Abstract
O6-alkylguanine-DNA alkyltransferase (AGT) is a ubiquitous enzyme with an amino acid sequence that is conserved in Eubacteria, Archaea and Eukarya. It repairs O6-alkylguanine and O4-alkylthymine adducts in single-stranded and duplex DNAs. In performing these functions, AGT must partition between adduct-containing sites and the large excess of adduct-free DNA distributed throughout the genome. Here we characterize the binding of human AGT to linear double-stranded, adduct-free DNAs ranging in length from 11 bp to 2686 bp. Moderately-cooperative binding (22.6 ± 3.7 ≤ ω ≤ 145.0 ± 37.0) results in an all-or-nothing association pattern on short templates. The apparent binding site size Sapp (mean = 4.39 ± 0.02 bp) oscillates with increasing template length. Oscillations in cooperativity factor ω have the same frequency but are of opposite phase to Sapp, with the result that the most stable protein-protein and protein-DNA interactions occur at the highest packing densities. The oscillation period (4.05 ± 0.02 bp/protein) is nearly identical to the occluded binding site size obtained at the highest measured binding density (4 bp/protein) and is significantly smaller than the contour length (~8 bp) occupied in crystalline complexes. A model in which protein molecules overlap along the DNA contour is proposed to account for these features. High AGT densities resulting from cooperative binding may allow efficient search for lesions in the context of chromatin remodeling and DNA replication.
O6-alkylguanine and O4-alkylthymine are mutagenic and carcinogenic adducts that are formed in DNA as a result of exposure to alkylating agents [1–3]. Both adducts are repaired by the ubiquitous repair protein, O6-alkylguanine-DNA alkyltransferase (AGT)1 [1,4,5]. Ironically, AGT also protects cells against chemotherapeutic drugs that methylate or chloroethylate DNA [6, 7]. Clinical trials of AGT-inhibitors are underway in an attempt to increase the efficacy of such drugs in cancer chemotherapy [8–10]. In addition, overexpression of AGT in hematopoietic stem cells is under study as a means to protect normal cells against the myelosuppressive effects of alkylating-agent chemotherapy [10, 11]. In spite of the interest focused on AGT as a result of its relevance to cancer, relatively little is known about its mechanisms of interaction with DNA and with proteins in its environment. A more complete characterization of these interactions is justified by the probability that they influence the repair activities of AGT in vivo as well as our ability to control DNA repair for therapeutic purposes.
Human AGT is a monomeric protein (207 amino acids, Mr = 21,519) that is expressed constitutively in normal cells [3, 5, 7] but may be overexpressed in some tumors that have been exposed to alkylating agents [12]. The protein binds DNA with modest affinity and little sequence specificity [13–15]. Studies with repair-inactive mutant proteins and with proteins in which the alkyltransfer function has been inactivated by prior alkylation of residue C145 (described below) suggest that binding affinities for alkylated DNA sites are only marginally greater than for the corresponding non-alkylated sequences ([15], Fried, unpublished results). While these results do not rule out preferential binding to O6-alkylguanine or O4-alkylthymine containing sequences, they raise the intriguing possibility that the lesion-search mechanism of AGT may not depend strongly on preferential binding. In repair reactions, each protein molecule mediates the transfer of a single alkyl group from the O6 position of guanine or O4 position of thymine to an active-site cysteine (C145 in the human protein). This reaction returns the DNA base to an unmodified state and permanently inactivates the alkyl-acceptor activity of the enzyme, which is ultimately degraded [4, 16]. As a result of this stoichiometric mechanism, the number of adducts that can be repaired at one time depends on the steady-state cellular concentration of the non-alkylated form of AGT [4, 5] and on its ability to partition between lesion sites and the large excess of genomic DNA in which they are embedded.
Several crystal structures of AGT-DNA complexes have been obtained in recent years. Those determined by Daniels et al. (PDB numbers 1T38 and 1T39) contain 1:1 AGT-DNA complexes [17] while that of Duguid et al. (PDB number 1YFH) contains two AGT molecules making separate contact with a single DNA molecule and a third binding a second DNA [18]. The isolated binding of single protein molecules that is a recurring theme in these structures contrasts with the close-packed, cooperative pattern of binding observed in free solution with both single-stranded and duplex templates ([15, 19], this work). These differences in binding mode suggest that binding in free solution may involve a different ensemble of molecular contacts than is present in the crystal structures. A more complete characterization of cooperative AGT-DNA interactions may allow us to account for this difference.
EXPERIMENTAL PROCEDURES
Enzymes and Reagents
T4 polynucleotide kinase and EcoRI endonuclease were purchased from New England Biolabs. [γ-32P]ATP was from ICN Radiochemicals. Electrophoresis grade polyacrylamide was from Fisher. All other chemicals were reagent-grade or better.
Human AGT Protein
Recombinant human AGT protein (tagged with His6 at its C-terminal end) was purified to apparent homogeneity according to published protocols [17]. The purity of the protein was verified by SDS-gel electrophoresis followed by silver-staining [20, 21]. Sedimentation equilibrium data were consistent with a single species of Mr = 21,860 ± 400 for [AGT] ≤ 100µM ([3] and M. Fried, unpublished results). This value is consistent with the value (Mr = 21,614) predicted from the sequence of this variant of the protein. The preparations used were >95% active in transfer of methyl groups from O6-[3H]methyl guanine-labeled calf-thymus DNA to AGT and >95% active in debenzylating O6-benzylguanine, as previously described [22, 23]. AGT concentrations were measured spectrophotometrically using ε280 = 3.93 × 104 M−1 cm−1 [23]. Samples were stored at −80°C until needed.
DNA substrates
Oligodeoxyribonucleotides of 11, 12, 14, 16, 18, 21, 22, 24, 26, 30 and 41 residues and their complements (sequences shown in Table 1) were synthesized by the Macromolecular Core Facility of the Penn State College of Medicine or were purchased from Midland Certified Reagent Company. Some DNA sequences were previously used in studies of DNA binding and/or repair by AGT (c.f., [14, 24]). Others were selected for particular sequence attributes (e.g., dG24•dC24 and dA24•dT24). Oligonucleotides were labeled at 5' termini with 32P as described by Maxam & Gilbert [25]. Single-stranded DNA concentrations were measured spectrophotometrically, using extinction coefficients calculated by the nearest-neighbor method [26, 27]. Depending on base composition and sequence, values of ε260 ranged from 8.45 × 103 M−1 cm−1 to 1.04 × 104 M−1 cm−1 (per base). Duplex DNAs were obtained by mixing a labeled oligonucleotide with a 1.05-fold molar excess of its unlabeled complement. Samples were heated to 90°C for 3 min and cooled to 20°C over 2h. Duplex formation was monitored by non-denaturing PAGE. Where necessary, samples were purified by preparative electrophoresis [28]. Duplex 147 bp and 207 bp DNA fragments containing the sea-urchin nucleosome positioning sequence were isolated by restriction digestion and preparative electrophoresis from plasmid p5S-207-18 [29], kindly provided by Dr. Sergei Grigoryev, Penn State University. Plasmid pUC19 DNA (New England Biolabs) was linearized with EcoRI endonuclease. Restriction fragments were deproteinized by phenol extraction and ethanol precipitation [30]. All DNAs were dialyzed against 10 mM Tris (pH 8.0), 0.1 mM EDTA prior to use or storage at −20°C. Duplex DNA concentrations were measured by absorbance using the average molar extinction coefficient ε260 = 1.3 × 104 M−1 cm−1 (per base pair).
Table 1.
