Abstract
An inflammatory and cytotoxic CD4+CD28– T cell subset infiltrates atherosclerotic plaques and is implicated in plaque rupture and myocardial infarctions. This pathologic subset develops with replicative stress and is found in patients with chronic inflammatory diseases such as RA as well as with aging. CD4+CD28– cells overexpress genes normally suppressed by DNA methylation in CD4+CD28+ T cells, such as KIR, perforin, and CD70. How this subset overexpresses methylation-sensitive genes is unknown. DNA methylation patterns are maintained in proliferating cells by Dnmts, which are up-regulated during mitosis by the ERK and JNK signaling pathways. We hypothesized that defects in these signaling pathways contribute to altered gene expression in human CD4+CD28– cells through effects on DNA methylation. We report that signaling through the ERK and JNK pathways is decreased in CD4+CD28– relative to CD4+CD28+ cells from the same individuals and that ERK and JNK pathway inhibition decreases Dnmt1 and -3a levels, which in turn, causes demethylation and overexpression of the TNFSF7 (CD70) gene. We also report that CD4+CD28– T cells overexpress PP5, a stress-induced inhibitor of the ERK and JNK signaling pathways that may contribute to the signaling defects. We conclude that decreased ERK and JNK signaling in the CD4+CD28– subset, arising with replicative stress, can lead to the overexpression of normally suppressed genes through effects on Dnmts and consequently, chromatin structure.
Keywords: senescence, DNA methylation, autoimmunity
Introduction
CD4+CD28– T cells are a heterogeneous subset found in elderly people as well as in patients with chronic inflammatory diseases such as RA [1, 2]. This subset develops with repeated mitogenic stimulation that results in damage to DNA and cellular components, referred to as replicative stress, and is characterized by decreased replicative potential, low telomerase levels, and shortened telomeres similar to other “senescent” cells [2, 3]. The CD4+CD28– subset expands in patients with acute coronary syndromes (unstable angina and myocardial infarctions) and is found in ruptured atherosclerotic plaques of people with fatal myocardial infarctions. These T cells secrete large amounts of IFN-γ, are cytotoxic to endothelial cells [1], and are implicated in atherosclerotic plaque development and rupture [4, 5].
CD4+CD28– T cells also overexpress genes such as KIR, perforin, and CD70, which contribute to the IFN-γ and cytotoxic responses [1, 6]. These genes are normally suppressed in CD4+ T cells by DNA methylation [7,8,9], a covalent modification promoting a transcriptionally repressive chromatin structure [10]. Our group reported that Dnmt1 and Dnmt3a are decreased in this subset, causing demethylation and overexpression of these genes [8]. How Dnmt levels are decreased in this subset is unknown. Dnmt1 is up-regulated following mitogenic stimulation by signals transmitted through the ERK and JNK pathways [11]. Dnmt3a is similarly up-regulated in part by signaling through the ERK pathway [12], although the role of JNK signaling in Dnmt3a regulation is unknown.
T cell signaling abnormalities develop with age [13]. We hypothesized that ERK and/or JNK signaling may be decreased in the CD4+CD28– subset, leading to decreased Dnmt levels and demethylation of methylation-sensitive genes. We therefore compared ERK and JNK signaling pathways in CD4+CD28+ and CD4+CD28– T cells generated by in vitro proliferative stress, RA, and aging and determined whether decreased ERK and/or JNK signaling could cause demethylation and overexpression of methylation-sensitive T cell genes. We also sought mechanisms contributing to the decreased signaling.
MATERIALS AND METHODS
Subjects
Subjects with RA (n=17, 55±14 years old, mean±sd) met revised criteria for RA [14] and were recruited from the University of Michigan Arthritis Clinics (Ann Arbor, MI, USA). Healthy subjects <60 years of age (n=15, 38±16 years old, mean±sd) were obtained by advertising. Healthy subjects >60 years of age (n=10, 72±7 years old) with osteoarthritis but without known chronic inflammatory diseases or other significant illnesses were similarly recruited from the University of Michigan Arthritis Clinics and the Human Subjects Core of the University of Michigan Claude D. Pepper Older Americans Independence Center. We and others [8] have found no relationship between medications received and gene expression alterations in RA subjects. This protocol was approved by the University of Michigan Institutional Review Board.
CD4+CD28– T cell quantitation, isolation, and culture
PBMC were isolated from healthy donors and RA patients by Ficoll-Hypaque density centrifugation, and CD4 and CD28 expression was measured by multi-color flow cytometry as described [15]. Total T cells, CD4+ T cells, or the CD4+CD28+ and CD4+CD28– subsets were purified using magnetic beads (Miltenyi Biotec, Auburn, CA, USA) and previously published protocols; purity is typically 95–98% by flow cytometry [8]. Where indicated, PBMC were stimulated with 1 μg/ml PHA and cultured in complete media (RPMI 1640/10% FBS supplemented with penicillin and streptomycin) also as described [15]. The signaling inhibitors PD98059 (Promega, Madison, WI, USA; 12.5 μM, 25 μM, 50 μM), U0126 (Promega; 10 μM, 20 μM, 40 μM), and/or SP600125 (Biosource, Carlsbad, CA, USA; 5 μM, 10 μM, 20 μM) were added where indicated. CD4+CD28– T cells were generated in vitro by stimulating PBMC from healthy, younger donors with PHA and then culturing bead-purified CD4+ cells in complete media, supplemented with 10 ng/ml rIL-2 (PeproTech, Rocky Hill, NJ, USA) and restimulating weekly with 106 irradiated (2500R) allogeneic PBMC/2.5 × 106-cultured CD4 T cells [16]. Fifty-two percent to 86% of the CD4+ cells were typically CD28– by Weeks 4–6.
