We used live imaging and EM to study migration of multiple nuclei in A. gossypii. Three types of nuclear movements, oscillation, rotation, and bypassing, depend on cytoplasmic microtubules while a fourth type, co-transport with the cytoplasmic stream, does not. Nuclear MTOCs emanating perpendicular and tangential cMTs lead cMT-dependent movements
Abstract
We investigated the migration of multiple nuclei in hyphae of the filamentous fungus Ashbya gossypii. Three types of cytoplasmic microtubule (cMT)-dependent nuclear movements were characterized using live cell imaging: short-range oscillations (up to 4.5 μm/min), rotations (up to 180° in 30 s), and long-range nuclear bypassing (up to 9 μm/min). These movements were superimposed on a cMT-independent mode of nuclear migration, cotransport with the cytoplasmic stream. This latter mode is sufficient to support wild-type-like hyphal growth speeds. cMT-dependent nuclear movements were led by a nuclear-associated microtubule-organizing center, the spindle pole body (SPB), which is the sole site of microtubule nucleation in A. gossypii. Analysis of A. gossypii SPBs by electron microscopy revealed an overall laminar structure similar to the budding yeast SPB but with distinct differences at the cytoplasmic side. Up to six perpendicular and tangential cMTs emanated from a more spherical outer plaque. The perpendicular and tangential cMTs most likely correspond to short, often cortex-associated cMTs and to long, hyphal growth-axis–oriented cMTs, respectively, seen by in vivo imaging. Each SPB nucleates its own array of cMTs, and the lack of overlapping cMT arrays between neighboring nuclei explains the autonomous nuclear oscillations and bypassing observed in A. gossypii hyphae.
INTRODUCTION
Nuclear migration is essential for normal growth and development of basically all eukaryotes (reviewed in Morris, 2000). In many cells, controlled nuclear migration depends on microtubules and their organizing centers (MTOCs). Despite being a morphologically diverse group of organelles, the function of MTOCs in nucleating and anchoring microtubules is highly conserved throughout all eukaryotes. Well-established and researched examples of MTOCs are the nuclear-associated centrosome of animal cells and the spindle pole body (SPB) of budding yeast (reviewed in Jaspersen and Winey, 2004; Luders and Stearns, 2007). A defining feature of all MTOCs is the presence of γ-tubulin, a phylogenetically conserved protein responsible for microtubule nucleation in vivo (Oakley and Oakley, 1989; Wiese and Zheng, 2006; Luders and Stearns, 2007). Based on the ability of γ-tubulin and its associated proteins to form a ring-shaped complex with an approximate diameter of a microtubule cylinder, it probably templates assembly of α- and β-tubulin heterodimers and serves as a cap to stabilize the microtubule minus end.
In multinucleated hyphae of filamentous fungi, nuclei are in continuous motion mainly, but not exclusively, in the direction of the growing hyphal tips. The mechanism of this long-range nuclear transport is not well understood. Cytoplasmic (cMTs) and MTOCs play an important role in this process as documented in several fungal species (Inoue et al., 1998; Minke et al., 1999a,b; Morris, 2000; Suelmann and Fischer, 2000; Requena et al., 2001; Xiang and Fischer, 2004; Veith et al., 2005; Mourino-Perez et al., 2006; Fischer et al., 2008), but it had also been argued that microtubule-independent mechanisms are essential for long-range nuclear migration (Meyer et al., 1988; Mourino-Perez et al., 2006). Of particular interest are studies showing that in filamentous fungi, including Aspergillus nidulans and Ustilago maydis, the γ-tubulin complex localizes not only to nuclear-associated MTOCs but also to other sites like hyphal tips and septa (Heitz et al., 2001; Straube et al., 2003; Konzack et al., 2005; Sawin and Tran, 2006; Zekert and Fischer, 2009). As shown by these authors, nonnuclear MTOCs have the ability to give rise to different populations of cMTs, including overlapping antiparallel microtubules that interconnect with each other to help regulate spatial positioning within the cytoplasm.
Hyphae of the filamentous fungus Ashbya gossypii contain many nuclei in the same cytoplasm. On hyphal extension, nuclei migrate toward the growing tip, showing not only long-range migration but also extensive oscillations and occasional bypassing of one another (Alberti-Segui et al., 2001). Nuclei do not divide synchronously but undergo asynchronous mitoses, pointing to a certain level of autonomy of individual nuclei within a common cytoplasm (Gladfelter et al., 2006). Nuclear division, bypassing, oscillation, and tip-directed movement are all assumed to be important for maintaining an approximately equidistant distribution of nuclei along the hypha (Alberti-Segui et al., 2001; Gladfelter et al., 2006; Kaufmann and Philippsen, 2009).
Unlike other filamentous fungi, the genome of A. gossypii is closely related to that of the budding yeast Saccharomyces cerevisiae; homologues of 95% of the 4720 A. gossypii genes have conserved gene order (synteny) with the S. cerevisiae genome, strongly indicating that both organisms derived from a common ancestor (Dietrich et al., 2004). Despite high similarity on the genome level, both organisms have a very distinct morphology. A. gossypii proliferates by sustained apical growth of multinucleated hyphae, which occasionally form septa between groups of eight to 10 nuclei but do not undergo cell divisions (Wendland and Walther, 2005; Kaufmann and Philippsen, 2009). In contrast, S. cerevisiae is a unicellular fungus that proliferates by growth of single daughter cells that separate from the mother cells after completion of mitosis. Its nuclear dynamics are relatively simple compared with A. gossypii; positioning of the nucleus at the mother-daughter bud neck before cytokinesis is the major nuclear movement required for mitotic growth. Efficient gene targeting is possible in A. gossypii just as it is in S. cerevisiae (Wendland et al., 2000; Kaufmann, 2009), making this organism a powerful system to study the mechanistic basis for nuclear migration in multinucleated hyphae, and, at the same time, to increase our knowledge about adaptive evolution due to its natural homology to budding yeast.
Indirect immunofluorescence microscopy of microtubules in A. gossypii suggests that they could be interconnected (Alberti-Segui et al., 2001), and this network of overlapping microtubules has been proposed to play a role in its complex nuclear dynamics, which was supported by early in vivo imaging data (Philippsen et al., 2005). Like other filamentous fungi, A. gossypii may use more than one type of MTOC to nucleate its microtubule cytoskeleton. However, based on the similarity between the A. gossypii and S. cerevisiae genomes, A. gossypii may solely use nuclear-associated SPBs to form the cMTs that regulate its nuclear dynamics. In such case, a nucleation capacity for longer, and probably also an increased number of cMTs, compared with S. cerevisiae, has to be hypothesized for A. gossypii SPBs to account for the differences in cell dimensions: an average haploid budding yeast cell is ∼10 μm long during mitosis, whereas a Ashbya hypha can extend approximately >50 μm between septa and the growing tip (Kaufmann and Philippsen, 2009). Modifications of tubulin and/or microtubule-associated proteins may also play a role in the differences in cMTs between the two organisms.
Here, we demonstrate by in vivo imaging that A. gossypii only forms nuclear-associated SPBs, which lead fast moving nuclei in the direction of migration, drive nuclear rotation to facilitate inversion of the nuclear migration direction, display micromovements in different directions sometimes following the direction of an attached cMT and nucleate up to six cMTs that often extend in opposite directions and bypass other SPBs without making connected networks. We find that three nuclear movements, oscillation, rotation, and bypassing, are dependent on cMTs, whereas the fourth and most basic type of long-range nuclear migration, which is driven by cytoplasmic streaming, is cMT-independent. To determine how cMTs are organized in A. gossypii to facilitate its unique lifestyle and pattern of nuclear dynamics, we analyzed SPB ultrastructure in serial thin sections by electron microscopy (EM). Comparison of A. gossypii and S. cerevisiae SPBs showed several important differences in the multilayered organelle between the two organisms. Most notably, the A. gossypii SPB has an altered outer plaque (OP) that can nucleate both perpendicular and tangential cMTs. This later class of cMTs has not been observed in budding yeast.
MATERIALS AND METHODS
A. gossypii Media and Growth Conditions
A. gossypii media and culturing are described in Ayad-Durieux et al. (2000) and Wendland et al. (2000), and strains are listed in Supplemental Table S1. To depolymerize microtubules, spores were grown for 16 h at 30°C in liquid AFM before nocodazole (Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 15 μg/ml. For microtubule regrowth experiments, cells were incubated 1 h at 30°C under shaking conditions before washing five times in AFM and incubation in AFM for 1 h at 30°C to allow microtubule repolymerization. Samples were taken before and after nocodazole treatment and every 10 min after nocodazole washout and immediately fixed for immunofluorescence staining. For time-lapse imaging of nocodazole treated hyphae, the cells were incubated 20 min in liquid AFM at a final concentration of 15 μg/ml nocodazole under shaking conditions before 70 μl of the cell suspension were transferred to a time-lapse agarose slide containing 15 μg/ml nocodazole.