Duplex Oligonucleotides
| Length, bp |
Sequence |
|---|---|
| 11 | 5’-TTT TTG TTT TT-3’ |
| 3’-AAA AAC AAA AA-5’ | |
| 12 | 5’-GAC TGA CTG ACT-3’ |
| 3’-CTG ACT GAC TGA-5’ | |
| 14 | 5’-GAC TGA CTG ACT GA-3’ |
| 3’-CTG ACT GAC TGA CT-5’ | |
| 16 | 5’-GAC TGA CTG ACT GAC T-3’ |
| 3’-CTG ACT GAC TGA CTG A-5’ | |
| 18 | 5’-GGA ACC TTG GAA CCT TGG-3’ |
| 3’-CCT TGG AAC CTT GGA ACC-5’ | |
| 21 | 5’-TGA AGT CCA AAG TTC AGT CCC-3’ |
| 3’-CT TCA GGT TTC AAG TCA GGG A-5’ | |
| 22 | 5’-CGC CAA CCC GCT GCC TAT CGT T-3’ |
| 3’-GCG GTT GGG CGA CGG ATA GCA A-5’ | |
| 24 | 5’-AAA AAA AAA AAA AAA AAA AAA AAA-3’ |
| 3’-TTT TTT TTT TTT TTT TTT TTT TTT-5’ | |
| 24 | 5’-GGG GGG GGG GGG GGG GGG GGG GGG-3’ |
| 3’-CCC CCC CCC CCC CCC CCC CCC CCC-5’ | |
| 26 | 5’-GAC TGA CTG ACT GAC TGA CTG ACT GA-3’ |
| 3’-CTG ACT GAC TGA CTG ACT GAC TGA CT-5’ | |
| 30 | 5’-GTG CCG CCA ACC CGC TGC CTA TCG TTA TAC-3’ |
| 3’-CAC GGC GGT TGG GCG ACG GAT AGC AAT ATG-5’ | |
| 41 | 5’-GCA ACG CAA TTA ATG TGA GTT AGC TCA CTC ATT AGG CAC CC-3’ |
| 3’-CGT TGC GTT AAT TAC ACT CAA TCG AGT GAG TAA TCC GTG GG-5’ |
Electrophoretic mobility shift assays (EMSA)
Mobility shift assays were carried out with DNAs of ≤ 41bp. Under the conditions used here, complexes formed with DNAs of 148 bp and larger were not sufficiently resolved from free DNA to allow quantitative binding analysis. Binding reactions were carried out at 20 ± 1 °C in 10mM Tris (pH 7.6), 100 mM NaCl, 1mM dithiothreitol, and 10µg/mL bovine serum albumin. Mixtures were equilibrated at 20 (±1) °C for 30 min. Duplicate samples incubated for longer periods gave identical results, indicating that equilibrium had been attained (result not shown). Electrophoresis was carried out in 10% polyacrylamide gels as previously described [19]. Autoradiographic images were captured on storage phosphor screens (type GP, GE Healthcare) detected with a Typhoon phosphorimager and quantitated with Image-Quant software (GE Healthcare). In a few cases autoradiographic data was captured on Kodak XL-1 film and band intensities were measured by excision and scintillation counting as previously described [15].
The serial dilution method [31, 32] was used to obtain estimates of binding stoichiometry (n). For the binding of n protein molecules to 1 DNA, the association constant is . Here P represents AGT protein, D represents DNA and PnD the AGT-DNA complex of protein-DNA ratio n. Separating variables and taking logarithms gives
| (1) |
Dilution of an AGT-DNA mixture changes [PnD]/[D] by mass action while maintaining the ratio of [P]total to [D] total. The free protein concentration at each dilution step can be estimated using [P]free = [P]input ∓ n[PnD] starting with an initial value of n; equation 1 was then used to calculate a new value of n from the DNA-binding distribution. This value was used to calculate a new estimate of [P]free and the calculations repeated recursively until values of n converged.
Analytical ultracentrifugation
Human AGT protein and duplex DNAs were dialyzed against 10mM Tris (pH 7.6), 1mM DTT, 1mM EDTA, 100mM NaCl. Analytical ultracentrifugation was performed at 20 ± 0.1°C in a Beckman XL-A centrifuge using an AN60Ti rotor. Most scans were obtained at 252 nm to minimize protein contribution to the DNA signal [33], although some data sets were obtained at 260 nm. Approach to equilibrium was considered to be complete when scans taken 6h apart were indistinguishable. Typically, equilibration times ≥24h met this criterion for AGT-DNA mixtures. Five scans were averaged for each sample at each wavelength and rotor speed.
For DNAs with small numbers of protein-binding sites, cooperative binding can be described by the simple mechanism nP + D ⇄ PnD in which free DNA (D) is in equilibrium with saturated complex (PnD) but intermediates with protein stoichiometries < n are not present at significant concentrations. At sedimentation equilibrium, the radial distribution of absorbance for such a system is given by equation 2.
| (2) |
Here A(r) is the absorbance at radial position r and αP, αD and αPnD are absorbances of protein, DNA and protein-DNA complex at the reference position, ro. and ε is a baseline offset that accounts for radial position-independent differences in the absorbances of different cell assemblies. The reduced molecular weights of AGT protein, DNA and protein-DNA complexes are given by σP = MP(1 − v̄Pρ)z2/(2RT), σD = MD(1 − v̄Dρ)z2/(2RT) and σPnD = (nMP + MD)(1 − v̄PnD ρ)z2/(2RT). Here MP and MD are the molecular weights of protein and DNA, n is the protein:DNA ratio of the complex; r is the solvent density, z, the rotor angular velocity, R is the gas constant and T the temperature (Kelvin). The partial specific volume of AGT (v̄ P = 0.744 mL/g) was calculated by the method of Cohn and Edsall [34], using partial specific volumes of amino acids tabulated by Laue and co-workers [35]. The partial specific volume of double-stranded NaDNA at 0.1M NaCl (0.540 ml/g) was estimated by interpolation of the data of Cohen and Eisenberg [36]. Partial specific volumes of each protein-DNA complex were estimated using Eq. 3.
| (3) |
Equation 2 was used in global analysis of multiple data sets obtained at different macromolecular concentrations and rotor speeds [37]. In this method, the values of αP, αD, αPnD and ε are unique to each sample but the value of n must be common to all data sets. Terms accounting for nonideal effects were not included because there was no evidence of nonideality (results not shown).
For large DNAs with many protein-binding sites, the concentration of protein-free DNA molecules becomes negligible long before available binding sites are saturated. In such systems, one observes mixtures of free protein and protein-DNA complex, with the weight-average molecular weight of the complex increasing smoothly until saturation is reached. These systems were analyzed using equation 4, with parameters defined as they were for Eq. 2, above.
| (4) |
Determination of equilibrium constants and cooperativity parameters
Association constants and cooperativity parameters were evaluated by direct titration of DNA with protein, with binding detected by EMSA or sedimentation equilibrium analysis. The total concentration of protein binding sites on DNA was always much less than that of the protein allowing the approximation [P]total = [P]free to be used. The dependence of binding density ν on the free protein concentration [P] was given by the McGhee-von Hippel isotherm [38] as modified by Record et al. [39] to account for finite lattice size (Eq.5).
| (5) |
Here ν is the binding density (protein molecules/nucleotide), K is the equilibrium association constant for binding a single site, ω is the cooperativity parameter, N is the length of the DNA in base pairs and s is the occluded site size (the size of the site, in base pairs, that one protein molecule occupies to the exclusion of others). The cooperativity parameter ω is equal to the equilibrium constant for moving a protein from an isolated site to a singly-contiguous one or from a singly-contiguous site to a doubly-contiguous one [38]. The model embodied in this equation is one in which proteins are assumed not to bind to fractional sites of length < s base pairs located within or at the ends of DNA molecules [39].
RESULTS
Determination of stoichiometries
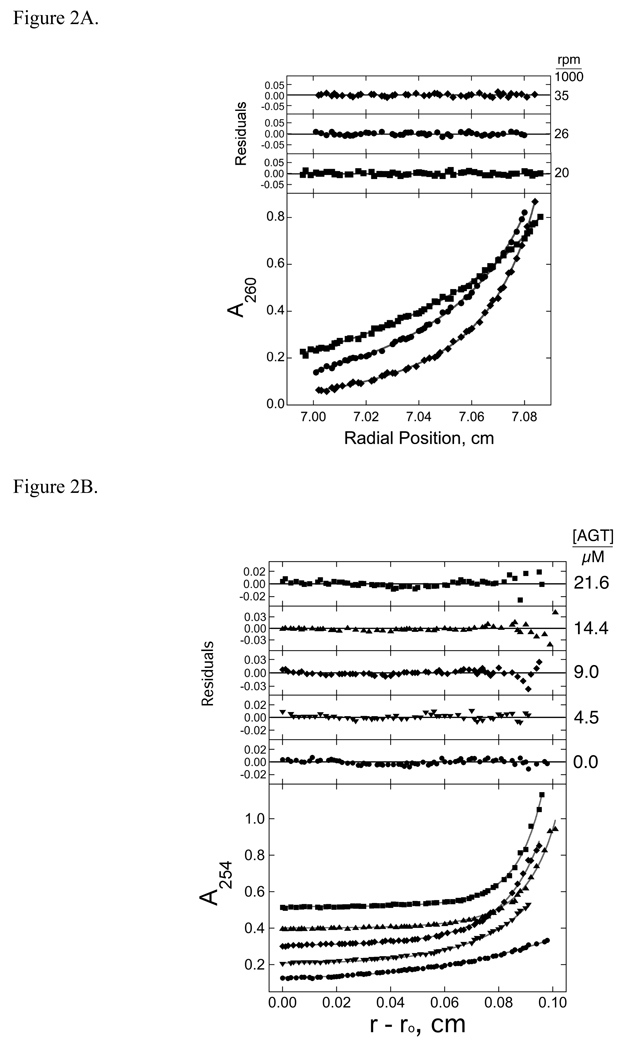
Sedimentation equilibrium and EMSA assays were used to determine binding stoichiometries. When short double-stranded DNAs (dsDNAs) were titrated with AGT, the major DNA species present at equilibrium were free DNA and the saturated protein-DNA complex (Figure 1). As previously discussed, the formation of a saturated multi-protein complex from free DNA without accumulation of significant concentrations of lower-stoichiometry species is a consequence of positively-cooperative binding [15]. Thus, for sedimentation equilibrium analysis of binding to templates ≤207 bp, data were fit using the sedimentation equation corresponding to the concerted binding model (nP + D ⇄ PnD; Eq. 2). For plasmid pUC19 DNA, the number of binding sites is much larger than the cooperativity parameter ω (described below) so over a wide range of [P], solutions contain only free protein and protein-DNA complexes, but no detectable free DNA. When this was the case, the data were fit using a two-species model (Eq. 4). The small, symmetrical residuals obtained from these fits (upper panels) indicate that these models account well for the mass distributions present in our samples. Molecular weights of protein-DNA complexes were obtained as fitting parameters and the stoichiometry of each complex was inferred from the known molecular weights of the DNA and AGT protein. The results of these analyses are summarized in Table 2.
