Abstract
The global rise of anti-microbial resistance, combined with the rapid rate of microbial evolution, and the slower development of novel antibiotics, underscores the urgent need for innovative therapeutics. We are facing a post-antibiotic era with a decreased armamentarium to combat infectious diseases. Development of novel drugs will rely on basic research aimed to increase our understanding of bacterial pathogenesis and the inter-cellular chemical signalling among bacterial cells. Such basic science, when combined with contemporary drug discovery technologies, may be translated into therapeutic applications to combat bacterial infections. In this review, we discuss many strategies aimed to interfere with bacterial cell-to-cell signalling via the quorum-sensing (QS) pathway to inhibit bacterial virulence and/or the development of microbial communities (known as biofilms), which are refractory to antibiotic treatment. QS antagonists should be viewed as blockers of pathogenicity rather than as anti-microbials and because QS is not involved in bacterial growth, inhibition of QS should not yield a strong selective pressure for development of resistance. QS inhibitors (QSIs) hold great expectations and we look forward to their application in fighting bacterial infections.
Keywords: enterohemorrhagic E. coli (EHEC), inter-kingdom signalling, Pseudomonas aeruginosa, quorum sensing, Staphylococcus aureus
Introduction
Bacteria were thought to live a simple existence lacking the cell-to-cell communication that highly evolved organisms possess. Theirs was deemed an uncomplicated repetition of cellular replication. But in the last three decades this concept has been challenged as bacteria were recognized to be equipped with the ability to sense chemical signals from other organisms in their surroundings, and capable of forming communities whose members interact with each other. This interaction can be intra- and/or inter-species or inter-kingdoms. Although most of these interactions probably evolved to allow co-existence of a bacterium and its host, pathogens seem to have ‘hijacked’ these communication systems to control their virulence traits.
Communication among bacteria is achieved by the release of chemical signals called autoinducers. These autoinducers allow bacteria to assess the density of the local bacterial population and coordinate critical gene expression. This mechanism, termed QS, was first observed in Vibrio fischeri, a bioluminescent bacterium that lives in the photophore, the light producing organ of the bobtail squid with which it has a symbiotic relationship (Eberhard, 1972; Eberhard et al, 1981; Kempner & Hanson, 1968; Stevens & Greenberg, 1997). By sensing autoinducers released by the continually growing bacterial population within the photophore, V. fischeri is able to discern when its density is high enough to trigger the transcription of luciferase, and the subsequent emission of light. Since this first discovery, many pathogens have been shown to utilize QS to determine the right time to express virulence related genes. Production of proteins and other gene products necessary for pathogenesis is an expensive endeavour that requires the bacteria to exert a lot of energy. Thus by releasing and/or sensing autoinducers, pathogens are able to optimally time the expression of their virulence factors, conserving energy and maximizing their survival, at the detriment of their host.
Although bacteria are capable of existing in a planktonic form, i.e. as single organisms floating in their fluid environment, there are instances in which it is advantageous for bacteria to live within a biofilm. These communities or biofilms are established when groups of bacteria synthesize hydrated polymeric matrices, within which they aggregate, and which they use for adhering to living or inert surfaces (Costerton et al, 1999). Biofilm development is coordinated by QS, and QS signalling has been shown to play a key role in the development of these living structures (Davies et al, 1998). Biofilms provide protection for the bacteria in a hostile environment while retaining nutrients for their inhabitants. These well protected, sessile communities endow bacteria with protection from the immune response, and an inherent antibiotic resistance that would not be present in their planktonic form (de Kievit, 2009). This resistance is responsible for many persistent and chronic bacterial infections.
Today QS signalling and biofilm formation have been identified in a growing list of medically relevant pathogens. These include Pseudomonas aeruginosa, the cause of several types of nosocomial infection and cystic fibrosis related lung infections, Staphylococcus aureus which causes a wide range of diseases from minor skin infections to toxic shock syndrome (TSS) and enteric bacteria. The last is a group of microbes that cause gut related infections as well as other complications including urinary tract infection (UTI) and haemolytic uraemic syndrome (HUS). The severity of disease caused by these pathogens, and the economic burden associated with prevention, treatment and control of infection, have compelled scientists and clinicians to invest substantial time and effort to not only understand how these mechanisms work, but also how they can interfere with them. In this review, we discuss several of these microbes' mechanisms that link their communication to human infections with emphasis on possible strategies that can be used to target them.
Glossary
- Antibiotics
Drugs used to kill or prevent bacterial growth.
- Autoinducers
Small organic compounds used by bacteria to achieve cell-to-cell signal. They can be seen as the bacterial version of hormones.
- Biofilms
Bacterial communities encased within a polysaccharide matrix. These communities are refractory to antibiotics and anti-bacterial treatments.
- Cellular replication
Mechanism by which one cell generates two daughter cells.
- Chemokines and cytokines
Soluble proteins that regulate the immune response, by serving as, e.g. chemoattractants or activators of immune cells.
- Cystic fibrosis
Inherited disease often accompanied by high susceptibility to bacterial infections in the lung.
- Enteric bacteria
Bacteria that inhabit the gastrointestinal tract of humans.
- Exotoxins
Toxins produced by bacteria that are excreted from the bacterial cell.
- Nosocomial infections
Hospital-acquired infections.
- Proteases
Enzymes that degrade proteins.
- Quorum sensing
The term used to depict bacteria cell-to-cell communication. This term was initially coined because the first bacterial signalling systems described were associated with bacterial density.
- Toxic shock syndrome
An overwhelming immune activation response caused by a bacterium toxin that leads to shock and in many cases death.
- Type III secretion system
Specialized bacterial secretion system that is used by bacteria to inject toxins (normally referred to as effectors) into the host cell.
- Virulence traits
Specific traits expressed by bacteria that render them virulent and able to cause disease.
