Summary
Attention-deficit hyperactivity disorder (ADHD) is a highly heritable disorder of impaired behavioral inhibition, increased motor activity, and inattention. The norepinephrine transporter (NET, SLC6A2) represents an important candidate gene for contribution to ADHD because it regulates catecholamine extracellular and tissue concentrations and contributes to executive functions disrupted in ADHD, and NET is a target for most effective ADHD therapeutics. We identified four NET coding single nucleotide polymorphisms (SNPs) in two ADHD sample sets, two SNPs produce protein variants (T283M, V245I), one of which, T283M, is a novel variant. Examination of the maternal family members through whom the T283M mutation was transmitted, provided no additional ADHD diagnoses. Given the previous identification of a NET mutation that contributes to a familial tachycardia syndrome, we examined autonomic function to reveal in the proband the highest standing-induced increase in heart rate among the ADHD subjects examined. We measured [3H]NE and [3H]dopamine transport for T283M, V245I, and a previously identified NET variant, T283R. T283M and V245I demonstrated decreased substrate transport, as did T283R, suggesting that the T283 residue is sensitive to mutation. Identification of polymorphic sites within NET, specifically those that produce functional consequences, is one critical step in elucidating the genetic variation contributing to the heritable component of diseases such as ADHD.
Keywords: norepinephrine, transporter, gene, SNP, polymorphism, attention-deficit hyperactivity disorder
Introduction
Attention Deficit Hyperactivity Disorder (ADHD) is a highly heritable disorder, characterized by impaired behavioral inhibition, increased motor activity, and inattention. Studies of concordance rates for ADHD between monozygotic versus dizygotic twins confirm genetic transmission, with an overall heritability estimate of 76% (Faraone et al., 2005). Meta-analysis of genome linkage scans reveals chromosomal regions with nominal linkage signals, but no genes have yet been identified via these analyses (Zhou et al., 2008). Candidate gene studies provide another route for discerning the genes that contribute to ADHD. Because of the role of catecholamines in attention and motor control, studies have focused on genes within dopaminergic and, to a lesser extent, noradrenergic systems, including receptors, enzymes involved in synthesis, inactivation and degradation, and regulators of vesicular release (Faraone et al., 2005; Waldman and Gizer, 2006).
Abnormal regulation of norepinephrine (NE) neurotransmission is hypothesized to contribute to ADHD (Beane and Marrocco, 2004; Pliszka, 2005). The prefrontal cortex (PFC) is a key anatomical substrate for working memory processes, attention, and organization of plans for behavior (Goldman-Rakic, 1996; Robbins, 1996) and its dysfunction is implicated in ADHD (Sullivan and Brake, 2003; Weinberger et al., 1986). Both NE and dopamine (DA) play a role in mediating attention and working memory in PFC, following the Yerkes-Dodson law, with an inverted U-shaped plot of level of performance dependent on concentration, predicting that intermediate levels of catecholamine are optimal (Arnsten, 1997; Aston-Jones et al., 1999).
NET is an important candidate gene for contributing to ADHD because it is a mediator of reuptake of both NE and DA (Gresch et al., 1995; Valentini et al., 2004), and is a target for nearly all effective ADHD therapeutics. Indeed, in addition to the psychostimulants that target NET and the dopamine transporter, atomoxetine, which selectively targets NET, is also an effective medication for ADHD (Spencer et al., 2002). Initial studies of NET genetic variation and ADHD reported neither association nor preferential transmission of SNPs in intron 7 (rs3785157), exon 9 (rs5569; the coding SNP 1287 G/A), intron 9 (rs998424), or intron 13 (rs2242447) (Barr et al., 2002; De Luca et al., 2004; McEvoy et al., 2002; Retz et al., 2008). However, two studies reported association of rs3785157, rs998424 and rs2242447 with ADHD (Bobb et al., 2005; Xu et al., 2005). Several more recent reports, while not replicating these previous findings, find preferential transmission of rs3785143 (intron 1) and rs11568324 (intron 5) SNPs (Brookes et al., 2006; Kim et al., 2008; Xu et al., 2008). A recent study found association of rs3785155 with performance on the continuous performance test (CPT) (Kollins et al., 2008). Medication response has also been examined in several studies. 1287 G/A has been associated with methylphenidate response on the hyperactive-impulsive symptoms (Yang et al., 2004). Haplotypes comprising regions intron 3 through intron 5 and exons 4–9 have been associated with response to amphetamine and atomoxetine, respectively (Dlugos et al., 2007; Ramoz et al., 2009).
A complementary approach to association studies with multiple or haplotypic markers is to focus on the evaluation of genetic variation having a functional impact on NET transcription, protein expression or regulation. Approximately 20 single nucleotide polymorphisms resulting in amino acid substitutions have been identified in NET. Our group previously identified a SNP that codes for a nonfunctional NET protein variant, A457P, that contributes to a familial form of a cardiovascular disorder, orthostatic intolerance (Hahn et al., 2003; Shannon et al., 2000). Other NET protein variants also have striking functional phenotypes, such as insensitivity to protein kinase C, which normally downregulates NET activity (Hahn et al., 2005). Recently, we identified a common, functional NET SNP (rs28386840) at position −3081 upstream of the transcription initiation site that is associated with Inattentive ADHD and phenotypes in major depression (Hahn et al., 2008; Kim et al., 2006a). This finding was replicated in a larger, independent sample of ADHD parent-offspring trios and siblings demonstrating preferential transmission of the T allele to affected children with the inattentive subtype of ADHD (Gizer et al., unpublished data). The sequence in the presence of the minor T allele is an E-box motif that binds members of the SNAIL family of transcription factors, and results in decreased transcriptional activity from NET promoter-reporter constructs (Kim et al., 2006a). In the current study we examine two ADHD cohorts for NET coding SNPs in an attempt to identify functional protein variants that impact ADHD.
2.0. Methods
2.1. Subjects
All procedures involving human subject participation were approved by the Vanderbilt University Medical Center Institutional Review Board or the University of Chicago, Children's National Medical Center, and the General Clinical Research Center Advisory Council. The ascertainment of these subject sets has been described previously (Mazei-Robison et al., 2005; Stein et al., 2005). Forty-nine Vanderbilt subjects of the inattentive or combined ADHD subtype were available at the time of the present study. The subjects were collected through the Center for Child Development and Clinical Trials Center at the Vanderbilt University Medical Center. The Kiddie-SADS-Present and Lifetime Version (Kaufman et al., 1997) was administered to determine that DSM-IV criteria were met for ADHD subtype; predominantly inattentive, predominantly hyperactive-impulsive, or combined inattentive and hyperactive-impulsive (Ambrosini, 2000). History of treatment for ADHD and other major medical conditions and family histories for the presence of other psychiatric and medical conditions were collected. In some cases, the Conner's Adult ADHD rating scale (CAARS-S:L) was administered to adult relatives of children with ADHD to ascertain attention deficit status (Conners et al., 1998). For the Vanderbilt sample set, including family members assessed, buccal cells were collected in Mouthwash samples and DNA was isolated using the Puregene Cell and Tissue DNA Isolation kit (Gentra systems, Minneapolis, MN) following the manufacturer's instructions. DNA was stored at −20°C. The University of Chicago cohort consisted of a subset of children who participated in a pharmacogenetic study (the exclusion of several subjects was due to lack of DNA sample) (Stein et al., 2005), and several additional subjects, for a total of 42 subjects of either inattentive or combined ADHD subtype. Chicago subjects completed a semistructured diagnostic interview and met DSM-IV criteria for ADHD (Stein et al., 2003). For the Chicago sample, genomic DNA was extracted from whole blood using a PureGene kit (Gentra systems).
