The protein chaperone Hsp90 and its co-chaperone Sba1/p23 are found to accumulate in the nucleus of haploid yeast cells as glucose is exhausted and in sporulating diploids. Novel and existing Hsp90 mutants exhibit defects in nuclear translocation and spore development, linking these two phenomena.
Abstract
The 90-kDa heat-shock protein (Hsp90) operates in the context of a multichaperone complex to promote maturation of nuclear and cytoplasmic clients. We have discovered that Hsp90 and the cochaperone Sba1/p23 accumulate in the nucleus of quiescent Saccharomyces cerevisiae cells. Hsp90 nuclear accumulation was unaffected in sba1Δ cells, demonstrating that Hsp82 translocates independently of Sba1. Translocation of both chaperones was dependent on the α/β importin SRP1/KAP95. Hsp90 nuclear retention was coincident with glucose exhaustion and seems to be a starvation-specific response, as heat shock or 10% ethanol stress failed to elicit translocation. We generated nuclear accumulation-defective HSP82 mutants to probe the nature of this targeting event and identified a mutant with a single amino acid substitution (I578F) sufficient to retain Hsp90 in the cytoplasm in quiescent cells. Diploid hsp82-I578F cells exhibited pronounced defects in spore wall construction and maturation, resulting in catastrophic sporulation. The mislocalization and sporulation phenotypes were shared by another previously identified HSP82 mutant allele. Pharmacological inhibition of Hsp90 with macbecin in sporulating diploid cells also blocked spore formation, underscoring the importance of this chaperone in this developmental program.
INTRODUCTION
The 90-kDa heat-shock protein (Hsp90) molecular chaperone is required for biogenesis and function of a variety of client proteins, most notably protein kinases and transcription factors (Picard, 2002). Hsp90 interacts with specific cochaperones that regulate substrate transfer from the Hsp70 chaperone system and subsequent maturation in an ATP-dependent manner (Pearl and Prodromou, 2006). In addition to operating as the center of a multichaperone complex, Hsp90 chaperones are capable of mediating protein folding independently of other cofactors. Hsp90 is an abundant cytosolic protein in all eukaryotes, whereas Hsp90 orthologues such as Grp94 and TRAP-1 are restricted to the endoplasmic reticulum and mitochondrial matrix, respectively (Felts et al., 2000; Richter et al., 2007). Hsp90 has been shown to shuttle between the nucleus and cytoplasm complexed with immature steroid receptor substrates. On steroid activation in metazoan cells, Hsp90 along with its late-stage cochaperones (FKBP52, TPR-domain immunophilins) and the client protein shift from a predominantly cytoplasmic localization into the nucleus before client dissociation (Pratt et al., 1999). In Saccharomyces cerevisiae, the Hsp90 proteins Hsp82 and heat-shock complex 82 (Hsc82) exhibit uniform localization throughout the cell during vegetative growth, consistent with involvement in both cytoplasmic and nuclear processes (Huh et al., 2003). It is unknown whether this distribution is altered under conditions of cellular stress.
Proliferation of S. cerevisiae in glucose is characterized by three distinct stages of growth (Herman, 2002). Cells in log phase proliferate at their maximal rate by using mixed respiro-fermentative metabolism. As glucose becomes limiting, cells progress through the diauxic shift and continue dividing at a slower rate based on respiration of ethanol derived from fermentation. On exhaustion of ethanol or other essential nutrients during proliferative growth, cells arrest at the G1 phase of the mitotic cell cycle and enter stationary phase, a period of reduced metabolic activity similar to nonmitotic, or quiescent, metazoan cells (Herman, 2002; Gray et al., 2004). Stationary phase/quiescent yeast cells are characterized by accumulation of storage carbohydrates such as glycogen and trehalose, increased stress resistance, thickening of the cell wall, and most importantly, the ability to survive extended periods without nutrients (Lillie and Pringle, 1980; Plesset et al., 1987; Gray et al., 2004). Simultaneous starvation of diploid S. cerevisiae cells for glucose and nitrogen leads to activation of the sporulation program and meiosis, resulting in production of four haploid progeny (Neiman, 2005). Spores share many of the characteristics of quiescent phase cells, rendering them capable of long-term survival (Smits et al., 2001).
In this report, we investigated whether the subcellular distribution of Hsp90 and its cochaperones responds to environmental conditions. We reveal for the first time a novel targeting event of Hsp90 into the nucleus upon glucose exhaustion in yeast. Nuclear accumulation of an Hsp90-green fluorescent protein (GFP) fusion protein was not observed in response to other stresses, including heat shock and ethanol toxicity, or acute depletion of glucose or other nutrients, suggesting that it is a specific response to transition through the diauxic shift into quiescence. The Hsp90 cochaperone Sba1/p23 also exhibited nuclear accumulation, and transport of both chaperones was largely autonomous. To understand the physiological relevance of Hsp90/Sba1 nuclear accumulation, we identified a novel nonlocalizing allele of HSP82, which along with an existing mutant allele of HSP82 were found to exhibit profound defects in sporulation at the level of spore formation. Hsp82 functional status correlated with sporulation defects and nuclear accumulation in quiescent cells, demonstrating that these diverse characteristics are linked. Pharmacological inhibition of Hsp90 in wild type diploids resulted in dose-dependent reduction in sporulation, underscoring the importance of this chaperone in this important developmental process.