Plasmid and Strain Construction
Plasmids generated and used in this study are described below. All DNA manipulations were carried out according to Sambrook and Russell (2001), with Escherichia coli DH5αF′ as host (Hanahan, 1983). Polymerase chain reaction (PCR) amplification was performed using standard methods with Taq DNA polymerase, Expand High Fidelity PCR system or the Expand Long Template PCR system (Roche Diagnostics, Indianapolis, IN). Oligonucleotides are listed in Supplemental Table S2 and were synthesized by Microsynth (Balgach, Switzerland). For recombination of plasmids and PCR products, both were cotransformed into the budding yeast host strain DY3 (MATα his3Δ200 trp1Δ63 leu2Δ1 ura3-52Δ) according to Gietz et al. (1995). Plasmids were isolated from yeast using the High Pure Plasmid Purification kit (Roche Diagnostics), with a modified protocol as described previously (Schmitz et al., 2006).
To generate the green fluorescent protein (GFP)-AgTub1 strain, the plasmid pCB2 was constructed as follows: the prom GFP-AgTUB1 fusion (764 base pairs upstream TUB1-opne reading frame [ORF], GFP-ORF without stop codon, 1347 base pairs TUB1-ORF plus downstream sequences) was isolated as BamHI/HindIII fragment form pFS197 (kindly provided by Florian Schaerer, Biozentrum University of Basel, Basel, Switzerland). Blunt ends were generated using the 3′–5′ exonuclease activity of T4 polymerase, and the 3923-base pair fragment was subcloned into pAIC opened at the ScaI site (Knechtle, 2002), thereby reconstituting a functional AgADE2 gene. pCB2 was digested with EcoRI and HindIII, and transformed into the partially deleted AgADE2 locus of the Agade2Δ1 strain (Knechtle, 2002). Transformants were obtained on minimal medium lacking adenine and verified by PCR analysis with primer pairs Agade2verfor_CB/Agade2_verup_CB and Agade2verrev_CB/Agade2_verds_1.
pAGT123 (Kaufmann, 2009) was used as a template to amplify YFP-LEU2 by using oligonucleotides AgTUB4_F1/F2. The resulting PCR product was cotransformed into yeast cells with pAG10748 that carries a genomic copy of AgTUB4 cloned into a pRS415 backbone (Dietrich et al., 2004) to generate pAgTUB4-YFP-LEU2. pAgTUB4-YFP-LEU2 was digested with AvaI and SacII to tag the endogenous AgTUB4 gene with YFP in the wild-type strain. Integration was verified with oligonucleotides pairs AgTUB4_A3/green2.2 and L3/AgTUB4-A4.
pAGT144 (Kaufmann, 2009) was used as a template to amplify RedStar2-GEN3 by using oligonucleotides AgTUB4_F1/F2. The resulting PCR product was cotransformed into yeast cells with pAG10748 to generate the plasmid pAgTUB4-RedStar2-GEN3. To generate AgH4-GFP Tub4-RFP the plasmid pAgTUB4-RedStar2-GEN3 was transformed into AgH4-GFP. This strain was always grown under selective conditions with 200 μg/ml G418 (Sigma-Aldrich) to maintain the plasmid.
Fluorescence Microscopy and Image Processing
Chitin (calcofluor white), DNA (Hoechst), and immunofluorescence stainings were performed as described previously (Ayad-Durieux et al., 2000; Gladfelter et al., 2006). Rat anti-α-tubulin (YOL1/34; Serotec, Oxford, United Kingdom) was used at a 1:25 dilution and Alexa Fluor 568 goat anti-rat immunoglobulin G (Invitrogen, Carlsbad, CA) at a 1:200 dilution.
An Axioplan2 microscope equipped with the objectives Plan-Apochromat 100×/1.40 numerical aperture (NA) Oil differential interference contrast (DIC) and Plan-Apochromat 63×/1.40 NA Oil DIC (Carl Zeiss, Feldbach, Switzerland) and appropriate filters (Carl Zeiss and Chroma Technology, Brattleboro, VT) was used for microscopy. The light source for fluorescence microscopy was either a 75-W XBO lamp (OSRAM, Augsburg, Germany), controlled by an MAC2000 shutter and filter wheel system (Ludl Electronics, Hawthorne, NY) or a Polychrome V monochromator (TILL Photonics, Gräfelfing, Germany). Images were acquired at room temperature using a cooled charge-coupled device camera CoolSNAP HQ (Photometrics, Tucson, AZ) with MetaMorph 6.2r5 software (Molecular Devices, Sunnyvale, CA). Out-of-focus shading references were used for DIC image acquisitions. For fluorescence images, multiple planes with a distance between 0.3 and 1 μm in the z-axis were taken. For time-lapse image acquisition, a glass slide was covered with 1 ml of A. gossypii minimal medium containing 1% agarose. Once the medium had solidified, 70 μl of young mycelia cultured in liquid medium was spotted onto the slides and covered by a coverslip.
Image processing was performed with MetaMorph 6.2r5 software. Z-stacks were deconvolved with Nearest Neighbor and compressed by maximum or average projection with Stack Arithmetic. Brightness and contrast were adjusted using Scale Image. Images were colored and overlaid by using Overlay Images and exported from MetaMorph as 8-bit grayscale or RGB TIFF files. Z-stacks were converted to QuickTime H.264 movies with QuickTime Player Pro (Apple Computer, Cupertino, CA).
Transmission Electron Microscopy
Spores were grown for 10–14 h in liquid AFM to give rise to small mycelia containing no >100 nuclei. Samples were frozen on EM-Pact (Leica, Wetzlar, Germany) at ∼2050 bar and then transferred under liquid nitrogen into 2% osmium tetroxide/0.1% uranyl acetate/acetone and transferred to an automated freeze-substitution apparatus (AFS) (Leica, Wetzlar, Germany). The freeze substitution protocol was as follows: −90°C for 16 h, up 4°C/h for 7 h, −60°C for 19 h, up 4°C/h for 10 h, −20°C for 20 h. Samples were removed from the AFS and placed in the refrigerator for 4 h and then allowed to incubate at room temperature for 1 h. Samples went through three changes of acetone over 1 h and were removed from the planchettes. Then, they were embedded in acetone/Epon mixtures to final 100% Epon over several days in a stepwise procedure as described previously (McDonald, 1999). Then, 60-nm serial thin sections were cut on a UC6 (Leica, Wetzlar, Germany), stained with uranyl acetate and Sato's lead and imaged on a Technai Spirit transmission electron microscope (FEI, Hillsboro, OR). Serial section images were aligned using AutoAligner (Bitplane, Zurich, Switzerland).
RESULTS
The SPB Is the Sole Site of Microtubule Nucleation in A. gossypii
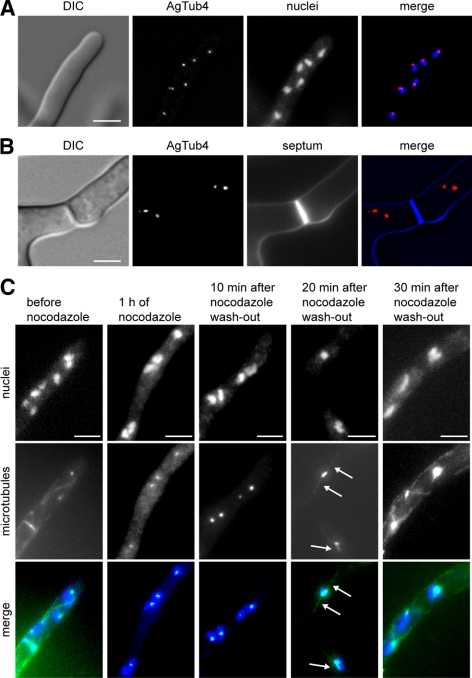
We first wanted to determine whether SPBs are the only site of microtubule nucleation in A. gossypii as they are in budding yeast or if other nonnuclear MTOCs also exist. To investigate the sites of microtubule nucleation, we fused the A. gossypii homologue of γ-tubulin with YFP because labeling of Tub4 with fluorophores has been used to visualize both nuclear and nonnuclear MTOCs in various fungi (Oakley et al., 1990; Horio et al., 1991; Stearns et al., 1991; Sobel and Snyder, 1995; Marschall et al., 1996; Spang et al., 1996; Heitz et al., 2001). AgTub4-yellow fluorescent protein (YFP) appeared as small foci fairly evenly distributed along the length of the hypha. Colabeling of nuclei with Hoechst revealed that AgTub4-YFP localizes exclusively to one or two spots at the nuclear periphery (Figure 1A). It was never observed at other locations such as hyphal tips or septa (Figure 1B).
Figure 1.