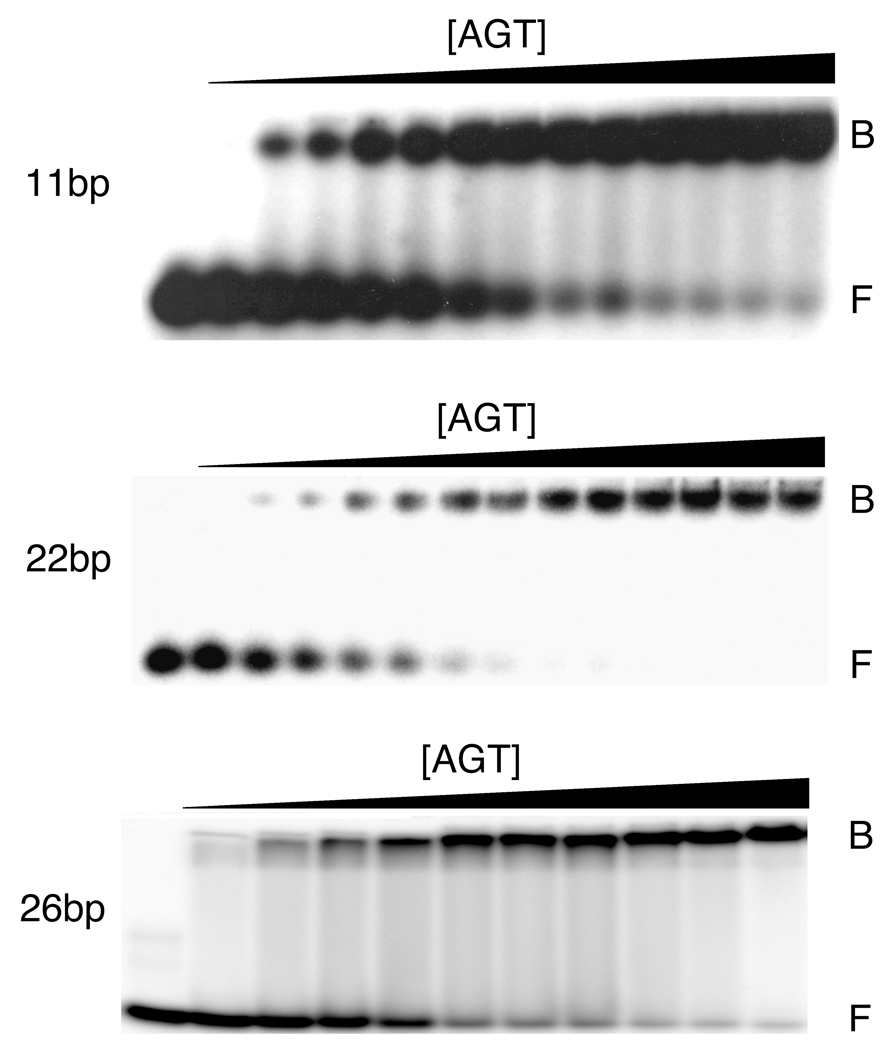
Fig. 1.
Titration of representative double-stranded DNAs with human AGT. Upper panel: 11-mer duplex DNA, 8.8 × 10−7 M with [AGT] increasing from 0M to 6.5 × 10−5 M from left to right. Middle panel: 22-mer duplex DNA, 3.6 × 10−7 M, with [AGT] increasing from 0 M to 1.5 × 10−5 M from left to right. Lower panel: 26-mer duplex DNA, 3.9 × 10−7 M, with [AGT] increasing from 0 M to 4.8 × 10−5 M, from left to right. Binding reactions were carried out at 20 ± 1 °C and samples were resolved on 10% polyacrylamide gels, as described in Experimental Procedures. Band designations B, bound DNA; F, free DNA. Although these images have been cropped and labeled for clarity, no additional bands were detectable between the origin of electrophoresis and the ionic front.
Table 2.
Properties of AGT-DNA Complexes
| DNA Length (bp) |
Mr (Saturated Complex)a |
Stoichiometry | Sapp bp/protein |
K/M | ω | K•ω |
|---|---|---|---|---|---|---|
| 11 | 5.08 ± 0.24 × 104 |
2.05 ± 0.11a 1.99 ± 0.15b |
5.36 ± 0.31a 5.52 ± 0.38b |
1.69 ± 0.19 × 103 c | 45.9 ± 6.0c | 7.75 ± 1.92 × 104 |
| 12 | - | 2.91 ± 0.14b | 4.13 ± 0.21b | 4.08 ± 1.12 × 103 c | 48.0 ± 5.5c | 1.96 ± 0.78 × 105 |
| 14 | - | 2.94 ± 0.10b | 4.75 ± 0.12b | 6.11 ± 0.96 × 103 c | 52.3 ± 5.1c | 3.18 ± 0.82 × 105 |
| 16 | 9.71 ± 0.17 × 104 |
3.94 ± 0.08a 3.98 ± 0.01b |
4.06 ± 0.08a 4.04 ± 0.02b |
1.25 ± 0.22 × 104 c | 105.2 ± 18.8c | 1.31 ± 0.47 × 106 |
| 18 | - | 3.82 ± 0.24b | 4.70 ± 0.18b | 4.23 ± 0.72 × 103 c | 85.7 ± 11.6c | 3.59 ± 1.10 × 105 |
| 21 | - | 5.06 ± 0.20b | 4.15 ± 0.17b | 6.07 ± 1.80 × 103 c | 92.6 ± 25.1c | 5.62 ± 3.21 × 105 |
| 22 | - | 4.58 ± 0.24b | 4.80 ± 0.24b | 3.80 ± 0.95 × 103 c | 68.3 ± 13.5c | 2.58 ± 1.16 × 105 |
| 24e | - | 5.93 ± 0.25b | 4.05 ± 0.18b | 2.31 ± 0.36 × 103 c | 83.1 ± 13.2c | 1.92 ± 0.61 × 105 |
| 24f | - | 6.01 ± 0.20b | 3.99 ± 0.14b | 4.01 ± 1.78 × 103 c | 110.1 ± 23.2c | 4.41 ± 2.89 × 105 |
| 26 | 1.37 ± 0.41 × 105 |
5.64 ± 0.18a 5.50 ± 0.16b |
4.60 ± 0.18a 4.72 ± 0.14b |
8.18 ± 2.52 × 103 c | 93.5 ± 26.5c | 7.64 ± 4.53 × 105 |
| 30 | 1.49 ± 0.34 × 105 |
6.08 ± 0.16a 6.43 ± 0.13b |
4.93 ± 0.13a 4.66 ± 0.11b |
9.71 ± 2.03 × 103 c | 64.7 ± 15.3c | 6.28 ± 2.79 × 105 |
| 41 | - | 9.01 ± 0.24b | 4.55 ± 0.13b | 1.87 ± 0.45 × 103 c | 145.0 ± 37.0c | 2.71 ± 1.35 × 105 |
| 148 | 5.66 ± 0.14 × 105 |
21.76 ± 0.65a | 6.80 ± 0.21a | 8.23 ± 1.51 × 103 d | 53.5 ± 22.1d | 4.40 ± 2.62 × 105 |
| 208 | 8.05 ± 0.33 × 105 |
31.04 ± 1.54a | 6.70 ± 0.35a | 7.87 ± 0.82 × 103 d | 63.6 ± 16.6d | 4.95 ± 1.82 × 105 |
| 2686 | 1.05 ± 0.51 × 107 |
406.3 ± 23.7a | 6.61 ± 0.41a | 8.52 ± 0.96 × 103 d | 22.6 ± 3.7d | 1.92 ± 0.53 × 105 |
Determined by sedimentation equilibrium analysis. The error ranges are 95% confidence limits.