MATERIALS AND METHODS
Strains and Growth Conditions
Standard yeast propagation and transformation procedures were used. Yeast strains and plasmids are described in Table 1. SWY519 and SWY3562 strains were a kind gift from Kathryn J. Ryan (Texas A&M University) (Ryan et al., 2007). JJ816 and JJ817 strains were a kind gift from Jill L. Johnson (University of Idaho) (Flom et al., 2005). Diploid strain D818 was constructed by mating JJ816 and JJ817 and confirmed by mating type testing. All experiments involving plasmid-borne alleles of HSP82-GFP or HSP82-yEm-red fluorescent protein (RFP) were carried out in strains JJ816 (haploid) or D818 (diploid) lacking endogenous HSC82 and HSP82 genes after counterselection of the wild-type HSP82 URA3 covering plasmid by using 5-fluoroorotic acid (5-FOA). Strains were grown in nonselective (YP, 1% yeast extract and 2% peptone) or selective (synthetic complete, SC) media containing 2% glucose. To induce sporulation, cells were first grown on presporulation plates consisting of 5% glucose, 3% Difco nutrient broth, and 1% yeast extract, on patches (3 cm2) that had been freshly grown on YPD for 1 d. Patches were transferred to presporulation plates for an additional day at 30°C. Cells were then transferred to supplemented liquid sporulation medium (1% potassium acetate, 0.01% zinc acetate, 1× uracil supplement, 1× histidine supplement, and 1× leucine supplement) and incubated with aeration at room temperature for 2 d followed by 2 d at 30°C. Pharmacological inhibition of Hsp90 in sporulating cultures was achieved by adding macbecin in dimethyl sulfoxide (DMSO) or DMSO alone to final concentrations between 0 and 25 μM, matching the total volume of DMSO in each case before placing cells into liquid sporulation medium. Cells were incubated with aeration at room temperature for 2 d followed by 2 d at 30°C and sporulation efficiency was quantified by determining the number of mature four-spore tetrads as a percentage of the total population. For quiescent phase/stationary cultures, cells were grown at 30°C in desired media for 3–4 d to allow cells to reach quiescence. Starvation media were made by using standard SC media (Sunrise Science Products, San Diego, CA) lacking glucose, NH4, PO4, or both NH4 and PO4. Spent media were obtained by growing cells in YPD for 3 d and removing the quiescent cells by centrifugation.
Table 1.
Strains and plasmids
| Strain/plasmid | Description | Reference |
|---|---|---|
| BY4741 | MATahis3Δ leu2Δ met15Δ ura3Δ | |
| SWY519 | MATα ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 ade2-1::ADE2:ura3 | Ryan et al. (2007) |
| SWY3562 | MATα kap95-E126K trp1-1 ura3-1 his3-11,15 leu2-3,112 can1-100ade2-1::ADE2:ura3 | Ryan et al. (2007) |
| JJ816 | MATahsc82::LEU2 hsp82::LEU2 ade2-1 trp1-1 ura3-1 leu2-3,112his3-11,15 met2 lys2 [Yep24-HSP82] | Flom et al. (2005) |
| JJ817 | MATαhsc82::LEU2 hsp82::LEU2 ade2-1 trp1-1 ura3-1 leu2-3,112his3-11,15 met2 lys2 [Yep24-HSP82] | Flom et al. (2005) |
| D818 | MATa/MATα hsc82::LEU2 hsp82::LEU2 ade2-1/ade2-1 trp1-1/trp1-1ura3-1/ura3-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 met2/met2lys2/lys2 [Yep24-HSP82] | This study |
| pHis6-Hsp82 | p414-GPD-6XHis-Hsp82 | Liu et al. (1999) |
| pHsp82-GFP | p414-GPD-6XHis-Hsp82-GFP(S65T)-His3MX6 | This study |
| pSba1-mCherry | p416-GPD-Sba1-mCherry | This study |
| pNL1 | pHsp82-GFP mutagenic PCR isolate | This study |
| pNL2 | pHsp82-GFP mutagenic PCR isolate | This study |
| pHsp82-I578F | pHsp82-GFP with single amino acid substitution at residue 578 (I->F) | This study |
| pTCA/Hsp82 | HSP82 driven from the GPD promoter, CEN/TRP1 vector | Bohen (1995) |
| pTCA/Hsp82-G313N | As pTCA/Hsp82, with G313N substitution | Bohen (1995) |
| pTCA/Hsp82-E431K | As pTCA/Hsp82, with E431K substitution | Bohen (1995) |
| pTCA/Hsp82-yEmRFP | Hsp82 tagged with y-EmRFP protein at C terminus | This study |
| pTCA/Hsp82-G313N-yEmRFP | Hsp82-G313N tagged with y-EmRFP protein at C terminus | This study |
| pTCA/Hsp82-E431K-yEmRFP | Hsp82-E431K tagged with y-EmRFP protein at C terminus | This study |
Plasmid Construction
pHsp82-GFP was constructed using recombinant cloning. In brief, p414GPD-6XHis-Hsp82 was digested with BamHI and SacII and used for gap repair via homologous recombination with pooled polymerase chain reaction (PCR) products amplified from the template plasmid pFA6a-GFP(S65T)-HIS3MX6, with homology to the gapped plasmid ends (Longtine et al., 1998; Liu et al., 1999). pSba1-mCherry was constructed using recombinant cloning. p416GPD (URA3) was digested with XhoI and SpeI and used for gap repair. Sba1 was amplified from genomic DNA incorporating a flanking SpeI site and homology to the gapped plasmid as well as homology to mCherry. mCherry plasmid was a kind gift from Ken Marians (Sloan-Kettering Institute) via William Margolin (University of Texas Medical School at Houston). mCherry was amplified from the original Escherichia coli codon-optimized mCherry expression vector incorporating a flanking XhoI site and homology to the 3′ end of SBA1 and the gapped plasmid. Plasmids pTCA-Hsp82, pTCA-Hsp82/G313N and pTCA-Hsp82/E431K were a kind gift from Avrom Caplan (City College of New York). Plasmids pTCA-Hsp82-yEm-RFP, pTCA-Hsp82/G313N-yEm-RFP, and pTCA-Hsp82/E431K-yEm-RFP were constructed using recombinant cloning. Original plasmids were digested with SacII and used for gap repair. yEmRFP was amplified from plasmid pRS316-Gap-Cherry (kind gift from Neta Dean, Stony Brook University), incorporating a flanking XhoI site and homology to Hsp90 and to the gapped plasmid. Colonies were visually screened for acquired purple fluorescence visible at the colony level and confirmed by Western blot analysis. Plasmid p416Gal-v-Src has been described previously (Xu and Lindquist, 1993).
