NTRAP is a novel protein that interacts with Trk receptors through its C2H2 zinc fingers in a kinase-dependent manner. It is associated with signaling endosomes in neurons. Down-regulation of NTRAP inhibits NGF-induced signaling within endosomes and neurite outgrowth in PC12 cells, and it also decreases retrograde neurotrophic signaling in cultured sensory neurons.
Abstract
Neurotrophins at axonal terminals signal to cell bodies to regulate neuronal development via signaling endosomes containing activated Trk receptor tyrosine kinases and mitogen-activated protein kinases (MAPKs). Requirements for the formation of signaling endosomes remain, however, poorly characterized. Here we show that a novel Trk-interacting protein, NTRAP (neurotrophic factor receptor–associated protein), plays a crucial role in this signaling process. NTRAP interacts with the Trk intracellular domain through its C2H2 zinc fingers in a kinase-dependent manner. It is associated with vesicles, some of which contain markers for signaling endosomes. Inhibition of NTRAP function suppresses neurotrophin-induced neurite outgrowth in PC12 cells by altering TrkA endocytic traffic, inhibiting the formation of endosomes containing persistently active MAPKs. In compartmentalized sensory neuron cultures, down-regulation of NTRAP abolishes the ability of neurotrophins applied to distal axons to activate the transcription factor adenosine 3′,5′-monophosphate response element-binding protein (CREB) and to promote neuronal survival. We propose that NTRAP regulates retrograde neurotrophic signaling by controlling the formation of signaling endosomes.
INTRODUCTION
The correct formation of neuronal connections is essential for the proper function of the nervous system. Such connections are generated through multiple steps that include specification and growth of axons and dendrites, target innervation, synaptogenesis, programmed cell death, and synaptic refinement. Neurotrophins secreted by innervated target tissues are crucial for these developmental processes (Zweifel et al., 2005; Cosker et al., 2008). They exert many biological effects by binding and activating specific Trk receptor tyrosine kinases at axonal terminals of presynaptic neurons; nerve growth factor (NGF) activates TrkA, brain-derived neurotrophic factor (BDNF) and neurotrophin 4/5 (NT4/5) activate TrkB, and neurotrophin 3 (NT3) activates TrkC (Reichardt, 2006). The activated Trk receptors then induce local responses at axonal terminals and retrograde responses in neuronal cell bodies residing at considerable distances from the terminals in many cases (Kuruvilla et al., 2004; Wickramasinghe et al., 2008).
Several models have been proposed to explain how the neurotrophic signal reaches cell bodies from axonal terminals. They include retrograde transport of internalized neurotrophin-receptor complexes in vesicles (signaling endosomes), retrograde propagation of signaling effectors, retrograde waves of Trk receptor activation along the plasma membrane, and retrograde calcium waves emanating from activated Trk receptors (Wu et al., 2008). Substantial evidence from in vivo and in vitro studies supports the signaling endosome model, which proposes that upon neurotrophin binding, neurotrophins and activated Trk receptors are internalized at axonal terminals, packaged into signaling endosomes, and retrogradely transported to neuronal cell bodies (Ehlers et al., 1995; von Bartheld et al., 1996; Howe et al., 2001; Delcroix et al., 2003; Ye et al., 2003; Heerssen et al., 2004). Signaling endosomes isolated from the sciatic nerve contain NGF, activated TrkA, phosphorylated mitogen-activated protein kinases (MAPKs), Rap1, and dynein (Delcroix et al., 2003). Because signaling endosomes have to move over long distances in some neurons, they should be specialized vesicles that have evolved to be long-lived by avoiding proteolytic degradation (Howe and Mobley, 2005; Zweifel et al., 2005). Recent studies using compartmentalized neuronal cultures have shown that retrograde transport of a neurotrophin-Trk receptor complex is necessary and sufficient for neuronal responses mediated by target-derived neurotrophins, including signaling events in cell bodies (Senger and Campenot, 1997; Watson et al., 2001), activation of the transcription factor cyclic AMP responsive element-binding protein (CREB; Riccio et al., 1999; Watson et al., 1999), and survival (Ye et al., 2003; Heerssen et al., 2004). However, the formation, sorting, and the exact composition of signaling endosomes remain unclear.
Here we report the identification of a novel Trk-interacting protein (named neurotrophic factor receptor–associated protein [NTRAP]), which contains one RING finger, five C2H2 zinc fingers, and two proline-rich regions. NTRAP interacts with Trk receptor tyrosine kinases in a kinase-dependent manner through the C2H2 zinc fingers. Results from PC12 cells and compartmentalized sensory neuron cultures indicate that NTRAP is essential for retrograde neurotrophic signaling, likely by controlling the formation of signaling endosomes.
MATERIALS AND METHODS
Reagents
Cell culture reagents were purchased from Invitrogen (Carlsbad, CA). Antibodies to Trk (sc-414, Santa Cruz Biotechnology, Santa Cruz, CA), phosphorylated CREB (p-CREB; Ser133), Akt, phosphorylated Akt (p-Akt), extracellular signal–regulated kinases 1 and 2 (ERK1/2), phosphorylated ERK1/2 (p-ERK1/2), phosphorylated TrkA (p-TrkA; Cell Signaling Technology, Danvers, MA), early endosome antigen 1 (EEA1; BD Biosciences, Franklin Lakes, NJ), Rap1 (BD Transduction Laboratories, San Jose, CA), Rab7, Myc, α-tubulin (Sigma-Aldrich, St. Louis, MO), Flag (Stratagene, Cedar Creek, TX), and hemagglutinin (HA; BabCO, Richmond, CA) were obtained commercially. Antibody against NGF (goat anti-mouse NGF) was a generous gift from Dr. Keith A. Crutcher (University of Cincinnati, Cincinnati, OH). Polyclonal TrkA antibodies (RTA) were previously described (Clary et al., 1994).
Yeast Two-Hybrid System
For library screening, the cDNA encoding the intracellular domain of rat TrkC was inserted into the plasmid pBTM116 (generated by Paul Bartel and Stan Fields, University of Washington, Seattle, WA) between the EcoRI and SmaI sites to generate pBTM-TrkC, which should express a LexA-TrkC fusion protein in yeast cells. To identify proteins that interact with the TrkC intracellular domain, we used a cDNA library generated from random cDNA fragments derived from mouse embryos at day E9.5-E10.5 (Vojtek et al., 1993). The library DNA (250 μg) was transformed into L40 Saccharomyces cerevisiae cells (Mata trp1 leu2 his3 LYS2::lexA-HIS3URA3::lexA-lacZ) containing pBTM-TrkC. Approximately 1 × 107 transformants were grown in synthetic medium lacking tryptophan, leucine, histidine, uracil, and lysine for 4 h at 30°C. After the recovery, the cells were plated onto synthetic medium lacking tryptophan, leucine, histidine, uracil, and lysine and containing 10 mM 3-amino-1,2,4-triazole (3-AT). The plates were incubated at 30°C for 2 wk. His+ colonies were grown on a synthetic medium lacking leucine and tryptophan at 30°C for 3 d and then assayed for β-galactosidase activity by a filter assay (Breeden and Nasmyth, 1985). Of 163 His+ colonies, 134 also expressed β-galactosidase. The library plasmids were recovered from His+LacZ+ colonies and sequenced.
For other yeast two-hybrid assays, cDNA for the intracellular domain of rat TrkA, mouse TrkB, or mouse Met was inserted into plasmid pBTM116 to create constructs that should express LexA-TrkA, LexA-TrkB, or LexA-Met fusion protein in yeast cells. Mouse NTRAP cDNA was inserted into vector pVP16 for the expression of a fusion protein containing NTRAP and the VP16 activation domain. To disrupt the third C2H2 zinc finger of NTRAP (NTRAP-ZFM), PCR-based site-directed mutagenesis was performed to change the two cysteine residues (C187 and C190) to two alanine residues using the Stratagene Mutagenesis kit (Stratagene). The PCR primers used to generate the mutant were as follows: 5′-catcggggacatccactcgctaagttcgctgacgagcgctacctggac-3′ (sense) and 5′-gtccaggtagcgctcgtcagcgaacttagcgagtggatgtccccgatg-3′ (anti-sense). To inactivate the tyrosine kinase of TrkC, mutagenesis was performed on pBTM116-TrkC to change the lysine residue at position 572 in the active site of the kinase to an asparagine residue (K572N). The PCR primers used for this mutagenesis were: 5′-cttgtggcagtgaacgccctgaaggatcc-3′ (sense) and 5′-ggatccttcagggcgttcactgccacaag-3′ (anti-sense). After yeast cells were digested with zymolyase, β-galactosidase activity was assayed as described (Ausubel et al., 2003).
Development and Purification of NTRAP Antibodies
To develop NTRAP antibodies, we synthesized a 50-amino acid peptide (CRRSEGVVSGEDYEEVDRYNRQGRAGRASGRGAQNRRGSWRYKREEEDRE) that corresponds to the sequence of NTRAP between the last C2H2 zinc finger and the first proline-rich domain and immunized rabbits with this peptide. We used ammonium sulfate to precipitate IgG from the anti-serum, dissolved IgG in PBS, and purified anti-NTRAP antibody with a column containing the peptide covalently linked to SulfoLink Coupling Gel (Pierce Biotechnology, Rockford, IL). The antibody was used for Western blots and immunohistochemistry at 0.5–1 μg/ml.
Constructs of NTRAP Short Hairpin RNA
To generate a construct expressing either NTRAP short hairpin RNA (shRNA) or a scrambled derivative of the NTRAP shRNA (control shRNA), two pairs of oligonucleotides were designed to produce DNA duplexes and subsequently cloned into the vector pSUPER (Brummelkamp et al., 2002): NTRAP shRNA, 5′-gatccccgatgcgggtgctgtgcaagttcaagagactcgcacagcacccgcatctttttggaaa-3′, 5′-agcttttccaaaaagatgcgggtgctgtgcgagtctcttgaactcgcacagcacccgcatcggg-3′; control shRNA, 5′-gatccccgtgaggcgttgccggatggttcaagagaccatccggcaacgcctcactttttggaaa-3′-3′, 5′-agcttttccaaaaatgaggcgttgccggatggtctcttgaaccatccggcaacgcctcacggg-3′. Two adeno-associated virus (AAV) constructs were developed: one expressing NTRAP shRNA and EGFP (NTRAP shRNA AAV) and another expressing the control shRNA and EGFP (control shRNA AAV). Enhanced green fluorescent protein (EGFP) was used to mark the infected cells. In these two constructs, shRNAs and EGFP are under the control of the H1 promoter and the cytomegalovirus promoter, respectively. The EGFP expression unit is downstream of the shRNA expression unit, and the two expression units are in the same orientation. Control shRNA AAV and NTRAP shRNA AAV viral particles were produced in AAV-293 cells according to the manufacturer's protocol (Stratagene).