Determined by EMSA-serial dilution analysis. The error ranges are 95% confidence limits.
Determined by scatchard analysis of EMSA data, using binding densities calculated from serial dilution analysis.
Determined by scatchard analysis of sedimentation equilibrium data, using binding densities calculated from Mw of the protein-DNA complex.
dG24•dC24
dA24•dT24
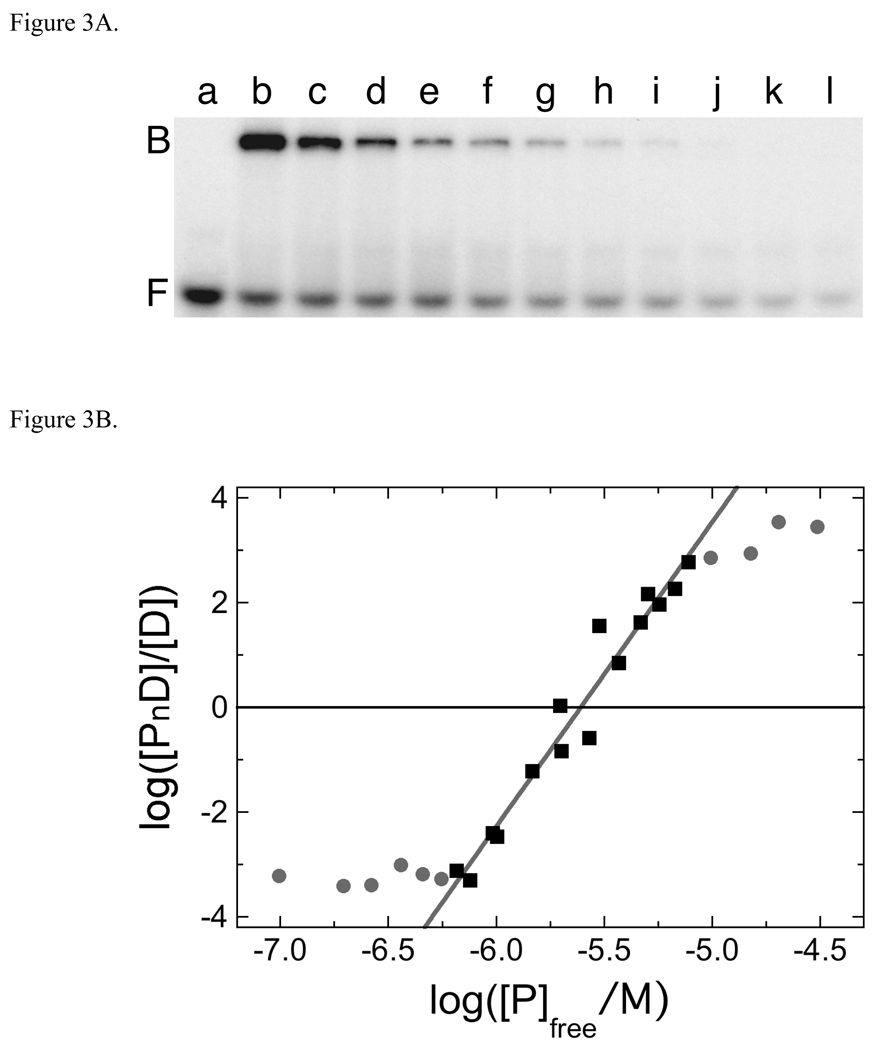
Additional stoichiometry values were determined by native gel electrophoresis (EMSA) using the serial dilution method [31, 32]. A representative experiment showing mass-action dissociation of the AGT complex formed with the dA24•dT24 duplex is shown in Fig. 3A. As with forward titrations, free DNA and an apparently saturated complex are the dominant species and stoichiometric intermediates are not detectably present. A graph of log[PnD]/[D] as a function of log[P] is shown in Fig. 3B. The slope of this graph near the mid-point of the binding reaction is a measure of the protein stoichiometry of the complex [40]. Stoichiometry values determined in this way are summarized in Table 2. The agreement of stoichiometry values determined by EMSA with those obtained by sedimentation equilibrium indicates that the same complexes are detected by both methods and demonstrates that dissociation of complexes during electrophoresis is not a significant factor under the conditions that were used here. These data also show that increasing DNA length results in higher limiting stoichiometries, indicating that for the DNA concentration range explored here and under conditions of AGT-excess, stoichiometry is limited by the number of binding sites available on each DNA.
Fig. 3.
Serial dilution analysis of the AGT complex formed with dA24•dT24. Panel A. Binding detected by EMSA. Sample a: 24-mer DNA (1.10 × 10−7 M) only. Sample b: 24-mer DNA (1.10 × 10−7 M) plus AGT (5.36 × 10−6 M). Samples c-l are sequential 1.33-fold dilutions of sample b. All samples were equilibrated in buffer consisting of 10mM Tris (pH 7.6), 100 mM NaCl, 1 mM DTT, 0.05 mg/mL bovine serum albumin for 30 min at 20 ± 1°C prior to resolution on native gels as described in Methods. Although this image has been cropped and labeled for clarity, no additional bands were detectable between the origin of electrophoresis and the ionic front. Panel B. Graph of the dependence of log[PnD]/[D] on log[P] for the AGT complex formed with dA24•dT24. Data from the experiment shown in Fig. 3A and others that provide additional [AGT] values. The line represents a least squares fit to the data ensemble for the range about the mid-point of the reaction (−6.18 ≤ log ([AGT]/M) ≤ −5.11), with [AGT]free calculated as described in Experimental Procedures. Symbols: the points used in the fit are indicated by (■) other points in the data set are indicated by closed circles (●). The slope equals 5.81 ± 0.34 for this subset of the data.
Apparent binding site size depends on DNA length
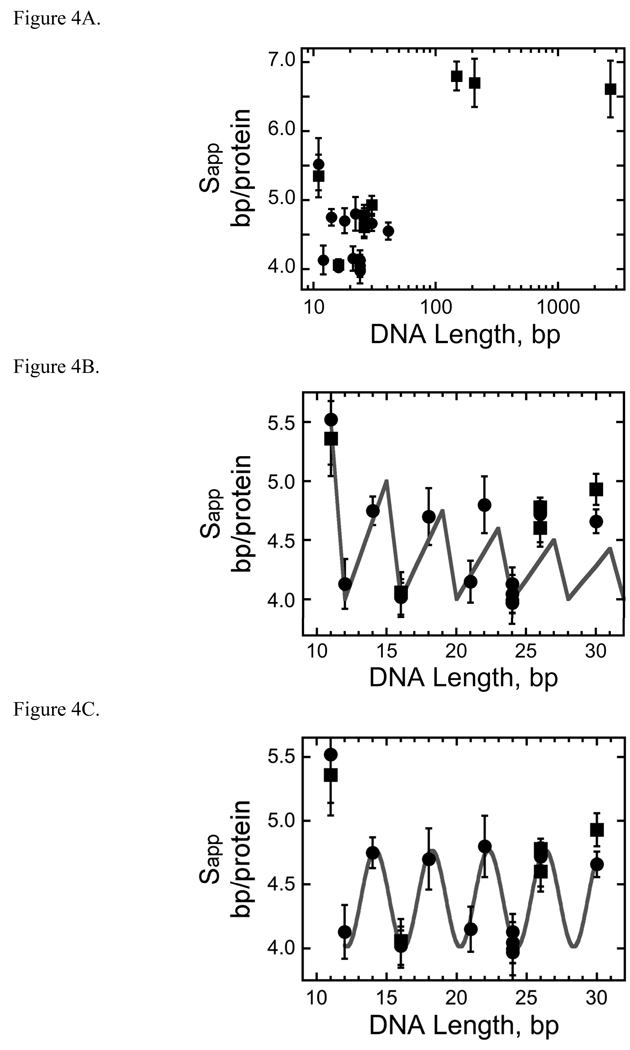
For a DNA of N bp, bound by a protein that occupies and occludes s bp/protein molecule, the maximum possible stoichiometry is the largest integer ≤ N/s, which we designate nmax. If binding-site utilization is efficient, i.e., if saturated complexes contain nmax proteins, we can define an apparent binding site size Sapp = N/nmax bp/protein. For short DNAs, values of Sapp are expected to fall between s and 2s-1 and oscillate with increasing N. Because minima in Sapp occur whenever N/s is an integer, this oscillation is expected to have a frequency equal to s. These properties, first described by Epstein [41], are characteristic of efficient, non-overlapping binding with occluded site size of s bp. Below we compare the properties of AGT-DNA complexes with the predictions of this simple model.