v-Src Activity Assay
Cells were prepared for immunoblot analysis by growing strains containing pGal-v-Src plasmid overnight at 23°C in media containing glucose. Early log phase cells were harvested and washed. Ten-milliliter cultures at an initial density of 0.4 A600 were grown for 10 h in media containing galactose for v-Src induction. Protein extracts were prepared following a modified alkaline lysis protocol to maintain tyrosine phosphorylation (Ooi et al., 1996). In brief, cells were harvested and resuspended in 150 μl of 1.85 M NaOH/1% β-mercaptoethanol and incubated on ice for 10 min. Then, 75 μl of 100% trichloroacetic acid (TCA) was added, followed by incubation on ice for 5 min. TCA precipitates were collected by centrifugation at 5000 × g for 2 min. The precipitate was then washed once with 1 ml of acetone and resuspended in 2× SDS loading buffer supplemented with 0.1 M Tris base. The sample was heated at 65°C for 10 min and subjected to SDS-polyacrylamide gel electrophoresis. Proteins were transferred onto nitrocellulose and blots were incubated overnight with anti-phosphotyrosine antibody (1 μg/ml; Millipore, Billerica, MA) in Tris-buffered saline containing 3% nonfat dry milk. Detection by enhanced chemiluminescence was performed after incubation with anti-mouse horseradish peroxidase-conjugated secondary. Load controls were obtained from the same blot with an antibody against glucose-6-phosphate dehydrogenase (GPD) (anti-GPD; Sigma-Aldrich, St. Louis, MO).
Microscopy
Cells were harvested by centrifugation, washed once with water, and resuspended in 70% ethanol for fixation for 1 min. Cells were once again harvested by centrifugation and resuspended in water for visualization. Nuclei of living cells were stained with the vital DNA dye Hoescht 33342 at a concentration of 10 μg/ml for 5 min. Cells were observed under an BX60 epifluorescence microscope (Olympus, Tokyo, Japan) equipped with a 100× immersion oil objective and 4,6-diamidino-2-phenylindole, GFP, or RFP filter cubes. Images were captured with a Photometrics CoolSNAP-fx cooled charge-coupled device camera driven by QED image-capturing software (Media Cybernetics, Bethesda, MD). All images were processed using Photoshop CS (Adobe Systems, San Jose, CA). A minimum sample number of 200 individual cells were counted for all experiments, chosen from multiple microscopic fields. Localization frequency was then calculated as a percent of total (i.e., cytoplasmic plus nuclear), with representative images are shown throughout.
Glucose and Ethanol Analysis
To determine medium ethanol concentrations, samples were taken at indicated time points during growth, and the ethanol concentration was measured using the EnzyChrom ethanol assay kit following the protocol recommended by the manufacturer (Bioassay Systems, Hayward, CA). For glucose concentrations, a fraction of the sample described above was tested using a glucose assay kit, as per the manufacturer's recommendations (Sigma-Aldrich). All measurements were done in duplicate.
PCR Random Mutagenesis and Mutation Analysis
We used PCR random mutagenesis to generate novel nonlocalizing alleles of HSP82. The HSP82 coding sequence was amplified from the pHsp82-GFP plasmid by using Taq polymerase in a reaction mixture spiked with different levels of dNTPs (0.5 mM dATP and dGTP and 1 mM dCTP and dTTP) and 0.1 mM MnCl2 to induce errors. pHsp82-GFP was digested with BamHI and SacII and used for gap repair via homologous recombination with pooled mutagenic PCR products, selecting for recombination after transformation into wild-type BY4741 cells on SC −HIS plates. Approximately 1500 colonies were visually screened for lack of nuclear accumulation in quiescent phase.
Spore Viability Stress Assay
Sporulated cultures of either wild type HSP82-GFP or hsp82-I578F-GFP were exposed to 55°C heat shock via incubation in 0.2-ml tubes placed in a thermal cycler for the indicated duration and plated on YPD in triplicate. Cells were allowed to grow for 2 d at 30°C.