siRNA “knockdown” and Dnmt levels
Purified T cells from healthy donors were transfected with 3 μg control or MEK- or JNK-specific siRNA (Dharmacon, Lafayette, CO, USA) using Amaxa (Walkersville, MD, USA) nucleofection, according to the manufacturer’s instructions. For double-knockdowns, 3 μg each siRNA was used. The target sequence for the down-regulation of both MEK isoforms was 5′-CAGAAGAAGCTGGAGGAGC-3′, found in MEK1 mRNA and MEK2 mRNA. Similarly, the target sequence for the down-regulation of both JNK isoforms was 5′-GAAAGAATGTCCTACCTTC-3′, found in JNK1 mRNA and JNK2 mRNA. BLAST searches were performed to verify the specificity of these sequences. Six hours after transfection, the medium was changed, as per the manufacturer’s instructions. Forty-eight hours following transfection, an aliquot of the cells was stimulated with PHA and Dnmt3a measured 6 h later, and Dnmt1 was measured 24 h later (vide infra) [12]. T cells express little Dnmt3b and so, were not studied [12]. At 72 h, the CD4+ cells were stimulated with PMA for 15 min and then purified, lysed, and protein isolated to verify suppression of MEK and JNK by Western blotting with MEK1/2 and JNK1/2 antibodies (Cell Signaling Technology, Beverly, MA, USA), performed as described [17]. RNA and DNA were also purified from the CD4+ cells at 72 h, as described for CD70 mRNA and DNA methylation analyses [18].
Flow cytometric signaling assays
For ERK and JNK signaling, PBMC were stained with FITC-labeled anti-CD28 (BD PharMingen, San Jose, CA, USA) and then cultured alone or with 50 ng/ml PMA and 50 ng/ml ionomycin for 15 min at 37°C. Treated and untreated cells were then fixed in prewarmed (37°C) Cytofix buffer (BD Bioscience, San Jose, CA, USA) for 10 min and then permeabilized in Phosflow Perm Buffer III (BD Bioscience) for 30 min on ice. The permeabilized cells were washed twice in PBS/3% FBS/0.1% sodium azide and then stained with PE-labeled anti-pERK1/2 (T202/Y204), purified rabbit anti-JNK (pT183/pY185), and PE-Cy5-labeled anti-CD4, respectively. For the detection of pJNK, the cells were reacted with PE-conjugated F(ab′)2 donkey anti-rabbit IgG. Isotype-matched control antibodies were used for surface and intracellular controls. All antibodies were obtained from BD PharMingen. The stained cells were then analyzed on a FACSCalibur cytometer (BD Bioscience) equipped with emission filters for FITC, PE, and Cy5-PE. Fold change was calculated by dividing the MFI of MFI stim by that of MFI unstim, i.e., MFI stim/MFI unstim. Serine/threonine PP5 was similarly quantitated using a 1/50 dilution of rabbit anti-PP5 antibody (Cell Signaling Technology). Isotype-matched control antibodies (BD PharMingen) were used for surface and intracellular controls [19].
Dnmt1, Dnmt3a, ERK, JNK, and PP5 immunoblotting
Where indicated, T cells were stimulated for 15 min with 50 ng/ml PMA + 50 ng/ml ionomycin and lysed, and then total and pERK and pJNK were measured by immunoblotting as described [17]. PP5 was quantitated using antibodies from Cell Signaling Technology.
Dnmt1, Dnmt3a, and CD70 quantitation
PBMC from healthy subjects were stimulated with PHA and treated with the MEK inhibitors PD98058 or U0126 and/or the JNK inhibitor SP600125, and then Dnmt3a mRNA and protein were measured 6 h later, and Dnmt1 mRNA and protein were measured 24 h later using protocols published previously [12]. Dnmt1 transcript levels are cell cycle-linked and so, were measured relative to histone H4, a similarly cell cycle-linked transcript, 24 h following stimulation, and Dnmt3a mRNA levels are maximal at 6 h following stimulation and so, were measured relative to the housekeeping gene β-actin [12]. CD70 transcripts were measured relative to β-actin 72 h following treatment with inhibitors also using protocols published previously [18].
Bisulfite sequencing
CD70 promoter methylation was determined by bisulfite sequencing as described previously [18], cloning, and sequencing 10 fragments/sample.
Statistical analysis
Statistical analyses were performed using Student’s t-test or ANOVA as appropriate and Systat (Evanston, IL, USA) software.
RESULTS
CD4+CD28– T cells increase in RA and in older, healthy individuals
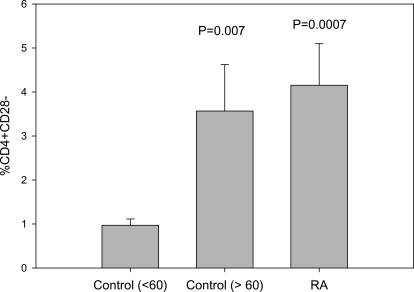
Figure 1 shows the percent of CD4+CD28– T cells relative to total CD4+ T cells in the control and RA cohorts used in this study and confirms that healthy older people [age >60, 72±7 (mean±sd) years of age, n=10] and patients with RA (55±14 years, n=17) have a significantly greater percentage of CD4+CD28– T cells than younger (age <60, 38±16 years, n=15) individuals as reported by others [1]. No significant difference in the percent CD4+CD28– was seen between the healthy, older cohort and the RA cohort.
Figure 1.
Percent CD4+CD28– T cells in younger and older, healthy subjects and in RA patients. PBMC from 15 healthy subjects, 38 ± 16 (mean±sd) years of age (controls <60); 10 healthy subjects, 72 ± 7 years of age (controls >60); and 17 RA patients, 55 ± 14 years of age, were stained with anti-CD4-FITC and anti-CD28-PE, and then the percent CD4+CD28– T cells was determined using two-color flow cytometry. P values refer to the difference in percent CD4+CD28– relative to the young cohort.