Values of Sapp measured for AGT oscillate with increasing DNA length (Figure 4). This is most evident for short templates (≤30 bp); above this length, the separation between tested values is too great for oscillation to be easily seen. Shown in Figure 4B, experimental Sapp values coincide with theoretical values calculated using s = 4bp, the occluded site size observed with 12, 16 and 24 bp DNAs. The agreement between theoretical and experimental Sapp values is best when N/4 is an integer. However, theoretical Sapp maxima decrease with increasing DNA length as the proportion of DNA length within an unoccupied partial binding site decreases and this decrease is not reproduced by the experimental data. This divergence may reflect increasingly degenerate binding as DNA length increases, that may result in population average stoichiometries < nmax. If this is the case, minimization of degeneracy whenever N/4 is an integer would still result in oscillations in Sapp with a period of 4bp. Fitting Sapp to a cosine function3 shows that the period and phase of the oscillation are strongly retained over the range 11 bp ≤ N ≤ 30bp (Figure 4C), with an oscillatory period of 4.05 ± 0.02 bp that is consistent with the s = 4 bp value inferred from minima in Sapp.
Fig. 4.
AGT forms a binding motif with a 4 bp periodicity. Panel A. Dependence of apparent binding site size (Sapp) on template length. Data from the entire set of AGT-dsDNA complexes. Sapp was calculated using Sapp = N/n, where N is DNA length in base pairs and n is the number of protein molecules bound to a DNA molecule. Sapp values determined from sedimentation equilibrium data are indicated by filled squares (■), values obtained from EMSA experiments are indicated by closed circles (●). The error bars correspond to 95% confidence limits. These data are also shown in Table 2. Panel B. Comparison of experimental and theoretical Sapp values as a function of DNA length. Experimental Sapp values determined from sedimentation equilibrium data are indicated by filled squares (■), values obtained from EMSA experiments are indicated by closed circles (●). The error bars correspond to 95% confidence limits. Theoretical Sapp values (solid line) were calculated using Sapp= N/nmax where nmax is the largest integer ≤ N/4. Panel C. Data for the subset of AGT-DNA complexes formed with DNAs of 41 bp or less. Data symbols are defined as in Panel A. The smooth curve is the least-squares fit of the equation3 Sapp = A cos (BL) + C in which A is the amplitude of the oscillation, B the displacement angle in degrees/bp and C is an offset equal to the mean value of Sapp. This fit returned A = −0.37 ± 0.04, B = 88.7 ± 0.5 degrees and C = 4.39 ± 0.02. This value of B indicates that successive binding sites are separated by 360/(88.7 ± 0.5) = 4.05 ± 0.02 bp along the DNA contour.
For DNAs of 148, 207 and 2686 bp, values of S are significantly larger than those found for shorter DNAs. Since incomplete occupancy of DNA ends should be equally possible for all DNAs, the less-efficient packing of AGT on these DNAs may reflect the presence of gaps between groups of tightly-packed protein molecules or a uniform, but less tightly-packed structure. The appearance of gaps between cooperative units is one of the features discussed by McGhee and von Hippel in their development of cooperative binding theory [38]. At the other extreme in template length, the value of S is significantly greater for the 11 bp DNA than for any of the short duplexes (up to and including the 41bp fragment). This is accounted for by the fact that with a site frequency of ~4bp/protein, only two AGT molecules can be accommodated on the 11mer and that the 3 residual base pairs are a larger proportion of the length of this DNA than of any larger template. Between these extremes, the dominant picture is one of a protein array in which the average separation between the start of two adjacent binding sites is ~4 bp. This spacing is significantly shorter than the ~8bp/protein occupied in crystalline AGT complexes [17, 18] and the optimum site size found for the rate of methyl-group transfer from single-stranded oligonucleotides (7 nt; [42]). As discussed below, our working hypothesis is that binding sites for AGT molecules overlap along the contour of dsDNA.
DNA-length dependence of K and ω
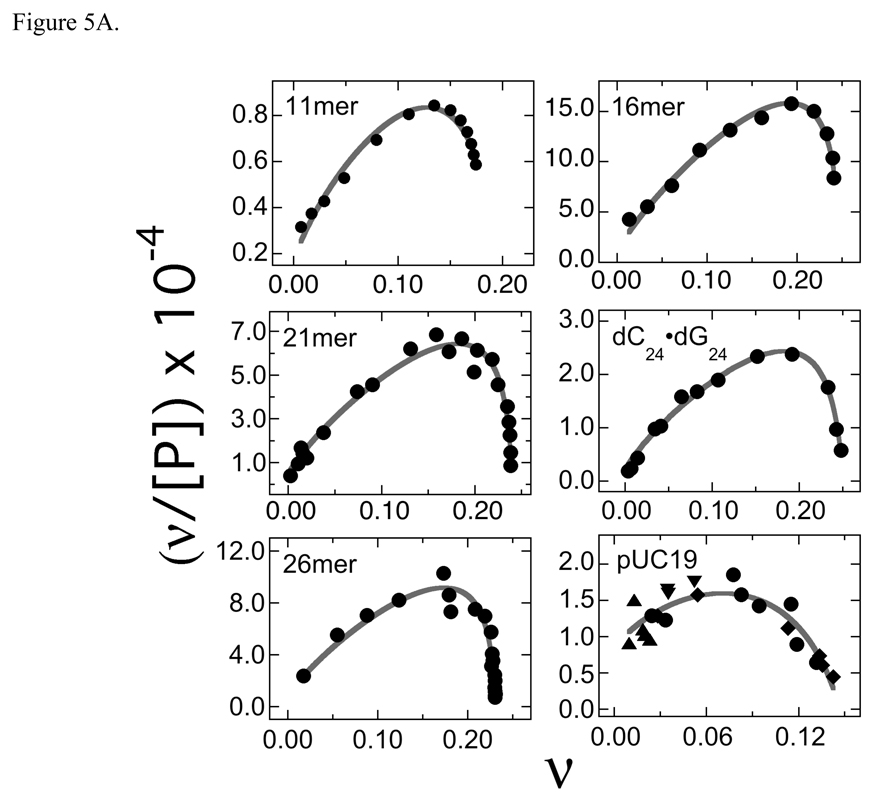
Changes in binding density may be accompanied by changes in the intrinsic association constant of AGT with DNA (K) or in the cooperativity with which AGT forms multi-protein complexes (ω). To explore these possibilities we recast our binding data according to the short-lattice variant of the McGhee-von Hippel relation (Eq. 5). This relates binding density ν to free protein concentration [P] in terms of K, ω and the occluded binding site size, s. Shown in Figure 5A are Scatchard plots of data obtained from serial dilution and direct titration experiments. The smooth curves represent fits of Eq. 5 to the data, in which values of s were calculated from stoichiometries determined independently (by sedimentation equilibrium or serial dilution as described above). The good correspondence of fits to data indicates that binding models that include neighbor exclusion and positive cooperativity can account for the binding activities in these systems.
Fig. 5.
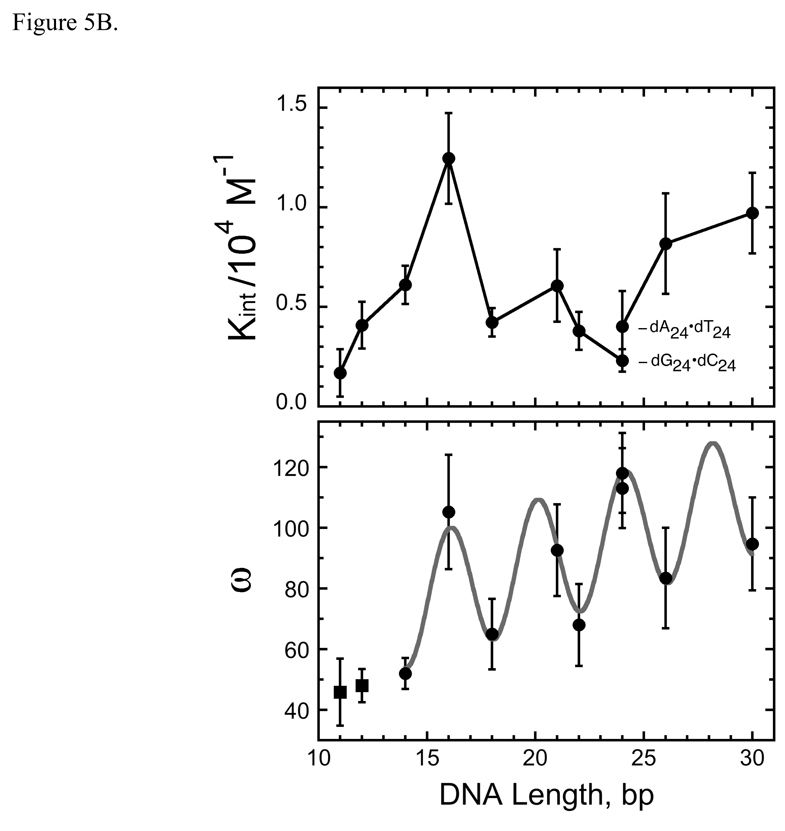
Analysis of binding affinities. Panel A: Scatchard plots. Forward titrations were carried out as described in Methods and binding was detected and quantitated by electrophoretic mobility shift assay. Each data set is a composite derived from 2 or 3 independent titrations. The smooth curves correspond to non-linear least squares fits of Eq. 2 to the data. The values of K and ω obtained as parameters of these fits are compiled in Table 2. Panel B: dependence of K and ω on DNA length (N). The data are derived in part from the experiments shown in Panel A. The error bars correspond to 95% confidence limits estimated for each parameter. K data for dA24-dT24 and dG24-dC24 templates are labeled Comparison with Fig. 4B shows that the oscillations of ω with increasing N have the same period but opposite phase as those of Sapp.