Pseudomonas aeruginosa
P. aeruginosa, is a Gram-negative, motile, ubiquitous bacillus found in soil, fresh and sea water. It is highly versatile, able to tolerate low oxygen conditions, grow at a wide range of temperatures (4–42 °C) and survive with minimal nutrients. It is this adaptability that allows the pathogen to adhere and survive on medical equipment and other hospital surfaces initiating outbreaks of nosocomial infections characterized by general inflammation and sepsis. These infections normally occur in compromised patients including burn victims and those with neoplasia or HIV. This opportunistic pathogen is also the major cause of chronic lung infections in cystic fibrosis patients and microbial keratitis (MK) in users of extended-wear contact lenses. Though unlikely to cross-healthy, intact anatomical barriers, Pseudomonas bacteria from poorly maintained community hot tubs and swimming pools have been linked to rashes, UTIs and external ear infections in immune-competent individuals.
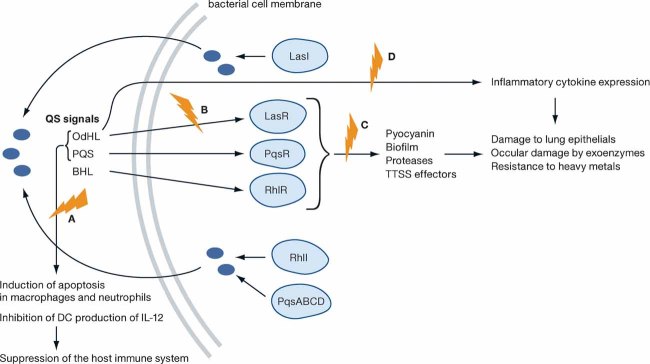
Research on P. aeruginosa has yielded much information on QS and biofilm formation. It uses several QS mechanisms to survive the harsh conditions on surfaces and within the host, as well as to circumvent the host immune system to cause disease. Figure 1 depicts several of these mechanisms. QS in P. aeruginosa depends on the release of a number of diffusible autoinducers which are divided into two groups based on their chemistry. The first group, the acyl homoserine lactones (AHLs) includes N-3-oxo-dodecanoyl-l-homoserine lactone (OdHL) and N-butanoyl-L-homoserine lactone (BHL), while the second group, the 4-quinolones (4Q) is represented by the Pseudomonas quinolone signal (PQS) (Brint & Ohman, 1995; Pearson et al, 1994, 1997; Pesci et al, 1999; Wilson et al, 1988). For these signals to be produced and sensed, the following QS systems are required: LasR–LasI and RhlR–RhlI for the AHLs, and PqsR/pqsABCDE for the PQS signal (Brint & Ohman, 1995; Farrow et al, 2008; Pearson et al, 1994; Pesci et al, 1999; Wade et al, 2005). LasI and RhlI synthesize OdHL and BHL, respectively, while the presence of pqsABCD is necessary for the synthesis of the PQS signal (Pesci et al, 1999; Wade et al, 2005). These signals bind to transcription regulators, and induce the expression of virulence genes such as exotoxins and proteases. OdHL binds to the LasR receptor, BHL binds to the RhlR receptor, and PQS to the PqsR (MvfR) LysR-like receptor (Brint & Ohman, 1995; Farrow et al, 2008; Pearson et al, 1994; Pesci et al, 1999; Wade et al, 2005). The activities of these QSIs are summarized in Fig 1 and Table 1.
Figure 1. The relationship between QS and virulence in P. aeruginosa.
The blue symbols indicate potential therapeutic targets. The green letters correspond to the targeted mechanisms in Table 1.
Table 1.
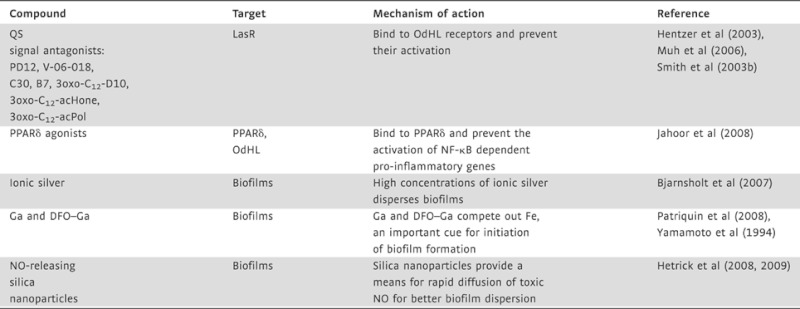
QS targeting and anti-biofilm compounds for P. aeruginosa
 |
The development of Pseudomonas biofilms relies on some of the QS signals described (Davies et al, 1998). The biofilm matrix provides a physical barrier that binds to or neutralizes host anti-microbials. Within the biofilm, Pseudomonas has a higher tolerance to heavy metals such as zinc, copper and lead than it would in its planktonic form (Whiteley et al, 2001). This tolerance prevents metal-based drugs from being able to clear chronic infections. The microenvironment in the biofilm is markedly different from that of the surrounding environment; low oxygen levels and low metabolic activity within the biofilm contribute to the disruption of the efficacy of anti-microbial drugs like tetracycline (Walters et al, 2003; Whiteley et al, 2001). Pseudomonas is capable of causing chronic (CF lung) and acute (ventilator associated/invasive) infections. Biofilms have been associated with chronic infections, while a mechanism relying on type III secretion (TSS) has been associated with acute infections. The type III secretion system (TTSS) provides bacterial pathogens, with a structure that channels effector proteins into the cytoplasm of eukaryotic host cells (Tampakaki et al, 2004). AHL-dependent QS has been reported to activate the expression of genes necessary for biofilm formation and repress genes encoding the TTSS (Bleves et al, 2005). However, a recent report by Mikkelsen et al (2009) showed that the type III effector exoenzyme S (ExoS) can also be detected in biofilm effluents but not in a planktonic cell supernatant. These results indicate that biofilms can also express the TTSS releasing effectors, in this case ExoS which is responsible for ocular damage in MK. The perceived contradictory reports by Bleeves and Mikkelsen could be reconciled by the possibility that while AHL-dependent QS may repress TTSS expression, it may not completely obliterate it, and other QS signals involved in biofilm formation could modulate the TTSS expression within biofilms.