ERK and JNK signaling is decreased in CD4+CD28– T cells
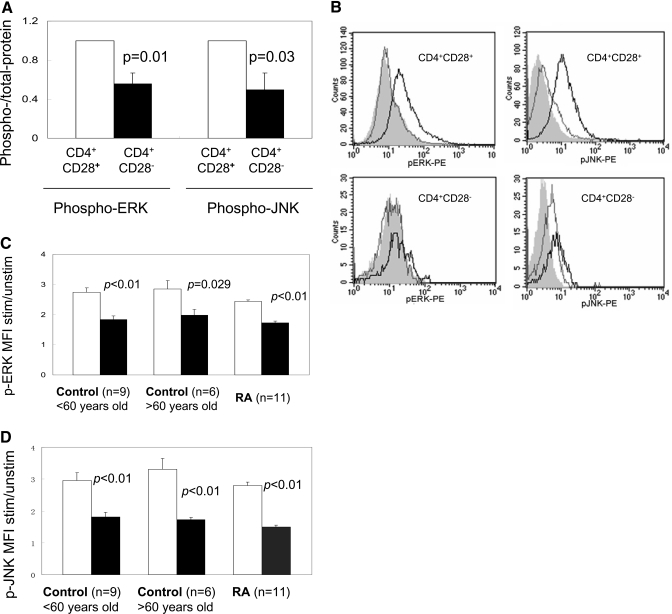
Initial studies compared ERK and JNK signaling pathways in CD4+CD28– T cells generated in vitro using repeated stimulation to cause replicative stress. PBMC from five healthy donors ages 23–62 years (45±16, mean±sd) were stimulated with PHA, and then CD4+ T cells were purified and maintained as long-term lines by weekly restimulation with irradiated, allogeneic PBMC and media supplemented with IL-2. After 4–6 weeks, the cells were fractionated into CD28+ and CD28– subsets, stimulated with PMA + ionomycin, and cytoplasmic pERK and pJNK measured. At the time of harvest, 51 ± 11% of the cells were CD4+CD28– (mean±SEM, n=5). Supplemental Figure 1 shows representative immunoblots comparing total and pERK and pJNK in CD4+CD28+ and CD4+CD28– T cells. pERK and pJNK are decreased in the CD28– subset. Figure 2A shows the mean ± sem of six similar experiments comparing pERK and pJNK in CD4+ CD28+ and CD4+CD28– T cells. pERK and pJNK are impaired significantly in the CD4+CD28– subset.
Figure 2.
ERK and JNK activation in CD4+CD28+ and CD4+CD28– T cells. (A) Mean ± sem of six experiments similar to those shown in Supplemental Figure 1 using immunoblotting to compare phosphorylated to total ERK and JNK in PMA-stimulated CD4+CD28+ T cells (open bars) and CD4+CD28– T cells (solid bars), generated by repeated stimulation in vitro. (B) Unstimulated (light lines) and PMA + ionomycin-stimulated (dark lines) T cells from a patient with RA were stained with PE-Cy5-labeled anti-CD4, FITC-anti-CD28–, and anti-pERK-PE or anti-pJNK-PE and then analyzed by flow cytometry as described in Materials and Methods. The shaded histograms represent isotype controls for the antibodies to the signaling molecules. The upper left panel shows pERK in CD4+CD28+ cells, the lower left pERK in CD4+CD28– cells, the upper right pJNK in CD4+ CD28+ cells, and the lower right pJNK in CD4+CD28– cells. (C) Bar graph showing the pERK MFI in CD4+CD28+ cells (open bars) and pERK MFI in CD4+CD28– cells (solid bars) in T cells from nine healthy controls age <60, 11 RA patients, and six controls age >60. (D) Bar graphs similarly showing pJNK MFI in CD4+CD28+ and CD4+CD28– T cells from the same subjects.
We then compared pERK and pJNK in CD4+CD28+ and CD4+CD28– T cells isolated from the peripheral blood of younger and older subjects without chronic inflammatory conditions and in patients with RA. The number of CD4+CD28– cells in peripheral blood is limiting (Fig. 1), so flow cytometric approaches described and validated by others [19] were used to compare pERK and pJNK levels in CD4+CD28+ and CD4+CD28– cells. Figure 2B shows representative histograms of pERK and pJNK staining in unstimulated and stimulated (PMA+ionomycin) CD4+CD28+ and CD4+CD28– cells from a 53-year-old woman with RA. There is less pERK and pJNK in the CD28– subset. Figure 2C shows the mean ± sem of similar pERK staining in stimulated CD4+CD28+ and CD4+CD28– T cells from nine younger, healthy people (age <60 years, 39±16 years old, mean±sd), six older, healthy subjects (age >60, 77±4 years old), and 11 RA patients (54±16 years old), and Figure 2D shows pJNK staining in stimulated CD4+CD28+ and CD4+CD28– T cells from the same subjects. pERK and pJNK were decreased by approximately the same degree in CD4+CD28– T cells from all three groups.