The least-squares “best” values of K and ω obtained as fitting parameters are summarized in Table 2. Tests using fixed values of K show that K and ω are weakly anti-correlated (an increase in one parameter produces a slight decrease in the other). On the other hand, the quality of the fits measured by the χ2 parameter decreased significantly if either parameter varied from its most probable value by more than the 95% confidence limits (results not shown). Thus, while the values of these parameters cannot be considered unique, the data quality is sufficient to allow us to parse binding affinity into reasonably well-defined estimates of K and ω.
Values of K and ω oscillate as DNA length increases (Figure 5B). The fact that these oscillations are larger than the confidence limits associated with individual parameters and the fact that they are not in phase argue that they are not consequences of fitting correlations between K and ω (as discussed above, K and ω are weakly anti-correlated). Oscillations of ω with DNA length are regular in amplitude and period; this oscillation has the same period but opposite phase as that for the oscillation of Sapp with length. As a result, the most compact complexes (with smallest Sapp) have relatively stronger protein-protein interactions than do less compact assemblies. One possible interpretation of these results is that optimal protein juxtaposition is lost as binding density decreases (Sapp increases). It is noteworthy that the baseline for the oscillation of ω increases uniformly for 11 bp ≤ DNA length ≤ 30 bp. This is an expected consequence of the growth of a cooperative array in which proteins bind adjacent sites on a linear substrate. Proteins at array ends interact with a single neighbor while proteins in the center of the array interact with two or more neighbors. As arrays increase in length, the proportion of proteins in the center increases, with corresponding increase in ω averaged over the whole assembly. A model of a cooperative assembly that unites these features is proposed below.
In contrast to the regular pattern seen with ω, oscillations in K are irregular in amplitude and phase, suggesting that factors other than DNA length and binding density contribute significantly to the strength of the AGT-DNA contact. The difference in K values for dG24•dC24 and dA24•dT24 templates (Fig 5B, Table 2) suggests that DNA sequence has an important influence on affinity. Consistent with this notion, the two DNAs that give local maxima in K (the 16mer and the 26mer) have repeating sequences that provide a guanine at every fourth residue on each strand (shown in Table 1). This frequency, which is the same as that of AGT in the cooperative complex, may allow the enzyme to make unusually favorable contacts with its substrate.
DISCUSSION
The data presented here are consistent with a binding mechanism in which weak affinity for duplex DNA (1.7 ± 0.2 × 103 M−1 ≤ K ≤ 1.3 ± 0.2 × 104 M−1) is paired with moderate binding cooperativity (22.6 ± 3.7 ≤ ω ≤ 145.0 ± 37.0). On short DNAs, this cooperativity produces tight packing that results in oscillation in the apparent binding site size Sapp with a period of 4 bp/protein molecule. On DNAs of length >41 bp, cooperativity is not sufficient to suppress binding degeneracy, so Sapp increases (Fig. 4), however our data for lengths greater than 41 bp is too sparse to allow us to determine whether oscillations in Sapp occur in much larger templates. The correlation of maxima in ω with minima in Sapp (Fig. 4B and Fig. 5B) indicates that the optimal juxtaposition for cooperativity occurs when neighboring binding sites are 4 bp apart. Since a single molecule of AGT occupies ~8bp with its center in the minor groove of double-stranded DNA [17, 18], this spacing requires that protein molecules overlap along the DNA contour, but it does not require overlap along the minor groove surface that provides the majority of DNA contacts4. Overlap along the DNA contour provides opportunities for protein-protein contacts that may contribute to cooperativity.
The cooperative binding mechanism described here for duplex DNA and previously for single-stranded DNAs [19] contrasts with the independent binding shown in currently-available crystal structures of AGT-DNA complexes [17, 18]. At present we cannot account for this difference. To date, cooperative binding has been observed with wild-type human AGT over a range of temperature (4°C ≤ T ≤ 40°C), salt concentrations (0M ≤ [KCl] ≤ 0.4M), magnesium ion concentrations (0 mM ≤ [Mg++]free ≤ 20 mM) and zinc availability (Zn-depleted and Zn-saturated proteins and Zn-saturated protein with 0 mM ≤ [Zn++]free ≤ 10mM). In addition, cooperative binding has been observed with active-site (C145) mutant- and C145-methylated and-benzylated proteins as well as C-terminal (His)6-tagged, N-terminal (His)6-tagged and untagged proteins [15, 19, 23, 43]. We conclude that cooperative binding is a bona-fide activity of human AGT.
The pattern described here for double-stranded DNA binding is remarkably similar to that found for single-stranded substrates [19]. Quantitative similarities include weak intrinsic affinity for substrate DNA (1 × 103 M−1 ≤ K (single stranded) ≤ 2 × 104 M−1), moderate cooperativity (35 ≤ ω (single stranded) ≤ 90) and a statistical binding site size that oscillates on short DNAs with a period of ~4nt/protein. Similar K values indicate that the free energies of protein-DNA interaction are almost the same for single stranded and duplex substrates, while comparison of K and ω values indicates that protein-protein contacts contribute a similar, large proportion (~33%) of the free energy of addition of a protein monomer to cooperative assemblies. Together, these results suggest why similar stabilities are observed for AGT complexes formed on single-stranded and duplex DNAs [15]: comparatively strong protein-protein interactions mask the contributions of relatively small differences in K to the overall product K•ω.
What role does cooperative binding play in sequence-specificity? Previous work has shown that overall affinity (K•ω) and DNA-repair efficiency are not strongly dependent on base composition or sequence of the substrate [14, 44, 45]. For the DNAs described here, values of K and ω vary by factors of 7.4 and 6.4, respectively. However, because changes in K and ω are not perfectly in phase, the largest difference in their product is only a factor of 17. These results suggest that differences in K may be offset by differences in ω, resulting in a very small sequence-dependence of overall binding affinity. The absence of a large dependence of binding affinity on base composition or sequence may be important to ensure that repair activity is available to all sequences. In addition, cooperative binding may contribute to the efficiency of lesion search and repair by concentrating repair activity at a higher density on the DNA contour than would be available in non-cooperative binding modes. With a new binding site starting every 4 bp, only modest protein displacement is needed for surveillance of every base-pair within a cooperative assembly. Preliminary kinetic results suggest that AGT complexes isomerize rapidly, as required for efficient surveillance according to this model (M. Fried, unpublished results). Although the moderate cooperativity detected here is insufficient to extend high-density binding over hundreds of base-pairs, it can generate densities approaching 1 protein every 4 bp for sections of template ≤50 bp long. Regions of this size are likely to become available between nucleosomes during the chromatin remodeling associated with transcription or DNA replication. The processive movements of transcription and replication complexes may allow systematic surveillance of large parts of the genome by AGT, while repair just upstream of a replication fork is likely to represent the last opportunity to prevent the conversion of a promutagenic lesion into an actual mutation.