Transmission Electron Microscopy (TEM)
TEM analysis was performed essentially as described previously (Wright, 2000), with modifications. Instead of using agar-embedded aggregates for selection of the yeast for electron microscopy, homogenous aliquots of yeast cultures were processed as suspensions. As needed during infiltration in epoxy resin, low-speed centrifugation (∼1500 × g) was used to recover and concentrate cells. After infiltration in low viscosity Spurr's resin, excess resin was removed from each of the specimen sediments, which were then transferred into Beem capsules and covered with appropriate aliquots of fresh resin which was subsequently polymerized at 60°C. After thin sectioning on a diamond knife, the stained specimens were transferred to copper grids and viewed using a CM12 electron microscope (JEOL, Tokyo, Japan) operated at 80 kV.
RESULTS
Quiescent Phase Nuclear Accumulation of Hsp90
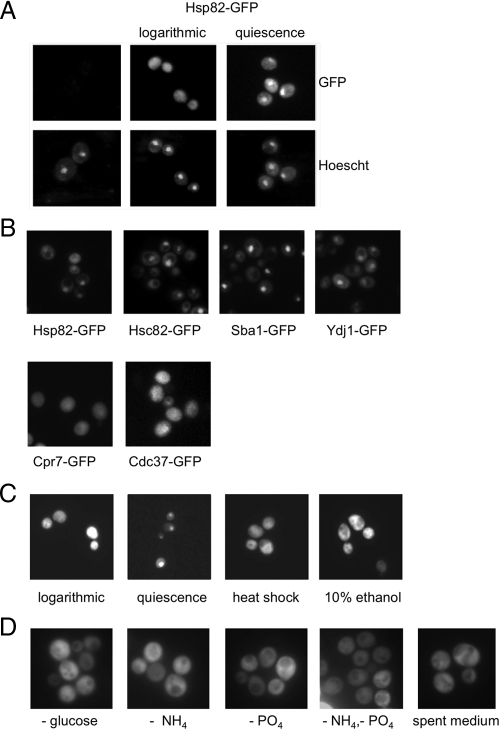
We examined localization dynamics of Hsp90 by using a strain with GFP inserted at the HSP82 genomic locus, creating a carboxy terminal-tagged Hsp82-GFP fusion protein (Huh et al., 2003). As shown in Figure 1A, during logarithmic phase growth in rich medium at 30°C, Hsp82-GFP was distributed uniformly throughout the cell. However, after 2 d of continued incubation a distinct concentration was observed, coincident with the nucleus as determined by Hoechst 33342 staining of DNA in live cells. The culture had by this time entered quiescence, characterized by the accumulation of large unbudded cells and cessation of proliferation as determined by optical density measurements. Western blot analysis verified that the Hsp82-GFP chimeric protein remained intact during growth, excluding the possibility that a GFP-containing proteolytic fragment was responsible for the observed nuclear localization (Supplemental Figure S1A). To confirm that the observed relocalization of Hsp82 reflected normal physiological activity and was not an artifact caused by addition of the GFP moiety, we asked whether Hsp82-GFP could function as the only copy of Hsp90 in the cell. Plasmid pHsp82-GFP bearing an Hsp82-GFP fusion was constructed using recombinant cloning and was able to sustain wild-type growth in the absence of both endogenous Hsp90 genes (Supplemental Figure S1B).
Figure 1.
Quiescent phase nuclear accumulation of Hsp90 and cochaperones. (A) A genomic Hsp82-GFP fusion was examined during logarithmic growth and after 3 d of growth and entry into quiescent phase. Representative photomicrographs are shown of GFP localization and nuclear position as assessed by Hoescht 33342 staining. (B) The indicated fusion proteins were localized as described in A. Only quiescent phase images are shown. (C) Log-phase cells bearing the genomic Hsp82-GFP were heat shocked for 1 h at 42°C or treated with 10% ethanol for 10 min. (D) Log phase cells were transferred to synthetic medium lacking the indicated nutrients, or spent medium collected from a quiescent phase culture, and images taken after 2 h.
We further examined logarithmic and quiescent phase localization of known Hsp90 cochaperones from the commercially available GFP collection of carboxy-terminal protein fusions: Sba1, Cdc37, Cpr7, Ssa1, Ydj1, Sti1, and Sse1 (Huh et al., 2003). In addition to both Hsp90 isoforms Hsp82 and Hsc82, we found that the yeast p23 orthologue Sba1 and the Hsp70 ATPase activating cochaperone Ydj1 also demonstrated quiescent phase nuclear accumulation (Figure 1B). Ydj1 has been reported previously to localize to the nucleus and perinuclear space due to posttranslational farnesylation that anchors a subpopulation in the contiguous endoplasmic reticulum (ER)/nuclear membrane (Caplan et al., 1992). Two Hsp90 cochaperones, Cpr7 and Cdc37, remained distributed throughout the cytoplasm regardless of growth phase (Figure 1B). A third pattern of localization into multiple dynamic puncta was observed with GFP-tagged Hsp70 (Ssa1), Hsp110 (Sse1), and HOP (Sti1), which will be reported elsewhere (data not shown). These findings demonstrate that Hsp90, along with the cochaperone Sba1, relocate to the nucleus after extended growth, exclusive of other known soluble Hsp90 partner proteins.