Inhibiting ERK and/or JNK signaling decreases Dnmt1 and Dnmt3a expression
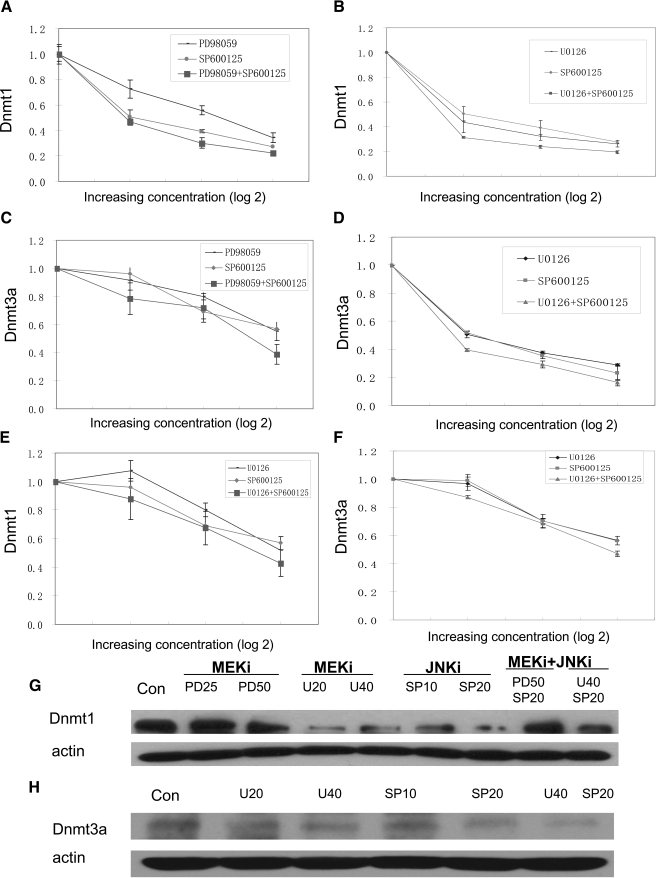
We next determined the effect of inhibiting ERK and JNK signaling, alone and in combination, on total T cell Dnmt1 and Dnmt3a levels. PBMC from young (18–30 years old), healthy controls were stimulated with PHA and treated with graded concentrations of the MEK inhibitors PD98059 or U0126 and/or the JNK inhibitor SP600125, and then Dnmt3a mRNA and protein were measured in T cells 6 h later, and Dnmt1 mRNA and protein were measured after 24 h [12]. Figure 3A shows the effects of the MEK inhibitor PD98059 and the JNK inhibitor SP600125 alone and in combination on Dnmt1 mRNA. PD98059 and SP600125 together decreased Dnmt1 levels to a slightly greater extent than PD98059 alone (P=0.03 by t-test; PD98059 50 μM alone vs. PD98059 50 μM+SP600125 20 μM). Figure 3B shows a similar experiment using the MEK inhibitor U0126 and SP600125, alone and in combination on Dnmt1. Each inhibitor alone suppressed T cell Dnmt1 transcripts, and again, there was a small additive effect when combined (P=0.02). Similar effects on Dnmt3a mRNA levels were observed when MEK and JNK pathways were inhibited. PD98059 and SP600125 (Fig. 3C) and U0126 and SP600125 (Fig. 3D) had effects on Dnmt3a transcripts resembling those observed on Dnmt1.
Figure 3.
Effect of ERK and JNK pathway inhibitors on T cell Dnmt1 and Dnmt3a. (A) PBMC from young, healthy controls were stimulated with PHA and treated with graded concentrations of the MEK inhibitor PD98059 (12.5 μM, 25 μM, or 50 μM), the JNK inhibitor SP600125 (5 μM, 10 μM, or 20 μM), or both, and then T cells were isolated and Dnmt1 transcripts measured relative to histone H4 24 h later. Results represent the mean ± sem of four independent experiments. (B) PBMC from healthy controls were similarly stimulated and treated with the MEK inhibitor U0126 (10 μM, 20 μM, 40 μM), SP600125 (5 μM, 10 μM, or 20 μM), or both, and T cell Dnmt1 mRNA was measured as in A. Results again represent the mean ± sem of four independent experiments. (C) PBMC from young, healthy controls were stimulated with PHA and treated with graded concentrations of PD98059, SP600125, or both as in A, and then T cells were purified, and Dnmt3a transcripts were measured relative to actin 6 h later. Results represent the mean ± sem of four independent experiments. (D) PBMC from healthy controls were similarly stimulated and treated with U0126, SP600125, or both as in B, and T cell Dnmt3a mRNA was measured as in C. Results again represent the mean ± sem of four independent experiments. (E) PBMC from young, healthy controls were stimulated with PHA and treated with graded concentrations of the MEK inhibitor U0126 (10 μM, 20 μM, or 40 μM), the JNK inhibitor SP600125 (5 μM, 10 μM, or 20 μM), or both as in A, and then CD4+ T cells were isolated and Dnmt1 transcripts measured relative to histone H4 24 h later. Results represent the mean ± sem of three independent experiments. (F) PBMC from healthy controls were similarly stimulated and treated with U0126, SP600125, or both, and then CD4+ T cell Dnmt3a mRNA was measured as in B. Results again represent the mean ± sem of three independent experiments. (G) PBMC were stimulated with PHA, treated with PD98059 (25 and 50 μM; PD25 and PD50, respectively), U0126 (20 and 40 μM; U20 and U40, respectively), SP600125 (10 and 20 μM; SP10 and SP20, respectively), or 50 μM PD98059 + 20 μM SP600125 (PD50 SP20) or 40 μM U0126 + 20 μM SP600125 (U40 SP20) as in A and B, and then T cell Dnmt1 and actin were measured by immunoblotting. (H) PBMC were stimulated with PHA and treated with U0126, SP600125, or U0126 + SP600125 as in G, and then T cell Dnmt3a and actin were measured by immunoblotting as in G. MEKi, MEK inhibitor; JNKi, JNK inhibitor; Con, control.
We then compared the effects of the MEK and JNK inhibitors in purified CD4+ T cells. Figure 3E shows suppressive effects of U0126 and SP600125 alone and in combination on Dnmt1 mRNA (P=0.007; U0126 40 μM vs. U0126 40 μM+SP600125 20 μM), and Figure 3F shows similar effects on Dnmt3a (P=0.05; U0126 40 μM vs. U0126 40 μM+SP600125 20 μM). The results again are additive and resemble those seen in unfractionated T cells.
