Abstract
Three articles from the early years of Molecular Biology of the Cell (MBoC) have had remarkably many citations in the literature since their publication ∼10 years ago. As a coauthor of these articles and the former editor of MBoC, I was asked for possible explanations. I believe the answer lies in the unusual nature of these articles: each presents and summarizes gene expression data for nearly every gene in the yeast or human genomes. Continuing interest in the data themselves by cell biologists, rather than results or conclusions drawn by the authors, best accounts for the citation history. The flatness of the numbers of citations over time, the continuing high rate of accesses to individual Web sites set up to allow searching and display of the underlying data, and the large fraction of citations in journals focused on mathematics and computation all support the same conclusion: it's the data.
Shortly after David Drubin accepted the position of editor-in-chief of Molecular Biology of the Cell (MBoC), he gave me a call. David wanted to talk about the early years of the journal, when Keith Yamamoto and I were responsible for its policies and editorial management. The discussion dwelt only briefly on the core values that drove the founding of the journal and its early management. After all, David had long served the journal as associate editor.
David was interested in understanding something quite different. He noticed that MBoC published quite a few articles in early years that appear to have attained notable and enduring influence, as measured by their citation history. I was a coauthor of a few of these. Why did I think these articles appeared at that time in MBoC? Is there a special niche, defined by such articles? Could MBoC do more to attract such manuscripts in the future? We agreed that I would think about this and potentially write a retrospective based on the articles I coauthored.
David chose three articles (citation statistics are from the Institute for Scientific Information [ISI; http://wokinfo.com/] database as of this writing): Spellman et al. (1998), “Comprehensive Identification of Cell Cycle-regulated Genes of the Yeast Saccharomyces cerevisiae by Microarray Hybridization” (2074 citations); Gasch et al. (2000), “Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes” (1384 citations); and Whitfield et al. (2002), “Identification of Genes Periodically Expressed in the Human Cell Cycle and Their Expression in Tumors” (395 citations). Among all the more than 34,000 articles MBoC had published by September, 2009, these three currently rank first, second, and thirteenth in citations in the ISI database.
These articles originated from a long and productive collaboration between my laboratory and Pat Brown's at Stanford. As their titles indicate, they each present genome-wide gene expression data of interest to cell biologists. Each article is connected to a large data set that is available as searchable supplementary tables and, more usefully, Web sites with more sophisticated search and display capacities that are maintained at Stanford (now also at Princeton) and linked to by more general databases such as SGD (Saccharomyces Genome Database).
What is it about our three articles that generates so many citations, year after year? None of them produced any major new principles; they were exploratory in nature. They were not designed to prove or to falsify any particular hypotheses, nor did they. They used, but they did not introduce, new technology or analytical methods: other articles were published to this end, some of which are also highly cited. I think the main reason these three articles are cited is for diverse data items others found useful in them. Each of the three articles contains information about the expression of thousands of individual genes of yeast or humans. Every time a scientist publishes something useful that they found in these data, a citation is generated. The title of this retrospective, a partial paraphrase of James Carville's famous political slogan from the Clinton era (“It's the economy, stupid”), says it all: “It's the data!”
The mass of the data presented a major challenge when we were writing these articles. We did our best to summarize the larger trends in the processes we had set out to study and to extract other generalities where we could. But we did not, and indeed we could not, foresee even a modest fraction of the specific uses and interpretations others ultimately found for data involving individual genes or subsets of genes. Our articles were consciously written to introduce potential users to the data and to provide experimental details that might aid users in the interpretation of the data in the context of their interests. The work of most cell biologists then, and still today, concerns only a small fraction of the total number of the functional genes of an organism; we wanted the data set to be useful to them. It is for this reason that we set up easily searchable Web sites with many display options for each article and asked MBoC to carrying a hyperlinked table of the primary data on their Web site and to allow anonymous download of some or all of the data.