Cell Cultures
PC12 cells were grown on collagen S–coated 96-well plates in DMEM medium supplemented with 2 mM l-glutamine, 5% horse serum, 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. HEK293 cells were cultured in DMEM medium containing 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM glutamine, and 10% FBS. Cells were maintained at 37°C under an atmosphere of 7.5% CO2 and saturated humidity.
Dorsal root ganglion (DRG) neurons were dissected from E18.5 rat embryos, enzymatically dispersed, and plated onto collagen-coated compartmentalized culture dishes. Compartmentalized cultures were established as described by Campenot, (1977). DRG cells were maintained in DMEM medium supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 μg/ml), and neurotrophins (NGF and BDNF at 100 ng/ml). Cytosine-β-d-arabinofuranoside (10 μM) was applied into the culture medium to block proliferation of nonneuronal cells during the first week. Medium was changed every 3 d. After neurons extended their axons to the side compartment over a period of 4–5 d, the concentration of neurotrophins in the central compartment was reduced to 10 ng/ml. On DIV3 (days in vitro), the cells were infected with NTRAP shRNA AAV or control shRNA AAV. For neuronal survival assays, neurotrophins were removed from the central compartment or the side compartment on DIV10. On DIV13 the cultures were incubated with 4′,6-diamidino-2-phenylindole (DAPI; 2 μg/ml) in PBS for 10 min and fixed. EGFP-positive cells with fragmented or condensed nuclei were counted as apoptotic cells. To examine CREB activation, neurotrophins were removed from both the central and side compartments on DIV12 for ∼16 h. BDNF and NGF were then added to the central compartment or the side compartment to a final concentration of 100 ng/ml for 20 min. The cultures were fixed and stained for p-CREB. For ERK1/2 activation assays, DRG neurons were cultured in 12-well plates and infected with AAV at DIV5 for 10 d in the presence of NGF. After overnight deprivation of serum and NGF, the infected cultures were treated with NGF (100 ng/ml) for 0–6 h. The amounts of total and activated ERK1/2 in cell lysates were determined using immunoblotting. In DRG cultures, the AAV infection efficiency ranged from 50 to 80%, depending on the timing of infection and culture conditions. The infection efficiency was similar among different groups in the same experiment.
Protein Expression
To generate mammalian expression constructs for NTRAP, TrkC, or TrkA, the cDNA encoding mouse NTRAP, rat TrkC or rat TrkA was inserted into the multiple cloning sites of mammalian expression vector pcDNA 3.1 (+) or pcDNA3.1/myc-HisA. In the shRNA-resistant NTRAP (NTRAPr) expressing construct, the NTRAP-shRNA–targeted sequence gatgcgggtgctgtgcgag was changed to gatgcgggtattatgcgag. The sequence change does not alter the amino acid sequence of the NTRAP protein. The Flag tag is located immediately after the signal peptide in the TrkA construct, whereas the Myc tag is at the C-terminus in the TrkC construct. To express ZF-HA, a portion of the NTRAP cDNA (corresponding to the protein region from A105 to Q243) tagged with HA sequence was generated by PCR and cloned into vector pLP-LNCX. The following PCR primers were used: 5′-actaatctcgaggccaccatggcattgtacaggcagctgctg-3′ (forward), 5′-actatcgattcaccgtgaaggtcctcctagcgatgcgtagtcagggacatcgtatgggtactgctcagtgctgcagcggccctcctcac-3′ (reverse). Cells were grown in 100-mm dishes and transfected with specific constructs using lipofectamine and PLUS reagent according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). For transient expression, cells were treated and analyzed 2–3 d after transfection. For stable expression of pLP-LNCX, PC12 cells were transfected and passed at a 1:10 dilution into selective medium (G418 at 300 μg/ml) 3 d after transfection. Eighty-five clones were isolated and examined for the expression of ZF-HA by Western blotting using anti-HA antibody. Two lines (ZF-HA1 and ZF-HA2) stably expressing ZF-HA were identified.
Cell Viability Assay
Cell viability was assessed by using a modified 3-(4,5-dimethylthiazol-2-yl)-2,5,-diphenyltetrazolium bromide (MTT) assay. Briefly, ∼10,000 PC12 cells per well were seeded in collagen S–coated 96-well plates with 100 μl of medium. After 6 h of incubation at 37°C, the medium was changed to serum-free medium. NGF (100 ng/ml) was added 45 min later, and cells were incubated for 24 h. At the end of the incubation period, 10 μl of MTT at 5 mg/ml was added to each well and then incubated for 1 h at 37°C. After 100 μl of solubilization solution (50% DMSO in 5% SDS) was added to stop the reaction, and absorption at 540 nm was measured with reference at 690 nm. The experiment was repeated four times, and averages of the four independent experiments were used in the analysis.
TUNEL Staining
PC12 cells were grown on collagen S–coated eight-well chambers (Nunc, Rochester, NY) in DMEM medium as described above. Approximately 100,000 PC12 cells were seeded onto each well in collagen S–coated eight-well chambers with 300 μl of growth medium. Cells were incubated at 37°C overnight. After serum starvation for 45 min, 100 ng/ml NGF was added to PC12 cells. Twenty-four hours later, cells were fixed and permeabilized with 0.2% Triton X-100 in PBS. Apoptotic cells were assessed by detection of DNA fragmentation using the standard TUNEL (terminal deoxynucleotidyl transferase–mediated biotinylated dUTP nick end labeling) method (DeadEnd Colorimetric TUNEL System, Promega, Madison, WI) according to the manufacturer's protocol. Observations were carried out using a Nikon microscope (Melville, NY). Apoptosis-positive or -negative cells were counted for each well.
Immunohistochemistry and Immunocytochemistry
We performed immunohistochemistry of sections and immunocytochemistry of cultured cells as described (Farinas et al., 1996; An et al., 2008). Mouse embryos at E12.5 were fixed in Carnoy's fixative (10% glacial acetic acid, 30% chloroform, and 60% ethanol) overnight. The embryos were dehydrated, embedded in paraffin, and sectioned at 7 μm on a rotary microtome. DRGs were dissected from E18.5 rat embryos, fixed immediately with 4% paraformaldehyde, and sectioned with a cryostat at 10 μm. Tissue sections or cultured cells on slides were washed with TBS (10 mM Tris, 150 mM NaCl, pH 7.5) twice. Cells and sections were permeabilized and blocked in a blocking buffer (0.4% Triton X-100, 1% glycine, 2–3% bovine serum albumin, 10% goat serum in TBS) for 1 h. Samples were incubated with primary antibodies overnight at 4°C. After washes (three times for 20 min in the blocking buffer), samples were incubated in dark for 1 h with FITC- or Texas Red–conjugated secondary antibodies in the blocking buffer. To label the actin cytoskeleton, cells were incubated with 50 μg/ml phalloidin-FITC for 40 min. Slides were mounted with Fluorescent Mounting Medium (DakoCytomation, Carpinteria, CA) and examined with a Zeiss 510 Meta confocal laser scanning microscope (LSM 510 META; Thornwood, NY). We quantified colocalization of two proteins within a cell by measuring percentage of intensity colocalized using ImageJ software (http://rsb.info.nih.gov/ij/).
Rap1 Activation Assay
To detect the active form of Rap1 (GTP-bound), the constructs containing glutathione S-transferase (GST) and Rap-binding domain of RalGDS (RalGDSRBD; kindly provided by Dr. Chenbiao Wu, Stanford University) were expressed in Escherichia coli strain BL21 LysE cells. The fusion proteins were purified as described (Castro et al., 2005). PC12 cells were treated with or without NGF (100 ng/ml) for different amounts of time after being starved in serum-free medium overnight. Cells were lysed on ice for 20 min in a lysis buffer (50 mM Tris-HCl, pH 7.5, 10% glycerol, 1% NP-40, 200 mM NaCl, 2.5 mM MgCl2, 2 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 1 μM pepstatin A, 2 μg/ml aprotinin). Cell lysates (2 mg) were incubated with 5 μg of RalGDSRBD/GST proteins prebound to agarose-glutathione conjugates at 4°C for 60 min with rotation. The beads were washed three times in cold lysis buffer and boiled in 2× SDS-PAGE sample buffer. The amounts of GTP-bound Rap1 were analyzed by Western blotting using SDS-PAGE gels and an antibody to Rap1 (Invitrogen).
Western Blotting
Cells were broken in a lysis buffer (20 mM Tris-Cl, 1% Triton X-100, 150 mM NaCl, 10% glycerol, 5 mM EDTA, 2 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 1 μM pepstatin A, 2 μg/ml aprotinin, pH 7.5) for 30 min on ice and centrifuged at 12,500 rpm for 20 min. Protein concentrations of lysates were measured using the BCA protein assay kit (Pierce Biotechnology). Protein lysates in the SDS loading buffer were denatured for 5 min at 100°C, separated on SDS polyacrylamide gels, and transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 5% nonfat milk in TBST (10 mM Tris, pH 7.5, 150 mM NaCl, 0.5% Tween-20) for 1 h and then incubated with primary antibodies overnight at 4°C. After two washes with TBST, the membranes were incubated with the appropriate secondary antibody conjugated to horseradish peroxidase (Pierce Biotechnology) for 1 h at room temperature. The membranes were then washed three times with TBST, and proteins were visualized using the ECL plus system (Amersham Biosciences, Piscataway, NJ). The membranes were then stripped and blotted with an antibody to α-tubulin.