In addition to being regulators of virulence genes and biofilm formation, the pathogen's QS signals can also modulate the host immune response. PQS and OdHL have been shown to induce apoptosis in neutrophils and macrophages during MK infections (Hoiby et al, 2001; Willcox et al, 2008; Zhu et al, 2002). Recent research into the effects of P. aeruginosa QS signals has also identified a role in the modulation of dendritic cell (DC) activity (Skindersoe et al, 2009). Skindersoe et al showed that PQS and OdHL divert TH1 cell differentiation towards TH2 cell differentiation ex vivo using cultured DCs.
Targeting QS and biofilm formation in P. aeruginosa
Conventional antibiotics work by either preventing bacterial cell division (bacteriostatic) or killing the cell (bacteridicial). However, this may increase the selective pressure towards antibiotic resistance. Targeting QS and biofilms provides an alternative that, in theory, applies a gentler evolutionary pressure towards development of drug resistance, given that QS does not control processes essential for cellular survival and/or growth. Although conventional antibiotics are used as anti-microbial drugs, some believe their function in nature at physiologically lower levels (subinhibitority anti-microbial levels) may be able to induce or interfere with QS signalling and even promote biofilm formation (Hoffman et al, 2005). Subinhibitory levels of aminoglicosides have been shown to promote biofilm development by Pseudomonas and Escherichia coli (Hoffman et al, 2005). Azithromycin (AZM) is an inhibitor of protein synthesis and is one of the few antibiotics that improves the clinical outcome of CF patients chronically infected with P. aeruginosa. Skindersoe et al showed that AZM and two other antibiotics, ceftazidime and ciprofloxacin also affect QS, possibly by altering the membrane permeability and affecting the OdHL flux. However, these antibiotics still have bacteridicial and bacteriostatic activities which may lead to the same problem of selective pressure and antibiotic resistance (Tramper-Stranders et al, 2007). These reports suggest that antibiotics may play a role in signalling in nature, as well as ‘killing’, and it will be advantageous to identify QSIs that while attenuating virulence do not affect essential bacterial processes.
Research into P. aeruginosa has provided vast information on QS and biofilms but also on potential drugs or molecules that could target these mechanisms (see Table 1). In 2006, Muh et al performed a high-throughput screening (HTS) on a library of 200,000 small compounds and identified two that were general inhibitors of QS (Muh et al, 2006). The compounds, PD12, a tetrazole with a 12-carbon alkyl tail and V-06-018, a phenyl ring with a 12-carbon alkyl tail, also inhibited the expression of the QS regulated virulence factor pyocyanin, by antagonizing LasR (the OdHL receptor). These QSIs added to the library of compounds previously identified by Smith et al (2003a, b; Suga & Smith, 2003) who showed that 2-aminocyclohexanone and 2-aminocyclopentanone are potent LasR antagonists. These QSIs and the QS signal OdHL share a chemical backbone but the QSIs have antagonistic activity on the OdHL receptor preventing the activation of downstream virulence factors. Another QSI, C-30, a derivative of a natural furanone was shown to inhibit QS and cause Pseudomonas biofilms to be susceptible to clearance by detergent and antibiotics when the biofilms were grown in its presence (Hentzer et al, 2003; Manefield et al, 2002). Although these molecules were shown to work in vitro to prevent pathogenesis, their activity in vivo in animal models remains to be addressed.
As discussed previously, QS signals can interfere with the host immune response, OdHL was shown to induce apoptosis of several cell types including macrophages and neutrophils by upregulating pro-inflammatory cytokines and chemokines (Shiner et al, 2006; Smith et al, 2001). However, how the bacterial signals were affecting the mammalian cells was unknown until 2008 when Jahoor showed that OdHL acted as an agonist to peroxisome proliferator-activated receptor β (PPARβ) and PPARδ while acting as a PPARγ antagonist. PPARγ is a trans acting repressor of the cytokine genes' transcription factor NF-κB (Pascual et al, 2005) and, by antagonizing PPARγ activity, OhDL was able to relieve the NF-κB transrepression, thus activating an inflammatory response and consequently causing tissue damage. The identification of the AHL mammalian receptors PPARβ/δ/γ also provided researchers and clinicians with potential therapeutic targets. Rosiglitazone, a PPARγ agonist was shown to block the pro-inflammatory effect of OdHL in lung epithelial cells (Jahoor et al, 2008). Identification of other PPARγ agonists may provide additional anti-inflammatory therapeutics for P. aeruginosa infections.
While targeting QS signalling is expected to have an impact on biofilm formation, several metals have direct anti-biofilm activity. Ionic silver has long been known to have anti-bacterial activity and when used in wound dressing clears planktonic P. aeruginosa infections (Melaiye et al, 2005). Chronically infected wounds, however, harbour mature biofilms that require higher silver concentrations than planktonic cells (Bjarnsholt et al, 2007). Therefore, it would be advisable that clinicians assess wounds to identify the form of colonizing Pseudomonas. If colonized by biofilms, dressing with higher ionic silver concentration should be used and chronic wounds should have their dressings frequently changed to allow for maximum silver dependent anti-microbial activity. Another metal, gallium (Ga), has also been shown to prevent the formation of biofilms that are characteristic of chronic infections (Yamamoto et al, 1994). Ga works by competing out iron (Fe) that is necessary for bacterial metabolism and also serves as a cue for biofilm formation (Banin et al, 2005, 2006). Recently, Banin et al showed that by coupling Ga to the siderophore desferrioxamine (DFO) they could form a complex (DFO–Ga) that acted like a Trojan horse (Banin et al, 2008; Patriquin et al, 2008). DFO–Ga is taken up by the bacterial cell and, once inside, it interferes with Fe metabolism and consequently biofilm formation.