Figure 3G shows a representative immunoblot confirming a decrease in Dnmt1 at the protein level caused by the MEK inhibitors, the JNK inhibitor, or both, and Figure 3H similarly shows an immunoblot confirming a decrease in Dnmt3a protein caused by the MEK inhibitors, the JNK inhibitor, or both. Together, these results indicate that defects in the ERK signaling pathway, the JNK signaling pathway, or both can decrease Dnmt1 and Dnmt3a levels.
Inhibiting ERK and/or JNK signaling causes TNFSF7 demethylation and overexpression
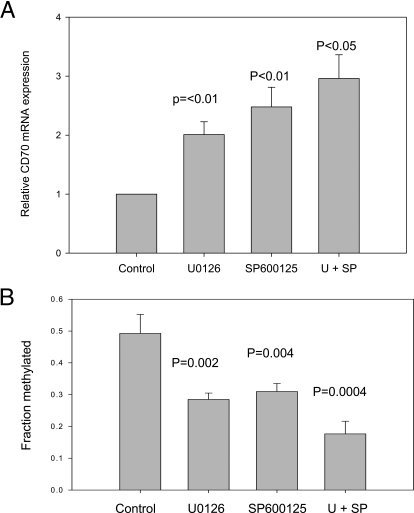
We reported previously that siRNA knockdowns of Dnmt1 and Dnmt3a were additive in causing demethylation and overexpression of KIR, perforin 1, and TNFSF7 (CD70), normally suppressed by DNA methylation in CD4+ T cells [8]. We therefore determined if decreasing signaling through the ERK and JNK pathways had similar effects on T cell DNA methylation and gene expression. CD70 is a B cell costimulatory molecule regulated in part by methylation of a sequence flanking the core promoter. Inhibiting T cell DNA methylation demethylates this sequence and increases CD70 expression, and methylation of this sequence in reporter constructs suppresses promoter function [18]. Figure 4A shows that U0126 and SP600125, used at the maximum concentrations shown in Figure 3, increased CD70 transcripts in CD4+ T cells and that their effects were additive. Supplemental Figure 2, A–D, shows the effects of treatment with U0126, SP600125, and U0126 + SP600125 on the methylation of each CG pair in the CD70 promoter using bisulfite sequencing performed on CD4+ T cells from four donors. Figure 4B depicts the average methylation across this region, calculated from the results shown in Supplemental Figure 2. The region shown previously to demethylate in drug-treated T cells and in human lupus T cells [18] also demethylated in T cells treated with MEK and JNK inhibitors, again, in an additive manner (P=0.002 overall by ANOVA).
Figure 4.
Effect of ERK and JNK pathway inhibitors on CD4+ T cell TNFSF7 (CD70) expression and methylation. (A) PBMC were stimulated with PHA and then left untreated (Control) or treated with 40 μM U0126, 20 μM SP600125, or both (U+SP), and 72 h later, CD70 mRNA was measured in purified CD4+ T cells. Results represent the mean ± sem of three independent experiments. (B) The overall methylation of the TNFSF7 promoter was determined for each experimental condition for each subject, and then the mean of the results for the four subjects was determined. The results are presented as the mean ± sem of the four determinations/experimental condition and significance determined relative to untreated cells using Student’s t-test.
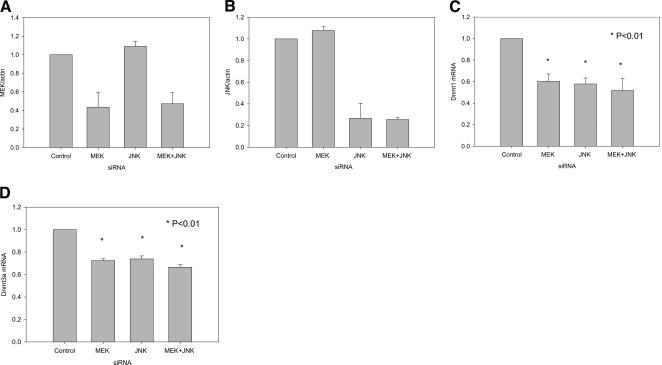
These results were confirmed using siRNA knockdowns to decrease MEK and JNK. Supplemental Figure 3 shows total and pMEK, pERK, and pJNK protein in PMA-stimulated T cells transfected with a MEK siRNA, a JNK siRNA, or both. The MEK siRNA decreased levels of total MEK and similarly decreased levels of pMEK and pERK, and JNK siRNA decreased total and pJNK. Figure 5A shows densitometric analysis of immunoblots, confirming the effects on MEK protein, and Figure 5B similarly shows densitometric analysis of immunoblots, confirming effects on JNK. The siRNAs demonstrated specificity and efficacy. Figure 5C shows that the MEK and JNK siRNAs decreased Dnmt1 mRNA and that their effects are modestly additive, and Figure 5D shows similar results on Dnmt3a transcripts (P<0.01 for all).
Figure 5.
Effects of T cell MEK and/or JNK RNA interference knockdowns on Dnmt levels. (A) Densitometric analysis of two experiments similar to those shown in Supplemental Figure 3 comparing control siRNA, MEK siRNA, JNK siRNA, or MEK + JNK siRNA on total MEK. Results represent the mean ± sd of the two experiments. (B) Densitometric analysis of two experiments similar to those shown in A comparing MEK siRNA, JNK siRNA, or both on total JNK. Results again represent the mean ± sd of the two experiments. (C) T cells from healthy donors were transfected with control siRNA, MEK siRNA, JNK siRNA, or MEK + JNK siRNA and then 48 h later, stimulated with PHA as in Figure 3. Dnmt1 mRNA was measured 24 h later as in Figure 3. Results represent the mean ± sem of three independent experiments. (D) T cells were similarly transfected with control siRNA, MEK siRNA, JNK siRNA, or MEK + JNK siRNA as in Supplemental Figure 3 and then 48 h later, stimulated with PHA and Dnmt3a transcripts, measured 6 h later as in Figure 3. Results again represent the mean ± sem of three independent experiments. *, P < 0.01.