We next investigated whether nuclear accumulation was a specialized consequence of entry into quiescent phase or a general response to cellular stress. To differentiate between the two possibilities, we applied a series of different stresses to cells including heat shock and toxic ethanol concentration and examined localization of Hsp82-GFP, as shown in Figure 1C. In contrast to stationary phase, neither heat shock (42°C; 10 min) nor 10% ethanol resulted in nuclear accumulation. Similar results were observed with the Sba1-GFP fusion (data not shown). These data support the interpretation that Hsp90 and Sba1 relocate to the nucleus specifically in response to growth status. To determine whether nuclear translocation was occurring in response to depletion of a specific nutrient, cells bearing Hsp82-GFP were grown to logarithmic phase and then switched to growth in minimal medium lacking glucose, a nitrogen source (−NH4), phosphate (−PO4), or both nitrogen and phosphate. In all cases, no translocation was observed, demonstrating that acute withdrawal of an essential nutrient is insufficient to drive nuclear accumulation of Hsp82-GFP (Figure 1D). In addition, it was possible that a metabolite was accumulating during growth and progression through the diauxic shift that could induce Hsp82 translocation. To test this theory, we allowed a parallel culture of Hsp82-GFP-bearing cells to reach quiescence through normal growth and collected the growth medium after centrifugation. Log phase Hsp82-GFP cells resuspended in this “spent” medium likewise failed to exhibit accumulation of the chaperone (Figure 1D).
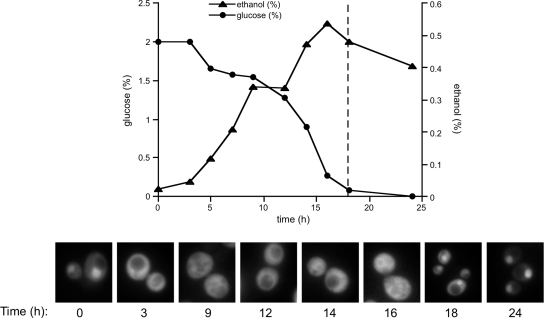
We next analyzed the kinetics of Hsp90 accumulation into the nucleus during growth in more detail. Quiescent cultures of Hsp82-GFP cells displaying nuclear accumulation were diluted into fresh medium and allowed to resume vegetative growth at 30°C (Figure 2A, time 0). Within three hours after the shift to glucose-rich media, Hsp90 was seen to redistribute throughout the cell (a representative image from each sample is shown). Glucose and ethanol concentrations were monitored as growth continued over 24 h. After ∼18 h, glucose was nearly exhausted from the media as determined by enzymatic assay, and ethanol produced during fermentation had reached its maximum. Coincident with this, Hsp90 was again observed to accumulate in the nucleus. Hsp90 therefore seems to respond to events connected to glucose depletion rather than the complete absence of a carbon source because at 18 h ethanol was still abundant (0.5%). Together, these data suggest that the Hsp90 chaperone in yeast cells responds to progression through the diauxic shift into the quiescent state by translocating to the nucleus. However, the precise signaling events required to initiate this change in subcellular localization are likely complex as acute depletion of glucose or other growth-limiting nutrients was unable to elicit the same response.
Figure 2.
Hsp90 nuclear accumulation is a consequence of glucose depletion. (A) Quiescent phase cells bearing the Hsp82-GFP fusion were shifted to fresh YPD growth medium, and the culture was followed for a growth period of 24 h. Samples were harvested at the indicated time points and processed for photomicroscopy or determination of ethanol (triangles) and glucose (circles) levels as described in Materials and Methods.
Hsp90 Transport to the Nucleus Is Independent of Sba1 and Requires the α/β Importin System
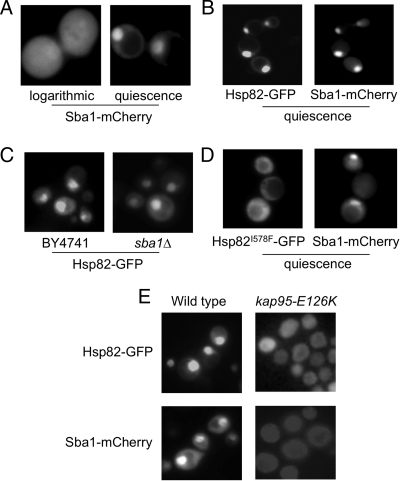
Sba1/p23 is a late-acting cofactor in the Hsp90 chaperoning cycle, binding to the amino-terminal nucleotide binding domains of both Hsp90 molecules in the functional dimer and stabilizing the chaperone in the ATP-bound state (Fang et al., 1998; Richter et al., 2004). This raised the possibility that nuclear accumulation of these two chaperones may be interdependent. Alternatively, Sba1 localization during quiescence may be regulated in parallel but independently of Hsp90. To better understand this relationship and distinguish between these two models, we constructed a plasmid-borne fusion of Sba1 with the dsRed variant mCherry. In the same manner as observed with the original GFP-tagged fusion, Sba1-mCherry was distributed uniformly throughout the cell during logarithmic growth and accumulated in the nucleus during quiescence (Figure 3A). Nuclear colocalization was evident upon coexpression of both fluorescently tagged proteins (Figure 3B). To directly test whether nuclear accumulation of Hsp82 required Sba1, we transformed BY4741 wild-type and isogenic sba1Δ cells with plasmid pHsp82-GFP. In the absence of Sba1, Hsp90 was observed to accumulate in the nucleus in a manner nearly indistinguishable from quiescent wild-type cells (Figure 3C).
Figure 3.