Immunoprecipitation
Cell lysates (1 mg protein) were precleared by incubation with 50 μl protein A/G agarose (Pierce Biotechnology). The supernatant was then incubated with primary antibodies at 4°C overnight with continuous rotation. Protein A/G agarose (50 μl) was added to the mixture in the next morning, and the tube was rotated at 4°C for 2 h. Protein A/G agarose was collected by centrifugation, washed five times in 1 ml of the lysis buffer, resuspended in 15 μl 2× SDS loading buffer, and heated at 100°C for 5 min. The immunoprecipitates were resolved on SDS polyacrylamide gels and blotted with specific antibodies.
Northern Blotting
Mouse adult tissue RNA blot (Mouse MTN blot) was purchased from Clontech Laboratories (Mountain View, CA). Probes were labeled with [α-32P]dCTP using the Rediprime II Random Prime Labeling System (GE Healthcare, Piscataway, NJ). Hybridization was performed in Rapid-hyb buffer according to the manufacturer's protocol (GE Healthcare).
Cell Fractionation
PC12 cells were fractionated using a previously described method (Wu et al., 2001). Equal numbers of cells were plated in 100-mm plates before NGF treatment. One plate of PC12 cells was used for one gradient. Membrane fractions were collected from the four interfaces of gradients after centrifugation. Proteins were precipitated from the membrane fractions using 13% trichloroacetic acid and washed with acetone. The air-dried pellets were dissolved in 2× SDS loading buffer and analyzed by immunoblotting.
RESULTS
Identification of a Novel Trk-interacting Protein
To identify proteins that are involved in neurotrophic signaling, we used a yeast two-hybrid system to search for novel proteins that interact with the Trk receptors. The intracellular domain of the TrkC receptor tyrosine kinase was fused to the LexA DNA-binding domain that contains a dimerization motif. The motif should induce dimerization of the fusion protein, which leads to activation of the TrkC tyrosine kinase. The fusion protein was used as a bait to screen a cDNA library constructed by fusing cDNAs prepared from mouse embryos to a DNA sequence encoding the VP16 transcription activation domain. The screen successfully pulled out the majority of known Trk-interacting proteins, including Shc, phospholipase C-γ (PLC-γ), Abl, and SH2-B (Reichardt, 2006). It also identified several novel Trk-interacting proteins.
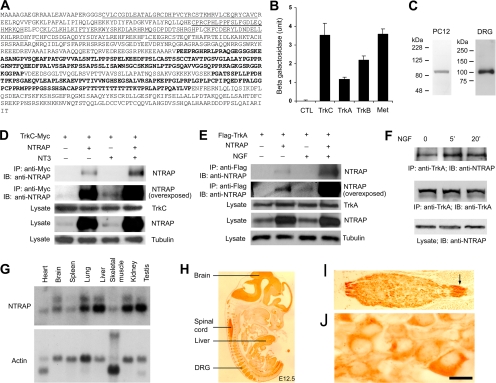
One of the novel Trk-interacting proteins is a 96-kDa protein that contains one RING finger, five tandem C2H2 zinc fingers, and two proline-rich regions (Figure 1A). This protein, which we named NTRAP, is identified as Zfp598 in the NCBI nucleotide database (Accession no. NM_183149) with no known functions and is conserved from yeast S. cerevisiae to humans. Yeast two-hybrid assays showed that NTRAP was also able to interact with TrkA, TrkB, and Met (the receptor for hepatocyte growth factor; Figure 1B) but not the receptor for epidermal growth factor (Supplemental Figure S1).
Figure 1.
NTRAP interacts with Trk receptor tyrosine kinases. (A) The amino acid sequence of NTRAP. The first underlined region represents a RING finger domain, and the next five underlined regions are five tandem C2H2 zinc finger motifs. The two proline-rich regions are shown in bold. (B) Interaction of NTRAP with the intracellular domain of TrkA, TrkB, TrkC, or Met in yeast two-hybrid assays. Laminin was used as a negative control (CTL). The interaction strength was determined by measuring the activity of the reporter β-galactosidase. (C) Specificity of NTRAP antibody. Protein extracts from PC12 cells or E18.5 rat DRGs were blotted with the affinity-purified NTRAP antibody. (D) Enhancement of interaction between NTRAP and TrkC by NT3. Lysates of HEK293 cells were immunoprecipitated with anti-Myc antibodies, and immunoprecipitates were blotted with the anti-NTRAP antibody. Cell lysates (25 μg) were also blotted with antibodies to Myc, NTRAP, and α-tubulin for examination of expression. (E) Enhancement of interaction between NTRAP and TrkA by NGF. HEK293 cells were transfected with constructs expressing Flag-tagged TrkA and NTRAP. Cell lysates were immunoprecipitated with anti-Flag antibodies, and immunoprecipitates were blotted with anti-NTRAP antibodies. Cell lysates (25 μg) were also blotted with antibodies to Flag, NTRAP, and α-tubulin. (F) Interaction of endogenous NTRAP and TrkA. PC12 cell lysates were immunoprecipitated with the mouse IgG against TrkA, and immunoprecipitates were blotted with the rabbit IgG against NTRAP or TrkA. Cell lysates were blotted with the antibody to NTRAP. NGF treatment enhanced the association of the two proteins. (G) Tissue distribution of NTRAP mRNA in adult mice, as revealed by Northern hybridization. (H) High levels of NTRAP in the nervous system and liver of an E12.5 mouse embryo, as revealed by immunohistochemistry. (I) NTRAP expression in DRG ganglia of an E12.5 mouse embryo. Note that axons of DRG neurons also contain NTRAP (arrow). (J) Localization of NTRAP in the cytoplasm of DRG neurons. Scale bar, 10 μm.
To confirm the association of NTRAP with TrkC in mammalian cells, we first raised rabbit antiserum against a synthesized 50-amino acid peptide corresponding to the sequence between the last C2H2 zinc finger and the first proline-rich domain and purified the serum with an affinity column. The purified antibody recognized only a single protein with the expected molecular weight of NTRAP in protein extracts prepared from PC12 cells and embryonic rat DRG neurons (Figure 1C), indicating that the antibody is highly specific to NTRAP. We expressed Myc-tagged TrkC (TrkC-Myc) alone or with NTRAP in HEK293 cells, treated cells with NT3 or vehicle for 20 min, precipitated protein extracts prepared from the treated cells with Myc antibodies, and performed immunoblotting to detect NTRAP in immunoprecipitates. A small amount of NTRAP was present in the immunoprecipitate in the absence of NT3. However, NT3 greatly increased the amount of NTRAP pulled down by Myc antibodies (Figure 1D). NTRAP was expressed in HEK293 cells, and overexposure indicated that the Myc antibody also pulled down endogenous NTRAP in an NT3-dependent manner (Figure 1D). In a similar experiment, we observed that 20-min NGF treatment greatly enhanced the association of NTRAP with TrkA (Figure 1E). Furthermore, NGF treatment also enhanced the association of endogenous NTRAP and TrkA in PC12 cells (Figure 1F). These results demonstrate that NTRAP is able to interact with the Trk receptors and that the activation of the tyrosine kinase greatly strengthens the interaction.
Expression Pattern of NTRAP
If the interaction between NTRAP and Trk receptors has physiological significance, NTRAP should be expressed in neurons. Northern hybridization analyses revealed that NTRAP mRNAs were widely expressed in adult mice with high levels of NTRAP mRNAs in lung, liver, kidney, testis, and brain (Figure 1G). However, NTRAP expression was mainly restricted to the nervous system in mouse embryos at E12.5, as revealed by immunohistochemistry (Figure 1H). At this stage, NTRAP was highly expressed in the brain, spinal cord, DRG, and liver. In DRG neurons, NTRAP was localized in axons and the cytoplasm (Figure 1, I and J). This expression pattern indicates that NTRAP and Trk receptors are expressed in the same DRG neurons because nearly every DRG neuron expresses one or more Trk receptors in E12.5 mouse embryos (Farinas et al., 1998).
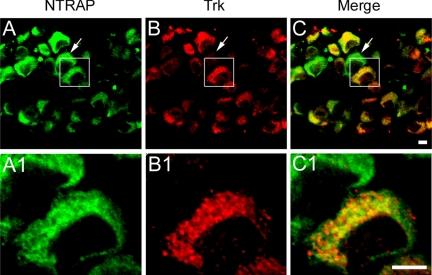
We used an antibody that recognizes all three Trk receptors for examining colocalization of NTRAP with Trk receptors in individual cells. In E18.5 rat DRG, all Trk-expressing neurons also expressed NTRAP (Figure 2, A–C). Nevertheless, a small number of DRG neurons expressed NTRAP but little Trk. This result is consistent with the observation that TrkA is down-regulated in nonpeptidergic Ret-expressing nociceptors during late embryogenesis and postnatally (Luo et al., 2007). Within the DRG neurons expressing both NTRAP and Trks, the majority of Trks (72 ± 3%, n = 56 neurons) were colocalized with NTRAP (Figure 2, A1–C1).
Figure 2.
Colocalization of NTRAP with Trk receptors in DRG neurons. Sections at 10 μm were obtained from dissected rat DRG at E18.5 and stained with the affinity-purified NTRAP antibody and an antibody that recognizes all three Trk receptors (A–C). The arrow indicates one neuron that expresses NTRAP but few Trk receptors. The boxed areas are shown in A1–C1 at higher magnification. Scale bars, 5 μm.