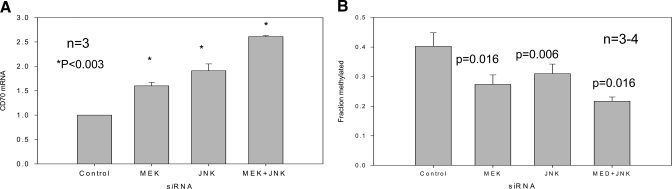
Figure 6A shows that the MEK and JNK siRNAs also increased CD70 expression and that their effects were additive, similar to the results seen with the pharmacologic inhibitors shown in Figure 4. Supplemental Figure 4, A–D, compares the effects of transfection with the MEK and JNK siRNA, alone and in combination on CD70 promoter methylation using bisulfite sequencing as in Figure 4, performed on CD4+ T cells from three to four donors, and Figure 6B shows the average methylation across this region for the control and transfected cells. The MEK and JNK siRNAs similarly demethylated the CD70 promoter, and their effects were again additive. Together, these experiments demonstrate that inhibiting the ERK and JNK signaling pathways, with pharmacologic inhibitors or siRNA, decreases Dnmt1 and Dnmt3a expression and causes demethylation and overexpression of CD70.
Figure 6.
Effects of MEK and/or JNK siRNA on CD4+ T cell TNFSF7 (CD70) expression and methylation. (A) T cells from three young, healthy controls were transfected with control siRNA, MEK siRNA, JNK siRNA, or MEK + JNK siRNA as in Figure 5, stimulated 48 h later with PHA, and 24 h later, CD70 mRNA was measured as described in Figure 4. Results represent the mean ± sem of the three independent experiments. *, P < 0.003. (B) The overall methylation of the TNFSF7 promoter was determined for each experimental condition for each subject, and then the mean of the results for the subjects was determined as in Figure 4. The results are presented as the mean ± sem of three to four determinations/experimental condition.
Mechanisms decreasing ERK and JNK signaling in CD28– T cells
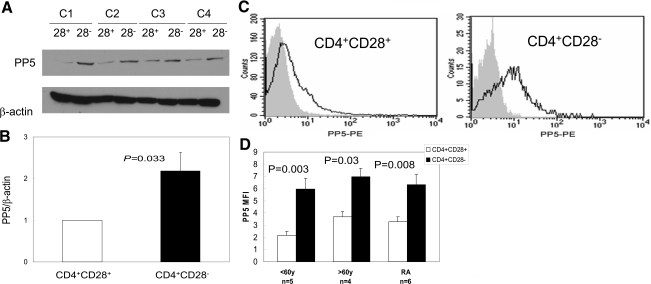
We next investigated mechanisms contributing to decreased signaling in the ERK and JNK pathways in CD4+CD28– T cells. Oxidative stress plays a major role in cellular senescence [20]. PP5 is a regulator of stress-induced signaling pathways, including oxidative stress, and inactivates ASK1, preventing JNK activation [21, 22]. PP5 also inactivates the ERK signaling pathway by dephosphorylating Raf-1 [23]. We therefore compared PP5 levels in CD4+CD28+ and CD4+CD28– T cells generated by repeated stimulation. Figure 7A shows PP5 protein in CD4+CD28+ and CD4+CD28– T cells from four young, healthy donors. PP5 expression was increased in the CD28– subset. Figure 7B shows the densitometric analyses of PP5 relative to β-actin in similar immunoblots performed on CD4+CD28+ and CD4+CD28– T cells from eight young, healthy controls. PP5 levels were significantly (P<0.01) higher in CD28– T cells. Figure 7C shows representative flow cytometric histograms of PP5 levels in CD4+CD28+ and CD4+CD28–, respectively, in T cells from a healthy control without chronic inflammatory disease. Again, the CD28– subset had higher PP5 levels. Figure 7D compares PP5 levels in CD4+CD28+ and CD4+CD28– T cells in five healthy subjects age <60 years (22–59, 37±19 years, mean±sd) 4 subjects >60 (61–74, 64±7 years) without known chronic inflammatory disease, and six RA patients (45–72, 55±9 years). Again, CD28– T cells have higher PP5 levels in all three groups. This suggests that the signaling regulator PP5 contributes to decreased signaling through the ERK and JNK pathways in the CD4+CD28– subset as in other cells subjected to oxidative stress [21,22,23].
Figure 7.
PP5 expression in cultured, senescent CD4+CD28– cells from normal donors and in CD4+CD28+ and CD4+CD28– T cells from RA patients and healthy controls. (A) CD4+CD28+ and CD4+CD28– T cells were generated by repeated stimulation of CD4+ T cells from four healthy controls (C1–C4) and then separated into CD28+ and CD28– subsets using magnetic beads. The cells were then lysed and PP5 levels measured by immunoblotting using β-actin as a control. (B) Densitometric analysis of PP5 relative to β-actin in similar immunoblots performed on chronically restimulated CD4+CD28+ and CD4+CD28– T cells from eight donors (mean±sem). (C) Representative flow cytometric histograms gating on PP5 levels in CD4+CD28+ and CD4+CD28– T cells from a healthy control. The dark lines represent anti-PP5 and the shaded portion, isotype control. (D) Similar analysis of PP5 in CD4+CD28– T cells from the same subject. PP5 MFI in CD4+CD28– (solid bars) and CD4+CD28+ (open bars) T cells from five healthy subjects age <60, four healthy subjects >60, and six RA patients. Results represent the mean ± sem.