Localization of MTOCs in A. gossypii. (A) Images of a tip compartment expressing AgTUB4-YFP to localize MTOCs and stained with Hoechst to visualize nuclei. (B) Images of a hyphal compartment with a calcofluor stained septum and YFP-marked MTOCs. No YFP fluorescence can be seen at the septum. (C) Images of A. gossypii hyphae before and after incubation with 15 μg/ml nocodazole for 1 h and at different times after washout of the drug. Nuclei are stained with Hoechst and microtubules are labeled with anti-α-tubulin antibodies. The untreated hypha shows cMTs, one metaphase spindle before orientation along the growth axis and fluorescent foci representing SPBs. After nocodazole treatment only SPB foci are detectable with anti-α-tubulin antibodies. Twenty minutes after nocodazole washout, faintly stained short cMTs reemerged at the nuclear SPBs (arrows). By 30 min, the microtubules seemed similar to untreated cells. Bars, 5 μm (A–C).
We also analyzed the sites of microtubule regrowth in A. gossypii after microtubule depolymerization using nocodazole. Newly formed microtubules that emerged after washout of nocodazole were associated with nuclei and were not seen at other sites (Figure 1C). Together, these results strongly suggest that similar to S. cerevisiae, perinuclear SPBs are the sole site of microtubule nucleation in A. gossypii. Thus, only microtubules nucleated at SPBs participate in nuclear dynamics of A. gossypii hyphae.
Dissection of Movements of SPBs and Nuclei in A. gossypii
Using time-lapse microscopy, we monitored hyphae expressing histone AgH4-GFP and AgTub4-RedStar2 to visualize possible connections between nuclear movements and the locations of SPBs. In total, 37 nuclei were monitored for 10 min in five hyphae growing at speeds from 0.15 to 1.28 μm/min. Table 1 summarizes the movements of all 37 nuclei examined, including the movement of nuclei and their SPBs in the slowest growing hypha (Figure 2) and hyphae with more rapid growth speeds (Supplemental Figure S1).
Table 1.
Nuclear movements in A. gossypii hyhae growing with different speeds
| Hypha 1 | Hypha 2 | Hypha 3 | Hypha 4 | Hypha 5 | |
|---|---|---|---|---|---|
| Hyphal speed (μm/min) | 0.15 | 0.53 | 0.83 | 0.94 | 1.28 |
| No. of nuclei screened | 11 | 5 | 8 | 5 | 6 |
| Time intervals analyzed | 110 | 68 | 168 | 100 | 120 |
| Forward movements (%)a | 34 (31) | 22 (33) | 60 (36) | 48 (48) | 75 (62) |
| Max continuous forward movement (min) | 4 | 3 | 4 | 4 | 7 |
| Backward movements (%)a | 30 (27) | 21 (30) | 23 (14) | 11 (11) | 14 (12) |
| Max continuous backward movement (min) | 5 | 2 | 2 | 1.5 | 1.5 |
| Fast bypassing eventsb | 4 | 1 | 0 | 0 | 5 |
| Max bypassing speed (μm/min) | 5.9 | 9.4 | 0 | 0 | 9.2 |
| Switch of direction in 10 min | 0–3 times | 1–4 times | 0–4 times | 2–3 times | 0–4 times |
| SPB-lead movements | 64/64 | 40/43 | 82/83 | 58/59 | 87/89 |
| 180° rotations in 1 min | 3 | 6 | 9 | 3 | 11 |
| Max speed of oscillation (μm/min)a | 2.8 | 4.2 | 4.5 | 4.9 | 4.8 |
| Tumbling nuclei (%)c | 46 (41) | 25 (37) | 85 (50) | 41 (41) | 31 (26) |
a Forward and backward refer to movements of nuclei during oscillation (no close contact to other nuclei) and to bypassing events. See Figure 2 and Supplemental S1.
b Bypassing is the movement of one nucleus past at least one adjacent nucleus. Fast bypassing events occur within 0.5–1 min when the nucleus moves up to 5 μm/30 s. Slow bypassing events, which last up to 3 min, are rare. See Figure 2 and Supplemental S1.
c Tumbling nuclei exhibit neither oscillation nor bypassing movements but undergo rotational movements of <45°/min. See Figure 2C.
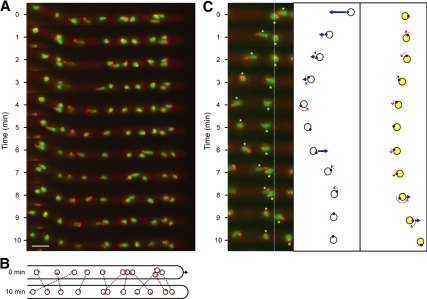
Figure 2.
Oscillation, rotation and bypassing of nuclei in an A. gossypii hypha. Hyphae expressing AgH4-GFP to mark nuclei (green) and AgTub4-RedStar2 to mark SPBs (red) were pregrown and mounted for fluorescence videomicroscopy at room temperature as described in Materials and Methods. GFP and red fluorescent protein (RFP) fluorescence was imaged for 500 ms each in five Z-planes, 0.75 μm apart at 1-min intervals and processed (Supplemental Movie S1). (A) Maximal projections at 11 time points showing frequent changes in the position and SPB orientation of the 11 nuclei monitored in Supplemental Movie S1. During the 110 analyzed 1-min time intervals, nuclei moved forward 34 times and backward 30 times, all of which were led by the SPB. During the remaining 46 intervals, nuclei moved only marginally. Nonmoving as well as moving nuclei rotated during 53 intervals by up to 180° as seen by changes of the SPB positions. The relative position of SPBs in all fast-moving nuclei was “head-first” (at the leading edge of nuclear movement), and inversions in the direction of nuclear movement were preceded by inversion of the SPB position. Bar, 5 μm. (B). Diagram of the position of nuclei (circles) and orientation of SPBs (red dots) at 0 and 10 min. Identical nuclei are connected by dotted lines highlighting the four bypassing events. The arrow marks the 1.5-μm elongation of the hypha during 10 min, which was measured using the overilluminated montage of A to visualize the background staining of the hyphal cytoplasm. (C). Detailed diagrammatic presentation of backward and forward as well as rotational movements of two nuclei (6, white asterisk and 7, yellow asterisk) within 10 min (movie frames 3–13 of Supplemental Movie S1). Nucleus 6 bypasses nucleus 7 by migrating 5 μm backward, inverts the position of the SPB by rotation at 4 and 5 min, and migrates 3 μm forward. Nucleus 7 represents a typical tumbling nucleus showing minor movements and small angle rotations until 8 min, at which time it undergoes a rotation and then a forward movement of 2 μm. Fast-moving nuclei in A and C seem to be stretched and sometimes display two SPBs. This does not reflect the real shape of fast nuclei or duplicated SPBs however; rather, it is due to the high migration speed and the fact that these images show projections of five focal planes taken within 6 s.
From our analysis of these nuclei, we found that the percentage of forward movements (in the direction of hyphal growth) increases with increasing hyphal speed and is concomitant with a decrease in the percentage of backward movements. In contrast, other types of nuclear movement were independent of hyphal speed, including rotations of nuclei and bypassing. One of the most striking observations was that 98% of all nuclear movements are led by the SPB. It is also remarkable that nuclei can invert their movement and reorient the position of their SPB by 180° within 30 s and that the frequency of such switches is up to 4 times in 10 min. Although bypassing was observed in hyphae of different growth speeds, an exceptional case of long-range nuclear migration was observed in the fastest hypha. The sixth nucleus bypassed four of the five more apical nuclei within 10 min and migrated 50 μm during that time (data not shown).
Nuclear Oscillations, Bypassing, and Rotation Require cMTs
When A. gossypii hyphae are treated with the microtubule-depolymerizing drug nocodazole, nuclei no longer show oscillations and bypassing as reported previously (Alberti-Segui et al., 2001). We repeated this experiment with the strain expressing the histone AgH4-GFP and Tub4-RedStar2 fusions to test whether all forms of nuclear migration in multinucleated hyphae depend on specific orientations of SPBs and an intact cMT cytoskeleton. In A. gossypii, the transport of secretory vesicles does not depend on cMTs (Köhli et al., 2008); thus, hyphae continue to grow even in the absence of cMTs.
After preincubation with 15 μg/ml nocodazole for 20 min no cMTs could be detected by immunofluorescence (Figure 1). Forty-eight nuclei in three hyphae were monitored at time intervals of 1 min; none showed oscillation, bypassing or rotation to orient SPBs in the direction of nuclear movements. However, all nuclei still migrated steadily in the direction of the growing hyphae. Figure 3A shows this type of migration in one hypha with 11 nuclei. SPBs assume apparently random positions with some tumbling rotations. The speeds of the nuclei followed a simple scheme: nuclei close to the tip migrated as fast as the tip elongation speed, whereas nuclei further subapically migrated with steadily decreasing speeds as illustrated in Figure 3B. The same pattern of position-dependent nuclear speed decrease was measured for two other hyphae with 21 and 16 nuclei, respectively (data not shown). Only forward movements were observed, with highest speeds close to the hyphal tips and lowest speeds for the most subapical nuclei.