2.2. Autonomic Function Tests
Supine and standing heart rates were measured in a subset of the Vanderbilt ADHD group in the Vanderbilt Clinical Trials Center. Subjects were asked to recline quietly for 5 minutes prior to assuming a standing position for 5 minutes. Heart rate was measured at the end of each period. Adult autonomic function tests were performed at the Elliot V. Newman Clinical Research Center at Vanderbilt University. For 3 days before testing, subjects consumed a diet that contained 150 milliEquivalents (mEq) of sodium per day and 70 mEq of potassium per day. The diet was free of caffeine-containing beverages. Heart rate, blood pressure, and plasma NE, epinephrine and DA were assessed after overnight rest with subject in the supine position and again after the subject had been standing for 30 minutes. The standing test was performed to assess the hemodynamic and biochemical responses to increased central hypovolemia (accentuated by the gravitational stress). For catecholamine measurements, blood was collected in plastic syringes and immediately transferred to chilled vacuum tubes with EGTA and reduced glutathione (Amersham International PLC) and immediately put on ice. The plasma was separated by refrigerated centrifugation at −4°C and stored at −70°C until the assay. Concentrations of NE, epinephrine and DA were measured by batch alumina extraction, followed by high-performance liquid chromatography for separation with electrochemical detection and quantification.
2.2. Genetic Analysis of NET
PCR amplification of NET exons
Oligonucleotide primers (Invitrogen, Carlsbad, CA) were designed to amplify exons 1 through 15 of NET. Forward and reverse primers were designed to flank and amplify the coding region and exon/intron junctions (Table 1). Positions are based on NCBI GenBank/EMBL accession numbers X91117-X91127 and AF061198 as described in Table 1. Polymerase chain reaction (PCR) was performed using 7.5 pmol of each forward and reverse primer, 7.5 nmol dNTPs (New England Biolabs, Beverly, MA), 2.5 U Amplitaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), 0.25 U pfu Turbo DNA polymerase (Stratagene, La Jolla, CA), and 50 ng of genomic DNA using the following amplification parameters: 95°C for 5 min followed by 30 cycles of 95°C for 30 s, 61°C for 45 s, 72°C for 1 min and a final extension of 72°C for 7 min. PCR products were verified on a 1% agarose gel.
Table 1.
Primers for amplification of human NET exons and splice junctions
| Name | Sequence (5'–3') | Location | Positiona | Amplicon Size (bp) |
|---|---|---|---|---|
| Exon 1-F | aaccacctcttttccctttatcca | 5' UTR | 4662–4685 | 441 |
| Exon 1-R | tccgtgtgtattccagctcctg | Intron 1 | 389–410 | |
| Exon 2-F | gattgctgcgcgtcgcctttg | Intron 1 | 79–100 | 353 |
| Exon 2-R | ccttagatctcaccactggag | Intron 2 | 411–431 | |
| Exon 3-F | catgcgacaggtcactggtg | Intron 2 | 1–20 | 466 |
| Exon 3-R | tagtgtttggctcaggtcatac | Intron 3 | 445–466 | |
| Exon 4-F | agagtggccaggtcctgtct | Intron 3 | 20–39 | 373 |
| Exon 4-R | cttgcacttccagctccatctt | Intron 4 | 371–392 | |
| Exon 5-F | tggcttcagggccttgcctagag | Intron 4 | 1–23 | 341 |
| Exon 5-R | acaagcctggcccaaggcttggt | Intron 5 | 319–341 | |
| Exon 6-F | ctgcccatctctggttcagaccat | Intron 5 | 11–34 | 333 |
| Exon 6-R | ggagagttggcttccagaccaga | Intron 6 | 321–343 | |
| Exon 7-F | gtatccatgtggcagcaggagc | Intron 6 | 6–27 | 355 |
| Exon 7-R | cacggaagagccatgcagccaa | Intron 7 | 339–360 | |
| Exon 8-F | ctatcatgtgcagctcagaccaatgg | Intron 7 | 70–95 | 321 |
| Exon 8-R | gtctgcaatttaaatagggccttctgg | Intron 8 | 364–390 | |
| Exon 9-F | caaggcagcctacatgagtcctgg | Intron 8 | 15–38 | 449 |
| Exon 9-R | taacagggctgaatggaatcctcag | Intron 9 | 439–463 | |
| Exon 10-F | cagttcccacgtttgaccaaagagg | Intron 9 | 646–670 | 209 |
| Exon 10-R | ggtgcaggattctaggaggactgg | Intron 10 | 831–854 | |
| Exon 11-F | catcttgcctcactgccctgctct | Intron 10 | 31–54 | 283 |
| Exon 11-R | atatcctcacccagctccatcctg | Intron 11 | 290–313 | |
| Exon 12-F | ctttgctgtgatgctcacttctcttca | Intron 11 | 602–628 | 409 |
| Exon 12-R | ttgaccctagtgtctgtgtccttctg | Intron 12 | 985–1010 | |
| Exon 13-F | gctgcaggatcaaatagcaggtgg | Intron 12 | 99–122 | 269 |
| Exon 13-R | gatgtgatgtctagccttgggcagt | Intron 13 | 343–367 | |
| Exon 14-F | cctttctgtccccaccatgtcatc | Intron 13 | 616–639 | 126 |
| Exon 14-R | tgcctgtgacctggacattggcat | Intron 14 | 718–741 | |
| Exon 15-F | gagaacatcacatttacgtctactc | Intron 14 | 3492–3516 | 233 |
| Exon 15-R | gcttcagtctcacattagcgagg | 3' UTR | 3702–3724 |
Primer positions are from NCBI Genbank accession numbers X91117–X91127, or, in the case of primer Exon 1-F, accession number AF061198.
F = forward, R = reverse.
Analysis of PCR products for polymorphic sites using temperature gradient capillary electrophoresis (TGCE)
To produce heteroduplex molecules, PCR products from subject DNA were mixed 1:1 with a reference DNA PCR product of the same amplicon whose sequence matches that of a reference NET sequence (NCBI GenBank accession numbers X91117-X91127, AF061198), and underwent the following protocol: 95°C for 3 min, 95°C for 1 min, −3°C/cycle for 5 cycles, 80°C for 1 min, −1°C/cycle for 30 cycles, 50°C for 20 min, 50°C for 1 min, −1°C/cycle for 5 cycles, 45°C for 1 min, −2°C/cycle for 10 cycles. The heteroduplexed products were then diluted and subjected to TGCE analysis by the Vanderbilt Center for Molecular Neuroscience Neurogenomic Core (REVEAL™, SpectruMedix, State College, PA) (Li et al., 2002). Analysis of electropherogram peak profiles between samples and reference DNA was performed using Reveal Revelation™ software (SpectruMedix, State College, PA). Samples that deviated from the reference DNA profile were sequenced using dideoxynucleotide terminator sequencing (ABI 310, Applied Biosystems, Center for Molecular Neuroscience Neurogenomics Core, Vanderbilt University, Nashville, TN).
Genotyping of the −3081 A/T polymorphism
A restriction fragment length polymorphism assay was used to genotype the −3081 A/T SNP. Primers flanking the polymorphic site were used to amplify a fragment that was then digested with the restriction enzyme Bsr 1. The presence of the A allele results in the Bsr 1 restriction site and digestion of the PCR product into two bands that were resolved on a 3% NuSieve agarose gel (Fisher Scientific, St. Louis, MO).
2.3. Plasmid Constructs
The expression vector pcDNA3 (Invitrogen, Carlsbad, CA) containing the coding sequence for NET, pcDNA3-hNET, described previously (Hahn et al., 2005), was used in the construction of the NET nonsynonymous SNPs, V245I, T283M, and T283R. Single-point mutations were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The following primers were used: V245I forward, 5'-CTG ATG GTC GTC ATC ATC GTC TTG TAT-3', and reverse, 5'-ATA CAA GAC GAT GAT GAC GAC CAT CAG-3'; T283M forward, 5'-GTC CAT GGC GTC ATG CTG CCC GGA GCC-3' and reverse, 5'-GGC TCC GGG CAG CAT GAC GCC ATG GAC-3'; T283R forward, 5'-GTC CAT GGC GTC AGG CTG CCC GGA GCC-3' and reverse, 5'-GGC TCC GGG CAG CCT GAC GCC ATG GAC-3'. Sequences were confirmed using dideoxynucleotide terminator sequencing (ABI 310, Applied Biosystems, Center for Molecular Neuroscience Neurogenomics Core, Vanderbilt University, Nashville, TN).