DISCUSSION
We and others [7, 18, 24,25,26] reported that some of the genes overexpressed in CD4+CD28– T cells, such as CD11a, perforin, CD70, IFN-γ, and the KIR gene family, are suppressed normally in T cells by DNA methylation and that inhibiting replication of DNA methylation patterns during mitosis is sufficient to increase their expression. Our group also reported that the CD70, perforin, and KIR2DL4 promoters are demethylated in CD4+CD28– T cells, that Dnmt1 and Dnmt3a levels are decreased in this subset, and that siRNA knockdown of Dnmt1, but not Dnmt3a, in CD4+CD28+ T cells caused similar demethylation and overexpression of KIR2DL4, perforin, and CD70, and simultaneous knockdown of Dnmt1 and Dnmt3a caused greater demethylation and overexpression of these genes than Dnmt1 alone [8]. T cells express little Dnmt3b, so this isoform was not studied [12].
The present studies extend this work by demonstrating that decreased ERK and JNK signaling contributes to demethylation of promoter sequences in CD4+CD28– T cells by decreasing Dnmt levels. Dnmt1 is up-regulated by signals transmitted through the ERK and JNK pathways [11]. Dnmt3a is similarly up-regulated, in part, by signaling through the ERK pathway [12], but the observation that JNK signaling regulates Dnmt3a is novel.
T cell signaling abnormalities develop with age [27]. However, subset specificity has not been studied, and this is the first report demonstrating ERK and JNK signaling abnormalities in CD4+CD28– cells. Functional consequences were confirmed by demonstrating that ERK and JNK inhibition or knockdown in CD4+ T cells replicated the Dnmt1 and Dnmt3a decreases observed in the CD4+CD28– subset [8] and caused demethylation and overexpression of the TNFSF7 (CD70) gene. This indicates that the signaling defects contribute to overexpression of some genes in CD4+CD28– cells through effects on DNA methylation and consequently, chromatin structure. Notably, the same signaling abnormalities were seen in CD4+CD28– T cells from RA patients, the elderly, and T cells subjected to replicative senescence, indicating similar mechanisms in all three conditions.
The decrease in JNK activation is not a result of loss of CD28, as CD28 is not required for JNK activation in T cells [28]. Rather, the present studies demonstrate that CD4+ CD28– T cells express PP5, which is a phosphatase that inhibits the JNK signaling pathway through effects on ASK1 [21, 22], and may contribute to the JNK pathway impairment in CD28– T cells. PP5 also inhibits the ERK signaling pathway through effects on Raf-1 [23]. PP5 is a regulator of stress-induced signaling networks [29], including oxidative stress [21], and its overexpression in the CD28– subset may similarly reflect a response to oxidative stress in these cells.
Others [2] have reported that the CD28– subset can be subdivided further based on markers such as CD27 and CD57. Although these subsubsets were not addressed specifically in the present studies, the fact that statistically significant differences were seen between the CD4+CD28+ and CD4+CD28– subsets, together with the flow cytometric studies showing signaling abnormalities in a majority of the CD4+CD28– T cells, suggests that a majority of the cells within the CD4+CD28– subset is affected.
In summary, our previous work demonstrated that maintenance Dnmt levels are decreased in CD4+CD28– T cells, resulting in demethylation and overexpression of genes that contribute to some of the autoreactive, inflammatory, and cytotoxic abnormalities characterizing this subset [8]. The present studies extend this work by demonstrating that impaired signaling through the ERK and JNK pathways plays an important role in the methyltransferase decrease. These studies also indicate that stress-induced PP5 expression may contribute to the signaling decreases. Further studies are now needed to define the role of PP5 in the generation of the CD4+CD28– subset and whether development of these cells can be prevented with inhibitors of oxidative stress. These studies are under way.
AUTHORSHIP
Dr. Chen designed and performed the majority of the experiments described in this manuscript. Dr. Gorelik provided guidance and data interpretation for the signaling studies, and Dr. Strickland provided guidance and data interpretation for the cell culture and flow cytometry studies. Dr. Richardson conceived and supervised the work and was responsible for writing the manuscript.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AG25877, AR42525, ES015214, and AG024824, and a merit grant from the Department of Veterans Affairs. The authors thank Ms. Cindy Bourke and Ms. Cheryl Glaser for their expert secretarial assistance and Ms. Ailing Wu, Ms. Sushma Yarlagadda, and Mr. Robert Hinderer for their expert technical assistance.