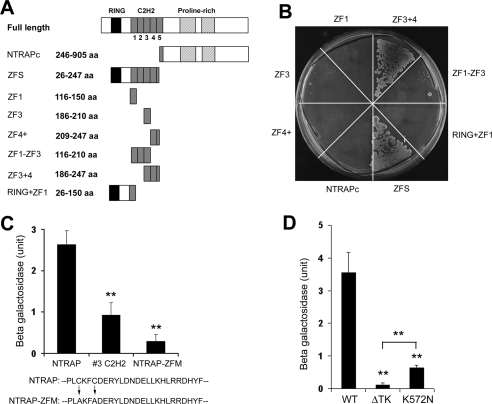
Identification of Interaction Domains in TrkC and NTRAP
To identify which domains of NTRAP are required to interact with the Trk receptors, we used yeast two-hybrid assays to examine interactions between the TrkC intracellular domain and NTRAP mutants. If two proteins interact, expression of the His3 reporter gene will be induced, which will allow host yeast cells to grow on plates lacking histidine. This assay showed that TrkC interacted with the N-terminal fragment containing the RING motif and C2H2 zinc fingers of NTRAP (ZFS), but not with its C-terminal half (NTRAPc; Figure 3, A and B). We next examined whether individual zinc fingers are capable of interacting with TrkC. As shown in Figure 3B, TrkC weakly interacted with the third C2H2 zinc finger (ZF3), but not with the RING motif, the first or the fourth C2H2 zinc finger (RING+ZF1, ZF1, and ZF4+). Interestingly, the addition of the first two (ZF1–ZF3) or the fourth (ZF3 + 4) C2H2 zinc finger enhanced the interaction of the third C2H2 zinc finger with TrkC. To demonstrate that the third C2H2 zinc finger is crucial for the interaction of NTRAP with the Trk receptors, we used a β-galactosidase activity assay to quantify the interaction of TrkC with the third C2H2 zinc finger alone (#3 C2H2) or an NTRAP mutant (NTRAP-ZFM) in which two cysteine residues of the third C2H2 zinc finger were replaced with two alanine residues (Figure 3C). The third C2H2 zinc finger interacted with TrkC well, whereas the cysteine-to-alanine replacement reduced the interaction strength between NTRAP and TrkC by 90%. These results show that the third C2H2 zinc finger is necessary and sufficient to mediate the interaction of NTRAP and TrkC.
Figure 3.
Identification of interaction domains in NTRAP and TrkC. (A) Schematic representation of NTRAP mutants. (B) Yeast cells expressing the intracellular domain of TrkC fused to LexA and VP16-NTRAP mutants were streaked onto a histidine-free plate containing 10 mM 3-AT and grown at 30°C for 8 d. The number of grown yeast cells is an indicator of the interaction strength between TrkC and an NTRAP mutant. (C) Interaction of TrkC with NTRAP mutants. The #3 C2H2 construct is comprised of the VP16 activation domain and the third C2H2 zinc finger motif. The NTRAP-ZFM construct is comprised of the VP16 activation domain and an NTRAP mutant in which the two cysteine residues in the third C2H2 zinc finger have been changed to two alanine residues. The β-galactosidase activity in yeast two-hybrid assays reflects the interaction strength between two proteins. Error bars, SEs. **p < 0.01 by Student's t test; n = 3. (D) Interaction of NTRAP with wild-type TrkC (WT) or TrkC mutants (ΔTK and K572N). Error bars, SEs. **p < 0.01 by Student's t test; n = 3.
Our immunoprecipitation results show the importance of the tyrosine kinase in the association of NTRAP with the Trk receptors (Figure 1, D–F). We further explored the role of the tyrosine kinase domain in the interaction using the yeast two-hybrid system. We inactivated TrkC tyrosine kinase by either deleting the tyrosine kinase domain and its trailing sequence (ΔTK) or substituting the lysine residue in the active site of the kinase with an asparagine residue (K572N). Using expression of β-galactosidase as an indicator, we found that in yeast cells ΔTK completely abolished, whereas the K572N substitution greatly reduced, the interaction (Figure 3D). Thus, NTRAP weakly interacts with inactive TrkC at the tyrosine kinase domain or its trailing sequence, and activation of the tyrosine kinase greatly enhances the interaction.
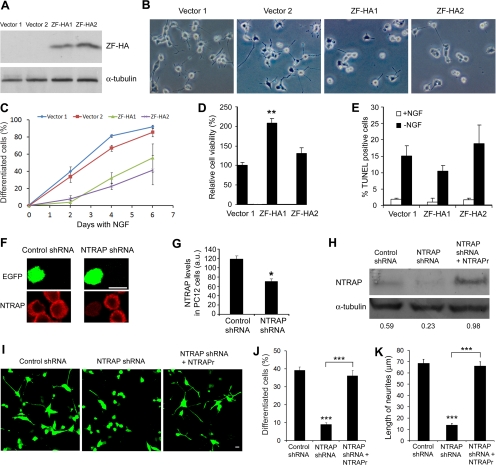
An Essential Role for NTRAP in NGF-dependent Differentiation of PC12 Cells
NGF promotes survival and differentiation of PC12 cells by activating the TrkA receptor. To determine the role of NTRAP in the action of neurotrophins, we used a retroviral vector to stably express its first four C2H2 zinc fingers (ZF) tagged with an HA epitope (ZF-HA) in PC12 cells. Because ZF-HA lacks the RING motif and the proline-rich domains of NTRAP, the expression of ZF-HA should interfere with the function of NTRAP. Two control lines transfected with the empty vector (vector 1 and vector 2) and two lines expressing ZF-HA (ZF-HA1 and ZF-HA2) were established. Western blots with an HA antibody revealed that line ZF-HA2 expressed a slightly higher amount of ZF-HA than line ZF-HA1 (Figure 4A). To assay the effect of ZF-HA on NGF-induced differentiation, we grew the four stable PC12 cell lines in a serum-free medium and treated cells with 100 ng/ml NGF. When a process was as long as or longer than two diameters of a cell, the cell was considered to be differentiated or have a neurite. The expression of ZF-HA reduced the number of cells with neurites after NGF treatment (Figure 4B). Quantification revealed that in the control lines neurites were observed in 34–40 and 85–92% of cells 2 d and 6 d after the addition of NGF, respectively. However, in the two lines expressing ZF-HA, neurites were only observed in 8–14 and 41–56% of cells 2 and 6 d after the addition of NGF, respectively (Figure 4C). To exclude the possibility that the differentiation phenotype might result from cell death, we carried out MTT and TUNEL assays to monitor NGF-dependent survival. The expression of ZF-HA did not decrease cell viability in the presence of NGF, as revealed by the MTT assay (Figure 4D). In fact, line ZF-HA1 cells had better viability than control cells in the presence of NGF. TUNEL staining showed that the expression of ZF-HA did not increase apoptosis in the presence or absence of NGF (Figure 4E). These results suggest that NTRAP is required for NGF-induced differentiation but not for NGF-dependent survival in PC12 cells.
Figure 4.
Inhibition of NTRAP blocks NGF-induced differentiation of PC12 cells. (A) Stable expression of HA epitope-tagged C2H2 zinc finger region of NTRAP (ZF-HA) in PC12 cells. Two control lines (vector 1 and vector 2) and two expression lines (ZF-HA1 and ZF-HA2) of PC12 cells were established. (B) Representative images showing inhibition of NGF-induced differentiation of PC12 cells by expression of ZF-HA. The images were taken after 4 d of NGF treatment. (C) Quantification of PC12 cell differentiation. The experiment was performed in triplicate. Percentage of cells with neurites was significantly lower in ZF-HA1 and ZF-HA2 lines compared with either of the two control lines at 2, 4, or 6 d of NGF treatment (p < 0.05 by Student's t test). Error bars, SEs. (D) Effect of ZF-HA expression on viability of PC12 cells in the presence of NGF. MTT assays were repeated four times. Error bars, SEs. **p < 0.01 by Student's t test. (E) No effect of HA-ZA expression on survival of PC12 cells in the presence or absence of NGF, as revealed by TUNEL staining (n = 4). Error bars, SEs. (F) Representative images showing reduced NTRAP immunoreactivity in PC12 cells expressing NTRAP shRNA 2 d after transfection. The shRNA constructs were cotransfected into PC12 cells with a construct expressing EGFP. (G) Quantification of NTRAP immunoreactivity in PC12 expressing either control shRNA or NTRAP shRNA. Error bars, SEs. *p < 0.05 by Student's t test. (H) Western blot analysis of extracts prepared from HEK293 cells transfected with adeno-associated viral (AAV) constructs expressing both EGFP and shRNA (either control shRNA or NTRAP shRNA). Some cells were also cotransfected with a construct expressing shRNA-resistant NTRAP mRNA (NTRAPr). The numbers represent the ratio of NTRAP amount to α-tubulin amount. (I) Representative images showing NGF-induced differentiation of PC12 cells expressing shRNA. PC12 cells were transfected with an AAV construct expressing EGFP and shRNA or cotransfected with the AAV construct expressing EGFP and NTRAP shRNA and a construct expressing NTRAPr mRNA. Two days later, the cells were treated with 100 ng/ml NGF for 3 d. Scale bar, 10 μm. (J) Quantification of NGF-induced differentiation of PC12 cells expressing shRNA after 3 d of NGF treatment. Error bars, SEs. ***p < 0.001 by Student's t test (n = 6–7 coverslips for each condition). (K) Measurement of neurite length in PC12 cells expressing shRNA after 3 d of NGF treatment. Error bars, SEs. ***p < 0.01 by Student's t test (n = 20–22 cells for each condition).
Because ZF-HA might block NGF-induced differentiation by sequestering proteins that are not involved in NGF signaling but that are important for neurite outgrowth, we further investigated the role of NTRAP in NGF signaling by using shRNAs. Plasmid pSUPER was used to express one shRNA that targets NTRAP mRNA (NTRAP shRNA) and one scrambled derivative of the shRNA (control shRNA). Each of the two shRNA constructs was transfected into PC12 cells along with a construct expressing EGFP. In control shRNA wells, NTRAP immunoreactivity was similar in EGFP-positive and negative cells. But expression of NTRAP shRNA greatly reduced NTRAP immunoreactivity in EGFP-positive cells (Figure 4, F and G), indicating that NTRAP shRNA is able to reduce NTRAP expression. This observation was further confirmed by immunoblot analysis of lysates prepared from HEK293 cells expressing either control shRNA or NTRAP shRNA (Figure 4H). To determine whether a reduction in NTRAP expression inhibits NGF-induced differentiation, we transfected PC12 cells with a construct expressing both EGFP and shRNA and 2 d after transfection treated the transfected cells with NGF. NGF-induced differentiation was severely impaired in PC12 cells expressing NTRAP shRNA compared with PC12 cells expressing control shRNA (Figure 4, I–K). The inhibitory effect was reversed by the expression of an shRNA-resistant NTRAP mRNA (NTRAPr) containing three silent single-nucleotide mutations at the NTRAP-shRNA–targeted site (Figure 4, H–K). These results further indicate that NTRAP is required for NGF-induced differentiation in PC12 cells.