Another anti-biofilm compound is nitric oxide (NO). NO is a reactive-free radical normally produced by phagocytes as part of the immune response to bacteria (Ghaffari et al, 2006). It has been shown that small molecules that release NO have anti-microbial properties and are capable of degrading biofilms (Barraud et al, 2006; De Groote & Fang, 1995). Recent studies presented a delivery system that provides a means to fine-tune the distribution of NO into biofilms (Hetrick et al, 2008, 2009). The authors use NO releasing silica nanoparticles to create a rapid diffusion that kills cells within established biofilms more effectively than a slow prolonged delivery.
Staphylococcus aureus
S. aureus is a Gram positive, coagulase and catalase positive coccus. It is responsible for a wide range of diseases from minor skin conditions like impetigo and abscesses to more serious conditions such as food poisoning, meningitis, endocarditis, pneumonia, septicaemia and TSS (Massey & Mangiafico, 1974). It is also one of the most common causes of nosocomial infections where it manifests itself as chronic wound infections. S. aureus colonizes approximately 30% of the population persistently, and close to 60% transiently. Most of the people with a healthy immune system, however, do not get infected, probably due to intact barriers that prevent invasive infection (Peterson et al, 2008). A major problem associated with S. aureus infections is the rise of methicillin resistant S. aureus (MRSA) as well as multidrug resistant strains (MDR) (Venter et al, 2004; Weinstein, 2001). New antibiotics such as linezolid and daptomycin are now being used to treat MRSA strains but some strains are already resistant to these drugs (Meka & Gold, 2004). It is therefore important to look for alternatives that would not engender the selective pressure to further increase drug resistance. One such alternative is to target QS cell-to-cell communication systems.
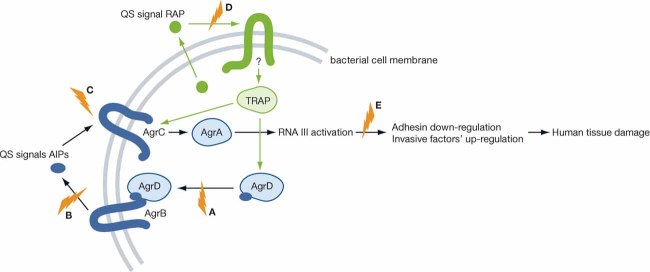
S. aureus has two phenotypes, an adhesive colonizer phenotype that is tolerated by the host, and a severe, invasive, infective phenotype that has high tissue damaging capabilities and is responsible for most of the manifestations of the disease (George & Muir, 2007). The switch in phenotypes is mediated by the agr QS system, the key regulator of gene expression in S. aureus and a vital determinant of species evolutionary diversification (Novick & Geisinger, 2008; Wright et al, 2005b). The agr QS system is depicted in Fig 2 and consists of a four gene operon (agrB, agrD, agrC, agrA) that synthesizes and secretes autoinducing cyclic thiolactone peptides (AIPs) (Novick et al, 1995). AIPs are produced and secreted by AgrBD and sensed through AgrC. AIPs bind to and activate AgrC, a membrane-bound histidine sensor kinase (the major environmental sensory system in prokaryotic cells), which in turn phosphorylates and activates AgrA, a transcriptional factor that regulates the production of the effector molecule RNAIII. RNAIII downregulates the expression of surface adhesins while upregulating the expression of invasive virulence factors such as proteases, secreted toxins and lipases (George & Muir, 2007; Peterson et al, 2008). There are four different types of AIPs with AIP1 being the most widespread signal (George & Muir, 2007; Peterson et al, 2008).
Figure 2. The relationship between QS and virulence in S. aureus.
The blue symbols indicate potential therapeutic targets. The green letters correspond to the targeted mechanisms in Table 2.
A study by Peterson et al (2008) recently identified an innate barrier that prevents the switch from the adhesive to the invasive phenotype. These authors showed that Apolipoprotein B, the major structural protein of lipoproteins, sequesters the QS signal AIP1, consequently inhibiting Agr-dependent virulence in MRSA isolates. In addition to Apolipoprotein B, another way to interfere with AIP AgrC signalling is the use of non-cognate AIPs. The four AIPs identified in S. aureus vary due to slight sequence differences, and have been shown to selectively bind to their cognate AgrC receptors with a natural QS inhibition occurring when AIPs bind to non-cognate receptors (Wright et al, 2005a). AIP4 differs from AIP1 by only a single amino acid but functions as an antagonist to AIP1 (Carmody & Otto, 2004; Chan et al, 2004). It has been proposed that identifying compounds with close sequence similarity to AIPs with antagonizing properties may provide an alternative way of treating S. aureus infections. Such compounds are summarized in Table 2.
Table 2.
QS targeting and anti-biofilm compounds for S. aureus
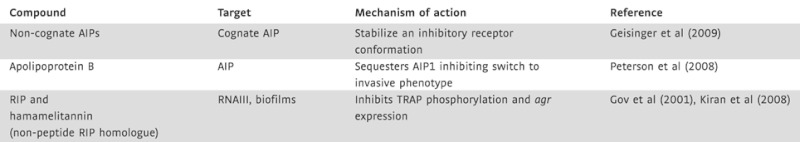 |
S. aureus survival in biofilms also depends on genes regulated by a second QS system RAP/TRAP, although its designation as a QS system is controversial (Balaban et al, 2001; Novick, 2003). The proposed autoinducer RNAIII-activating protein (RAP) is believed to be secreted and by an as yet unknown mechanism reenters the cell and activates the Target of RAP (TRAP). Activated TRAP up-regulates agr expression and promotes cellular adherence, an integral part of biofilm formation (Harraghy et al, 2007; Kiran et al, 2008; Korem et al, 2005). This system can be exploited as another alternative to antibiotics, e.g. by the use of a heptapeptide that inhibits TRAP activity (Gov et al, 2001; Kiran et al, 2008). The heptapeptide RNAIII-inhibiting peptide (RIP) has been shown to inhibit TRAP phosphorylation and agr expression. Kiran et al (2008) showed that RIP is effective against some strains of MRSA and identified a RIP non-peptide analogue, hamamelitannin, that prevents device-associated MRSA infections in a concentration-dependent manner (Kiran et al, 2008).