2.4. Cell Culture and Transfection
All experiments were performed in transiently transfected Cath.a-differentiated (CAD) cells (American Type Culture Collection, Manassas, VA). CAD cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 8% fetal bovine serum (HyClone Laboratories, Logan, UT), L-Glutamine (2 mM) (Invitrogen), and 100 U/ml penicillin/100 μg/ml streptomycin (Invitrogen) in a humidified incubator at 37°C and 5% CO2. One day before transfection, cells were plated in individual wells of 24-well plates at a density of 5 × 104 cells/well. Transfection was performed using FuGene 6 reagent as described by the manufacturer (Roche Applied Science, Indianapolis, IN). All experimental manipulations were begun ~24 h after transfection. Observations in all experiments were made using multiple DNA stocks to control for any variability in DNA plasmid preparation.
2.5. [3H]NE and [3H]DA Transport Assays
Transport was assayed essentially as described previously (Hahn et al., 2005). In brief, cells were washed twice with Krebs-Ringer-HEPES buffer (KRH; 120 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 2.2 mM CaCl2, and 10 mM HEPES, pH 7.4) and preincubated with assay buffer (KRH, 10 mM D-glucose, 100 μM ascorbic acid, 100 μM pargyline, and 1 mM tropolone) for 10 min at 37°C, with 1 μM desipramine added to a subset of wells to assess nonspecific accumulation, followed by the addition of radiolabeled substrate for a 10-min transport assay. Saturation kinetics to determine KM and VMAX values were carried out using serial dilutions of NE or DA (100 nM to 10 μM) with [3H]NE or [3H]DA of constant specific activity DA (~36 or ~49 Ci/mmol, respectively; Amersham Biosciences AB, Uppsala, Sweden), respectively. In experiments assessing the effects of phorbol-12-myristate-13-acetate ({beta}-PMA; Sigma-Aldrich) on transport, PMA (1 nM-10 μM) was added 10 min before the addition of [3H]NE. After 10 min of uptake, cells were washed three times in KRH, incubated for 2 h in Microscint 20 scintillation fluid (PerkinElmer Life and Analytical Sciences, Boston, MA), and accumulated radiolabeled substrate was quantified in a Topcount plate scintillation counter (PerkinElmer).
2.6. Statistical Analyses
Logistic regression was used to test for effect of the 1287 G/A SNP in ADHD (SPSS v.16 for Mac). The ability of genotype to predict ADHD subtype was tested in the entire sample set and in Caucasians only. The effect of race on genotype was also tested. For transport studies, KM and VMAX values were calculated by nonlinear regression analysis according to a single-site hyperbolic model (Prism version 5; GraphPad Software Inc., San Diego, CA). KI values for the inhibition of [3H]NE uptake by β-PMA were calculated for data expressed as the percentage inhibition of total uptake versus the log of drug concentration by nonlinear regression analysis according to a single-site competition model (Prism). One-way ANOVA followed by Dunnett's post-hoc test were used.
3.0. Results
3.1. Vanderbilt and University of Chicago Sample Demographics
Two sample sets of ADHD subjects were examined in these studies, one from Vanderbilt University and one from the University of Chicago (Mazei-Robison et al., 2005; Stein et al., 2005). Forty-nine subjects from Vanderbilt and 42 subjects from the University of Chicago were included in the present study. Both populations showed a predominance of male subjects, with 84 and 74% males in the Vanderbilt and Chicago cohorts, respectively. This is consistent with the reported higher incidence of ADHD in males. We examined subjects with either the combined or inattentive subtype of ADHD. Subtype diagnosis was 76% combined and 24% inattentive for the Vanderbilt and 57% combined and 31% inattentive (5 patients were lacking subtype designation) for the Chicago samples, consistent with literature that the combined subtype is the most prevalent. Both samples were also predominantly Caucasian, with 84 and 76% for the Vanderbilt and Chicago group, respectively. We combined the Vanderbilt and Chicago samples for analyses, thus producing similar demographics; the combined sample set was 79% male, 87% white and 67% combined subtype (Table 2).
Table 2.
NET 1287 G/A genotypes in Vanderbilt and University of Chicago ADHD subjects
| Genotype | Subjects | Gender | Race/Ethnicity | ADHD Diagnosis | ||||
|---|---|---|---|---|---|---|---|---|
| Male | Female | Caucasian | A.A. | Other | Inattentive | Combined | ||
| Total | 91 | 72 (79%) | 19 (21%) | 73 (80%) | 13 (14%) | 2 (2%) | 25 (27%) | 61 (67%) |
| GG | 58 (64%) | 43 (60%) | 15 (79%) | 42 (58%) | 12 (92%) | 1 (50%) | 17 (68%) | 36 (59%) |
| GA | 26 (29%) | 12 (32%) | 14 (16%) | 25 (34%) | 1 (8%) | 0 | 5 (20%) | 21 (34%) |
| AA | 7 (8%) | 6 (8%) | 1 (5%) | 6 (8%) | 0 | 1 (50%) | 3 (12%) | 4 (7%) |
A.A., African can American. Three subjects had unknown ethnicity and five subjects lacked ADHD subtype diagnosis.
3.2. Vanderbilt and University of Chicago Polymorphism Discovery
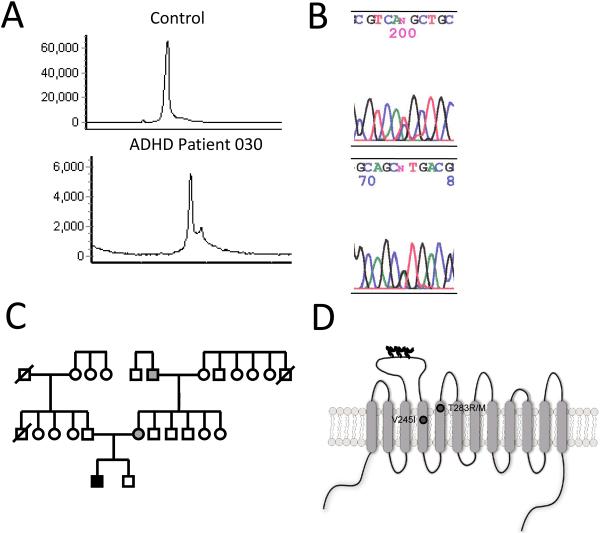
SNP discovery was performed on DNA isolated from buccal cells using temperature gradient capillary electrophoresis (TGCE) followed by dideoxynucleotide sequencing to determine the identity of polymorphisms. Primers juxtaposed to the exon-intron boundaries were used to identify coding polymorphisms (Table 1). Four coding region SNPs were identified in the samples (Table 3). Two were synonymous (rs1805068, rs5569) and two were nonsynonymous (rs1805066, unassigned rs number). Three have been observed in the NET gene previously, whereas one is a novel variant. The novel SNP is nonsynonymous, generating the protein variant T283M, present as a heterozygote present in 1 of 91 subjects. This is the first identification of this variant in an individual. The SNP was identified in exon 5 of the gene by observation of an additional peak on the TCGE chromatogram and sequencing revealed a C to T substitution the converts an ACG to an ATG codon (Fig. 1A, B). T283M is located in the top of TM5. Rs1805066, or V245I, was present as a heterozygote, in 1 of our 91 samples, for an allele frequency less than 1%, similar to what has been reported for this variant (Stöber et al., 1996). V245I is located in TM4 of NET. Rs1805068 was present in 4 of the subjects, all heterozygote, for an allele frequency of 2.2%, similar to that reported (NCBI dbSNP). Of these polymorphisms, rs5569 had a minor allele frequency sufficient to allow statistical analyses (Table 2, 3). Thus, we examined 1287 G/A (rs5569) for association with ADHD subtype in the combined sample set. Logistic regression analysis revealed no association of genotype with combined versus inattentive ADHD. We also tested if genotype was influenced by race, as SNPs in the NET gene, including 1287 G/A, demonstrate ethnic heterogeneity. Genotype differed by race (p < 0.05). Thus, we reexamined the genotype-subtype interaction in the Caucasian only subgroup, with no significant effects.