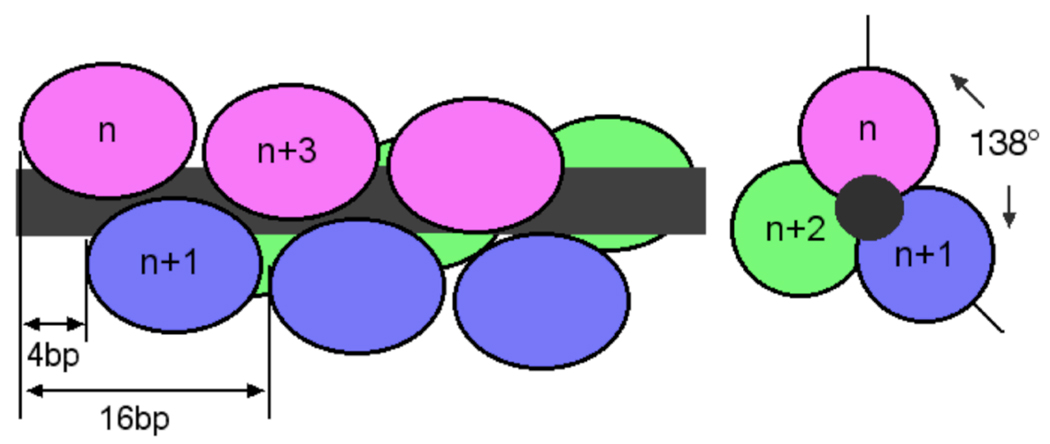
How are AGT proteins arranged in the cooperative array? Our working model is based on the binding density of 1 protein/4 base-pairs characteristic of complexes formed on short DNAs. This frequency allows identical protein contacts with the minor groove a B-DNA duplex if each protein is rotated clockwise approximately (4 bp•protein−1/10.4 bp•turn−1) × 360 deg•turn−1 = 138.4 deg•protein−1 with respect to its immediate neighbor. This arrangement would allow each AGT protein to maintain the minor-groove contacts that are seen in the crystal structures of AGT-dsDNA complexes [17, 18] and it also produces a 3-start helical array of protein molecules, with the nth protein molecule positioned to interact with protein n + 3 (shown schematically in Fig. 6). The ability to form an n-to-n+3 contact may account for the significantly greater cooperativity seen in the 4:1 complex formed with 16mer DNA than in 2:1 and 3:1 complexes formed with 11, 12 and 14 bp DNAs (Fig. 5B). The resulting tight-packing of proteins accounts for the dramatic phasing of Sapp and cooperativity (ω) with DNA length that we have observed. It also allows us to make testable predictions about the protein interactions that maintain the complex in vitro and are likely to dominate its functions in vivo. A few of these are outlined below.
Fig. 6.
A model of binding topology for the cooperative array. Proteins are shown schematically as filled ovals and the DNA as a dark grey rod. Drawings are not to scale. Left, overlapping binding viewed from perpendicular to the DNA axis. Proteins are arranged as a three-start helix with individual arrays colored red, blue and green. Each protein is rotated ~138° with respect to its nearest neighbors, as predicted if all AGT molecules make identical contacts with the minor groove of B-form DNA and binding sites are separated by four base pairs. This view emphasizes the n-to-n+3 juxtaposition predicted for proteins along the same face of the DNA cylinder. Right: view down the long axis of the DNA. Only the first three proteins are shown for clarity. This view emphasizes the rotational juxtaposition of proteins n, n+1 and n+2.
First, AGT is likely to bind relaxed-duplex DNA with greater affinity than it binds positively or negatively supercoiled molecules. If locally-stiff duplex DNA constrains the protein-protein contacts to a sub-optimal geometry compared to those formed on torsionallymore-flexible single-stranded DNA, the free energy cost should be reflected in a reduction in ω on duplex templates compared to the value on single-stranded DNAs. Since experimental values of ω are nearly the same for single-stranded and duplex DNAs ([19], this work), it seems unlikely that the helical period of the DNA that is optimal for protein-protein contacts differs much from that of relaxed B-DNA. If correct, this prediction is likely to be functionally relevant. Since chromatin imposes topological constraints on genomic DNA and since transcription and DNA replication have the potential to impose torsional stress, we predict that AGT- and topoisomerase-activities will be found to co-segregate within chromosomal domains.
Second, cooperative interactions are likely to influence the rates of AGT binding and dissociation from target DNAs and hence, the kinetics of lesion search and repair. The portion of a cooperative array of bound proteins that is active in protein binding and/or dissociation is likely to influence the kinetics of these processes. Our current thinking is influenced by the notion that the addition or removal of a protein unit from the middle of a cooperative assembly is likely to be slow compared to the addition or removal of a unit from one end. If this is the case, the rate of transfer of AGT molecules between DNA segments may depend more strongly on the concentration of cooperative assemblies (and hence the concentration of end-monomers) than on the concentration of DNA-bound AGT monomers, themselves.
Finally, unless the rate of alkyltransfer greatly exceeds the rate of forming a cooperative complex, DNA-repair rates are likely to depend on the length of the cooperative complex. This is because repair requires the correct juxtaposition of an active AGT monomer and the damaged base. If the correct juxtaposition of AGT and lesion is not achieved during formation of the protein-DNA complex, repair may require repositioning of the entire protein array. If this is the case, productive binding and repair might be more rapidly achieved with short protein arrays than with long. Tests of these predictions are currently underway.
Fig. 2.
Sedimentation equilibrium of solutions containing AGT and double-stranded DNAs at 20 ± 1 °C. Panel A: Representative data for binding to 26 bp DNA. Samples contained DNA (5 × 10−7 M) and AGT (1.45 × 10−5 M) in buffer consisting of 10 mM Tris (pH 7.6), 1 mM DTT, 1 mM EDTA, 100 mM NaCl. Radial scans taken at 20,000 rpm (■), 26,000 rpm (●), and 35,000 rpm (♦) are shown with vertical offsets for clarity. The smooth curves correspond to a global fit of Eq. 3 to a data set that includes these scans and scans obtained at 2 additional AGT concentrations (1.82 × 10−5 M and 2.18 × 10−5 M). The small residuals, nearly symmetrically-distributed about zero (upper panels) indicate that the cooperative nP + D ⇄ PnD model is consistent with the mass distributions of DNA in these samples, and that n = 5.65 ± 0.21. Panel B: Representative data for binding to linear pUC19 DNA (2686 bp). Samples contained DNA (5 × 10−7 M) and AGT in buffer consisting of 10 mM Tris (pH 7.6), 1 mM DTT, 1 mM EDTA, 100 mM NaCl. AGT concentrations were: 0M (●), 4.5 × 10−6 M (▼), 9.0 × 10−6 M (♦), 1.44 × 10−5 M (▲) and 2.16 × 10−5 M (■). Radial scans shown here were taken at 3,000 rpm. The smooth curves are fits of Eq. 5 to each data set.
Footnotes
Supported by NIH grants GM-070662 (to M.G.F.) and CA-97209 (to A.E.P.) and Medical Scientist Training Program Grant 5 T32 GM-08601-05 (J.J.R.).
Also called O6-methylguanine DNA methyltransferase, MGMT; EC 2.1.1.63. Homologous proteins have been identified in Eubacteria, Archaea and Eukarya [3].
This is the occluded site size obtained for complexes with the highest binding densities (formed with DNAs of 12bp, 16 bp and 24bp).
This corresponds to the first terms of a Fourier cosine series [46]. We chose this simple expression because it adequately models the oscillation and because the fit was not significantly improved when additional sine or cosine terms were added (results not shown).
In the structures of Duguid et al. [18] and Daniels et al. [17], AGT contacts 4bp in the minor groove of slightly-distorted B-DNA. The occluded site size s = 4bp results in binding sites that do not overlap on the minor groove surface. This justifies the use of Eq. 5, which describes non-overlapping cooperative binding, in our analyses.