Hsp90 and Sba1 colocalize to the nucleus during quiescence via the Kap95 importin system. (A) BY4741 transformed with pSba1-mCherry was analyzed for subcellular localization at the indicated growth phases. (B) BY4741 harboring both pHsp82-GFP and pSba1-mCherry was examined during quiescence. Control strains transformed with only one chaperone fusion protein were used to confirm lack of signal bleedthrough to other channels. (C) BY4741 and sba1Δ cells transformed with plasmid pHsp82-GFP were examined in quiescence as in Figure 1B. (D) JJ816 cells bearing plasmid pHsp82-I578F as the sole Hsp90 (Hsp82I578-GFP) and pSba1-mCherry were grown to quiescence and analyzed for localization of both chaperone fusions. (E) Strain kap95-E126K transformed with either pHsp82-GFP or pSba1-mCherry was grown to quiescence at the semipermissive temperature of 30°C and analyzed as described in D.
Proteins larger than 60 kDa require several soluble factors to facilitate transport into and out of the nucleus through nuclear pore complexes. These factors, known as karyopherins, recognize nuclear localization signals (NLSs) within transport substrates that are required for the targeting of proteins into the nucleus (Mosammaparast and Pemberton, 2004). Nuclear egress requires a distinct nuclear export signal (NES), recognized by different karyopherins (Mosammaparast and Pemberton, 2004). To further dissect the molecular mechanisms contributing to Hsp90 nuclear accumulation during quiescence, we transformed plasmid pHsp82-GFP into known karyopherin gene deletions and examined localization of Hsp82-GFP during both vegetative growth and quiescence. Cells lacking Kap104, Kap108, Kap114, Kap119, Kap120, Kap122, Kap123, and Kap142 all exhibited Hsp82-GFP localization patterns identical to wild-type cells, indicating either a lack of involvement or redundant transport capabilities (data not shown). The yeast SRP1/KAP95 gene encodes the essential orthologue of the classical α/β importin system (Enenkel et al., 1995; Rexach and Blobel, 1995). We obtained a strain bearing a temperature-sensitive allele of KAP95 (kap95-E126K) to test the involvement of this system in Hsp90 nuclear accumulation (Ryan et al., 2007). kap95-E126K cells demonstrated impaired accumulation of both Hsp82-GFP (6% nuclear accumulation in quiescence vs. 88.4% for isogenic wild type) and Sba1-mCherry (10.2% nuclear accumulation in quiescence vs. 73.3%) under all growth conditions tested, suggesting that even at growth-permissible temperatures, nuclear transport is significantly affected in this mutant (30°C; Figure 3E). Thus, both Hsp90 and Sba1 accumulate within the nucleus via the SRP1/KAP95 importin system, and Hsp90 localization does not require Sba1.
Reduced Hsp90 Nuclear Accumulation Is Coincident with Growth and Developmental Defects
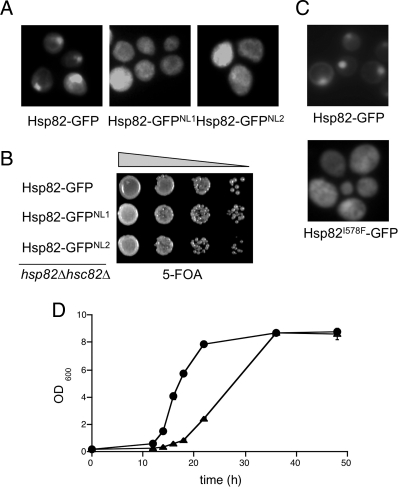
To address the phenotypic relevance of Hsp90 nuclear accumulation in response to glucose exhaustion and entry into quiescence, we sought to generate a nonlocalizing HSP82 allele by using error-prone PCR and a manual microscopic screen. From ∼1500 independent recombinants, two nonlocalizing (NL) mutants were isolated and subsequently determined to be capable of conferring viability when present as the sole source of Hsp90 in hsp82Δ hsc82Δ cells (Figure 4, A and B). Complete amino acid sequences for the two surviving mutants were acquired and analyzed. Both alleles possessed multiple amino acid substitutions distributed along the entire length of the protein with no common mutations between them. The allele with the least number of mutations (NL1) was chosen to parse the substitutions to determine whether a single point mutation was capable of conferring the nonaccumulating phenotype. Of the five amino acid changes found initially, substitution of isoleucine 578 to phenylalanine in the C-terminal domain of the protein was sufficient to block nuclear accumulation of Hsp90 (Figure 4C). This isoleucine residue is highly conserved within the fungal lineage as well as in human Hsp90, but no obvious nuclear localization signals could be distinguished in the neighboring region. Based on the fact that we were able to identify at least two nonlocalizing HSP82 alleles, quiescent phase nuclear localization is apparently not required for survival. However, more detailed analysis of the growth characteristics of the hsp82-I578F mutant revealed a slower overall growth rate and a pronounced lag in the transition to logarithmic phase growth from quiescence, possibly reflecting a defect in reentry into the cell cycle from G0 (Figure 4D). In addition, identification of this allele allowed us to ask whether Sba1 required association with Hsp90 for nuclear accumulation. As shown in Figure 3D, when Hsp90 accumulation is reduced from ∼95.4% in the wild type to 17.4% in the hsp82-I578F mutant, nuclear localization of Sba1-mCherry is reduced from 85.6 to 38.9%, but not eliminated. These data suggest that Sba1 nuclear accumulation in quiescence is a consequence of both association with translocating Hsp90 as well as an Hsp90-independent mechanism.