Table 3.
Coding Single Nucleotide Polymorphisms in NET
| rs number | Exon | Positiona | Nucleotide substitution | Amino acid | Minor Allele Frequency | |
|---|---|---|---|---|---|---|
| wt | variant | |||||
| rs1805066 | Exon4 | 733 | gtc - atc | Val 245 | Ile | 1/182 (0.005) |
| - | Exon 5 | 848 | acg - atg | Thr283 | Met | 1/182 (0.005) |
| rs1805068 | Exon 6 | 955 | ttg - ctg | Leu 319 | 4/182 (0.022) | |
| rs5569 | Exon 9 | 1287 | acg - aca | Thr429 | 40/182 (0.22) |
Nucleotide positions are designated from the coding sequence of hNET, with the first coding nucleotide designated as 1.
Figure 1.
Identification of the T283M coding variant in an ADHD proband. Electropherograms from TCGE from control (upper panel) and subject (lower panel) (A). Chromatogram of dideoxynucleotide sequencing of T283M mutation using forward (upper panel) and reverse primers (lower panel) (B). Pedigree of the T283M proband (C). The black box indicates the proband carrying the T283M heterozygous variant and diagnosed with ADHD. Grey boxes represent family members carrying T283M but unaffected by ADHD. Schematic illustration of NET indicating the positions of T283M protein variant in TM5 (D). V245I, in TM4, is also represented.
3.3. T283M Proband and Family Characterization
We identified a novel, nonsynonymous SNP that results in the T283M variant. The T283M subject is diagnosed with inattentive ADHD and was effectively treated with Adderall™ at the time of the study. Medical and psychiatric histories and DNA samples were collected from family members of this subject. DNA sequencing revealed that the T283M mutation was inherited through the maternal side, with the mother and maternal grandfather heterozygous carriers of the mutant allele (Fig. 1C). Although self-report indicated that the maternal grandfather experienced depression and the mother was a poor student, no family members met DSM-IV criteria for psychopathology or were diagnosed with ADHD based on the Conner's Adult ADHD rating scale (CAARS-S:L).
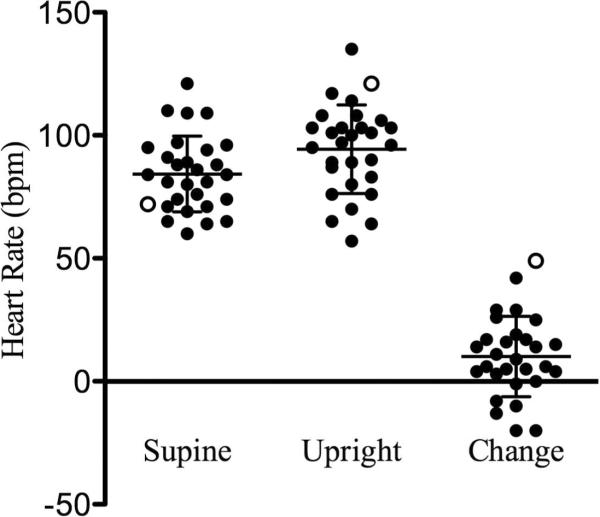
We have previously identified and characterized a NET mutation, A457P, that associates with a familial form of orthostatic intolerance, characterized by an abnormally elevated and sustained heart rate displayed upon standing (Hahn et al., 2003; Shannon et al., 2000). In order to assess the influence of T283M on tachycardia, we collected supine and standing heart rates following five minutes of standing in a subset of our samples in the Vanderbilt group. Group mean heart rates for supine, standing, and change upon standing were 84.3+15.4, 94.4+18.0 and 10.1+16.4 bpm, respectively (Fig. 2). The T283M proband demonstrated an increase in heart rate upon standing of 49 bpm (Fig. 2). This change in heart rate was the highest of all subjects and was greater than two standard deviations from the mean. We also administered autonomic function tests to the mother of the proband. We measured heart rate and plasma norepinephrine levels following 30 minutes of upright posture. In adults, the diagnostic criteria for orthostatic intolerance used at Vanderbilt are a standing heart rate change of greater than 30 bpm and a plasma NE level of greater than 600 pg/ml. The heart rate of the mother increased 26 bpm, whereas her plasma NE levels were elevated to 595 pg/ml, very close to the cutoff for orthostatic intolerance diagnosis. Epinephrine levels were 91 pg/ml. Our group also recently identified a functional promoter SNP, −3081 A/T that associates with ADHD. The T283M proband and family members were assessed for this SNP. All maternal family members were homozygous for the, T, minor allele with the exception of the sibling of the T283M proband, who was heterozygous for this polymorphism (Table 4).
Figure 2.
Heart rate measurements in T283M proband and ADHD children. Heart rates were measured following 5 min supine posture, and 5 min standing, and the change in heart rate was calculated. Individual data points are shown and the mean ± S.D (N = 29) is given for each measure. T283M proband is represented by the open circle. Bpm, beats per minute.
Table 4.
Genotype of T283M Proband Pedigree
| T283M | 1287 G/A | −3081 A/T | |
|---|---|---|---|
| Affected Proband | T283M het. | GA | TT |
| Brother | T283 | GA | TA |
| Mother | T283M het. | GA | TT |
| Maternal Uncle | T283 | AA | TT |
| Maternal Uncle | T283 | AA | TT |
| Maternal Aunt | T283 | AA | TT |
| Maternal Aunt | T283 | AA | TT |
| Maternal Grandfather | T283M het. | GA | TT |
| Maternal Grandmother | T283 | AA | TT |
Het, heterozygous
3.4. Functional Properties of NET Protein Variants Identified in ADHD
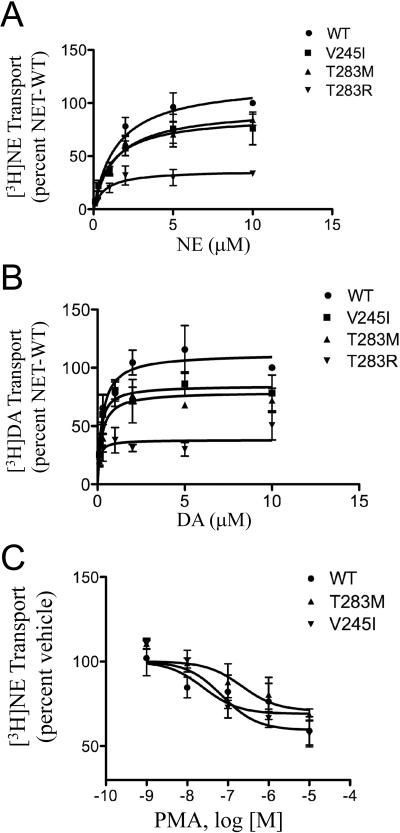
The T283M and V245I mutations were engineered into a plasmid vector and transfected in CAD cells to examine functional properties. Additionally, we generated and characterized T283R, a NET SNP deposited in dbSNP and as yet uncharacterized. We performed transport assays, generating saturation curves with VMAX and KM values for both NE and DA uptake (Fig. 3A,B, Table 5). Significant changes were observed for NE transport (F(3,8) = 17.8; p < 0.001). T283R was significantly different from wild-type NET (WT) on post-hoc testing, p < 0.05. Significant changes were also observed for DA transport (F(3,8) = 20.9, p < 0.001. The VMAX for DA transport for V245I, T283M and T283R all differed from WT. There were no significant changes in KM, despite an apparent 2–3-fold decrease in KM values for T283R (Table 5).