REFERENCES
- 1.Loveless A. Nature (London) Vol. 223. 1969. Possible relevance of O6 alkylation of dexoyguanosine to he mutagenicity and carcinogenicity of nitrosamines and nitrosamides; pp. 206–207. [DOI] [PubMed] [Google Scholar]
- 2.Pegg AE. Methylation of the O6 position of guanine in DNA is the most likely initiating event in carcinogenesis by methylating agents. Cancer Invest. 1984;2:223–231. doi: 10.3109/07357908409104376. [DOI] [PubMed] [Google Scholar]
- 3.Pegg AE, Dolan ME, Moschel RC. Structure, function and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog. Nucl. Acid Res. and Mol. Biol. 1995;51:167–223. doi: 10.1016/s0079-6603(08)60879-x. [DOI] [PubMed] [Google Scholar]
- 4.Pegg AE. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogens and therapeutic agents. Cancer Res. 1990;50:6119–6129. [PubMed] [Google Scholar]
- 5.Margison GP, Santibáñez-Koref MF. O6-alkylguanine-DNA alkyltransferase: role in carcinogenesis and chemotherapy. BioEssays. 2002;24:255–266. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- 6.Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J. Clin. Oncol. 2002;20:2388–2399. doi: 10.1200/JCO.2002.06.110. [DOI] [PubMed] [Google Scholar]
- 7.Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutat. Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 8.Rabik CA, Njoku MC, Dolan ME. Inactivation of O6-alkylguanine DNA alkyltransferase as a means to enhance chemotherapy. Cancer Treat. Rev. 2006;32:261–276. doi: 10.1016/j.ctrv.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Ranson M, Middleton MR, Bridgewater J, Lee SM, Dawson M, Jowle D, Halbert G, Waller S, McGrath H, Gumbrell L, McElhinney RS, Donnelly D, McMurry TB, Margison GP. Lomeguatrib, a potent inhibitor of O6-alkylguanine-DNA-alkyltransferase: phase I safety, pharmacodynamic, and pharmacokinetic trial and evaluation in combination with temozolomide in patients with advanced solid tumors. Clin. Cancer Res. 2006;12:1577–1584. doi: 10.1158/1078-0432.CCR-05-2198. [DOI] [PubMed] [Google Scholar]
- 10.Verbeek B, Southgate TD, Gilham DE, Margison GP. O6-Methylguanine-DNA methyltransferase inactivation and chemotherapy. Br. Med. Bull. 2008;85:17–33. doi: 10.1093/bmb/ldm036. [DOI] [PubMed] [Google Scholar]
- 11.Zaboikin M, Srinivasakumar N, Schuening F. Gene therapy with drug resistance genes. Cancer Gene Ther. 2006;13:335–345. doi: 10.1038/sj.cgt.7700912. [DOI] [PubMed] [Google Scholar]
- 12.Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, Greenman C, Edkins S, Bignell G, Davies H, O'Meara S, Parker A, Avis T, Barthorpe S, Brackenbury L, Buck G, Butler A, Clements J, Cole J, Dicks E, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Richardson D, Shepherd R, Small A, Solomon H, Tofts C, Varian J, West S, Widaa S, Yates A, Easton DF, Riggins G, Roy JE, Levine KK, Mueller W, Batchelor TT, Louis DN, Stratton MR, Futreal PA, Wooster R. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan CL, Wu Z, Ciardelli T, Eastman A, Bresnick E. Kinetic and DNA-binding properties of recombinant human O6-methylguanine-DNA methyltransferase. Arch. Biochem. Biophys. 1993;300:193–200. doi: 10.1006/abbi.1993.1027. [DOI] [PubMed] [Google Scholar]
- 14.Fried MG, Kanugula S, Bromberg JL, Pegg AE. DNA binding mechanisms of O6-alkylguanine-DNA alkyltransferase: stoichiometry and effects of DNA base composition and secondary structures on complex stability. Biochemistry. 1996;35:15295–15301. doi: 10.1021/bi960971k. [DOI] [PubMed] [Google Scholar]
- 15.Rasimas JJ, Pegg AE, Fried MG. DNA-binding mechanism of O6-alkylguanine-DNA alkyltransferase. Effects of protein and DNA alkylation on complex stability. J. Biol. Chem. 2003;278:7973–7980. doi: 10.1074/jbc.M211854200. [DOI] [PubMed] [Google Scholar]
- 16.Xu-Welliver M, Pegg AE. Degradation of the alkylated form of the DNA repair protein, O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2002;23:823–830. doi: 10.1093/carcin/23.5.823. [DOI] [PubMed] [Google Scholar]
- 17.Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat. Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 18.Duguid EM, Rice PA, He C. The structure of the human AGT protein bound to DNA and its implications for damage detection. J. Mol. Biol. 2005;350:657–666. doi: 10.1016/j.jmb.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 19.Rasimas JJ, Kar SR, Pegg AE, Fried MG. Interactions of human O6-alkylguanine-DNA alkyltransferase (AGT) with short single-stranded DNAs. J. Biol. Chem. 2007;282:3357–3366. doi: 10.1074/jbc.M608876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Nesterenko MV, Tilley M, Upton SJ. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods. 1994;28:239–242. doi: 10.1016/0165-022x(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 22.Kanugula S, Goodtzova K, Edara S, Pegg AE. Alteration of arginine-128 to alanine abolishes the ability of human O6-alkylguanine-DNA alkyltransferase to repair methylated DNA but has no effect on its reaction with O6-benzylguanine. Biochemistry. 1995;34:7113–7119. doi: 10.1021/bi00021a024. [DOI] [PubMed] [Google Scholar]
- 23.Rasimas JJ, Kanugula S, Dalessio PM, Ropson IJ, Fried MG, Pegg AE. Effects of zinc occupancy on human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2003;42:980–990. doi: 10.1021/bi026970b. [DOI] [PubMed] [Google Scholar]
- 24.Fang Q, Kanugula S, AE P. Function of domains of human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2005;44:15396–15405. doi: 10.1021/bi051460d. [DOI] [PubMed] [Google Scholar]
- 25.Maxam A, Gilbert WS. A new method for sequencing DNA. Proc. Natl. Acad. Sci. U.S.A. 1977;74:560–565. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantor CR, Tinoco I., Jr Absorption and optical rotatory dispersion of seven trinucleoside diphosphates. J. Mol. Biol. 1965;13:65–77. doi: 10.1016/s0022-2836(65)80080-8. [DOI] [PubMed] [Google Scholar]
- 27.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5'-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandelkern M, Elias JG, Eden D, Crothers DM. The dimensions of DNA in solution. J. Mol. Biol. 1981;152:153–161. doi: 10.1016/0022-2836(81)90099-1. [DOI] [PubMed] [Google Scholar]
- 29.Simpson RT, Thoma F, Brubaker JM. Chromatin reconsituted from tandemly repeated cloned DNA fragments and core histones: A model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 30.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:449–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 31.Fried MG, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucl. Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeman SM, Crothers DM. Simultaneous measurement of binding constants and unwinding angles by gel electrophoresis. Methods Enzymol. 2001;340:51–68. doi: 10.1016/s0076-6879(01)40417-4. [DOI] [PubMed] [Google Scholar]
- 33.Daugherty MA, Fried MG. Protein-DNA interactions at sedimentation equilibrium. In: Scott D, editor. Modern Analytical Ultracentrifugation: Techniques and Method. Oxford: Royal Society of Chemistry; 2005. pp. 195–209. [Google Scholar]
- 34.Cohn EJ, Edsall JT. Proteins, amino acids and peptides as ions and dipolar ions. In: Cohn EJ, Edsall JT, editors. Proteins, Amino Acids and Peptides as Ions and Dipolar Ions. Reinhold, New York: 1943. pp. 370–381.pp. 428–431. [Google Scholar]
- 35.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Harding JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, England: The Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 36.Cohen G, Eisenberg H. Deoxyribonucleate solutions: sedimentation in a density gradient, partial specific volumes, density and refractive density increments and preferential interactions. Biopolymers. 1968;6:1077–1100. doi: 10.1002/bip.1968.360060805. [DOI] [PubMed] [Google Scholar]
- 37.Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Analysis of data from the analytical ultracentrifuge by non-linear least squares techniques. Biophys. J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGhee J, von Hippel PH. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 39.Tsodikov OV, Holbrook JA, Shkel IA, Record MT., Jr Analytic binding isotherms describing competitive interactions of a protein ligand with specific and nonspecific sites on the same DNA oligomer. Biophys. J. 2001;81:1960–1969. doi: 10.1016/S0006-3495(01)75847-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams C, Fried MG. analysis of protein-DNA equilibria by native gel electrophoresis. In: Schuck P, editor. Protein Interactions: Biophysical Approaches For The Study of Multicomponent Systems. Academic Press: New York; 2007. pp. 417–446. [Google Scholar]
- 41.Epstein IR. Cooperative and non-cooperative binding of large ligands to a finite one-dimensional lattice A model for ligand-oligonucleotide interactions. Biophys. Chem. 1978;8:327–339. doi: 10.1016/0301-4622(78)80015-5. [DOI] [PubMed] [Google Scholar]
- 42.Liem LK, Wong CW, Lim A, Li BFL. Factors influencing the repair of the mutagenic lesion O6-methylguanine in DNA by human O6-methylguanine-DNA methyl-transferase. J. Mol. Biol. 1993;231:950–959. doi: 10.1006/jmbi.1993.1344. [DOI] [PubMed] [Google Scholar]
- 43.Rasimas JJ, Dalessio PA, Ropson IJ, Pegg AE, Fried MG. Active-site alkylation destabilizes human O6-alkylguanine DNA alkyltransferase. Protein Sci. 2004;13:301–305. doi: 10.1110/ps.03319404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pegg AE, Dolan ME. Investigation of sequence specificity in DNA alkylation and repair using oligodeoxynucleotide substrates. In: Lambert MW, Laval J, editors. DNA Repair Mechanisms and their Biological Implications in Mammalian Cells. New York: Plenum Press; 1989. pp. 45–59. [Google Scholar]
- 45.Bender K, Federwisch M, Loggen U, Nehls P, Rajewsky MF. Binding and repair of O6-ethylguanine in double-stranded oligodeoxynucleotides by recombinant human O6-alkylguanine-DNA alkyltransferase do not exhibit significant dependence on sequence context. Nucl. Acids Res. 1996;24:2087–2094. doi: 10.1093/nar/24.11.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burington RS. Handbook of Mathematical Tables and Formulas. 5th ed. New York: McGraw-Hill; 1973. [Google Scholar]