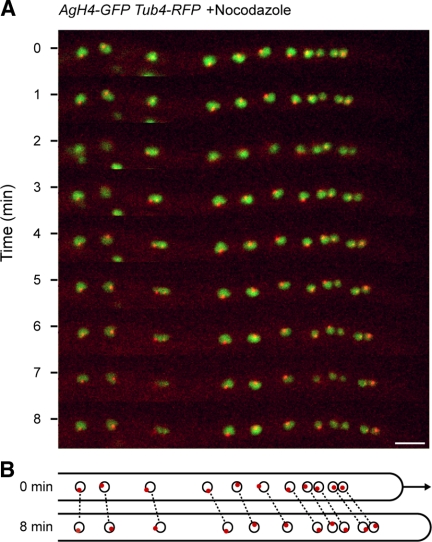
Figure 3.
Migration of nuclei in the absence of cMTs. The strain expressing AgH4-GFP (green) and AgTub4-RedStar2 (red) was preincubated in the presence of 15 μg/ml nocodazole for 20 min and mounted for fluorescence videomicroscopy on agar slices containing the same concentration of nocodazole. GFP and RFP fluorescence was imaged for 500 ms each in three Z-planes, 0.75 μm apart, at 30-s intervals and processed (Supplemental Movie S2). (A) Nine maximal projections of Supplemental Movie S2 at 1-min intervals showing 11 evenly moving nuclei with SPBs only rarely oriented in the direction of migration. Bar, 5 μm. (B) Diagram of positions of nuclei (circles) and orientation of SPBs (red dots) at 0 and 8 min. Identical nuclei are connected by dotted lines. The arrow marks the elongation of the hypha within 8 min.
This clearly indicates one main mechanism for nuclear migration: cotransport with cytoplasmic streaming. Porins, responsible for water influx are most likely evenly distributed along the hyphal cortex. The incoming water molecules are directed toward the growing tip, the only site of surface expansion in a fungal hypha. Thus, cytoplasmic streaming is slowest in the most subapical region, for example close to a septum, and highest in the tip region. Cotransport by cytoplasmic streaming is cMT-independent and can be viewed as the basic mechanism for nuclear migration in A. gossypii hyphae. Superimposed are three cMT-dependent movements of nuclei: oscillation, bypassing and rotation, all very likely dependent on the ability of SPBs to organize the cMTs.
In Vivo Imaging of cMTs and Micromovements of SPBs in A. gossypii
The mobility of SPBs in Figures 2 and 3 was monitored with a time resolution of 0.5–1 min. To get a more refined picture about mobilities of SPBs in multinucleated hyphae and the arrangement of cMTs, we wanted to perform time-lapse studies with a resolution of a few seconds using a strain expressing GFP-AgTub1. These experiments turned out to be extremely challenging, as A. gossypii seems to tolerate only weakly expressed GFP-AgTub1 in the presence of normal expression levels of untagged α-tubulin, which results in a low signal-to-noise ratio. To visualize the very thin cMTs, exposure times of 1.5 s are necessary, preventing acquisition of a sufficiently large number of images (e.g., for 4-dimensional movies) before bleaching of the GFP-AgTub1 signal. However, by focusing only on one Z-plane in the middle of hyphae, we could visualize micromovements of SPBs at a time resolution of 6 s (Supplemental Movie S3).
As an example, 11 consecutive frames of Supplemental Movie S3 are presented in Figure 4A. Eighty percent of the SPBs changed their locations within 6 s by an average of 0.32 μm. In consecutive intervals, SPBs rarely moved in one contiguous direction and pausing was often seen. This raises the question how these heterogeneous micromovements of SPBs can lead to the observed nuclear movements in the micrometer range. We therefore determined the net movement of SPBs during the 7 min of Supplemental Movie S3. For example, the net movement of the second SPB of Figure 4A is 2 μm backward during the first min, 4.5 μm forward during the next 4 min, and 1 μm backward during the final 3 min, an overall movement typical for nuclear oscillation (Table 1 and Supplemental Figure S1).
Figure 4.
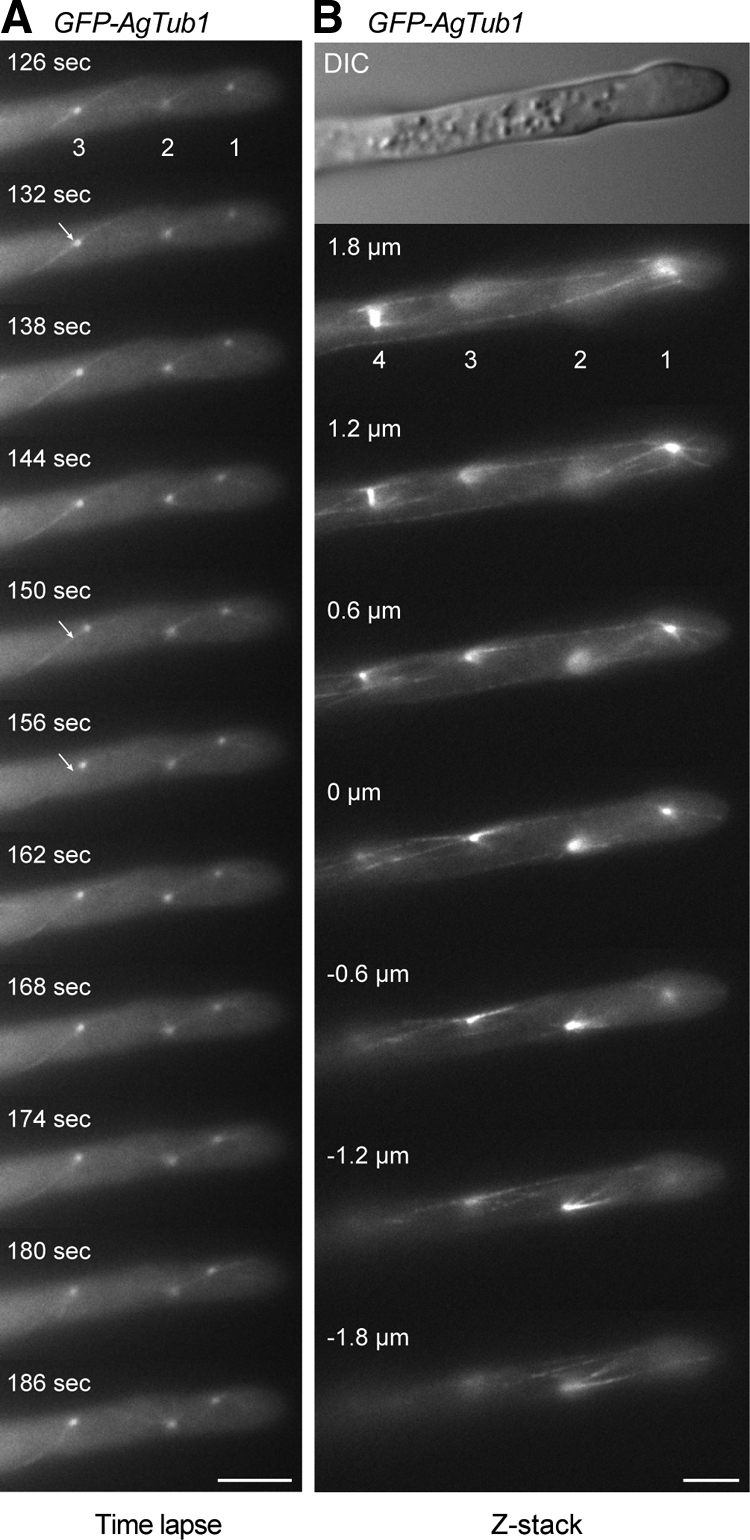
Motility of SPBs and visualization of cMT arrays. Hyphae expressing GFP-tagged and nontagged AgTub1 were pregrown and mounted for fluorescence microscopy on thin agar slices. Images were taken from a single Z-plane every 6 s with 1.5-s exposure time for 7 min (Supplemental Movie S3) or as Z-stack with 18 planes each 0.3 μm apart and 1.5-s exposure time (Supplemental Movie S4). (A) 11 representative frames of Movie S3 showing three SPBs as distinct dots and attached cMTs as thin, weakly fluorescent filaments. The apparent connection of SPBs 1 and 2 (frames 162–180 s) represents two cMTs superimposed by chance because both are visible as two independent cMTs in frame 186 s. Bar, 5 μm. (B) Images of selected focal planes of the Z-stack (Supplemental Movie S4) of a hypha expressing GFP-AgTub1. Four SPBs and attached cMTs are seen in the different focal planes. Short and long cytoplasmic microtubules emanate from both sides of bright foci representing SPBs (1–3) and from SPBs of a metaphase spindle (4). Short cytoplasmic microtubules frequently point toward the cell cortex; long cytoplasmic microtubules run along the longitudinal axis of the hyphae and can often be followed over several focal planes. They either terminate in the cytoplasm or seem to glide along the cell cortex. The SPB from nucleus 1 is connected to the cell cortex of the tip region by three short cytoplasmic microtubules, whereas at least three longer cytoplasmic microtubules run toward the distal part of the hypha. Bar, 5 μm.