Figure 3.
Functional analysis of NET protein variants. Saturation kinetics of [3H]NE (A) and [3H]DA (B) uptake of NET and variant NETs. CAD cells were transiently transfected with NET or variant cDNAs. Twenty-four hours later, saturation transport assays were performed as described in Materials and Methods. Transport was determined by incubating cells for 10 min with 100 nM to 10 μM [3H]NE or [3H]DA, and nonspecific activity was defined by 1 μM desipramine at each concentration. Data are expressed as the percentage of NET transport at 10 μM concentration of substrate and are the mean ± S.E.M. values of three experiments. KM and VMAX values were determined using Prism as described in Materials and Methods. Data were analyzed using one-way ANOVA followed by Dunnett's test. Effects of μ-PMA treatment on [3H]NE transport by NET and variants (C). CAD cells were transfected with NET or variant cDNAs. Twenty-four hours later, μ-PMA treatment was performed as described in Materials and Methods. Cells were treated for 10 min with 1 nM-10 μM β–-PMA followed by a 10 min incubation with 50 nM [3H]NE. Data points are mean ± S.E.M. of percent inhibition of total uptake (vehicle) for each NET construct versus the log of drug concentration for three experiments. KI values for the inhibition of [3H]NE uptake by β-PMA were calculated with nonlinear regression analysis according to a single-site competition model (Prism). One-way ANOVA followed by Dunnett's post-hoc test were used.
Table 5.
Transport Kinetics of NET Variants
| hNET Variant | [3H]NE Transport | [3H]DA Transport | ||
|---|---|---|---|---|
| KM(μM) | VMAX(%NET) | KM (μM) | VMAX(%NET) | |
| hNET | 1.5 ± 0.4 | 100.0 ± 0.0 | 0.3 ± 0.1 | 100.0 ± 0.0 |
| V245I | 1.1 ± 0.4 | 76.2 ± 15.4 | 0.2 ± 0.1 | 78.2 ± 15.6* |
| T283M | 1.4 ± 0.4 | 84.3 ± 5.2 | 0.2 ± 0.1 | 72.1 ± 10.3* |
| T283R | 0.7 ± 0.4 | 33.6 ± 3.6* | 0.09 ± 0.07 | 50.6 ± 17.1* |
, significantly different from NET, p < 0.05.
Stimulation of PKC via phorbol ester treatment or receptor-mediated activation is well-described to down-regulate NET in cell culture and native tissue (Apparsundaram et al., 1998a; Apparsundaram et al., 1998b; Bauman et al., 2000). Additionally, we have identified NET protein variants that have altered sensitivity to phorbol ester-induced NET down-regulation (Hahn et al., 2005). Thus, we next measured the response of these variants to PKC-mediated down-regulation, induced by phorbol-12-myristate-13-acetate ({beta}-PMA).
Transfected CAD cells were treated for 10 min with 1–10 μM β-PMA followed by a 10 min incubation with [3H]NE. WT NET demonstrated a 30% decrease in [3H]NE transport consistent with previous observations (Fig. 3C) (Hahn et al., 2005). The NET variants showed similar reductions, in the range of 30–40% (F(3,8) = 0.898). The IC50 values for the variants were 2.5 × 10−8, 2.2 × 10−7, and 7.2 × 10−8 for WT, T283M, and V245I, respectively. Despite a 10-fold shift to the right in the IC50 of T283M for the effect of β-PMA, this did not reach significance.
4.0. Discussion
The current study examined coding variation in two sample sets of ADHD patients to identify novel, functional variants that may contribute to ADHD. Examining the phenotypes of individuals and families that bear such mutations can provide invaluable knowledge regarding the physiological processes mediated by NET and the consequences of disrupting such control. Indeed, we previously identified in a family a rare yet functional NET variant that correlated with orthostatic tachycardia and elevated plasma norepinephrine levels (Hahn et al., 2003; Shannon et al., 2000). Such rare variants with a prominent effect on function of that gene product may in fact, in combination with other variants of large effect, be responsible for complex traits. Indeed, it has been proposed that a summation of rare variants with low frequency but high penetrance are responsible for common, complex traits, as opposed to the common variant-common disease model (Bodmer and Bonilla, 2008). Indeed, candidate gene approaches in the study of phenotypes such as lipid metabolism have revealed rare variants with moderate to large effects that contribute to those traits (Cohen et al., 2004). A similar point is made by the observation that genome-wide association studies fail to detect most of the heritable factors contributing to complex traits in part because they do not capture information about rare variants (Frazer et al., 2009).
Previous work has demonstrated association of 1287 G/A (rs5569) with ADHD or response to medication. The exon 9 SNP has been associated with methylphenidate response on the hyperactive-impulsive symptoms (Yang, 2004). Interestingly, the G/G genotype was also reported to be associated with response to the NET blocker nortriptyline in depressed patients (Kim et al., 2006b). A number of studies of ADHD and NET association were restricted to the combined subtype or did not perform subtype by genotype analyses. Our previous work indicates an association of −3081 A/T with the inattentive subtype in the Vanderbilt ADHD group and in a large set of ADHD families. Thus, we examined the frequency of 1287 G/A in inattentive versus combined subjects in our sample. It is important to consider that synonymous coding SNPs may be functional through such mechanisms as influencing protein folding during translation (Kimchi-Sarfaty et al., 2007). We did not observe an association of this SNP with ADHD in the current study, indicating that even when accounting for subtype, there is no association of this SNP with ADHD. However, our small sample size may preclude us from observing effects.
The TCGE SNP discovery analysis identified a novel protein variant, T283M, in an ADHD subject and family members. The variant was present in the maternal grandfather and mother, although these family members did not meet criteria for ADHD. The NET variant A457P that contributes to a familial form of tachycardia also diminishes transport, albeit in that case nearly completely. Thus, we reasoned that T283M might also produce a cardiovascular phenotype. The ADHD proband demonstrated a standing-induced heart rate elevation that was higher than all other subjects for which heart rate was measured. Autonomic functions tests demonstrated that the mother had high levels of standing-induced plasma NE. These initial observations suggest cardiovascular phenotypes are present in the T283M family, and analysis of family members under more extensive testing conditions is warranted. Other NET variants identified in cardiovascular phenotypes also demonstrate functional impact, including altered expression, transport and regulation by signal transduction pathways (Hahn et al., 2005). These findings may have relevance for cardiac side-effects of medications used to treat ADHD. Cardiovascular side-effects of stimulant medications, as well as atomoxetine, have been reported, and although the clinical relevance remains controversial, studies show that a small percentage of ADHD patients demonstrate changes in heart rate and blood pressure that are clinically significant (Vitiello, 2008). Thus, genetic variation in genes such as NET, that contribute to both central mechanisms of attention and motor activity as well as cardiovascular control, may contribute to increased sensitivity to the cardiovascular side-effects of medication in a subset of patients.
Expression of T283M in cell culture demonstrated decreased transport activity. Our discovery of T283M in an ADHD sample prompted us to examine the function of the T283R variant, demonstrating a decrease in transport of both NE and DA. T283R is a variant identified by the Pharmacogenomics Knowledge Base Consortium at a frequency of 0.42% and was present in an Affymetrix screen. Thus, while falling into the rare mutation category, this variant has been found in more than one individual or family. The sensitivity of T283, which is located in TM5, to both substitutions of methionine and arginine on transport suggest that this residue lies in a region of the transporter important to its function. A high resolution structure of a SLC6 bacterial family member, the leucine transporter from Aquifex aeolicus (LeuTAa), was recently achieved (Yamashita, 2005). Whereas TMs 1, 3, 6 and 8 contain residues that are critical to the permeation pathway, TM5, with TM4, forms one of two V-like structures that may act as pincers to facilitate conformational change to open and close a gating mechanism to allow neurotransmitter movement through the pore (Henry et al., 2006).