Figure 4.
Generation and analysis of nonlocalizing HSP82 alleles. (A) Identification of two novel HSP82-GFP mutants that fail to accumulate in the nucleus in quiescence. (B) Mutants were transformed into strain JJ816 and serial dilutions plated on medium containing 5-FOA to select for loss of the Yep24-HSP82 plasmid, resulting in the indicated pHsp82-GFP plasmid as the sole source of Hsp90. (C) The Hsp82-GFPNL1 mutant was sequenced and of the five amino acid mutations detected, I578F was sufficient to confer the nonaccumulating phenotype. (D) JJ816 cells harboring either wild-type HSP82-GFP (closed circles) or the hsp82-I578F mutant (closed triangles) were diluted from a quiescent phase culture into fresh YPD medium and growth rates followed for 50 h.
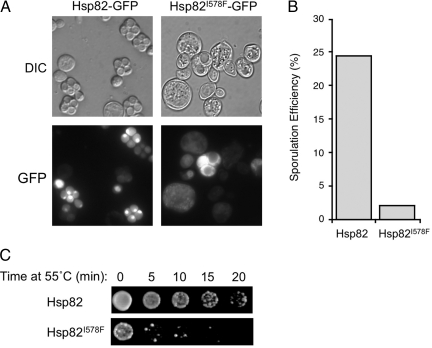
Simultaneous starvation of diploid S. cerevisiae cells for a fermentable carbon source and nitrogen leads to activation of the sporulation program and meiosis, resulting in production of four haploid spore progeny (Neiman, 2005). If localization of Hsp90 to the mitotic nucleus occurs during glucose starvation, then we reasoned that the chaperone should likewise localize to the nuclei within the developing spores. Diploid strains were generated expressing either wild type HSP82-GFP or the hsp82-I578F-GFP allele as the sole copy of Hsp90 and meiosis was induced through growth on sporulation medium. We indeed observed localization of Hsp90 to all four spore nuclei, (Figure 5A), consistent with the idea that nuclear localization of Hsp90 is not a general stress response, but a specialized response to nutrient starvation. A striking phenotype was observed upon examining spore formation in the hsp82-I578F mutant; very few mature four-spore asci were found. Instead, a panoply of sporulation defects ranging from aberrantly large cells apparently devoid of spores to cells containing compartments suggestive of immature (nonbirefringent) or disorganized spores were observed (Figure 5A). Quantitation of sporulation efficiency as scored by the acquisition of mature four-spore asci revealed a nearly complete failure to produce spores (Figure 5B). Importantly, this defect is likely not a result of failure to initiate or proceed through meiosis, as multiple disorganized nuclear bodies were detectable by Hoescht staining of sporulated diploids bearing hsp82-I578F-GFP (Supplemental Figure S2). Nuclei not encapsulated within spores in the hsp82-I578F-GFP mutant seemed to be highly unstable and could only be detected early in sporulation, precluding precise quantitation of meiotic progression.
Figure 5.
The hsp82-I578F mutant displays sporulation defects. (A) D818 diploid cells carrying either pHsp82-GFP or pHsp82-I578F-GFP as the sole copy of Hsp90 were sporulated and visualized using differential interference contrast (Nomarksi; differential interference contrast [DIC]) optics and epifluorescence. (B) Quantitation of sporulation efficiency of cells from A. (C) Sporulated cultures of D818 bearing pHsp82-GFP or pHsp82-I578F-GFP were subjected to 55°C heat shock and plated for viability at 30°C as described in Materials and Methods.
Spore formation in S. cerevisiae leads to the production of highly stress-resistant cells that can withstand severe environmental insults such as heat shock and desiccation. Cultures of sporulated HSP82-GFP and hsp82-I578F-GFP diploids were subject to a lethal 55°C heat shock and subsequently plated for viability at 30°C (Figure 5C). Although HSP82-GFP cells retained significant viability up to 20-min heat shock, cultures of hsp82-I578F-GFP exhibited drastically reduced survival after only 5 min of treatment, consistent with the marked reduction in spore production observed in this strain. These findings demonstrate that in addition to accumulation in the nucleus of cells transitioning into G0, diploid cells deficient in nuclear accumulation of Hsp90 exhibit sporulation defects.
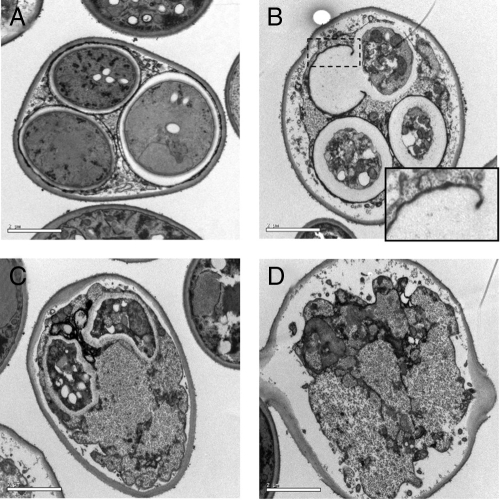
Similar to metazoan differentiation programs, spore formation is induced in response to a nutrient stimulus in specific cell types and is characterized by the ordered progression of morphogenic stages that lead to a differentiated state (Neiman, 2005). The spore wall is composed of two inner spore wall layers, which resemble a vegetative cell, and two outer layers containing spore-specific materials, chitosan and dityrosine (Neiman, 2005). To examine the structural and morphological defects associated with loss of Hsp90 nuclear accumulation at high resolution, sporulated HSP82-GFP and hsp82-I578F-GFP diploid cells were examined by transmission electron microscopy. The typical ultrastructure of a wild-type spore is exemplified by the ascus shown in Figure 6A. In contrast, hsp82-I578F-GFP asci exhibited multiple defects in spore wall construction. A possible defect in assembly of the outer dityrosine layer is exhibited in Figure 6B, where the spore wall seems to have ruptured, releasing its contents but retaining an incomplete dityrosine ghost structure. Figure 6, C and D, illustrates asci with partial to complete failure of spore assembly, resulting in catastrophic sporulation. Nuclear-localized Hsp90 may therefore be required for one or more critical steps in spore morphogenesis, including but not limited to spore wall construction.