Supplementary Material
Footnotes
Abbreviations: ASK1=apoptosis signal-regulating kinase 1, Dnmt=DNA methyltransferase, KIR=killer cell Ig-like receptor, MFI=mean fluorescence intensity, MFI stim=stimulated MFI, MFI unstim=unstimulated MFI, pERK=phospho-ERK, pJNK=phospho-JNK, pMEK=phospho-MEK, PP5=protein phosphatase 5, RA=rheumatoid arthritis, siRNA=small interfering RNA, TNFSF7=TNF superfamily 7
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
References
- Weyand C M, Fulbright J W, Goronzy J J. Immunosenescence, autoimmunity, and rheumatoid arthritis. Exp Gerontol. 2003;38:833–841. doi: 10.1016/s0531-5565(03)00090-1. [DOI] [PubMed] [Google Scholar]
- Weng N P, Akbar A N, Goronzy J. CD28(–) T cells: their role in the age-associated decline of immune function. Trends Immunol. 2009;30:306–312. doi: 10.1016/j.it.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goronzy J J, Fujii H, Weyand C M. Telomeres, immune aging and autoimmunity. Exp Gerontol. 2006;41:246–251. doi: 10.1016/j.exger.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Liuzzo G, Goronzy J J, Yang H, Kopecky S L, Holmes D R, Frye R L, Weyand C M. Monoclonal T-cell proliferation and plaque instability in acute coronary syndromes. Circulation. 2000;101:2883–2888. doi: 10.1161/01.cir.101.25.2883. [DOI] [PubMed] [Google Scholar]
- Gerli R, Schillaci G, Giordano A, Bocci E B, Bistoni O, Vaudo G, Marchesi S, Pirro M, Ragni F, Shoenfeld Y, Mannarino E. CD4+CD28– T lymphocytes contribute to early atherosclerotic damage in rheumatoid arthritis patients. Circulation. 2004;109:2744–2748. doi: 10.1161/01.CIR.0000131450.66017.B3. [DOI] [PubMed] [Google Scholar]
- Lee W W, Yang Z Z, Li G, Weyand C M, Goronzy J J. Unchecked CD70 expression on T cells lowers threshold for T cell activation in rheumatoid arthritis. J Immunol. 2007;179:2609–2615. doi: 10.4049/jimmunol.179.4.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kuick R, Hanash S, Richardson B. DNA methylation inhibition increases T cell KIR expression through effects on both promoter methylation and transcription factors. Clin Immunol. 2009;130:213–224. doi: 10.1016/j.clim.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Richardson B. Decreased DNA methyltransferase levels contribute to abnormal gene expression in “senescent” CD4+CD28– T cells. Clin Immunol. 2009;132:257–265. doi: 10.1016/j.clim.2009.03.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Weyand C M, Goronzy J J. Epigenetic mechanisms of age-dependent KIR2DL4 expression in T cells. J Leukoc Biol. 2008;84:824–834. doi: 10.1189/jlb.0807583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B. Primer: epigenetics of autoimmunity. Nat Clin Pract Rheumatol. 2007;3:521–527. doi: 10.1038/ncprheum0573. [DOI] [PubMed] [Google Scholar]
- Rouleau J, MacLeod A R, Szyf M. Regulation of the DNA methyltransferase by the Ras-AP-1 signaling pathway. J Biol Chem. 1995;270:1595–1601. doi: 10.1074/jbc.270.4.1595. [DOI] [PubMed] [Google Scholar]
- Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, Richardson B. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- Hasler P, Zouali M. Immune receptor signaling, aging, and autoimmunity. Cell Immunol. 2005;233:102–108. doi: 10.1016/j.cellimm.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Arnett F C, Edworthy S M, Bloch D A, McShane D J, Fries J F, Cooper N S, Healey L A, Kaplan S R, Liang M H, Luthra H S. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179:6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- Michel J J, Turesson C, Lemster B, Atkins S R, Iclozan C, Bongartz T, Wasko M C, Matteson E L, Vallejo A N. CD56-expressing T cells that have features of senescence are expanded in rheumatoid arthritis. Arthritis Rheum. 2007;56:43–57. doi: 10.1002/art.22310. [DOI] [PubMed] [Google Scholar]
- Gorelik G, Fang J Y, Wu A, Sawalha A H, Richardson B. Impaired T cell protein kinase C δ activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol. 2007;179:5553–5563. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- Lu Q, Wu A, Richardson B C. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol. 2005;174:6212–6219. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- Perez O D, Nolan G P. Phospho-proteomic immune analysis by flow cytometry: from mechanism to translational medicine at the single-cell level. Immunol Rev. 2006;210:208–228. doi: 10.1111/j.0105-2896.2006.00364.x. [DOI] [PubMed] [Google Scholar]
- Muller M. Cellular senescence: molecular mechanisms, in vivo significance, and redox considerations. Antioxid Redox Signal. 2009;11:59–98. doi: 10.1089/ars.2008.2104. [DOI] [PubMed] [Google Scholar]
- Morita K, Saitoh M, Tobiume K, Matsuura H, Enomoto S, Nishitoh H, Ichijo H. Negative feedback regulation of ASK1 by protein phosphatase 5 (PP5) in response to oxidative stress. EMBO J. 2001;20:6028–6036. doi: 10.1093/emboj/20.21.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Golden T, Aragon I V, Honkanen R E. Ser/Thr protein phosphatase 5 inactivates hypoxia-induced activation of an apoptosis signal-regulating kinase 1/MKK-4/JNK signaling cascade. J Biol Chem. 2004;279:46595–46605. doi: 10.1074/jbc.M408320200. [DOI] [PubMed] [Google Scholar]
- von Kriegsheim A, Pitt A, Grindlay G J, Kolch W, Dhillon A S. Regulation of the Raf-MEK-ERK pathway by protein phosphatase 5. Nat Cell Biol. 2006;8:1011–1016. doi: 10.1038/ncb1465. [DOI] [PubMed] [Google Scholar]
- Lu Q, Ray D, Gutsch D, Richardson B. Effect of DNA methylation and chromatin structure on ITGAL expression. Blood. 2002;99:4503–4508. doi: 10.1182/blood.v99.12.4503. [DOI] [PubMed] [Google Scholar]
- Lu Q, Wu A, Ray D, Deng C, Attwood J, Hanash S, Pipkin M, Lichtenheld M, Richardson B. DNA methylation and chromatin structure regulate T cell perforin gene expression. J Immunol. 2003;170:5124–5132. doi: 10.4049/jimmunol.170.10.5124. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Larbi A, Kempf J, Pawelec G. Oxidative stress modulation and T cell activation. Exp Gerontol. 2007;42:852–858. doi: 10.1016/j.exger.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Rivas F V, O'Herrin S, Gajewski T F. CD28 is not required for c-Jun N-terminal kinase activation in T cells. J Immunol. 2001;167:3123–3128. doi: 10.4049/jimmunol.167.6.3123. [DOI] [PubMed] [Google Scholar]
- Golden T, Swingle M, Honkanen R E. The role of serine/threonine protein phosphatase type 5 (PP5) in the regulation of stress-induced signaling networks and cancer. Cancer Metastasis Rev. 2008;27:169–178. doi: 10.1007/s10555-008-9125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.