The proband, mother and maternal grandfather who carry the T283M allele were all homozygous for the decreased function T allele of the NET −3081 A/T polymorphism (Kim et al., 2006a). Thus, in addition to a decrease in transporter due to the presence of a protein variant, these individuals are homozygous for a common polymorphism that decreases NET gene transcription. The sibling of the proband who is not diagnosed with ADHD both lacks the T283M variant and is heterozygous for the hypofunctioning −3081 allele. The pedigree shows that the T283M variant is carried on the T allele. This suggests that to the extent that other individuals in the population are identified who harbor this mutation, it will be on the background of this lower functioning NET allele. The presence of multiple, functional NET SNPs in an individual would presumably magnify the consequences to altered NET capacity and noradrenergic system signaling.
The NET variant, V245I, was present in our ADHD sample and this NET variant decreased substrate transport. Stöber et al. first identified this SNP in 1996 in the first paper to describe NET protein variants, with a reported frequency of 0.43% (Runkel et al., 2000; Stöber et al., 1996). Consistent with the present findings, characterization of V245I by this group revealed a 50% reduction in VMAX compared to WT(Runkel et al., 2000). This substitution may appear a conservative one; however, variants identified in the serotonin transporter that substitute either an isoleucine or leucine for the valine at amino acid 425 result in striking altered transporter velocity and kinase/phosphatase regulation and are associated with autism-spectrum disorders (Ozaki et al., 2003; Prasad et al., 2009).
V245I has been validated by several other large-scale genome SNP discovery efforts. Although not a common SNP by standard definitions, it is present in a proportion of the population, i.e., not a single, familial occurrence. One of the replicated findings for associations of a NET SNP with ADHD is that of rs11568324, located in intron 5. Two groups reported its association, along with rs3785143, with ADHD (Brookes et al., 2006; Kim et al., 2008). A third replication of the finding for rs11568324 was published, when combining the new subject pool with those of the previous studies (Xu et al., 2008). V245I, is located approximately 7000 bp from rs11568324 and HapMap data report a D' value of 1.0 for this SNP pair, suggesting that there is high linkage disequilibrium (LD) between the two SNPs. If it should be verified that these 2 SNPs are in high LD, it is possible that V245I is the functional SNP that drives the association in these ADHD populations. Although the scope of the current work was to identify and characterize coding variation in NET, even if those variants occur at a low frequency, a much larger sample set could now be used to assess the association of V245I with ADHD. Assembly of a catalogue of functional NET SNPs, both common and rare, will be the first important step in studies that sequence multiple variants in candidate genes for their contribution to complex traits and disease.
The variants characterized in the present study would presumably enhance extracellular NE and DA levels in cortical brain regions. The influence of catecholamine levels on attentional processes and ADHD is complex, working through multiple dopaminergic and noradrenergic receptor subtypes on different target neurons in cortex. The effects of NE and DA on attention and WM follow the Yerkes-Dodson law, an inverted U-shaped plot of level of performance dependent on concentration, predicting that intermediate levels of catecholamine are optimal (Arnsten, 1997; Aston-Jones et al., 1999). Indeed, alpha-2 agonists improve working memory in nonhuman primates and alpha-1 agonists impair working memory in monkeys, rats and mice (Arnsten et al., 1999; Franowicz et al., 2002) and D1 agonists can alternately disrupt or enhance performance, depending on prefrontal cortex DA levels at the time of drug administration (Floresco and Phillips, 2001; Phillips et al., 2004). Additionally, stress disrupts working memory (Arnsten, 2004), and may be due to elevating the levels of catecholamine into the descending portion of the inverted-U curve of function. Thus, the influence of hypofunctioning transporter variants on ADHD (and other disorders) must be considered in the face of this complex physiology.
Acknowledgments
The authors thank the families who generously volunteered to participate in these studies. We thank Denise Malone and Tracy-Jarrett Moore of the Vanderbilt Center for Molecular Neuroscience Neurogenomics Core for Reveal and sequencing. We also thank Jill Janssen, Elizabeth Roof, and Geri Rice of the Clinical Trials Center at Vanderbilt for recruiting and ascertaining subjects and Michelle-Mazei Robison for Vanderbilt DNA isolation. We thank David Robertson and the Vanderbilt Clinical Research Center for providing autonomic function testing. We would like to acknowledge Randy Blakely in whose lab these studies were initiated. This work was supported by NIH awards K01 MH076018-03 (M.K.H.) PO1 HL56693 (D. Robertson), MO1 RR-00095 to the General Clinical Research Center at Vanderbilt, K24MHO1823 (M.A.S.) and MO1-RR13297 from the General Clinical Research Center Program of the National Center for Research Resources (M.A.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambrosini PJ. Historical development and present status of the schedule for affective disorders and schizophrenia for school-age children (K-SADS) J Am Acad Child Adolesc Psychiatry. 2000;39:49–58. doi: 10.1097/00004583-200001000-00016. [DOI] [PubMed] [Google Scholar]
- Apparsundaram S, Galli A, DeFelice LJ, Hartzell HC, Blakely RD. Acute regulation of norepinephrine transport: I. protein kinase C- linked muscarinic receptors influence transport capacity and transporter density in SK-N-SH cells. J Pharmacol Exp Ther. 1998a;287:733–743. [PubMed] [Google Scholar]
- Apparsundaram S, Schroeter S, Giovanetti E, Blakely RD. Acute regulation of norepinephrine transport: II. PKC-modulated surface expression of human norepinephrine transporter proteins. J Pharmacol Exp Ther. 1998b;287:744–751. [PubMed] [Google Scholar]
- Arnsten AF. Catecholamine regulation of the prefrontal cortex. J Psychopharmacol. 1997;11:151–162. doi: 10.1177/026988119701100208. [DOI] [PubMed] [Google Scholar]
- Arnsten AF. Adrenergic targets for the treatment of cognitive deficits in schizophrenia. Psychopharmacology (Berl) 2004;174:25–31. doi: 10.1007/s00213-003-1724-3. Epub 2003 Dec 2019. [DOI] [PubMed] [Google Scholar]
- Arnsten AF, Mathew R, Ubriani R, Taylor JR, Li BM. Alpha-1 noradrenergic receptor stimulation impairs prefrontal cortical cognitive function. Biol Psychiatry. 1999;45:26–31. doi: 10.1016/s0006-3223(98)00296-0. [DOI] [PubMed] [Google Scholar]
- Aston-Jones G, Rajkowski J, Cohen J. Role of locus coeruleus in attention and behavioral flexibility. Biol Psychiatry. 1999;46:1309–1320. doi: 10.1016/s0006-3223(99)00140-7. [DOI] [PubMed] [Google Scholar]
- Barr CL, Kroft J, Feng Y, Wigg K, Roberts W, Malone M, Ickowicz A, Schachar R, Tannock R, Kennedy JL. The norepinephrine transporter gene and attention-deficit hyperactivity disorder. Am J Med Genet. 2002;114:255–259. doi: 10.1002/ajmg.10193. [DOI] [PubMed] [Google Scholar]