Adjacent SPBs did not seem to coordinate their micromovements in a sense that they comigrated in the same direction. This indicates a high level of autonomy for SPB mobility in A. gossypii hyphae. In rare cases, two SPBs seem to be connected by a cMT. Such an intriguing connection can be seen between the first and the second SPB in Figure 4A (162–180 s). However, the next frame (186 s) indicates two separate cMTs emerging from the first and second SPB, respectively, in the direction of the other SPB. Thus, it does not seem that cMTs from adjacent or distant SPBs form overlapping networks (see below).
The images presented in Figure 4A show one to three cMTs emanating in different directions from each SPB. This number is an underestimation because only those cMTs are visible, which lie within or close to the focal plane of this time-lapse movie. Despite this experimental limitation, we asked whether the direction of the SPB micromovements is determined by the orientation of an attached cMT. For example, the third SPB (Figure 4A, white arrow) is attached to two cMTs that emanate in opposite orientations and both reach the hyphal cortex (132 s). This SPB migrates 0.9 μm, two steps with one pause, in the direction of the upper cMT (150 s) and then 0.4 μm back in the direction of the lower cMT (156 s), indicating that in this specific instance, force generated by single cMTs probably guided the movement of the SPB. This may be pulling forces similarly as described for SPB movements in meiotic nuclear oscillations of Schizocaccharomyces pombe (Vogel et al., 2009). If this is indeed the case, the pulling force for SPB movements in A. gossypii will most likely result from several cMTs emanating from SPBs in different orientations.
Arrangement of cMTs Attached to Individual SPBs
To overcome the limitations of single plane movies to determine numbers and arrangements of cMTs, we tracked SPBs and microtubules in the GFP-AgTub1 strain through Z-stacks of 18 focal planes with 300-nm distances for nine apical hyphal compartments each carrying four to eight nuclei. Representative images from one Z-stack documenting this analysis for four SPBs are shown in Figure 4B. Arrays of up to 6 short and long cMTs emanate from bright foci representing SPBs 1–3. Array 4 consists of two SPBs of a metaphase spindle, each connected to three cMTs. In total, we analyzed 55 SPBs and found on average 4.25 ± 1.4 microtubules emanating per SPB. Short cMTs (<5 μm) radiated from the SPB in different directions into the cytoplasm often reaching the cortex. Long cMTs (>5 μm) were oriented along the polarity-axis and frequently bypassed one or several other SPBs (e.g., top four images of Figure 4B). By tracking long cMTs through several planes, some could be followed over distances up to 17 μm. Of the 55 analyzed SPBs, 50 nucleated both short and long cMTs, and 37 SPBs nucleated long cMT in opposite orientations.
We also searched for cMTs connecting two SPBs, that is microtubules emanating from different SPBs and potentially aligning with their plus ends to form antiparallel bundles. When maximal projections of Z-planes indicated a potential cMT connection between two SPBs, we tracked the two microtubules through three-dimensional space and found that they were running close to each other but did not align (for one example see Supplemental Figure S2 and Supplemental Movie S5). Thus, it is highly unlikely that arrays of anti-parallel cMTs form throughout the mycelium and control the dynamic behavior of nuclei. Rather, our results strongly suggest that both long and short microtubules form a regional microtubule network to guide nuclear oscillations, rotations and bypassing of every nucleus independently. This can explain why nuclei behave autonomously in a contiguous cytoplasm.
The A. gossypii SPB Is Embedded in the Nuclear Membrane
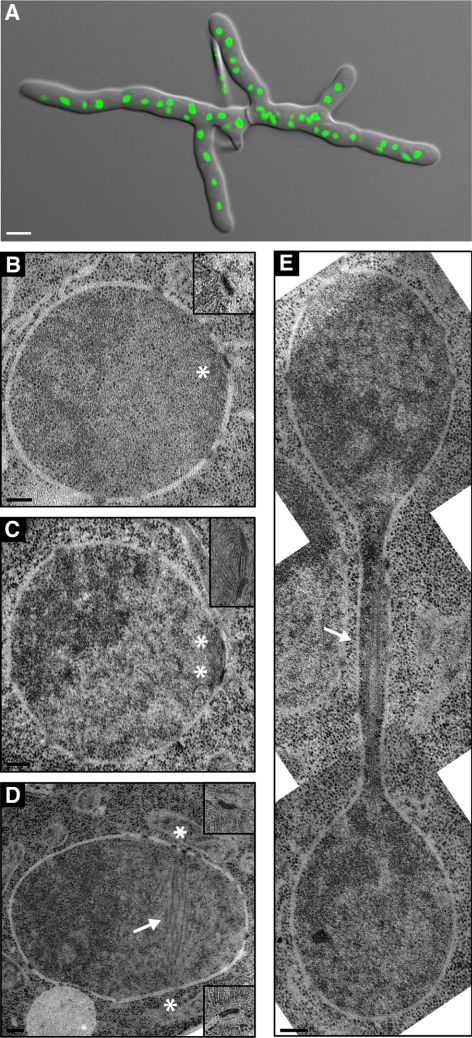
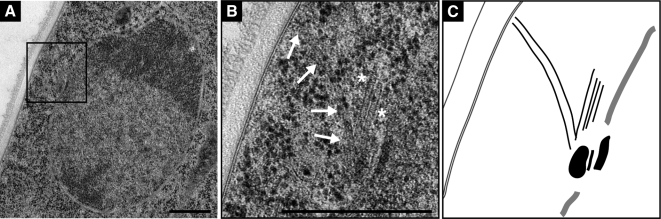
How is it possible that nuclear-associated SPBs organize such complex arrays of cMTs? To better understand the structural basis for the cMT arrays we decided to determine the ultrastructure of A. gossypii SPBs. Electron microscopy (EM) has provided valuable insight not only into SPB structure and assembly but also into microtubule organization in a variety of organisms, including budding and fission yeast. Here, we report the first EM study of nuclei in multinucleate hyphae of A. gossypii. To ensure that all nuclei analyzed by EM were actively dividing, we prepared young mycelium that had no >10 growing tips and contained no >100 nuclei (Figure 5A). These samples were fast-frozen and then freeze-substituted to preserve the structural integrity of the SPBs and microtubules, and serial thin sections (∼60 nm) of nuclei were examined. We found that the nuclear envelope is continuous throughout the nuclear cycle (Figure 5, B–E), indicating that A. gossypii undergoes a closed mitosis. Single SPBs (Figure 5B) or duplicated SPBs (Figure 5, C and D) were embedded in the nuclear envelope throughout the life cycle like in budding yeast.
Figure 5.
EM analysis of nuclei in multinucleated hyphae. (A) Overlay of a DIC and a fluorescence image of a young A. gossypii mycelium that was stained with Hoechst to visualize nuclei. Such mycelia with five to 10 tips and 50–100 nuclei were prepared for thin section EM analysis as described in Materials and Methods. Bar, 5 μm. (B–E) EM of nuclei in different nuclear cycle stages. The continuous nuclear membrane and nuclear pore complexes within the nuclear envelope can be seen in all images. Bars, 200 nm. (B) A single SPB (asterisk) is embedded in the nuclear envelope. A higher magnification is shown in the top right corner. (C) Duplicated SPBs (asterisks) connected by a bridge are embedded in the nuclear envelope. A higher magnification is presented in the top right corner. (D) A nucleus with spindle microtubules (arrow) and continuous nuclear membrane. The two SPBs were observed in the adjacent sections at positions marked by the asterisks. Magnifications are shown in the top and bottom right corners. (E) Montage of three EM images showing a nucleus in anaphase. The continuous nuclear envelope and spindle microtubules (arrow) are visible. The SPBs are in other sections.