Interference with inter-kingdom signalling: enterohemorrhagic Escherichia coli (EHEC) O157:H7: a case study
Ingestion of food and water contaminated by EHEC leads to gastroenteritis of varying severity, and also other non-gastro related symptoms including fever, meningitis and septicaemia. EHEC is a Gram negative, rod-shaped, facultative anaerobe whose infection in humans is characterized by a usually self-resolving bloody diarrhoea (Kaper et al, 2004). It colonizes the large intestine and forms attaching and effacing (AE) lesions on intestinal epithelial cells. Most of the genes required to form this lesion are encoded on a chromosomal pathogenicity island termed the locus of enterocyte effacement, LEE. EHEC expresses Shiga toxin (Stx) in the intestine and this potent inhibitor of protein synthesis can be absorbed systemically where it binds to receptors found in the kidneys and central nervous system (CNS), causing HUS, seizures, cerebral oedema and/or coma. The genes encoding Stx are located within the late genes of a λ bacteriophage, and are transcribed only when the phage enters its lytic cycle upon induction of an SOS response in EHEC (Wagner et al, 2001). There are few, if any, good treatment options for HUS. Antibiotics and anti-motility agents are contraindicated for EHEC infections. In fact, antibiotics promote the expression and release of Stxs, thereby increasing the occurrence and severity of HUS and CNS involvement (Kimmitt et al, 1999, 2000). Consequently, innovative, cost-effective EHEC treatments are urgently needed to address a significant unmet healthcare need.
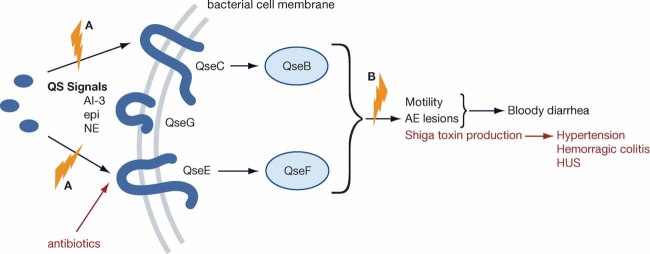
As shown in Fig 3, EHEC senses three signals to activate transcription of virulence genes: a bacterial aromatic autoinducer (AI-3) produced by the normal GI flora, and two hormones (epinephrine/norepinephrine) produced by the host (Sperandio et al, 2003). Any of these signalling molecules can trigger the QseC membrane bound sensor kinase thereby relaying the presence of these chemical signals to a complex regulatory cascade leading to transcription of key virulence genes (Clarke et al, 2006; Walters & Sperandio, 2006). QseC will also activate expression of yet a second sensor, QseE, which discriminates between AI-3 and epinephrine/NE, further fine-tuning this signalling cascade (Reading et al, 2009). These transcription events ultimately enable the organism to form AE lesions and to produce Stx, thereby leading to the clinical manifestations of infection. These observations suggest that an inhibitor of the AI-3/epi/NE-induced pathogenesis could be a valuable treatment for EHEC infections. Strictly speaking, such an inhibitor would be a ‘pathogenesis blocker’ rather than an anti-microbial, which causes bacterial stasis or death. This signalling cascade and its interference strategies are summarized in Fig 3 and Table 3.
Figure 3. The relationship between QS and virulence in EHEC.
The red shows the effect of antibiotics on virulence and disease. The blue symbols indicate potential therapeutic targets. The green letters correspond to the targeted mechanisms in Table 3.
Table 3.
QS targeting compound for EHEC
 |
This QSI is effective in animal models of infection.
Rasko et al (2008) carried out a HTS using a library of 150,000 small organic compounds to identify a lead structure (N-phenyl-4-[[(phenylamino)thioxomethyl]amino]-benzenesulfonamide) LED209 which selectively blocked binding of signals (AI-3/epinephrine and NE) to QseC, preventing QseC's autophosphorylation, and consequently inhibiting QseC-mediated activation of virulence gene expression, LED209 inhibited EHEC pathogenesis but did not block transcription generally nor kill EHEC cells. These properties are critical since cell damage could initiate Stx production. In addition, LED209 was not toxic to host cells, but inhibited expression of key virulence traits of EHEC pathogenesis (AE lesions and Stx production) (Rasko et al, 2008).
QseC homologues are present in at least 25 important human and plant pathogens (Rasko et al, 2008), and qseC mutants of EHEC (Clarke et al, 2006), Salmonella typhimurim (Spector et al, 1999) and Francisella tularensis (Wagner et al, 2007) have been shown to be attenuated in animal models of infection. These studies suggested a central role of the AI-3/epinephrine/NE QseC receptor signalling system in the virulence of several important pathogens. In agreement with these studies, LED209 also inhibited virulence of Salmonella typhimurium and F. tularensis in vitro and in vivo (animal models of infections) (Rasko et al, 2008). This study revealed that inter-kingdom cell-to-cell signalling pathways and QseC in particular provide an attractive target for novel therapeutics. Since qseC is present in many important animal and plant pathogens, drugs that target this sensor kinase have the potential to be broad spectrum. Furthermore, since the QseC-dependent inter-kingdom signalling system does not directly influence bacterial growth, inhibition of this signalling pathway may not exert strong selective pressure towards development of drug resistance. LED290 provides proof of principle that a strategy to develop drugs that obscure inter-kingdom signalling is a feasible approach towards development of novel therapeutics to combat bacterial infections.
Pending issues
Need to evaluate the efficacy of most QSIs identified so far in animal models of infection.
Detailed studies on how rapidly resistance arises to QSIs in comparison to conventional antibiotics are missing.
Critical analyses of spectrum, interference with normal microbial flora and cost/benefits of QSIs are needed.
We are still scratching the tip of the iceberg in terms of how many classes of signals bacteria use to communicate. Expanding our knowledge of the ‘bacterial language’ will also expand the repertoire of drug targets.