Inhibition of NTRAP Function Abolishes NGF-induced Persistent Activation of MAPKs
Binding of NGF to the TrkA receptor in PC12 cells activates the ERK family of MAPKs via two small GTPases, Ras and Rap1. Ras induces the initial and transient ERK activation on the plasma membrane, whereas Rap1 induces persistent ERK activation in endosomes (York et al., 1998; Wu et al., 2001). Persistent ERK activity is required for NGF-induced differentiation in PC12 cells (Cowley et al., 1994; Traverse et al., 1994; Hisata et al., 2007). In agreement with these findings, TrkA endocytosis is essential for NGF-dependent differentiation but not survival in PC12 cells (Zhang et al., 2000). To investigate the possibility that NTRAP regulates differentiation through the Rap1-ERK pathway, we first measured levels of activated ERK1/2 in PC12 cells that express ZF-HA using antibodies specific to p-ERK1/2. Levels of activated ERK1/2 in the two lines expressing ZF-HA were significantly lower at 60 and 120 min but not 5 min after the addition of NGF when compared with those in a control line (Figure 5, A and B). To examine the effect of ZF-HA expression on Rap1 activation, we used the Rap1-binding domain of RalGDS to pull down the active form of Rap1 (GTP-bound) from cell lysates and then used immunoblotting to measure the amount of active Rap1. As shown in Figure 5C, the expression of ZF-HA did not affect levels of total Rap1 in PC12 cells, but delayed NGF-induced Rap1 activation. In support of this observation, the expression of ZF-HA altered subcellular localization of Rap1 (Figure 5D). It has been shown that endosomes containing Rap1 move to the perinuclear region from the cell periphery after TrkA is activated (Mochizuki et al., 2001; Wu et al., 2001). NGF-induced Rap1 relocation was significantly attenuated in cells of line ZF-HA2 compared with cells of line vector 1 (Figure 5, D and E). In contrast, the expression of ZF-HA did not affect NGF-induced Akt activation in PC12 cells (Figure 5F), which is consistent with the observation that the expression of ZF-HA did not interfere with NGF-dependent survival (Figure 4E). To investigate if NTRAP is also required for persistent activation of ERK1/2 in neurons, we knocked down NTRAP expression by infecting cultured DRG neurons with adeno-associated virus expressing shRNA and then examined activation of ERK1/2 (Figure 5G). NGF-induced ERK1/2 activation lasted for more than 6 h in DRG neurons, and the expression of NTRAP shRNA decreased levels of p-ERK1/2 at later time points but not at earlier time points when compared with the expression of control shRNA (Figure 5G). Taken together, these results suggest that NTRAP is required for the sustained TrkA signaling, likely in endosomes, in NGF-treated PC12 cells or neurons.
Figure 5.
NTRAP affects the Rap1-ERK1/2 pathway. (A) Representative Western blots indicating that the sustained activation of ERK1/2 by NGF is diminished in ZF-HA2 line of PC12 cells. After overnight serum starvation, PC12 cells were incubated with NGF (100 ng/ml) for 0, 5, 20, 60, or 120 min. Lysates were blotted with antibodies to p-ERK1/2. The same membrane was also blotted with antibodies to ERK1/2 for examination of expression. (B) Quantification of p-ERK1/2 levels. Intensities of p-ERK1/2 bands were measured using ImageJ software and normalized to levels of total ERK1/2 (n = 3). Error bars, SEM. *p < 0.05 by Student's t test when compared with line vector 1. (C) Representative Western blots showing that Rap1 activation is delayed in ZF-HA2 line of PC12 cells. After overnight starvation in serum-free medium, cells were treated with NGF (100 ng/ml) for 0, 30, or 60 min. (D) Representative images of Rap1 immunocytochemistry in vector 1 and ZF-HA2 lines of PC12 cells. The outline of PC12 cells and location of nucleus are revealed by phaloidin staining of actin filaments (green) and DAPI staining of DNA (blue), respectively. Scale bar, 5 μm. (E) Distribution of Rap1 in the cytoplasm of PC12 cells. Rap1 immunofluorescence was measured at 0.5-μm intervals from the edge of the nucleus to the cell periphery. Error bars, SEM. *p < 0.05 by Student's t test (n = 30 cells for each condition). (F) Normal Akt activation in PC12 cells expressing ZF-HA. After overnight serum starvation, PC12 cells were incubated with NGF (100 ng/ml) for 0, 5, 20, or 60 min. Lysates were blotted with antibodies to p-Akt. The same membrane was also blotted with antibodies to Akt for examination of expression. (G) Representative Western blots indicating that the sustained activation of ERK1/2 by NGF is decreased in DRG neurons expressing NTRAP shRNA. AAV-infected DRG neuronal cultures were incubated with NGF (100 ng/ml) for 0, 1, 2, 4, or 6 h. Lysates were blotted with antibodies to p-ERK1/2. The same membrane was then blotted with antibodies to ERK1/2 for examination of expression.
NTRAP Regulates Endocytic Trafficking of the Activated TrkA Receptor
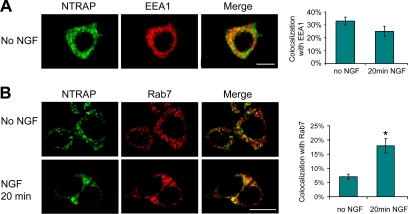
After the addition of NGF, activated TrkA bound to NGF is internalized into vesicles, which are converted to early endosomes and then late endosomes (Howe and Mobley, 2005). Deficits in either TrkA internalization or TrkA trafficking along the endosome differentiation pathway could diminish NGF-induced persistent ERK1/2 activation in PC12 cells expressing ZF-HA. We found that the expression of ZF-HA in PC12 cells or NTRAP shRNA in DRG neurons did not affect internalization of the TrkA receptor (Supplemental Figure S2). To investigate the role of NTRAP in endocytic trafficking of Trk receptors, we first examined the distribution of NTRAP in early and late endosomes. Confocal microscopy revealed that NTRAP was localized in vesicles in PC12 cells (Figure 6A). Some NTRAP-positive vesicles contained EEA1, a marker for early endosomes (Figure 6A), but very little Rab7, a late endosome component (Figure 6B). A 20-min NGF treatment caused a trend of reduction in colocalization of NTRAP with EEA1 (Figure 6A) and significantly increased the presence of NTRAP in late endosomes (Figure 6B), suggesting that NTRAP moved along with activated TrkA from early to late endosomes, perhaps through association.
Figure 6.
NGF-induced localization of NTRAP in late endosomes. (A) Colocalization of NTRAP with the early endosome marker, EEA1. The graph shows percentage of NTRAP immunoreactivity that was colocalized with EEA1. Scale bar, 10 μm. (B) Colocalization of NTRAP with the late endosome marker, Rab7. PC12 cells were starved for serum overnight and then incubated with/without NGF (100 ng/ml) for 20 min. NGF treatment increased colocalization of NTRAP and Rab7. Scale bar, 10 μm.
To further examine the role of NTRAP in endocytic trafficking of TrkA, we used discontinuous iodixanol density gradients to separate different subcellular membranes. Membrane fractions were collected from the four interfaces after centrifugation and analyzed on immunoblots for their protein contents (Figure 7A). EEA1 and NTRAP were enriched in interface 4, whereas Rab7 was enriched in interfaces 2 and 3 (Figure 7A). Consistent with the result shown in Figure 6B, some NTRAP protein was shifted from interface 4 to interfaces 2 and 3 where late endosomes were enriched after 20-min NGF treatment. This result suggests that NTRAP is primarily localized in early endosomes in the absence of NGF, but NGF treatment relocates some NTRAP protein from early endosomes to late endosomes.
Figure 7.
NTRAP controls TrkA trafficking and ERK activation in endosomes. (A) Fractionation of membranous organelles. Membranous components isolated from PC12 cells were separated in iodixanol density gradients. The numbers inside the diagram indicate the concentration of iodixanol. Immunoblots show the distribution of EEA1, Rab7, and NTRAP isolated from vector 1 line of cells. (B) Distribution of the TrkA receptor among the four interfaces before and after NGF treatment. The numbers show the distribution of TrkA among the four interfaces. (C) Distribution of ERK1/2 among the four interfaces before and after NGF treatment. The numbers show the distribution of ERK1/2 among the four interfaces.
In the absence of NGF, the TrkA receptor was similarly distributed among interfaces 1–4 in the samples prepared from cells of vector 1 line and ZF-HA2 line (Figure 7B). It was enriched in interfaces 3 and 4 with its highest level in interface 4. However, TrkA appeared to follow different trafficking routes in vector 1 line and ZF-HA2 line after NGF treatment (Figure 7B). In vector 1 cells, the TrkA receptor moved to lower density of membrane compartments after 10-min NGF treatment and become enriched in interfaces 2 and 3, which is consistent with the flow of TrkA from the plasma membrane to early endosomes and then late endosomes. It moved back to interfaces 3 and 4 with its highest level in interface 3 after 20-min NGF treatment, which might result from a partial recycle of the internalized TrkA back to the plasma membrane. In ZF-HA2 cells, the TrkA receptor also moved to interfaces 2 and 3 after 10-min NGF treatment and remained there after 20-min NGF treatment. These results suggest that TrkA is either stuck in late endosomes or moved to a different type of membrane compartment that has a similar density to late endosomes. The distribution of activated TrkA (p-TrkA) among the four interfaces was similar to the total TrkA after NGF treatment (Figure 7B). Taken together, these results suggest that NTRAP plays a crucial role in normal trafficking of TrkA through early endosomes and late endosomes after NGF activation.
