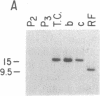
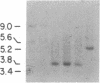
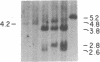
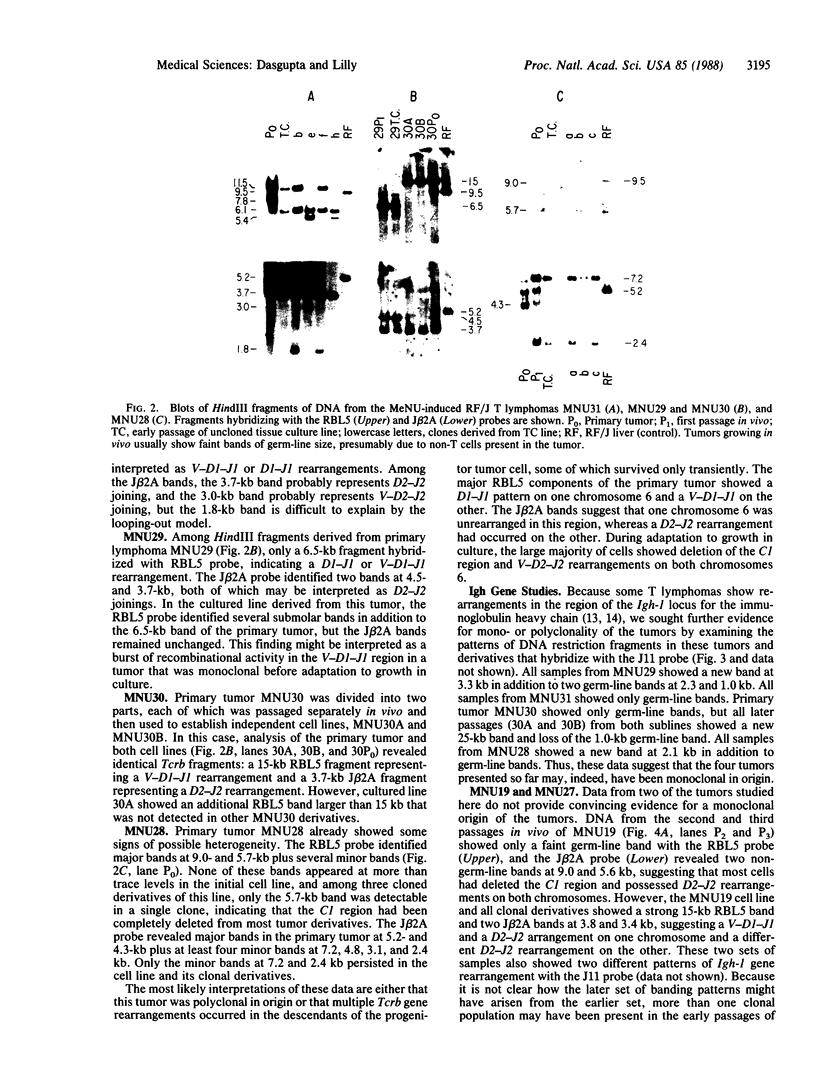
Abstract
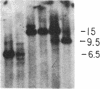
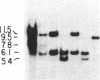
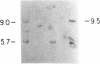
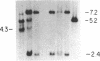
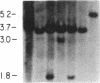
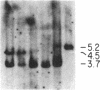
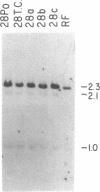
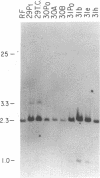
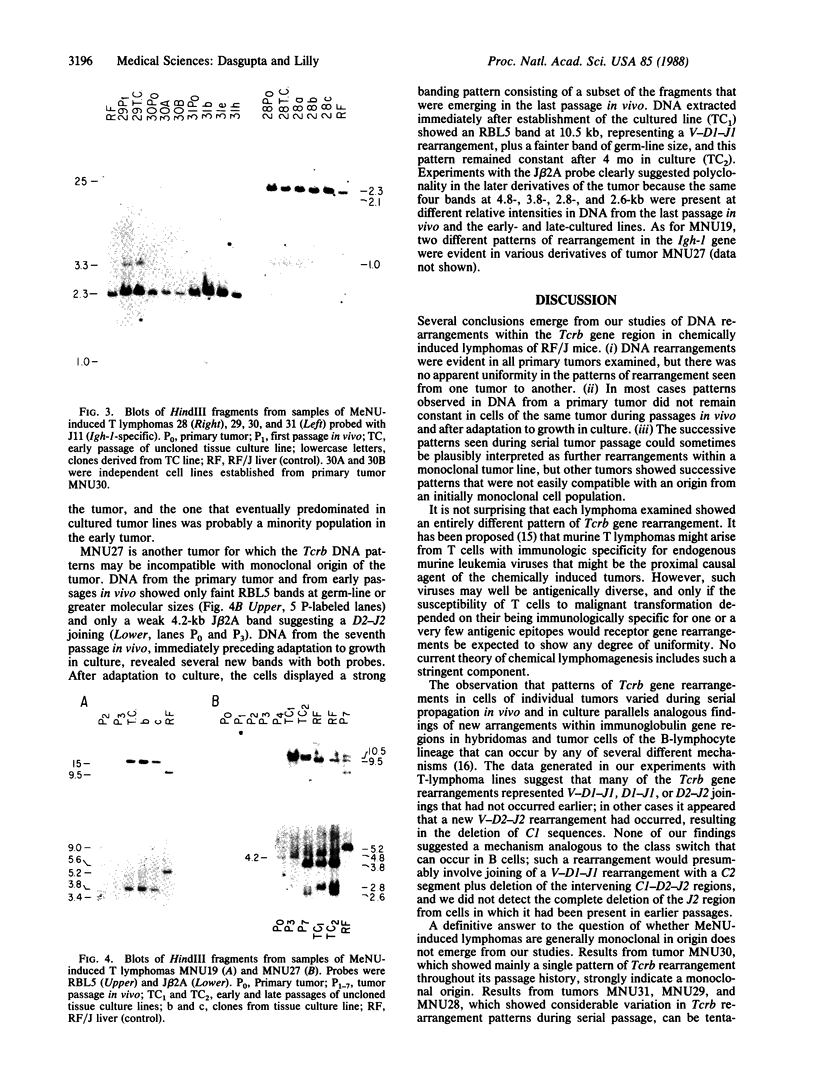
The first constant region of the Tcrb gene was completely deleted from the DNA of 8/10 mouse cell lines established from 3-methylcholanthrene-induced RF/J thymic lymphomas, but 6/7 primary lymphomas contained the first constant region sequences. DNA from RF/J thymic lymphomas induced by N-methyl-N-nitrosourea was then examined serially as the tumors were passaged in vivo and adapted to growth in culture as uncloned and, in some cases, cloned lines. Patterns of Tcrb-specific restriction fragments from most tumors changed extensively during continued propagation. Analysis of the patterns often suggested that initial DNA rearrangements within the Tcrb complexes of monoclonal tumors had been followed by further rearrangements within the same genes. However, these different patterns may alternatively have represented successive outgrowth of separate lineages from lymphomas that were polyclonal in origin.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Caccia N., Kronenberg M., Saxe D., Haars R., Bruns G. A., Goverman J., Malissen M., Willard H., Yoshikai Y., Simon M. The T cell receptor beta chain genes are located on chromosome 6 in mice and chromosome 7 in humans. Cell. 1984 Jul;37(3):1091–1099. doi: 10.1016/0092-8674(84)90443-4. [DOI] [PubMed] [Google Scholar]