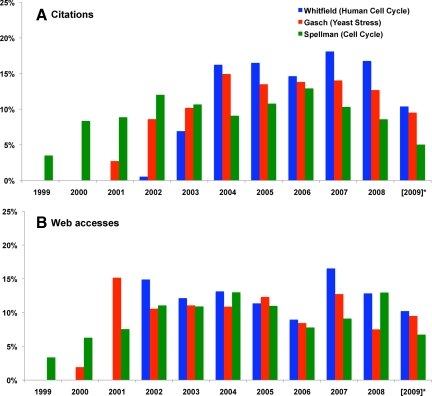
Citations to our three articles have an interesting and unusual property: they are minimally dependent on what our group intended to study, and indeed they only rarely refer to the results and conclusions of our article. Figure 1 shows that the citation history is remarkably flat over time, with citations continuing at a high rate year after year. Usage of the supplementary tables and the companion Web sites (assessed independently from the server logs) is similarly high and flat. To me, these observations are a strong indication that the citation rate is driven by the uses readers make of the data themselves. If results and conclusions were responsible for the citations, I would have anticipated that number of citations would rise over the years as the results and conclusions become accepted and then would fall as they appear in reviews and texts and ultimately become common knowledge taken for granted. They certainly would not be expected to increase every year, a decade later. If our articles were being cited for results and conclusions, searches of the data in the databases would be expected to fall in frequency much earlier than citations to the article. Only if, as I believe, the interest in our work is for the data themselves would the citations increase and the database accesses continue in parallel.
Figure 1.
The citation history of three articles from the early years of MBoC. Source: ISI database.
A bit of further research in the ISI citation databases produced another remarkable statistic that fortifies my belief that the citations are for the data themselves. For Spellman et al. (1998), half (49.8%) of the citations are in journals categorized by ISI as being devoted to mathematics or computation (20.5%, mathematical and computational biology; 14.8%, probability and statistics; and 14.5%, computer science), leaving only 50.2% of the citations in journals devoted primarily to biological subjects, i.e., biochemistry and molecular biology, cell biology, genetics and heredity, and biotechnology and applied microbiology. The statistics for Gasch et al. (2000) and Whitfield et al. (2002) are somewhat lower, but no less remarkable; 22 and 34%, respectively, of the citations are to the journals in fields devoted mainly to mathematics and computation. The citing articles reflect the explosion of interest by nonbiologists from the physical and mathematical sciences and engineering in applications to biology (e.g., molecular evolution, biostatistics, computer modeling, and regulatory networks) on the one hand, and the rise of what appear to be robust new disciplines at the interface of biology and mathematics and computation (bioinformatics and systems biology), on the other. I think the three MBoC articles attracted all these citations because the data sets themselves were both readily available in a useful form and coherently described. They have been used, literally hundreds of times, as test beds for new algorithms, statistical tests, and bioinformatic computational systems.
We published these three articles in MBoC because MBoC offered a permanent venue for housing, in a convenient and accessible way, the entire primary data (simple tab-delimited and hyperlinked tables of expression levels), whereas at the same time allowing us, through its flexibility with respect to manuscript length, to describe what we did fully enough so that both biologist and computational and statistical communities could get and use the data well.
Ten years ago, Pat Brown and I concluded an early review by stressing the necessity of open and continued access to all the primary data underlying articles such as the three MBoC articles. I cannot improve on what we wrote then (Brown and Botstein, 1999). After introducing the challenge to publishers of dealing with data-rich exploratory manuscripts useful mainly for their data, we wrote:
For the moment, our own group has been addressing this problem in three ways: first, we provide complete data tables whenever we submit publications so that the journals can provide them to readers; second, we “self-publish” by means of our own Web sites, which provide searchable databases and visualization tools so that anybody can find out what we learned about any gene of interest; and third, we provide all the data to the relevant genomic databases, such as the Saccharomyces Genome Database. We believe that publishing these descriptive data are as essential a part of the process of genomic exploration as the publication of maps and journals was to the lasting value of the expedition of Lewis and Clarke. A fresh approach to scientific publication may be one of the next critical advances in the post-genome era.
The final lesson from the three articles may well be that MBoC should continue to play a leading role in addressing the problems of large-data-set integrity, completeness, usability, and availability in ways that help scientists everywhere, now and in the future. If it can be developed, a simple and robust database schema on MBoC's Web site that would allow better searching and display of data would be especially valuable, because it would obviate the need for housing data on author's individual Web sites. In this way the journal could serve to archive the feature of these articles that readers find most valuable over time: it's the data!
ACKNOWLEDGMENTS
I am grateful to Mike Cherry for providing the usage data at Stanford and Kara Dolinski and Mike Livstone for help in preparing the illustration.
REFERENCES
- Brown P. O., Botstein D. Exploring the new world of the genome with DNA microarrays. Nat. Genet. 1999;21(1 Suppl):33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., Eisen M. B., Brown P. O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M. L., et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]