Because persistent ERK activation occurs in endosomes (York et al., 1998; Wu et al., 2001), the expression of ZF-HA may affect persistent ERK activation by altering TrkA trafficking through endosomes. We found that the distribution of total ERK1/2 in the four interfaces was similar among the lysates prepared from cells of vector 1 and ZF-HA2 lines (Figure 7C). In the gradient of the vector 1 lysate, p-ERK1/2 was more abundant in the interfaces enriched in p-TrkA; however, amounts of p-ERK1/2 were low in interfaces enriched in p-TrkA in the gradient of the ZF-HA2 lysate although ERK1/2 was present there (Figure 7C). These results suggest that NTRAP is required for TrkA to activate ERK1/2 in endosomes.
NTRAP Is Associated with Signaling Endosomes in Sensory Neurons
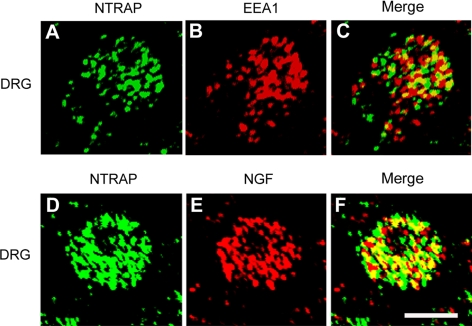
To determine whether NTRAP is also localized in early endosomes in neurons as in PC12 cells, we performed immunohistochemistry on DRG sections from E18 rat embryos with antibodies to NTRAP, EEA1, and NGF. In the DRG small-sized neurons are dependent on NGF for support and are TrkA-expressing nociceptors (Snider and Silos-Santiago, 1996). In these neurons, NTRAP and EEA1 immunoreactivity displayed a punctate pattern and a great fraction of EEA1 (43 ± 3%, n = 51 cells) was colocalized with NTRAP (Figure 8, A–C), indicating that some NTRAP is associated with early endosomes. To determine whether some of the NTRAP+ early endosomes are signaling endosomes, we examined colocalization of NGF and NTRAP. Because embryonic DRG neurons do not express NGF, the NGF immunoreactivity should come from target-derived NGF and thus marks signaling endosomes. In these neurons, 41 ± 3% (n = 50 cells) of NGF immunoreactivity was colocalized with NTRAP (Figure 8, D–F). These observations indicate that NTRAP is associated with signaling endosomes in DRG neurons.
Figure 8.
Colocalization of NTRAP with signaling endosome markers in DRG neurons. Dissected dorsal root ganglia from E18.5 rat embryo were sectioned at 10 μm, stained with antibodies to NTRAP and EEA1 (A–C) or to NTRAP and NGF (D–F), and examined with a confocal microscope. Scale bars, 5 μm.
NTRAP Is Required for Retrograde Signaling of Neurotrophins
Because NTRAP is important for trafficking of activated TrkA receptors along endosomes in PC12 cells and is localized in signaling endosomes in DRG neurons, we asked whether NTRAP plays a role in retrograde neurotrophic signaling. To this end, we used compartmentalized cultures of DRG neurons to mimic the action of a target-derived neurotrophin. It has been shown that neurotrophins on distal axons support the survival of cultured neurons and activate the transcription factor CREB (Tsui-Pierchala and Ginty, 1999; Watson et al., 1999; Ye et al., 2003). If NTRAP is essential for retrograde neurotrophic signaling, down-regulation of NTRAP with shRNA should reduce the effect of NGF and BDNF applied to distal axons on neuronal survival and CREB activation.
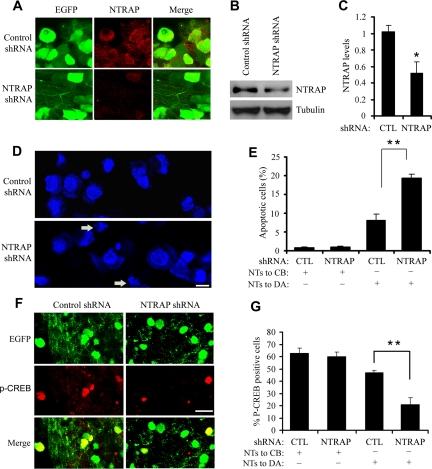
AAV was used to express NTRAP shRNA (NTRAP shRNA AAV) or a scrambled derivative of the shRNA (control shRNA AAV). Each AAV construct also expresses EGFP to mark infected neurons. Rat DRG neurons at E18.5 were plated into the central compartment and cultured in the presence of BDNF and NGF. They were infected with NTRAP shRNA AAV or control shRNA AAV on the third day in vitro (DIV3). AAV started to express shRNA ∼5–7 d after infection, as indicated by appearance of EGFP. Infection of NTRAP shRNA AAV or control shRNA AAV did not interfere with DRG cells extending axons into the side compartment (Supplemental Figure S3). Immunocytochemistry at DIV13 revealed that NTRAP expression in NTRAP shRNA AAV-infected neurons was diminished (Figure 9A). To quantify the reduction, we prepared protein extracts from AAV-infected DRG neuronal cultures at DIV13 and performed Western blots to determine levels of NTRAP (Figure 9B). The amount of NTRAP was reduced by 48% in NTRAP shRNA AAV-infected cultures in comparison with control shRNA AAV-infected cultures (Figure 9C; p = 0.03). Because not every cell in the cultures was infected, the actual reduction in the level of NTRAP in NTRAP shRNA AAV-infected neurons should have been larger than 48%. These results indicate that viral expression of NTRAP shRNA efficiently reduces levels of NTRAP in DRG neurons.
Figure 9.
Down-regulation of NTRAP reduces neuronal survival and CREB activation promoted by retrograde neurotrophic signaling. (A) Reduced NTRAP immunoreactivity in cultured DRG neurons infected with NTRAP shRNA AAV compared with those infected by control shRNA AAV. (B) Reduced levels of NTRAP in cultured DRG cells infected with NTRAP shRNA AAV, as revealed by Western blots. (C) Quantification of NTRAP levels. NTRAP signals on Western blots were normalized to those of α-tubulin. Error bars, SEs. p < 0.05 by Student's t test, n = 3. (D) DAPI staining of cultured DRG neurons. Arrows indicate apoptotic cells with fragmented or condensed nuclei. Scale bar, 5 μm. (E) Quantification of apoptotic neurons in compartmentalized neuronal cultures. Neurotrophins (NTs) were applied to distal axons (DA) or cell bodies (CB) as indicated. EGFP-positive cells with fragmented or condensed nuclei were counted as apoptotic cells. Error bars, SEs. **p < 0.01 by Student's t test; n = 3. (F) Activation of CREB by neurotrophins applied to distal axons, revealed by immunocytochemistry with an antibody specific to p-CREB. AAV-infected neurons were marked by EGFP. P-CREB immunoreactivity was lower in neurons infected by NTRAP shRNA AAV than in neurons infected by control shRNA AAV. Scale bar, 10 μm. (G) Reduced CREB activation by neurotrophins applied to distal axons in DRG neurons expressing NTRAP shRNA. Neurons positive for EGFP and neurons positive for both EGFP and p-CREB were counted and the numbers were used to calculate percentage of p-CREB–positive cells. Error bars, SEs. **p < 0.01 by Student's t test; n = 3.
To determine whether down-regulation of NTRAP blocks the ability of neurotrophins applied to distal axons to promote survival of DRG neurons, we infected DRG neurons with NTRAP shRNA AAV or control shRNA AAV on DIV3, deprived cell bodies in the central compartment or distal axons in the side compartment of neurotrophins for 3 d on DIV10, and performed DAPI staining to detect neurons undergoing apoptosis. Cells with fragmented or condensed nuclei were counted as apoptotic cells (Figure 9D, arrows). Expression of NTRAP shRNA significantly increased apoptosis by 135% in comparison with control shRNA when NGF and BDNF were removed from cell bodies (Figure 9E, p < 0.01). Importantly, NTRAP shRNA did not affect survival of DRG neurons when NGF and BDNF remained in the central compartment (Figure 9E). These results indicate that neurotrophins at distal axons require NTRAP to promote neuronal survival.
To further demonstrate that NTRAP is required for retrograde neurotrophic signaling, we examined CREB activation by BDNF and NGF applied to distal axons. Previous studies demonstrate that activation of the MAPK signaling cascade by Trk receptors leads to phosphorylation of CREB (p-CREB) at Ser133, which promotes a nuclear response to neurotrophins (Ginty et al., 1994; Bonni et al., 1995; Finkbeiner et al., 1997). DRG cells in compartmentalized cultures were infected with AAV expressing shRNAs on DIV3, deprived of neurotrophins on DIV12 overnight, and treated with BDNF and NGF at distal axons for 20 min. The p-CREB immunoreactivity in neurons expressing NTRAP shRNA was greatly reduced in comparison with neurons expressing control shRNA (Figure 9, F and G). The percentage of NTRAP shRNA AAV-infected neurons expressing p-CREB was reduced by 55% in comparison with control shRNA AAV-infected neurons (Figure 9G, p < 0.01). No significant differences in CREB activation were observed between DRG neurons expressing NTRAP shRNA and control shRNA when neurotrophins were applied to cell bodies (Figure 9G). Taken together, the results from the apoptosis and CREB phosphorylation studies show that NTRAP is required for retrograde signaling of neurotrophins from distal axons to cell bodies.
DISCUSSION
NTRAP interacts with receptor tyrosine kinases through a novel mechanism. Most signaling proteins interact with receptor tyrosine kinases through their SH2 domain (Src homology domain 2) or PTB domain (phosphotyrosine-binding domain), such as Shc and PLC-γ, which is dependent on receptor activation and phosphorylation of key tyrosine residues (Schlessinger, 2000). However, a small number of signaling proteins interact with receptor tyrosine kinases independent of activation status of the receptors. For example, ARMS (an ankyrin-rich membrane-spanning protein) is required for sustained neurotrophin signaling and interacts with the transmembrane domain of Trk receptors (Arevalo et al., 2004). Our results indicate that NTRAP does not belong to either one of the two classes. First, NTRAP interacts with inactive Trk receptors in both yeast and mammalian cells. Second, neurotrophin activation greatly enhances the interaction of NTRAP with Trk receptors. Although we have not identified the Trk sequence that interacts with NTRAP, the interaction site must include the tyrosine kinase domain and/or the short carboxy terminus of a Trk receptor because deletion of these two regions completely abolishes the interaction of TrkC with NTRAP.