Conclusions
A great challenge of the 21st century is the identification of new anti-microbial targets, and/or the development of novel anti-microbial/anti-virulence drugs. We are facing a post-antibiotic era with limited capability to combat microbial infections. The rapid bacterial evolution combined with the slower process of drug development leads to an unsettling post-antibiotic era.
The development of QSIs offers a promising strategy to tackle the anti-microbial resistance issue. It is perceived that if one targets non-essential bacterial processes, which do not compromise bacterial growth and/or survival, the evolutionary pressure engendered towards development of drug resistance is milder. Bacterial virulence can be controlled by many cell-to-cell signalling systems and in this review we discussed and highlighted the QS systems more prone to drug development. The majority of these systems, specifically the ones targeting AHLs and peptides, are very specific to certain bacterial species (in some cases receptors), offering the possibility of developing highly specific ‘designer’ drugs. This could be viewed as an advantageous strategy, because one would infer that it may not interfere with the normal microbial flora of the host. However, the spectrum of such drugs would be smaller, making their development more expensive and less profitable. Some systems, such as the QseC system, are more broadly distributed and therefore inhibitors of this system have an increased spectrum but are likely to interfere with the normal microbial flora.
It is also worth noting that most QSIs are still in the basic science level of development. Although, some pharmaceutical companies have been interested in developing these drugs none are at the clinical stage. This may be due to the fact that this is a young and growing field in biology, and also that most of these inhibitors showed efficacy ‘in vitro’, but have not been successfully tested ‘in vivo’ in animal models. The one exception is LED209 (QseC inhibitor), which has been shown to work in two different animal models of infection (Rasko et al, 2008).
Although it is assumed that anti-QS drugs will engender a milder evolutionary pressure for bacterial anti-microbial resistance, resistance may eventually develop overtime. The rapid rate of bacterial evolution provides a strong driving force for a pathogen to overcome the mechanisms of action of a drug. Constant vigilance and basic research on the processes associated with bacterial virulence and inter-cellular signalling will continue to aid the development of novel and effective therapeutics. This will likely remain an ongoing ‘battle’ between microbes and their hosts in pathogenic associations. The anti-QS strategies developed so far have not yet been applied in a broad scale clinical trial and hence, it is difficult to assess their true potential and drawbacks at this stage. It is however certain that we need to expand our anti-microbial/anti-virulence targets and strategies, and interference with inter-cellular signalling appears as a viable and promising avenue to conduct drug development.
Acknowledgments
The authors declare that they have no conflict of interest.
References
- Balaban N, Goldkorn T, Gov Y, Hirshberg M, Koyfman N, Matthews HR, Nhan RT, Singh B, Uziel O. Regulation of Staphylococcus aureuspathogenesis via target of RNAIII-activating protein (TRAP) J Biol Chem. 2001;276:2658–2667. doi: 10.1074/jbc.m005446200. [DOI] [PubMed] [Google Scholar]
- Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosabiofilm formation. Proc Natl Acad Sci USA. 2005;102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E, Brady KM, Greenberg EP. Chelator-induced dispersal and killing of Pseudomonas aeruginosacells in a biofilm. Appl Environ Microbiol. 2006;72:2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E, Lozinski A, Brady KM, Berenshtein E, Butterfield PW, Moshe M, Chevion M, Greenberg EP, Banin E. The potential of desferrioxamine-gallium as an anti-Pseudomonastherapeutic agent. Proc Natl Acad Sci USA. 2008;105:16761–16766. doi: 10.1073/pnas.0808608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T, Kirketerp-Moller K, Kristiansen S, Phipps R, Nielsen AK, Jensen PO, Hoiby N, Givskov M. Silver against Pseudomonas aeruginosabiofilms. APMIS. 2007;115:921–928. doi: 10.1111/j.1600-0463.2007.apm_646.x. [DOI] [PubMed] [Google Scholar]
- Bleves S, Soscia C, Nogueira-Orlandi P, Lazdunski A, Filloux A. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosaPAO1. J Bacteriol. 2005;187:3898–3902. doi: 10.1128/JB.187.11.3898-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brint JM, Ohman DE. Synthesis of multiple exoproducts in Pseudomonas aeruginosais under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody AB, Otto M. Specificity grouping of the accessory gene regulator quorum-sensing system of Staphylococcus epidermidisis linked to infection. Arch Microbiol. 2004;181:250–253. doi: 10.1007/s00203-003-0644-2. [DOI] [PubMed] [Google Scholar]
- Chan WC, Coyle BJ, Williams P. Virulence regulation and quorum sensing in staphylococcal infections: competitive AgrC antagonists as quorum sensing inhibitors. J Med Chem. 2004;47:4633–4641. doi: 10.1021/jm0400754. [DOI] [PubMed] [Google Scholar]
- Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci USA. 2006:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- De Groote MA, Fang FC. NO inhibitions: antimicrobial properties of nitric oxide. Clin Infect Dis. 1995;21:S162–S165. doi: 10.1093/clinids/21.supplement_2.s162. [DOI] [PubMed] [Google Scholar]
- de Kievit TR. Quorum sensing in Pseudomonas aeruginosabiofilms. Environ Microbiol. 2009;11:279–288. doi: 10.1111/j.1462-2920.2008.01792.x. [DOI] [PubMed] [Google Scholar]