- Cory S., Adams J. M., Kemp D. J. Somatic rearrangements forming active immunoglobulin mu genes in B and T lymphoid cell lines. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4943–4947. doi: 10.1073/pnas.77.8.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Reynals M. L., Lilly F., Bosch A., Blank K. J. The genetic basis of susceptibility to leukemia induction in mice by 3-methylcholanthrene applied percutaneously. J Exp Med. 1978 Feb 1;147(2):459–469. doi: 10.1084/jem.147.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt L. A., Birshtein B. K. Independent immunoglobulin class-switch events occurring in a single myeloma cell line. Mol Cell Biol. 1985 Apr;5(4):856–868. doi: 10.1128/mcb.5.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenow M. M., Lilly F. Expression of differentiation and murine leukemia virus antigens on cells of primary tumors and cell lines derived from chemically induced lymphomas of RF/J mice. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7612–7616. doi: 10.1073/pnas.81.23.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W., Gilbert W. Free and integrated recombinant murine leukemia virus DNAs appear in preleukemic thymuses of AKR/J mice. J Virol. 1984 Apr;50(1):155–162. doi: 10.1128/jvi.50.1.155-162.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W., Perlmutter A. P., Gilbert W. Monoclonal AKR/J thymic leukemias contain multiple JH immunoglobulin gene rearrangements. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7433–7436. doi: 10.1073/pnas.80.24.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaka S. T., Lilly F. Genetically determined resistance to 3-methylcholanthrene-induced lymphoma is expressed at the level of bone marrow-derived cells. J Exp Med. 1987 Aug 1;166(2):565–570. doi: 10.1084/jem.166.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi V. V., Frei J. V. Effects of dose and schedule of methylnitrosourea on incidence of malignant lymphoma in adult female mice. J Natl Cancer Inst. 1970 Aug;45(2):335–339. [PubMed] [Google Scholar]