The A. gossypii SPB Is a Multilayered Structure
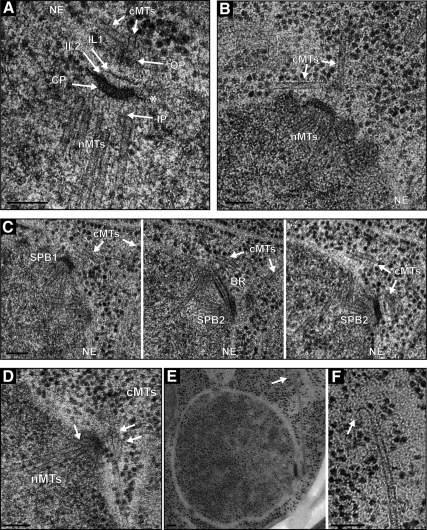
Detailed ultrastructural analysis revealed that A. gossypii SPBs are laminar structures (Figure 6A), which is similar to the SPB of S. cerevisiae (Byers and Goetsch, 1974; Bullitt et al., 1997; Adams and Kilmartin, 1999; O'Toole et al., 1999). An electron-dense central plaque (CP) seems to anchor the SPB in the nuclear membrane via hook-like appendages. The inner plaque (IP) at the nuclear side of the budding yeast SPB lacks a distinctive staining pattern, and this is what we also observe in the region below the CP of A. gossypii SPBs. This ill-defined IP serves as the site of nuclear microtubule formation. Directly above the CP are two sharply staining plaques that correspond to intermediate layer (IL)1 and IL2 of the budding yeast SPB. A region of amorphous material follows, from which cMTs emanate (Figure 6, A–C), indicating it is the outer plaque (OP). Two types of cMTs emerge from the OP, one in a perpendicular manner and a second in a tangential direction. Perpendicular and tangential microtubules were observed emanating from unduplicated, duplicated and separated SPBs, indicating that their formation is very likely not regulated during the mitotic division cycle (Figure, 6 A–D). The existence of two types of OP microtubules can explain the arrangement of cMTs observed by in vivo imaging.
Figure 6.
High-resolution EM analysis of A. gossypii SPBs and microtubules. (A) Electron micrograph showing five discrete SPB layers: the inner plaque (IP), central plaque (CP), intermediate layer 2 (IL2), intermediate layer 1 (IL1) and outer plaque (OP). Hook-like appendages (asterisk) extending from the CP anchor the SPB in the nuclear envelope (NE). Nuclear microtubules (nMTs) emanate from the IP and cMTs (cMTs) from the OP. (B) Two types of cMTs, one extending from the OP in a perpendicular direction and the other in a tangential direction. (C) Three serial sections of a duplicated SPB (SPB1 and SPB2) connected by a bridge (BR). Again, perpendicular and tangential cMTs nucleate at OPs. (D) SPB with capped nuclear and cMTs (arrows). The rounded caps at the microtubule ends are associated with electron dense material. (E) Low- and (F) high-magnification image of a cMT that ends in a distinct flare at its plus end (arrow). Bars, 100 nm (A–F).
A structure equivalent to the S. cerevisiae half-bridge was found in only two of >50 analyzed SPBs. This structure may be present only during a short period of the nuclear cycle and/or may not be detected easily by EM in A. gossypii. However, because we observed duplicated SPBs connected by a bridge (Figures 5C and 6C), it seems likely that A. gossypii uses the half-bridge mechanism for SPB duplication. The bridge is associated with the nuclear envelope and appears as three distinct layers throughout its length (Figure 6C). Interestingly, we observed an unexpectedly high number of nuclei (11 of 39) containing SPBs in this so-called “side-by-side” configuration compared with what is observed in asynchronously growing budding yeast. In S. cerevisiae, the side-by-side configuration of SPBs is only observed in wild-type cells that are at the end of G1 phase of the cell cycle (Byers and Goetsch, 1974, 1975).
Dimensions of A. gossypii SPBs
Even though the general structure of the A. gossypii SPB is similar to that of S. cerevisiae, careful analysis of the size and spacing between A. gossypii SPB layers revealed distinct differences compared with budding yeast SPBs. One of the most notable features of A. gossypii SPBs is the distance between the OP and IL2, which is ∼25 nm greater than it is in S. cerevisiae. There is also a slight increase in the spacing between IL2 and IL1, whereas the distance between the remaining plaques is equivalent (Figure 7). A second major distinction is the shape of the OP. In A. gossypii, the OP does not form a distinctive plaque but rather appears as an amorphous sphere with a diameter significantly reduced compared with the other layers of the SPB (Figure 7). These features result in a considerable degree of variability in the height measurement of the A. gossypii SPBs (Figure 7).
Figure 7.
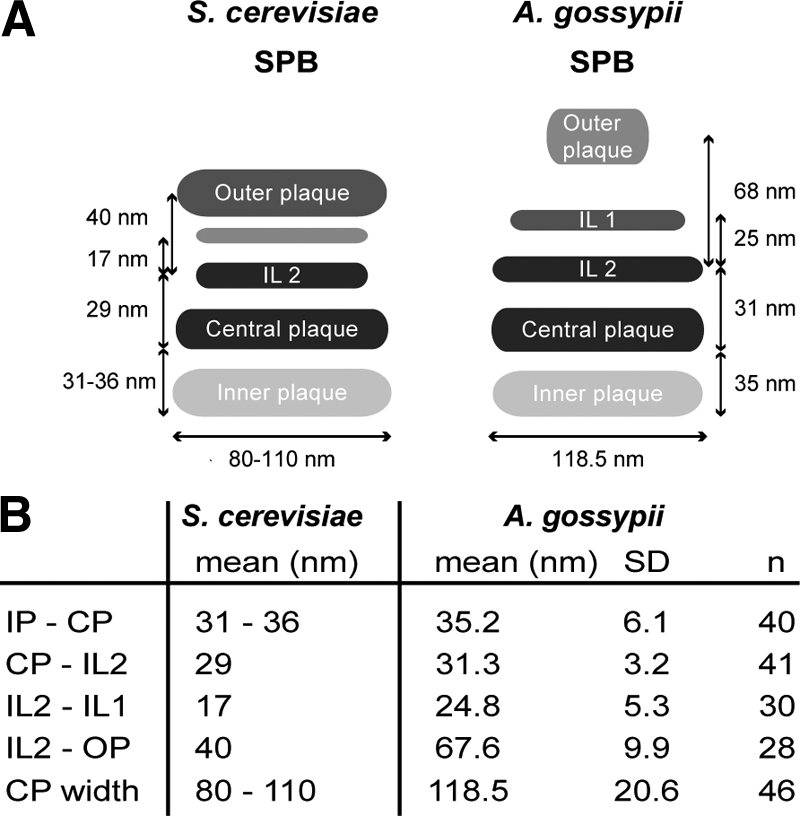
Comparison of A. gossypii and S. cerevisiae SPB structure based on EM analysis. (A) Schematic depicting the S. cerevisiae and A. gossypii SPB layers and distances between SPB layers. Although the CP and IL2 are similar in size and structure in A. gossypii and S. cerevisiae, the IL1 and OP of A. gossypii are considerably smaller and the spacing between those layers is increased compared with budding yeast. The A. gossypii OP appears amorphous rather than electron dense like in S. cerevisiae. (B) Quantitation of distances between A. gossypii SPB layers with SD and number of plaques used for the measurements. The data for S. cerevisiae SPB plaques were compiled from published work (Byers and Goetsch, 1974; Bullitt et al., 1997; O'Toole et al., 1999; Schaerer et al., 2001).
The average diameter of the A gossypii SPB based on the width of the CP was 119 ± 21 nm (n = 46) (Figure 7). This finding has interesting implications in terms of the number of spindle microtubules A. gossypii SPBs are capable of nucleating to ensure segregation of its seven chromosomes. In budding yeast, the diameter of the CP grows from 80 nm in G1-110 nm in mitosis, which is sufficient to nucleate ∼19–20 nuclear microtubules in haploid cells: one microtubule for each of its 16 chromosomes and three to four microtubules to interdigitate with microtubules from the other half spindle (Byers and Goetsch, 1974; Winey et al., 1995).
Structure of the Ends of Microtubules
The microtubule minus ends formed at the IP and OP seemed rounded and often associated with electron dense material (Figure 6D). Based on the similarity to capped microtubules described previously in S. cerevisiae (Byers et al., 1978; Rout and Kilmartin, 1990; Bullitt et al., 1997; O'Toole et al., 1999), these probably are the closed ends of nuclear and cMTs formed at the SPB by the γ-tubulin complex. No difference between the minus ends of nuclear and cMTs was observed. In some cases where a distinct microtubule plus end could be visualized in the cytoplasm, we observed a “flaring” or “peeling” of its tip: one side of the microtubule end extended and curved outward (Figure 6, E and F). These flared microtubule ends were rarely observed on nuclear microtubules in wild-type cells. This structure is highly similar to that described previously for cMTs in budding yeast and probably represents a depolyermizing microtubule end (Byers et al., 1978; Mandelkow et al., 1991; O'Toole et al., 1999).
Types of A. gossypii Microtubules
We observed cMTs that are nucleated perpendicularly to the SPB OP. At least some of these perpendicular cMTs could be tracked to a region near the cell cortex (Figure 8). We suspect that these microtubules correspond to the short microtubules we observed by live cell imaging of GFP-AgTub1–labeled cells. In addition we also observed tangential microtubules extending into the hyphal cytoplasm parallel to the growth axis (Figure 8). This later class of microtubules has not been observed in S. cerevisiae and might account for some of the differences in nuclear dynamics and cytoskeletal organization between the two organisms. The observation that tangential cMTs run in parallel to the hyphal cortex is consistent with the idea that they correspond to the long microtubules that elongate along the polarity-axis as observed by GFP-AgTub1-labeled strains.
Figure 8.