How does neurotrophin activation enhance the interaction of NTRAP and Trk receptors? C2H2 zinc finger proteins comprise a large family of regulatory proteins, many of which are transcription factors, bind to specific DNA sequences, and participate in a variety of cellular activities (Iuchi, 2001). Some C2H2 zinc fingers bind to RNAs or mediate self-association or selective association with other proteins (Iuchi, 2001; Edelstein and Collins, 2005). NTRAP contains five tandem C2H2 zinc fingers, and the third C2H2 zinc finger motif is required for the interaction with Trk receptor. Because a majority of C2H2 zinc fingers bind to nucleic acids, phosphorylation of tyrosine residues may bestow some properties of nucleic acids on Trk receptors. TrkC contains five key tyrosine residues at positions 485, 674, 678, 679, and 789, which serve as the docking sites for signaling proteins Shc, Frs2, SH2-B, and PLC-γ when they are phosphorylated (Reichardt, 2006). However, we found that substitution of these tyrosine residues with phenylalanine residues did not weaken the interaction of the two proteins (Supplemental Figure S4), indicating that phosphotyrosine residues are not required for binding of NTRAP to Trk receptors. Therefore, it is more likely that neurotrophin activation alters the conformation of the Trk intracellular domain, which enhances the association of the two proteins. These observations also suggest that the active site and the three autophosphorylation sites (tyrosine residues at 674, 678, and 679 in the rat TrkC) in the tyrosine kinase domain have distinct roles in neurotrophic signaling. Although the third C2H2 zinc finger is critical for the interaction of NTRAP and Trk receptors, other C2H2 zinc fingers of NTRAP enhance the interaction. These C2H2 zinc fingers may affect the interaction by either interacting weakly with Trk receptors or mediating dimerization of NTRAP.
Our results from both PC12 cells and DRG neurons indicate that NTRAP plays a crucial role in retrograde neurotrophic signaling. Inhibiting NTRAP function with expression of its N-terminal fragment diminished NGF-dependent differentiation without affecting survival in PC12 cells. Because signaling cascades activated by TrkA receptor on cell surface are sufficient to mediate NGF-induced survival, but NGF-induced differentiation requires internalization of the Trk receptor in PC12 cells (Zhang et al., 2000), Our observation suggests that NTRAP may be involved in the internalization and/or subsequent endocytic sorting of activated Trk receptors. The observation that expression of NTRAP shRNA or ZF-HA did not affect internalization of Trk receptors in PC12 cells and DRG neurons indicates that NTRAP is likely required for intracellular sorting of internalized Trk receptors. Our several observations support this argument. First, a significant fraction of NTRAP is localized to early endosomes and, after neurotrophin stimulation, to late endosomes. Second, membrane fractionation analysis showed that endocytic TrkA trafficking was normal in the first 10 min of NGF treatment, which should include the internalization step, and aberrant in the second 10 min of NGF treatment in cells expressing ZF-HA. Finally, expression of ZF-HA impaired NGF-induced persistent ERK activation in PC12 cells, which is dependent on TrkA signaling in Rap1-associated endosomes (York et al., 1998; Wu et al., 2001). In light of substantial evidence indicating that retrograde neurotrophic signaling is mediated by endosomes carrying internalized Trk receptors (Cosker et al., 2008; Wu et al., 2008), these observations further suggest that NTRAP may play a critical role in retrograde neurotrophic signaling. Indeed, NTRAP is localized to signaling endosomes in DRG neurons, as indicated by colocalization of NTRAP and NGF in small-sized DRG neurons. Importantly, we show that NTRAP knockdown inhibits the ability of neurotrophins applied to the side compartment to promote neuronal survival and to activate CREB without affecting the action of neurotrophins applied to cell bodies in compartmentalized cultures of DRG neurons. Taken together, these results support the notion that NTRAP regulates retrograde neurotrophic signaling by controlling endocytic trafficking of internalized Trk receptors.
In the endocytic pathway, GTPases Rab5 and Rab7 act in a sequential manner by controlling targeting and fusion of clathrin-coated vesicles to early endosomes and regulating progression from early to late endosomes, respectively (Pfeffer, 2003; Rink et al., 2005; Deinhardt et al., 2006). Internalized molecules in early and late endosomes can be recycled back to the plasma membrane. They can also be stored in endosomes for a long time or transported to lysosomes in which they are degraded (Sorkin and Von Zastrow, 2002). Several studies indicate that Trk receptors at axonal terminals are internalized upon neurotrophin activation via clathrin-mediated (Grimes et al., 1996; Grimes et al., 1997; Howe et al., 2001) or pincher-dependent endocytosis (Valdez et al., 2005, 2007). Characterization of endosomes localized in axons suggests that the retrograde neurotrophic signal is carried in either early endosomes (Delcroix et al., 2003) or late endosomes (Deinhardt et al., 2006). Activated TrkA receptors in either early or late endosomes are able to induce sustained activation of ERK1/2 (Delcroix et al., 2003; Hisata et al., 2007). However, it is unknown whether signaling endosomes are a specialized population of early or late endosomes and, if so, which protein controls their generation. Biochemical fractionation analyses revealed that in PC12 cells with disrupted NTRAP function the internalized TrkA receptor was stuck in fractions enriched in late endosomes and was not associated with activated ERK1/2. These results suggest that signaling endosomes are indeed specialized ones and require NTRAP for their formation or maturation. This argument is further supported by our observation that inhibition of NTRAP reduced the sustained activation of ERK1/2, Rap1 activation, and differentiation in PC12 cells after NGF treatment. Future live imaging of Trk trafficking along different membrane compartments is necessary to reveal the precise role of NTRAP in the formation of signaling endosomes.
NTRAP likely regulates endocytic trafficking of Trk receptors by linking Trk receptors to other factors important for sorting of endosomes. NTRAP has a RING finger motif at its N-terminus, which is commonly found in E3 ubiquitin ligases (Jackson et al., 2000). Although we failed to detect any effect of NTRAP on ubiquitination or sumoylation of Trk receptors (data not shown), NTRAP may mediate posttranslational modifications of other associated proteins. It can recruit proteins to Trk-containing endosomes with its two proline-rich regions and modify these proteins. Posttranslational modifications can change the property of the endosomes and specify the endosomes as retrograde cargoes. As an example, the RNA-binding protein La is anterogradely transported into axons by kinesin motors, and sumoylation in axons triggers retrograde transport of La because sumoylated La exclusively binds to dynein (van Niekerk et al., 2007). Rab5 is regulated through the binding of guanine nucleotides and is active when bound to GTP. Its activation is required to complete vesicle targeting and fusion processes and is positively regulated by several guanine exchange factors, which stimulate the release of GDP, allowing GTP to enter the guanine-binding site (Carney et al., 2006). It is intriguing to speculate that NTRAP may control the endocytic pathway by posttranslationally modifying the regulators of Rab5. Further investigation of the ubiquitination activity and interacting partners of NTRAP should provide new insights into the molecular mechanism of retrograde neurotrophic signaling.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rejji Kuruvilla (Johns Hopkins University) for demonstration of compartmentalized cultures, Keith Crutcher for goat anti-NGF antibody, David Kaplan (Toronto Medical Discovery Tower) for tagged TrkA cDNAs, Daniel Pak (Georgetown University) for sharing rat embryos, Chenbiao Wu (Stanford University) for the RalGDSRBD expression construct, and Hui Li for technical assistance. Recombinant human BDNF was generously provided by Regeneron Pharmaceuticals (Tarrytown, NY). This work was supported by grants from the March of Dimes Foundation (1-FY07-532) and the U.S. National Institutes of Health (NS050596) to B.X.
Abbreviations used:
- AAV
adeno-associated virus
- BDNF
brain-derived neurotrophic factor
- CREB
adenosine 3′,5′-monophosphate response element-binding protein
- DRG
dorsal root ganglion
- EEA1
early endosome antigen 1
- ERK1/2
extracellular signal–regulated kinases 1 and 2
- MAPK
mitogen-activated protein kinase
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5,-diphenyltetrazolium bromide
- NTRAP
neurotrophic factor receptor–associated protein
- NTRAPr
shRNA-resistant NTRAP
- shRNA
short hairpin RNA
- TrkA
tropomyosin-related kinase A
- TrkC
tropomyosin-related kinase C
- TUNEL
terminal deoxynucleotidyl transferase–mediated biotinylated dUTP nick end labeling
- ZF-HA
HA-tagged zinc figure fragment of NTRAP.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-04-0321) on October 28, 2009.