- Kronenberg M., Goverman J., Haars R., Malissen M., Kraig E., Phillips L., Delovitch T., Suciu-Foca N., Hood L. Rearrangement and transcription of the beta-chain genes of the T-cell antigen receptor in different types of murine lymphocytes. Nature. 1985 Feb 21;313(6004):647–653. doi: 10.1038/313647a0. [DOI] [PubMed] [Google Scholar]
- Kronenberg M., Siu G., Hood L. E., Shastri N. The molecular genetics of the T-cell antigen receptor and T-cell antigen recognition. Annu Rev Immunol. 1986;4:529–591. doi: 10.1146/annurev.iy.04.040186.002525. [DOI] [PubMed] [Google Scholar]
- Lee N. E., D'Eustachio P., Pravtcheva D., Ruddle F. H., Hedrick S. M., Davis M. M. Murine T cell receptor beta chain is encoded on chromosome 6. J Exp Med. 1984 Sep 1;160(3):905–913. doi: 10.1084/jem.160.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath M. S., Weissman I. L. AKR leukemogenesis: identification and biological significance of thymic lymphoma receptors for AKR retroviruses. Cell. 1979 May;17(1):65–75. doi: 10.1016/0092-8674(79)90295-2. [DOI] [PubMed] [Google Scholar]
- Pelicci P. G., Knowles D. M., 2nd, Dalla Favera R. Lymphoid tumors displaying rearrangements of both immunoglobulin and T cell receptor genes. J Exp Med. 1985 Sep 1;162(3):1015–1024. doi: 10.1084/jem.162.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu G., Kronenberg M., Strauss E., Haars R., Mak T. W., Hood L. The structure, rearrangement and expression of D beta gene segments of the murine T-cell antigen receptor. 1984 Sep 27-Oct 3Nature. 311(5984):344–350. doi: 10.1038/311344a0. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G. D., Alt F. W. Regulation of the assembly and expression of variable-region genes. Annu Rev Immunol. 1986;4:339–368. doi: 10.1146/annurev.iy.04.040186.002011. [DOI] [PubMed] [Google Scholar]
- Yoshikai Y., Anatoniou D., Clark S. P., Yanagi Y., Sangster R., Van den Elsen P., Terhorst C., Mak T. W. Sequence and expression of transcripts of the human T-cell receptor beta-chain genes. Nature. 1984 Dec 6;312(5994):521–524. doi: 10.1038/312521a0. [DOI] [PubMed] [Google Scholar]