Two types of cMTs in A. gossypii. (A) EM micrograph of a hyphal thin section showing a nucleus with its SPB near the cell cortex. (B) Magnified view of the cortex region with a perpendicular cMT (arrows) ending at the cell cortex and two cMTs (asterisks) tangential to the nucleus and parallel to the hyphal axis (center). (C) Graphic presentation of the SPB and the emanating cMTs shown in B. Bars, 500 nm.
DISCUSSION
Our studies were aimed at understanding the mechanistic basis of nuclear migration in multinucleated fungal hyphae as a model for long-range nuclear transport that occur in a variety of cell types and during development. We could experimentally verify four types of nuclear motility. Three were dependent on SPB-emanating cMTs: back and forth oscillation, rotation to keep the SPB in a “head-first” position and bypassing of other nuclei. In addition, one cMT-independent mechanism exists for basic nuclear migration: cotransport of nuclei with the cytoplasmic stream. If hyphae can grow perfectly well in the presence of microtubule-destabilyzing drugs such as benomyl (Alberti-Segui et al., 2001) and rely solely on cotransport of nuclei by cytoplasmic streaming as documented in this article, why did A. gossypii evolve three cMT-dependent mechanisms for nuclear mobility?
Oscillatory movements may help to keep similar distances between nuclei. SPB-emanating cMTs are also probably needed to actively guide nuclei into emerging branches and to support elongation and orientation of anaphase spindles. Nuclear bypassing facilitates “mixing” of nuclei, which will prevent misfunctioning of a growing hypha if, for example, the front nucleus gains and then spreads through its mitotic division a detrimental mutation for an essential polarity factor. Nuclear mixing may also be beneficial for a fungus such as A. gossypii that carries haploid nuclei and seems to lack a diploid phase; having nonsister nuclei as neighbors could enhance meiotic recombination and the generation of genetic diversity before spore formation. Nuclear rotation seems to be important to keep the SPB head-first before directional change of migration. Alternatively, it could be the direct consequence of an oscillation in which cMTs emanating in different directions from an SPB exert alternating pulling forces.
Earlier studies of fixed A. gossypii cells stained with anti-α-tubulin suggested a complex microtubule network within hyphae (Alberti-Segui et al., 2001). Long cMTs anchored in the nuclear membrane were proposed to interact with microtubules from other SPBs, the cell wall or nonnuclear MTOCs to provide forces that could explain the distinct nuclear dynamics of A. gossypii. Our data suggest that a nuclear-associated SPB in growing A. gossypii hyphae is the sole site of microtubule nucleation with each SPB forming its own cytoskeletal subdomain consisting of short and long cMTs, the short ones often reaching the cortex and the long ones frequently growing far beyond adjacent nuclei (Figure 9A). Interactions between microtubules from adjacent nuclei are, if anything, transient. The ability of nuclei in multinucleate hyphae to form distinct cytoskeletal domains is likely essential for their autonomous dynamic behavior, which is independent from the dynamics of adjacent nuclei. A unique cytoskeletal domain for each nucleus would also permit nonsynchronized oscillatory movements and facilitate orientation of the spindle axis and nuclear bypassing. If we assume that long microtubules are important for nuclear migration along a hypha, the different growth-mode of budding yeast could explain why the ability to nucleate this class of microtubules may have disappeared in the 100 million y between the divergence of A. gossypii and S. cerevisiae. However, our current work does not address the relative contribution of long and short cMTs to nuclear dynamics. Nor does it exclude the possibility that short and long microtubules can interconvert through microtubule growth or shrinkage. Analysis of proteins involved in microtubule nucleation and anchoring will shed light on these questions.
Figure 9.
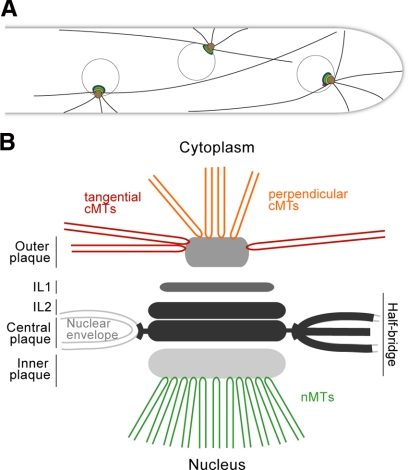
Model of A. gossypii SPB and cMT arrays emanating at SPBs. (A) Model of three nuclei with independent cMT arrays in a hypha. The most apical nucleus has close contact to the growing tip via its short cMTs. Loss of cMTs will increase the distance between the first nucleus and the tip as shown in Figure 3. The two other nuclei are connected with the hyphal cortex via short cMTs and most likely also long cMTs. Growth and shrinkage of these cMTs provides pushing and pulling forces for short-range nuclear oscillations. The most likely pulling force in the direction of a single short cortex-connected cMT was discussed in Figure 4A. Nuclear bypassing is a long-range movement and is very likely achieved by pulling forces of a long cMT when the cortex connection of short cMTs is reduced or absent. Nuclei cannot oscillate or bypass in hyphae lacking short and long cMTs as shown in Figure 3. Nuclei with mutant SPBs able to emanate mainly/only long cMTs cannot oscillate but are still able to bypass other nuclei (unpublished observations). (B) Schematic representation of an A. gossypii SPB and associated microtubules.
We have shown that SPBs are the only MTOCs in the multinucleate hyphae of A. gossypii. SPBs of this filamentous fungus assemble into a multi-layered organelle that is permanently embedded in the nuclear membrane, similar to S. cerevisiae SPBs (Figure 9B). However, the A. gossypii SPB is structurally distinct, particularly on the cytoplasmic side. In our EM images, the OP of A. gossypii appears as an amorphous sphere rather than an electron-dense plaque like in S. cerevisiae. Also, the distance between IL2 and the OP is increased. In budding yeast, Cnm67 controls the spacing between these layers: decreasing or increasing its size by removal or addition of 270 amino acids of protein-internal sequences decreases or increases the IL2-OP distance by ∼20 nm, respectively (Schaerer et al., 2001). Interestingly, AgCnm67 is 281 amino acids longer than ScCnm67 whereas most other pairs of SPB orthologues have similar sizes (Lang et al., 2009). Why would the size of this spacer protein and thus the distance between IL2 and the OP, which nucleates cMTs, have evolved so differently? One striking difference between S. cerevisiae and A. gossypii is the types of cMTs that form at the SPB OP (Figure 9B). In A. gossypii we found both perpendicular and tangential cMTs emanating from the SPB OP. Perhaps an increased distance between IL2 and the OP correlates with the ability to nucleate perpendicular as well as tangential cMTs.
The ability to form cMTs that associate tangentially with the SPB is not unique to A. gossypii and has been observed in other fungi. EM analysis of S. pombe showed cMTs that were oblique with respect to the SPB, similar to the tangential microtubules we observed in A. gossypii. Often the oblique microtubules were not attached to the SPB and may have originated at one of fission yeast's nonnuclear MTOCs (Ding et al., 1997). Tangential cMTs can also be observed in EM preparations of A. nidulans SPBs (Oakley and Morris, 1980, 1983). How these cMTs are formed, including the involvement of the nonnuclear MTOCs, has not been addressed, so it is unclear whether they play a similar function in A. nidulans nuclear dynamics as in A. gossypii hyphae. Neurospora crassa also has two classes of cMTs, one of which is reported to connect adjacent nuclei. The second type of cMT has no obvious association with the nucleus and is probably not nucleated by its SPB (Minke et al., 1999a). In contrast to these organisms, the ability to nucleate cMTs at different angles, including both perpendicular and tangential, must be intrinsic to the SPB in A. gossypii as it is the only MTOC in this organism. Therefore, SPB components or tightly associated proteins with the SPB OP are most likely involved in formation of both types of cMTs.
In conclusion, the multinucleate growth mode of A. gossypii results in unique requirements for microtubule nucleation. Through our analysis of the A. gossypii microtubule cytoskeleton and comparison to that of S. cerevisiae, we can better understand the adaptive properties of the cytoskeleton involved in growth and development of many eukaryotic cells. Our studies provide the groundwork for future analysis of nuclear and microtubule dynamics in the A. gossypii system and illustrate the power of combining high-resolution EM and live cell image analysis.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dominic Hoepfner for guidance in the early stage of this project and to Mark Winey and Tom Giddings for advice and suggestions at later stages. We also thank Jenny Friederichs, Teri Johnson, and Fengli Guo for assistance with EM and Katie Perko for help with AutoAligner. This work was supported by a grant from the Swiss National Science Foundation grant 3100A0-112688 (to P. P.). S.L.J. is supported by a March of Dimes Basil O'Connor Award and the Stowers Institute for Medical Research.
Abbreviations used:
- cMT
cytoplasmic microtubule
- CP
central plaque
- EM
electron microscopy
- IL
intermediate layer
- IP
inner plaque
- MEN
mitotic exit network
- MTOC
microtubule-organizing center
- OP
outer plaque
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0063) on November 12, 2009.