REFERENCES
- An J. J., et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo J. C., Yano H., Teng K. K., Chao M. V. A unique pathway for sustained neurotrophin signaling through an ankyrin-rich membrane-spanning protein. EMBO J. 2004;23:2358–2368. doi: 10.1038/sj.emboj.7600253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., editors. New York: John Wiley & Sons; 2003. Current Protocols in Molecular Biology. [Google Scholar]
- Bonni A., Ginty D. D., Dudek H., Greenberg M. E. Serine 133-phosphorylated CREB induces transcription via a cooperative mechanism that may confer specificity to neurotrophin signals. Mol. Cell Neurosci. 1995;6:168–183. doi: 10.1006/mcne.1995.1015. [DOI] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harb. Symp. Quant. Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T. R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Campenot R. B. Local control of neurite development by nerve growth factor. Proc. Natl. Acad. Sci. USA. 1977;74:4516–4519. doi: 10.1073/pnas.74.10.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney D. S., Davies B. A., Horazdovsky B. F. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 2006;16:27–35. doi: 10.1016/j.tcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Castro A. F., Rebhun J. F., Quilliam L. A. Measuring Ras-family GTP levels in vivo—running hot and cold. Methods. 2005;37:190–196. doi: 10.1016/j.ymeth.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Clary D. O., Weskamp G., Austin L. R., Reichardt L. F. TrkA cross-linking mimics neuronal responses to nerve growth factor. Mol. Biol. Cell. 1994;5:549–563. doi: 10.1091/mbc.5.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker K. E., Courchesne S. L., Segal R. A. Action in the axon: generation and transport of signaling endosomes. Curr. Opin. Neurobiol. 2008;18:270–275. doi: 10.1016/j.conb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley S., Paterson H., Kemp P., Marshall C. J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- Deinhardt K., Salinas S., Verastegui C., Watson R., Worth D., Hanrahan S., Bucci C., Schiavo G. Rab5 and Rab7 control endocytic sorting along the axonal retrograde transport pathway. Neuron. 2006;52:293–305. doi: 10.1016/j.neuron.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Delcroix J. D., Valletta J. S., Wu C., Hunt S. J., Kowal A. S., Mobley W. C. NGF signaling in sensory neurons: evidence that early endosomes carry NGF retrograde signals. Neuron. 2003;39:69–84. doi: 10.1016/s0896-6273(03)00397-0. [DOI] [PubMed] [Google Scholar]
- Edelstein L. C., Collins T. The SCAN domain family of zinc finger transcription factors. Gene. 2005;359:1–17. doi: 10.1016/j.gene.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Ehlers M. D., Kaplan D. R., Price D. L., Koliatsos V. E. NGF-stimulated retrograde transport of trkA in the mammalian nervous system. J. Cell Biol. 1995;130:149–156. doi: 10.1083/jcb.130.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas I., Wilkinson G. A., Backus C., Reichardt L. F., Patapoutian A. Characterization of neurotrophin and Trk receptor functions in developing sensory ganglia: direct NT-3 activation of TrkB neurons in vivo. Neuron. 1998;21:325–334. doi: 10.1016/s0896-6273(00)80542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas I., Yoshida C. K., Backus C., Reichardt L. F. Lack of neurotrophin-3 results in death of spinal sensory neurons and premature differentiation of their precursors. Neuron. 1996;17:1065–1078. doi: 10.1016/s0896-6273(00)80240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S., Tavazoie S. F., Maloratsky A., Jacobs K. M., Harris K. M., Greenberg M. E. CREB: a major mediator of neuronal neurotrophin responses. Neuron. 1997;19:1031–1047. doi: 10.1016/s0896-6273(00)80395-5. [DOI] [PubMed] [Google Scholar]
- Ginty D. D., Bonni A., Greenberg M. E. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Grimes M. L., Beattie E., Mobley W. C. A signaling organelle containing the nerve growth factor-activated receptor tyrosine kinase, TrkA. Proc. Natl. Acad. Sci. USA. 1997;94:9909–9914. doi: 10.1073/pnas.94.18.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes M. L., Zhou J., Beattie E. C., Yuen E. C., Hall D. E., Valletta J. S., Topp K. S., LaVail J. H., Bunnett N. W., Mobley W. C. Endocytosis of activated TrkA: evidence that nerve growth factor induces formation of signaling endosomes. J. Neurosci. 1996;16:7950–7964. doi: 10.1523/JNEUROSCI.16-24-07950.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerssen H. M., Pazyra M. F., Segal R. A. Dynein motors transport activated Trks to promote survival of target-dependent neurons. Nat. Neurosci. 2004;7:596–604. doi: 10.1038/nn1242. [DOI] [PubMed] [Google Scholar]
- Hisata S., Sakisaka T., Baba T., Yamada T., Aoki K., Matsuda M., Takai Y. Rap1-PDZ-GEF1 interacts with a neurotrophin receptor at late endosomes, leading to sustained activation of Rap1 and ERK and neurite outgrowth. J. Cell Biol. 2007;178:843–860. doi: 10.1083/jcb.200610073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe C. L., Mobley W. C. Long-distance retrograde neurotrophic signaling. Curr. Opin. Neurobiol. 2005;15:40–48. doi: 10.1016/j.conb.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Howe C. L., Valletta J. S., Rusnak A. S., Mobley W. C. NGF signaling from clathrin-coated vesicles: evidence that signaling endosomes serve as a platform for the Ras-MAPK pathway. Neuron. 2001;32:801–814. doi: 10.1016/s0896-6273(01)00526-8. [DOI] [PubMed] [Google Scholar]
- Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol. Life Sci. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P. K., Eldridge A. G., Freed E., Furstenthal L., Hsu J. Y., Kaiser B. K., Reimann J. D. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- Kuruvilla R., Zweifel L. S., Glebova N. O., Lonze B. E., Valdez G., Ye H., Ginty D. D. A neurotrophin signaling cascade coordinates sympathetic neuron development through differential control of TrkA trafficking and retrograde signaling. Cell. 2004;118:243–255. doi: 10.1016/j.cell.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Luo W., Wickramasinghe S. R., Savitt J. M., Griffin J. W., Dawson T. M., Ginty D. D. A hierarchical NGF signaling cascade controls Ret-dependent and Ret-independent events during development of nonpeptidergic DRG neurons. Neuron. 2007;54:739–754. doi: 10.1016/j.neuron.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Mochizuki N., Yamashita S., Kurokawa K., Ohba Y., Nagai T., Miyawaki A., Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- Pfeffer S. Membrane domains in the secretory and endocytic pathways. Cell. 2003;112:507–517. doi: 10.1016/s0092-8674(03)00118-1. [DOI] [PubMed] [Google Scholar]
- Reichardt L. F. Neurotrophin-regulated signalling pathways. Philos. Trans. R Soc. Lond. B Biol. Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio A., Ahn S., Davenport C. M., Blendy J. A., Ginty D. D. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science. 1999;286:2358–2361. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Senger D. L., Campenot R. B. Rapid retrograde tyrosine phosphorylation of trkA and other proteins in rat sympathetic neurons in compartmented cultures. J. Cell Biol. 1997;138:411–421. doi: 10.1083/jcb.138.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider W. D., Silos-Santiago I. Dorsal root ganglion neurons require functional neurotrophin receptors for survival during development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1996;351:395–403. doi: 10.1098/rstb.1996.0034. [DOI] [PubMed] [Google Scholar]
- Sorkin A., Von Zastrow M. Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 2002;3:600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- Traverse S., Seedorf K., Paterson H., Marshall C. J., Cohen P., Ullrich A. EGF triggers neuronal differentiation of PC12 cells that overexpress the EGF receptor. Curr. Biol. 1994;4:694–701. doi: 10.1016/s0960-9822(00)00154-8. [DOI] [PubMed] [Google Scholar]
- Tsui-Pierchala B. A., Ginty D. D. Characterization of an NGF-P-TrkA retrograde-signaling complex and age-dependent regulation of TrkA phosphorylation in sympathetic neurons. J. Neurosci. 1999;19:8207–8218. doi: 10.1523/JNEUROSCI.19-19-08207.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez G., Akmentin W., Philippidou P., Kuruvilla R., Ginty D. D., Halegoua S. Pincher-mediated macroendocytosis underlies retrograde signaling by neurotrophin receptors. J. Neurosci. 2005;25:5236–5247. doi: 10.1523/JNEUROSCI.5104-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez G., Philippidou P., Rosenbaum J., Akmentin W., Shao Y., Halegoua S. Trk-signaling endosomes are generated by Rac-dependent macroendocytosis. Proc. Natl. Acad. Sci. USA. 2007;104:12270–12275. doi: 10.1073/pnas.0702819104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niekerk E. A., Willis D. E., Chang J. H., Reumann K., Heise T., Twiss J. L. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc. Natl. Acad. Sci. USA. 2007;104:12913–12918. doi: 10.1073/pnas.0611562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek A. B., Hollenberg S. M., Cooper J. A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- von Bartheld C. S., Williams R., Lefcort F., Clary D. O., Reichardt L. F., Bothwell M. Retrograde transport of neurotrophins from the eye to the brain in chick embryos: roles of the p75NTR and trkB receptors. J. Neurosci. 1996;16:2995–3008. doi: 10.1523/JNEUROSCI.16-09-02995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson F. L., Heerssen H. M., Bhattacharyya A., Klesse L., Lin M. Z., Segal R. A. Neurotrophins use the Erk5 pathway to mediate a retrograde survival response. Nat. Neurosci. 2001;4:981–988. doi: 10.1038/nn720. [DOI] [PubMed] [Google Scholar]
- Watson F. L., Heerssen H. M., Moheban D. B., Lin M. Z., Sauvageot C. M., Bhattacharyya A., Pomeroy S. L., Segal R. A. Rapid nuclear responses to target-derived neurotrophins require retrograde transport of ligand-receptor complex. J. Neurosci. 1999;19:7889–7900. doi: 10.1523/JNEUROSCI.19-18-07889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe S. R., Alvania R. S., Ramanan N., Wood J. N., Mandai K., Ginty D. D. Serum response factor mediates NGF-dependent target innervation by embryonic DRG sensory neurons. Neuron. 2008;58:532–545. doi: 10.1016/j.neuron.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Cui B., He L., Chen L., Mobley W. C. The coming of age of axonal neurotrophin signaling endosomes. J Proteomics. 2009;72:46–55. doi: 10.1016/j.jprot.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Lai C. F., Mobley W. C. Nerve growth factor activates persistent Rap1 signaling in endosomes. J. Neurosci. 2001;21:5406–5416. doi: 10.1523/JNEUROSCI.21-15-05406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Kuruvilla R., Zweifel L. S., Ginty D. D. Evidence in support of signaling endosome-based retrograde survival of sympathetic neurons. Neuron. 2003;39:57–68. doi: 10.1016/s0896-6273(03)00266-6. [DOI] [PubMed] [Google Scholar]
- York R. D., Yao H., Dillon T., Ellig C. L., Eckert S. P., McCleskey E. W., Stork P. J. Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature. 1998;392:622–626. doi: 10.1038/33451. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Moheban D. B., Conway B. R., Bhattacharyya A., Segal R. A. Cell surface Trk receptors mediate NGF-induced survival while internalized receptors regulate NGF-induced differentiation. J. Neurosci. 2000;20:5671–5678. doi: 10.1523/JNEUROSCI.20-15-05671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel L. S., Kuruvilla R., Ginty D. D. Functions and mechanisms of retrograde neurotrophin signalling. Nat. Rev. Neurosci. 2005;6:615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.