- Bauman AL, Apparsundaram S, Ramamoorthy S, Wadzinski BE, Vaughan RA, Blakely RD. Cocaine and antidepressant-sensitive biogenic amine transporters exist in regulated complexes with protein phosphatase 2A. J Neurosci. 2000;20:7571–7578. doi: 10.1523/JNEUROSCI.20-20-07571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane M, Marrocco RT. Norepinephrine and acetylcholine mediation of the components of reflexive attention: implications for attention deficit disorders. Prog Neurobiol. 2004;74:167–181. doi: 10.1016/j.pneurobio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Bobb AJ, Addington AM, Sidransky E, Gornick MC, Lerch JP, Greenstein DK, Clasen LS, Sharp WS, Inoff-Germain G, Wavrant-De Vrieze F, Arcos-Burgos M, Straub RE, Hardy JA, Castellanos FX, Rapoport JL. Support for association between ADHD and two candidate genes: NET1 and DRD1. Am J Med Genet B Neuropsychiatr Genet. 2005;134:67–72. doi: 10.1002/ajmg.b.30142. [DOI] [PubMed] [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes K, Xu X, Chen W, Zhou K, Neale B, Lowe N, Anney R, Franke B, Gill M, Ebstein R, Buitelaar J, Sham P, Campbell D, Knight J, Andreou P, Altink M, Arnold R, Boer F, Buschgens C, Butler L, Christiansen H, Feldman L, Fleischman K, Fliers E, Howe-Forbes R, Goldfarb A, Heise A, Gabriels I, Korn-Lubetzki I, Johansson L, Marco R, Medad S, Minderaa R, Mulas F, Muller U, Mulligan A, Rabin K, Rommelse N, Sethna V, Sorohan J, Uebel H, Psychogiou L, Weeks A, Barrett R, Craig I, Banaschewski T, Sonuga-Barke E, Eisenberg J, Kuntsi J, Manor I, McGuffin P, Miranda A, Oades RD, Plomin R, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Taylor E, Thompson M, Faraone SV, Asherson P. The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol Psychiatry. 2006;11:934–953. doi: 10.1038/sj.mp.4001869. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Conners C, Erhardt D, Sparrow E. Connor's Adult ADHD Rating Scales (CAARS) NCS Pearson Inc.; Minneapolis: 1998. [Google Scholar]
- De Luca V, Muglia P, Jain U, Kennedy JL. No evidence of linkage or association between the norepinephrine transporter (NET) gene MnlI polymorphism and adult ADHD. Am J Med Genet. 2004;124B:38–40. doi: 10.1002/ajmg.b.20075. [DOI] [PubMed] [Google Scholar]
- Dlugos A, Freitag C, Hohoff C, McDonald J, Cook EH, Deckert J, de Wit H. Norepinephrine transporter gene variation modulates acute response to D-amphetamine. Biol Psychiatry. 2007;61:1296–1305. doi: 10.1016/j.biopsych.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, Sklar P. Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Floresco SB, Phillips AG. Delay-dependent modulation of memory retrieval by infusion of a dopamine D1 agonist into the rat medial prefrontal cortex. Behav Neurosci. 2001;115:934–939. [PubMed] [Google Scholar]
- Franowicz JS, Kessler LE, Borja CM, Kobilka BK, Limbird LE, Arnsten AF. Mutation of the alpha2A-adrenoceptor impairs working memory performance and annuls cognitive enhancement by guanfacine. J Neurosci. 2002;22:8771–8777. doi: 10.1523/JNEUROSCI.22-19-08771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Regional and cellular fractionation of working memory. Proc Natl Acad Sci U S A. 1996;93:13473–13480. doi: 10.1073/pnas.93.24.13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresch PJ, Sved AF, Zigmond MJ, Finlay JM. Local influence of endogenous norepinephrine on extracellular dopamine in rat medial prefrontal cortex. J Neurochem. 1995;65:111–116. doi: 10.1046/j.1471-4159.1995.65010111.x. [DOI] [PubMed] [Google Scholar]
- Hahn MK, Blackford JU, Haman K, Mazei-Robison M, English BA, Prasad HC, Steele A, Hazelwood L, Fentress HM, Myers R, Blakely RD, Sanders-Bush E, Shelton R. Multivariate permutation analysis associates multiple polymorphisms with subphenotypes of major depression. Genes Brain Behav. 2008;7:487–495. doi: 10.1111/j.1601-183X.2007.00384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MK, Mazei-Robison MS, Blakely RD. Single nucleotide polymorphisms in the human norepinephrine transporter gene affect expression, trafficking, antidepressant interaction, and protein kinase C regulation. Mol Pharmacol. 2005;68:457–466. doi: 10.1124/mol.105.011270. [DOI] [PubMed] [Google Scholar]
- Hahn MK, Robertson D, Blakely RD. A mutation in the human norepinephrine transporter gene (SLC6A2) associated with orthostatic intolerance disrupts surface expression of mutant and wild-type transporters. J Neurosci. 2003;23:4470–4478. doi: 10.1523/JNEUROSCI.23-11-04470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry LK, Defelice LJ, Blakely RD. Getting the message across: a recent transporter structure shows the way. Neuron. 2006;49:791–796. doi: 10.1016/j.neuron.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Birmaher B, Brent D, Rao U, Flynn C, Moreci P, Williamson D, Ryan N. Schedule for Affective Disorders and Schizophrenia for School-Age Children-Present and Lifetime Version (K-SADS-PL): initial reliability and validity data. J Am Acad Child Adolesc Psychiatry. 1997;36:980–988. doi: 10.1097/00004583-199707000-00021. [DOI] [PubMed] [Google Scholar]
- Kim CH, Hahn MK, Joung Y, Anderson SL, Steele AH, Mazei-Robinson MS, Gizer I, Teicher MH, Cohen BM, Robertson D, Waldman ID, Blakely RD, Kim KS. A polymorphism in the norepinephrine transporter gene alters promoter activity and is associated with attention-deficit hyperactivity disorder. Proc Natl Acad Sci U S A. 2006a;50:19164–19169. doi: 10.1073/pnas.0510836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lim SW, Kim S, Kim JW, Chang YH, Carroll BJ, Kim DK. Monoamine transporter gene polymorphisms and antidepressant response in koreans with late-life depression. JAMA. 2006b;296:1609–1618. doi: 10.1001/jama.296.13.1609. [DOI] [PubMed] [Google Scholar]
- Kim JW, Biederman J, McGrath CL, Doyle AE, Mick E, Fagerness J, Purcell S, Smoller JW, Sklar P, Faraone SV. Further evidence of association between two NET single-nucleotide polymorphisms with ADHD. Mol Psychiatry. 2008;13:624–630. doi: 10.1038/sj.mp.4002090. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kollins SH, Anastopoulos AD, Lachiewicz AM, FitzGerald D, Morrissey-Kane E, Garrett ME, Keatts SL, Ashley-Koch AE. SNPs in dopamine D2 receptor gene (DRD2) and norepinephrine transporter gene (NET) are associated with continuous performance task (CPT) phenotypes in ADHD children and their families. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1580–1588. doi: 10.1002/ajmg.b.30876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Liu Z, Monroe H, Culiat CT. Integrated platform for detection of DNA sequence variants using capillary array electrophoresis. Electrophoresis. 2002;23:1499–1511. doi: 10.1002/1522-2683(200205)23:10<1499::AID-ELPS1499>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Mazei-Robison MS, Couch RS, Shelton RC, Stein MA, Blakely RD. Sequence variation in the human dopamine transporter gene in children with attention deficit hyperactivity disorder. Neuropharmacology. 2005;49:724–736. doi: 10.1016/j.neuropharm.2005.08.003. [DOI] [PubMed] [Google Scholar]