- Eberhard A. Inhibition and activation of bacterial luciferase synthesis. J Bacteriol. 1972;109:1101–1105. doi: 10.1128/jb.109.3.1101-1105.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Structural identification of autoinducer of Photobacterium fischeriluciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- Farrow JM, III, Sund ZM, Ellison ML, Wade DS, Coleman JP, Pesci EC. PqsE functions independently of PqsR-Pseudomonasquinolone signal and enhances the rhl quorum-sensing system. J Bacteriol. 2008;190:7043–7051. doi: 10.1128/JB.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Muir TW, Novick RP. Agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc Natl Acad Sci USA. 2009;106:1216–1221. doi: 10.1073/pnas.0807760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George EA, Muir TW. Molecular mechanisms of agr quorum sensing in virulent staphylococci. Chembiochem. 2007;8:847–855. doi: 10.1002/cbic.200700023. [DOI] [PubMed] [Google Scholar]
- Ghaffari A, Miller CC, McMullin B, Ghahary A. Potential application of gaseous nitric oxide as a topical antimicrobial agent. Nitric Oxide. 2006;14:21–29. doi: 10.1016/j.niox.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Gov Y, Bitler A, Dell'Acqua G, Torres JV, Balaban N. RNAIII inhibiting peptide (RIP), a global inhibitor of Staphylococcus aureuspathogenesis: structure and function analysis. Peptides. 2001;22:1609–1620. doi: 10.1016/s0196-9781(01)00496-x. [DOI] [PubMed] [Google Scholar]
- Harraghy N, Kerdudou S, Herrmann M. Quorum-sensing systems in staphylococci as therapeutic targets. Anal Bioanal Chem. 2007;387:437–444. doi: 10.1007/s00216-006-0860-0. [DOI] [PubMed] [Google Scholar]
- Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, et al. Attenuation of Pseudomonas aeruginosavirulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick EM, Shin JH, Stasko NA, Johnson CB, Wespe DA, Holmuhamedov E, Schoenfisch MH. Bactericidal efficacy of nitric oxide-releasing silica nanoparticles. ACS Nano. 2008;2:235–246. doi: 10.1021/nn700191f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick EM, Shin JH, Paul HS, Schoenfisch MH. Anti-biofilm efficacy of nitric oxide-releasing silica nanoparticles. Biomaterials. 2009;30:2782–2789. doi: 10.1016/j.biomaterials.2009.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- Hoiby N, Krogh Johansen H, Moser C, Song Z, Ciofu O, Kharazmi A. Pseudomonas aeruginosaand the in vitro and in vivo biofilm mode of growth. Microbes Infect. 2001;3:23–35. doi: 10.1016/s1286-4579(00)01349-6. [DOI] [PubMed] [Google Scholar]
- Jahoor A, Patel R, Bryan A, Do C, Krier J, Watters C, Wahli W, Li G, Williams SC, Rumbaugh KP. Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosaautoinducer. J Bacteriol. 2008;190:4408–4415. doi: 10.1128/JB.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kempner ES, Hanson FE. Aspects of light production by Photobacterium fischeri. J Bacteriol. 1968;95:975–979. doi: 10.1128/jb.95.3.975-979.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmitt PT, Harwood CR, Barer MR. Induction of type 2 Shiga toxin synthesis in Escherichia coliO157 by 4-quinolones. Lancet. 1999;353:1588–1589. doi: 10.1016/s0140-6736(99)00621-2. [DOI] [PubMed] [Google Scholar]
- Kimmitt PT, Harwood CR, Barer MR. Toxin gene expression by shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg Infect Dis. 2000;6:458–465. doi: 10.3201/eid0605.000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran MD, Giacometti A, Cirioni O, Balaban N. Suppression of biofilm related, device-associated infections by staphylococcal quorum sensing inhibitors. Int J Artif Organs. 2008;31:761–770. doi: 10.1177/039139880803100903. [DOI] [PubMed] [Google Scholar]
- Korem M, Gov Y, Kiran MD, Balaban N. Transcriptional profiling of target of RNAIII-activating protein, a master regulator of staphylococcal virulence. Infect Immun. 2005;73:6220–6228. doi: 10.1128/IAI.73.10.6220-6228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M, Rasmussen TB, Henzter M, Andersen JB, Steinberg P, Kjelleberg S, Givskov M. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology. 2002;148:1119–1127. doi: 10.1099/00221287-148-4-1119. [DOI] [PubMed] [Google Scholar]
- Massey ED, Mangiafico JA. Microagglutination test for detecting and measuring serum agglutinins of Francisella tularensis. Appl Microbiol. 1974;27:25–27. doi: 10.1128/am.27.1.25-27.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meka VG, Gold HS. Antimicrobial resistance to linezolid. Clin Infect Dis. 2004;39:1010–1015. doi: 10.1086/423841. [DOI] [PubMed] [Google Scholar]
- Melaiye A, Sun Z, Hindi K, Milsted A, Ely D, Reneker DH, Tessier CA, Youngs WJ. Silver(I)-imidazole cyclophane gem-diol complexes encapsulated by electrospun tecophilic nanofibers: formation of nanosilver particles and antimicrobial activity. J Am Chem Soc. 2005;127:2285–2291. doi: 10.1021/ja040226s. [DOI] [PubMed] [Google Scholar]
- Mikkelsen H, Bond NJ, Skindersoe ME, Givskov M, Lilley KS, Welch M. Biofilms and type III secretion are not mutually exclusive in Pseudomonas aeruginosa. Microbiology. 2009;155:687–698. doi: 10.1099/mic.0.025551-0. [DOI] [PubMed] [Google Scholar]
- Muh U, Schuster M, Heim R, Singh A, Olson ER, Greenberg EP. Novel Pseudomonas aeruginosaquorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob Agents Chemother. 2006;50:3674–3679. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriquin GM, Banin E, Gilmour C, Tuchman R, Greenberg EP, Poole K. Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol. 2008;190:662–671. doi: 10.1128/JB.01473-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. Structure of the autoinducer required for expression of Pseudomonas aeruginosavirulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosalas and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MM, Mack JL, Hall PR, Alsup AA, Alexander SM, Sully EK, Sawires YS, Cheung AL, Otto M, Gresham HD. Apolipoprotein B is an innate barrier against invasive Staphylococcus aureusinfection. Cell Host Microbe. 2008;4:555–566. doi: 10.1016/j.chom.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Moreira CG, Li de R, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading NC, Rasko DA, Torres AG, Sperandio V. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc Natl Acad Sci USA. 2009;106:5889–5894. doi: 10.1073/pnas.0811409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiner EK, Terentyev D, Bryan A, Sennoune S, Martinez-Zaguilan R, Li G, Gyorke S, Williams SC, Rumbaugh KP. Pseudomonas aeruginosaautoinducer modulates host cell responses through calcium signalling. Cell Microbiol. 2006;8:1601–1610. doi: 10.1111/j.1462-5822.2006.00734.x. [DOI] [PubMed] [Google Scholar]