REFERENCES
- Adams I. R., Kilmartin J. V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti-Segui C., Dietrich F., Altmann-Johl R., Hoepfner D., Philippsen P. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 2001;114:975–986. doi: 10.1242/jcs.114.5.975. [DOI] [PubMed] [Google Scholar]
- Ayad-Durieux Y., Knechtle P., Goff S., Dietrich F., Philippsen P. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 2000;113:4563–4575. doi: 10.1242/jcs.113.24.4563. [DOI] [PubMed] [Google Scholar]
- Bullitt E., Rout M. P., Kilmartin J. V., Akey C. W. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb. Symp. Quant. Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Shriver K., Goetsch L. The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae. J. Cell Sci. 1978;30:331–352. doi: 10.1242/jcs.30.1.331. [DOI] [PubMed] [Google Scholar]
- Dietrich F. S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Ding R., West R. R., Morphew D. M., Oakley B. R., McIntosh J. R. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R., Zekert N., Takeshita N. Polarized growth in fungi–interplay between the cytoskeleton, positional markers and membrane domains. Mol. Microbiol. 2008;68:813–826. doi: 10.1111/j.1365-2958.2008.06193.x. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gladfelter A. S., Hungerbuehler A. K., Philippsen P. Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 2006;172:347–362. doi: 10.1083/jcb.200507003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Heitz M. J., Petersen J., Valovin S., Hagan I. M. MTOC formation during mitotic exit in fission yeast. J. Cell Sci. 2001;114:4521–4532. doi: 10.1242/jcs.114.24.4521. [DOI] [PubMed] [Google Scholar]
- Horio T., Uzawa S., Jung M. K., Oakley B. R., Tanaka K., Yanagida M. The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J. Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- Inoue S., Turgeon B. G., Yoder O. C., Aist J. R. Role of fungal dynein in hyphal growth, microtubule organization, spindle pole body motility and nuclear migration. J. Cell Sci. 1998;111:1555–1566. doi: 10.1242/jcs.111.11.1555. [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Kaufmann A. A plasmid collection for PCR-based gene targeting in the filamentous ascomycete Ashbya gossypii. Fungal Genet. Biol. 2009;46:595–603. doi: 10.1016/j.fgb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Kaufmann A., Philippsen P. Of bars and rings: Hof1-dependent cytokinesis in multiseptated hyphae of Ashbya gossypii. Mol. Cell Biol. 2009;29:771–783. doi: 10.1128/MCB.01150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knechtle P. Ph.D. Thesis. Basel, Switzerland: Biozentrum Universität Basel; 2002. AgSPA2 and AgBOI Control Landmarks of Filamentous Growth in the Filamentous Ascomycete Ashbya gossypii. [Google Scholar]
- Köhli M., Galati V., Boudier K., Roberson R. W., Philippsen P. Growth-speed-correlated localization of exocyst and polarisome components in growth zones of Ashbya gossypii hyphal tips. J. Cell Sci. 2008;121:3878–3889. doi: 10.1242/jcs.033852. [DOI] [PubMed] [Google Scholar]
- Konzack S., Rischitor P. E., Enke C., Fischer R. The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol. Biol. Cell. 2005;16:497–506. doi: 10.1091/mbc.E04-02-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J., Stearns T. Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- Mandelkow E. M., Mandelkow E., Milligan R. A. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J. Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall L. G., Jeng R. L., Mulholland J., Stearns T. Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J. Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K. High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol. Biol. 1999;117:77–97. doi: 10.1385/1-59259-201-5:77. [DOI] [PubMed] [Google Scholar]
- Meyer S. L., Kaminskyj S. G., Heath I. B. Nuclear migration in a nud mutant of Aspergillus nidulans is inhibited in the presence of a quantitatively normal population of cytoplasmic microtubules. J. Cell Biol. 1988;106:773–778. doi: 10.1083/jcb.106.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke P. F., Lee I. H., Plamann M. Microscopic analysis of Neurospora ropy mutants defective in nuclear distribution. Fungal Genet. Biol. 1999a;28:55–67. doi: 10.1006/fgbi.1999.1160. [DOI] [PubMed] [Google Scholar]
- Minke P. F., Lee I. H., Tinsley J. H., Bruno K. S., Plamann M. Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol. Microbiol. 1999b;32:1065–1076. doi: 10.1046/j.1365-2958.1999.01421.x. [DOI] [PubMed] [Google Scholar]
- Morris N. R. Nuclear migration. From fungi to the mammalian brain. J. Cell Biol. 2000;148:1097–1101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourino-Perez R. R., Roberson R. W., Bartnicki-Garcia S. Microtubule dynamics and organization during hyphal growth and branching in Neurospora crassa. Fungal Genet. Biol. 2006;43:389–400. doi: 10.1016/j.fgb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- O'Toole E. T., Winey M., McIntosh J. R. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B. R., Morris N. R. Nuclear movement is β-tubulin dependent in Aspergillus nidulans. Cell. 1980;19:255–262. doi: 10.1016/0092-8674(80)90407-9. [DOI] [PubMed] [Google Scholar]
- Oakley B. R., Morris N. R. A mutation in Aspergillus nidulans that blocks the transition from interphase to prophase. J. Cell Biol. 1983;96:1155–1158. doi: 10.1083/jcb.96.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B. R., Oakley C. E., Yoon Y., Jung M. K. gamma-Tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Oakley C. E., Oakley B. R. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Kaufmann A., Schmitz H. P. Homologues of yeast polarity genes control the development of multinucleated hyphae in Ashbya gossypii. Curr. Opin. Microbiol. 2005;8:370–377. doi: 10.1016/j.mib.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Requena N., Alberti-Segui C., Winzenburg E., Horn C., Schliwa M., Philippsen P., Liese R., Fischer R. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol. Microbiol. 2001;42:121–132. doi: 10.1046/j.1365-2958.2001.02609.x. [DOI] [PubMed] [Google Scholar]
- Rout M. P., Kilmartin J. V. Components of the yeast spindle and spindle pole body. J. Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sawin K. E., Tran P. T. Cytoplasmic microtubule organization in fission yeast. Yeast. 2006;23:1001–1014. doi: 10.1002/yea.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerer F., Morgan G., Winey M., Philippsen P. Cnm67p is a spacer protein of the Saccharomyces cerevisiae spindle pole body outer plaque. Mol. Biol. Cell. 2001;12:2519–2533. doi: 10.1091/mbc.12.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz H. P., Kaufmann A., Kohli M., Laissue P. P., Philippsen P. From function to shape: a novel role of a formin in morphogenesis of the fungus Ashbya gossypii. Mol. Biol. Cell. 2006;17:130–145. doi: 10.1091/mbc.E05-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel S. G., Snyder M. A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J. Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Geissler S., Grein K., Schiebel E. gamma-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J. Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T., Evans L., Kirschner M. gamma-Tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Straube A., Brill M., Oakley B. R., Horio T., Steinberg G. Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis. Mol. Biol. Cell. 2003;14:642–657. doi: 10.1091/mbc.E02-08-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suelmann R., Fischer R. Nuclear migration in fungi–different motors at work. Res. Microbiol. 2000;151:247–254. doi: 10.1016/s0923-2508(00)00151-0. [DOI] [PubMed] [Google Scholar]
- Veith D., Scherr N., Efimov V. P., Fischer R. Role of the spindle-pole-body protein ApsB and the cortex protein ApsA in microtubule organization and nuclear migration in Aspergillus nidulans. J. Cell Sci. 2005;118:3705–3716. doi: 10.1242/jcs.02501. [DOI] [PubMed] [Google Scholar]
- Vogel S. K., Pavin N., Maghelli N., Julicher F., Tolic-Nørrelykke I. M. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 2009;7:918–928. doi: 10.1371/journal.pbio.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland J., Ayad-Durieux Y., Knechtle P., Rebischung C., Philippsen P. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene. 2000;242:381–391. doi: 10.1016/s0378-1119(99)00509-0. [DOI] [PubMed] [Google Scholar]
- Wendland J., Walther A. Ashbya gossypii: a model for fungal developmental biology. Nat. Rev. 2005;3:421–429. doi: 10.1038/nrmicro1148. [DOI] [PubMed] [Google Scholar]
- Wiese C., Zheng Y. Microtubule nucleation: gamma-tubulin and beyond. J. Cell Sci. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
- Winey M., Mamay C. L., O'Toole E. T., Mastronarde D. N., Giddings T. H., Jr, McDonald K. L., McIntosh J. R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X., Fischer R. Nuclear migration and positioning in filamentous fungi. Fungal Genet. Biol. 2004;41:411–419. doi: 10.1016/j.fgb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Zekert N., Fischer R. The Aspergillus nidulans kinesin-3 UncA motor moves vesicles along a subpopulation of microtubules. Mol. Biol. Cell. 2009;20:673–684. doi: 10.1091/mbc.E08-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.