- McEvoy B, Hawi Z, Fitzgerald M, Gill M. No evidence of linkage or association between the norepinephrine transporter (NET) gene polymorphisms and ADHD in the Irish population. Am J Med Genet. 2002;114:665–666. doi: 10.1002/ajmg.10416. [DOI] [PubMed] [Google Scholar]
- Ozaki N, Goldman D, Kaye WH, Plotnicov K, Greenberg BD, Lappalainen J, Rudnick G, Murphy DL. Serotonin transporter missense mutation associated with a complex neuropsychiatric phenotype. Mol Psychiatry. 2003;8:933–936. doi: 10.1038/sj.mp.4001365. [DOI] [PubMed] [Google Scholar]
- Phillips AG, Ahn S, Floresco SB. Magnitude of dopamine release in medial prefrontal cortex predicts accuracy of memory on a delayed response task. J Neurosci. 2004;24:547–553. doi: 10.1523/JNEUROSCI.4653-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliszka SR. The neuropsychopharmacology of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57:1385–1390. doi: 10.1016/j.biopsych.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Prasad HC, Steiner JA, Sutcliffe JS, Blakely RD. Enhanced activity of human serotonin transporter variants associated with autism. Philos Trans R Soc Lond B Biol Sci. 2009;364:163–173. doi: 10.1098/rstb.2008.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramoz N, Boni C, Downing AM, Close SL, Peters SL, Prokop AM, Allen AJ, Hamon M, Purper-Ouakil D, Gorwood P. A Haplotype of the Norepinephrine Transporter (Net) Gene Slc6a2 is Associated with Clinical Response to Atomoxetine in Attention-Deficit Hyperactivity Disorder (ADHD) Neuropsychopharmacology. 2009 doi: 10.1038/npp.2009.39. [DOI] [PubMed] [Google Scholar]
- Retz W, Rosler M, Kissling C, Wiemann S, Hunnerkopf R, Coogan A, Thome J, Freitag C. Norepinephrine transporter and catecholamine-O-methyltransferase gene variants and attention-deficit/hyperactivity disorder symptoms in adults. J Neural Transm. 2008;115:323–329. doi: 10.1007/s00702-007-0822-5. [DOI] [PubMed] [Google Scholar]
- Robbins TW. Dissociating executive functions of the prefrontal cortex. Philos Trans R Soc Lond B Biol Sci. 1996;351:1463–1470. doi: 10.1098/rstb.1996.0131. [DOI] [PubMed] [Google Scholar]
- Runkel F, Bruss M, Nothen MM, Stober G, Propping P, Bonisch H. Pharmacological properties of naturally occurring variants of the human norepinephrine transporter. Pharmacogenetics. 2000;10:397–405. doi: 10.1097/00008571-200007000-00003. [DOI] [PubMed] [Google Scholar]
- Shannon JR, Flattem NL, Jordan J, Jacob G, Black BK, Biaggioni I, Blakely RD, Robertson D. Clues to the origin of orthostatic intolerance: a genetic defect in the cocaine- and antidepressant sensitive norepinephrine transporter. New Eng J Med. 2000;342:541–549. doi: 10.1056/NEJM200002243420803. [DOI] [PubMed] [Google Scholar]
- Spencer T, Heiligenstein JH, Biederman J, Faries DE, Kratochvil CJ, Conners CK, Potter WZ. Results from 2 proof-of-concept, placebo-controlled studies of atomoxetine in children with attention-deficit/hyperactivity disorder. J Clin Psychiatry. 2002;63:1140–1147. doi: 10.4088/jcp.v63n1209. [DOI] [PubMed] [Google Scholar]
- Stein MA, Maher CS, Waldman ID, Robb AS, Conlon C, Pearl PL, Black DO, Seymour KE, Newcorn JH. A dose-response study of OROS methylphenidate in children with attention-deficit/hyperactivitydisorder. Pediatrics. 2003;112:e404. doi: 10.1542/peds.112.5.e404. [DOI] [PubMed] [Google Scholar]
- Stein MA, Waldman ID, Sarampote CS, Seymour KE, Robb AS, Conlon C, Kim SJ, Cook EH. Dopamine transporter genotype and methylphenidate dose response in children with ADHD. Neuropsychopharmacology. 2005;30:1374–1382. doi: 10.1038/sj.npp.1300718. [DOI] [PubMed] [Google Scholar]
- Stöber G, Nothen MM, Porzgen P, Bruss M, Bonisch H, Knapp M, Beckmann H, Propping P. Systematic search for variation in the human norepinephrine transporter gene: identification of five naturally occurring missense mutations and study of association with major psychiatric disorders. Am J Med Genet. 1996;67:523–532. doi: 10.1002/(SICI)1096-8628(19961122)67:6<523::AID-AJMG3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Sullivan RM, Brake WG. What the rodent prefrontal cortex can teach us about attention-deficit/hyperactivity disorder: the critical role of early developmental events on prefrontal function. Behav Brain Res. 2003;146:43–55. doi: 10.1016/j.bbr.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Valentini V, Frau R, Di Chiara G. Noradrenaline transporter blockers raise extracellular dopamine in medial prefrontal but not parietal and occipital cortex: differences with mianserin and clozapine. J Neurochem. 2004;88:917–927. doi: 10.1046/j.1471-4159.2003.02238.x. [DOI] [PubMed] [Google Scholar]
- Vitiello B. Understanding the risk of using medications for attention deficit hyperactivity disorder with respect to physical growth and cardiovascular function. Child Adolesc Psychiatr Clin N Am. 2008;17:459–474. xi. doi: 10.1016/j.chc.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman ID, Gizer IR. The genetics of attention deficit hyperactivity disorder. Clin Psychol Rev. 2006;26:396–432. doi: 10.1016/j.cpr.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Weinberger DR, Berman KF, Zec RF. Physiologic dysfunction of dorsolateral prefrontal cortex in schizophrenia. I. Regional cerebral blood flow evidence. Arch Gen Psychiatry. 1986;43:114–1124. doi: 10.1001/archpsyc.1986.01800020020004. [DOI] [PubMed] [Google Scholar]
- Xu X, Hawi Z, Brookes KJ, Anney R, Bellgrove M, Franke B, Barry E, Chen W, Kuntsi J, Banaschewski T, Buitelaar J, Ebstein R, Fitzgerald M, Miranda A, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Sonuga-Barke E, Steinhausen HC, Faraone SV, Gill M, Asherson P. Replication of a rare protective allele in the noradrenaline transporter gene and ADHD. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1564–1567. doi: 10.1002/ajmg.b.30872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Knight J, Brookes K, Mill J, Sham P, Craig I, Taylor E, Asherson P. DNA pooling analysis of 21 norepinephrine transporter gene SNPs with attention deficit hyperactivity disorder: no evidence for association. Am J Med Genet B Neuropsychiatr Genet. 2005;134:115–118. doi: 10.1002/ajmg.b.30160. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl–dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- Yang L, Wang YF, Li J, Faraone SV. Association of norepinephrine transporter gene with methylphenidate response. J Am Acad Child Adolesc Psychiatry. 2004;43:1154–1158. doi: 10.1097/01.chi.0000131134.63368.46. [DOI] [PubMed] [Google Scholar]
- Zhou K, Dempfle A, Arcos-Burgos M, Bakker SC, Banaschewski T, Biederman J, Buitelaar J, Castellanos FX, Doyle A, Ebstein RP, Ekholm J, Forabosco P, Franke B, Freitag C, Friedel S, Gill M, Hebebrand J, Hinney A, Jacob C, Lesch KP, Loo SK, Lopera F, McCracken JT, McGough JJ, Meyer J, Mick E, Miranda A, Muenke M, Mulas F, Nelson SF, Nguyen TT, Oades RD, Ogdie MN, Palacio JD, Pineda D, Reif A, Renner TJ, Roeyers H, Romanos M, Rothenberger A, Schafer H, Sergeant J, Sinke RJ, Smalley SL, Sonuga-Barke E, Steinhausen HC, van der Meulen E, Walitza S, Warnke A, Lewis CM, Faraone SV, Asherson P. Meta-analysis of genome-wide linkage scans of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1392–1398. doi: 10.1002/ajmg.b.30878. [DOI] [PMC free article] [PubMed] [Google Scholar]