- Skindersoe ME, Zeuthen LH, Brix S, Fink LN, Lazenby J, Whittall C, Williams P, Diggle SP, Froekiaer H, Cooley M, et al. Pseudomonas aeruginosaquorum-sensing signal molecules interfere with dendritic cell-induced T-cell proliferation. FEMS Immunol Med Microbiol. 2009;55:335–345. doi: 10.1111/j.1574-695X.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- Smith RS, Fedyk ER, Springer TA, Mukaida N, Iglewski BH, Phipps RP. IL-8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N-3-oxododecanoyl homoserine lactone is transcriptionally regulated by NF-kappa B and activator protein-2. J Immunol. 2001;167:366–374. doi: 10.4049/jimmunol.167.1.366. [DOI] [PubMed] [Google Scholar]
- Smith KM, Bu Y, Suga H. Induction and inhibition of Pseudomonas aeruginosaquorum sensing by synthetic autoinducer analogs. Chem Biol. 2003a;10:81–89. doi: 10.1016/s1074-5521(03)00002-4. [DOI] [PubMed] [Google Scholar]
- Smith KM, Bu Y, Suga H. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosaautoinducer. Chem Biol. 2003b;10:563–571. doi: 10.1016/s1074-5521(03)00107-8. [DOI] [PubMed] [Google Scholar]
- Spector MP, Garcia del Portillo F, Bearson SM, Mahmud A, Magut M, Finlay BB, Dougan G, Foster JW, Pallen MJ. The rpoS-dependent starvation-stress response locus stiA encodes a nitrate reductase (narZYWV) required for carbon-starvation-inducible thermotolerance and acid tolerance in Salmonella typhimurium. Microbiology. 1999;145:3035–3045. doi: 10.1099/00221287-145-11-3035. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens AM, Greenberg EP. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J Bacteriol. 1997;179:557–562. doi: 10.1128/jb.179.2.557-562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Smith KM. Molecular mechanisms of bacterial quorum sensing as a new drug target. Curr Opin Chem Biol. 2003;7:586–591. doi: 10.1016/j.cbpa.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Tampakaki AP, Fadouloglou VE, Gazi AD, Panopoulos NJ, Kokkinidis M. Conserved features of type III secretion. Cell Microbiol. 2004;6:805–816. doi: 10.1111/j.1462-5822.2004.00432.x. [DOI] [PubMed] [Google Scholar]
- Tramper-Stranders GA, Wolfs TF, Fleer A, Kimpen JL, van der Ent CK. Maintenance azithromycin treatment in pediatric patients with cystic fibrosis: long-term outcomes related to macrolide resistance and pulmonary function. Pediatr Infect Dis J. 2007;26:8–12. doi: 10.1097/01.inf.0000247109.44249.ac. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Wade DS, Calfee MW, Rocha ER, Ling EA, Engstrom E, Coleman JP, Pesci EC. Regulation of Pseudomonasquinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol. 2005;187:4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner PL, Neely MN, Zhang X, Acheson DW, Waldor MK, Friedman DI. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia colistrain. J Bacteriol. 2001;183:2081–2085. doi: 10.1128/JB.183.6.2081-2085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Zimmermann S, Brenner-Weiss G, Hug F, Prior B, Obst U, Hansch GM. The quorum-sensing molecule N-3-oxododecanoyl homoserine lactone (3OC12-HSL) enhances the host defence by activating human polymorphonuclear neutrophils (PMN) Anal Bioanal Chem. 2007;387:481–487. doi: 10.1007/s00216-006-0698-5. [DOI] [PubMed] [Google Scholar]
- Walters M, Sperandio V. Quorum sensing in Escherichia coliandSalmonella. Int J Med Microbiol. 2006;296:125–131. doi: 10.1016/j.ijmm.2006.01.041. [DOI] [PubMed] [Google Scholar]
- Walters MC, III, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosabiofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein RA. Controlling antimicrobial resistance in hospitals: infection control and use of antibiotics. Emerg Infect Dis. 2001;7:188–192. doi: 10.3201/eid0702.010206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosabiofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- Willcox MD, Zhu H, Conibear TC, Hume EB, Givskov M, Kjelleberg S, Rice SA. Role of quorum sensing by Pseudomonas aeruginosain microbial keratitis and cystic fibrosis. Microbiology. 2008;154:2184–2194. doi: 10.1099/mic.0.2008/019281-0. [DOI] [PubMed] [Google Scholar]
- Wilson R, Sykes DA, Watson D, Rutman A, Taylor GW, Cole PJ. Measurement of Pseudomonas aeruginosaphenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect Immun. 1988;56:2515–2517. doi: 10.1128/iai.56.9.2515-2517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JS, III, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci USA. 2005a;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JS, III, Traber KE, Corrigan R, Benson SA, Musser JM, Novick RP. The agr radiation: an early event in the evolution of staphylococci. J Bacteriol. 2005b;187:5585–5594. doi: 10.1128/JB.187.16.5585-5594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Kaneko M, Changchawalit S, Serichantalergs O, Ijuin S, Echeverria P. Actin accumulation associated with clustered and localized adherence in Escherichia coliisolated from patients with diarrhea. Infect Immun. 1994;62:2917–2929. doi: 10.1128/iai.62.7.2917-2929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Thuruthyil SJ, Willcox MD. Determination of quorum-sensing signal molecules and virulence factors of Pseudomonas aeruginosaisolates from contact lens-induced microbial keratitis. J Med Microbiol. 2002;51:1063–1070. doi: 10.1099/0022-1317-51-12-1063. [DOI] [PubMed] [Google Scholar]