Figure 6.
Spore wall construction defects in the hsp82-I578F mutant. (A) D818 cells carrying pHsp82-GFP as the sole copy of Hsp90 exhibit normal spore formation as shown by transmission electron microscopy. (B–D) D818 cells carrying pHsp82-I578F-GFP as the sole copy of Hsp90 exhibit diverse sporulation defects. Inset in B is a 2× digital zoom of dashed rectangle. Bar, 2 μm.
Hsp90 Activity and Localization Are Intertwined and Required for Sporulation
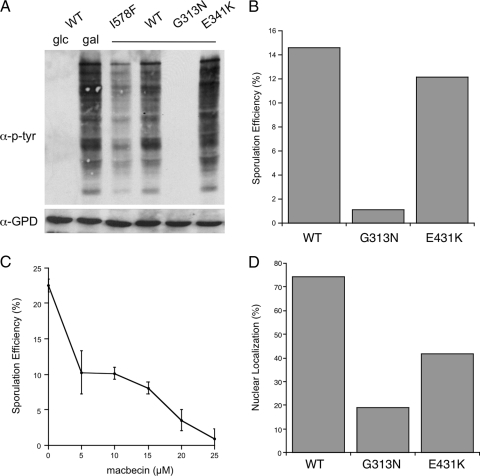
The preceding results raise the possibility that functional Hsp90 is required in the nucleus before initiation of sporulation to generate intact, viable spores. An alternative explanation is that the hsp82-I578F allele, in addition to blocking nuclear translocation, is hypomorphic for Hsp90 activity and that loss of chaperone function is sufficient to cause the defects we have observed. We tested these two scenarios by assaying the ability of hsp82-I578F to support activation and function of the mammalian Hsp90 client tyrosine kinase v-Src in yeast (Xu and Lindquist, 1993). When expressed from a galactose-inducible promoter, v-Src promiscuously phosphorylates several unknown endogenous yeast proteins, detectable by immunoblot with anti-phosphotyrosine antibodies (Xu and Lindquist, 1993). As shown in Figure 7A, the characteristic banding pattern was observed in cells bearing wild-type HSP82 and the v-Src plasmid grown in galactose. No signal was observed in glucose medium, confirming the specificity of the antibody. However hsp82-I578F cells exhibited substantial reduction in tyrosine phosphorylation, indicative of reduced Hsp90 chaperone activity. To place these results in context, we also examined two previously characterized HSP82 mutant alleles, hsp82-G313N and hsp82-E431K (Bohen and Yamamoto, 1993). When compared directly with a strain bearing the appropriate HSP82 wild-type allele (see Materials and Methods), hsp82-G313N displayed complete loss of v-Src activity, whereas hsp82-E431K seemed normal (Figure 7A). These results are consistent with a previous functional analysis using steroid receptor clients, demonstrating that the G313N substitution results in severe diminution of Hsp90 activity, whereas E431K behaves essentially as wild type with the exception of glucocorticoid signaling (Bohen and Yamamoto, 1993; Bohen, 1995). We next investigated the ability of the G313N and E431K mutants to support sporulation as the sole source of Hsp90 in diploid cells. Strikingly, whereas hsp82-E431K and the corresponding wild-type diploids exhibited similar sporulation, the hsp82-G313N cells displayed a dramatic sporulation defect in line with that observed with the nonlocalizing hsp82-I578F mutant (Figure 7B). These findings suggest that reduction in Hsp90 chaperone activity is sufficient to inhibit proper sporulation. To further test this hypothesis, we asked whether pharmacological ablation of endogenous Hsp90 in wild-type cells with the Hsp90-specific inhibitor macbecin would have the same effect (Whitesell et al., 1994). Indeed, as shown in Figure 7C, macbecin treatment resulted in dose-dependent inhibition of overall sporulation efficiency, completely blocking sporulation at established concentrations (∼20 μM) known to inhibit Hsp90 activities in yeast cells (Bohen, 1998).
Figure 7.
Reduction in Hsp90 activity is sufficient to cause defective sporulation and is coincident with loss of nuclear accumulation. (A) D818 cells bearing pHsp82-GFP or pHsp82-I578F-GFP, or plasmids pTCA-Hsp82, pTCA-Hsp82/G313N and pTCA-Hsp82/E431K and p416Gal-v-Src were induced by growth in the presence of galactose for 10 h (gal), or maintained in glucose (glc), and protein extracts were assayed by immunoblot for bulk tyrosine phosphorylation (α-p-tyr). Levels of glucose phosphate dehydrogenase were assessed as a gel loading control (α-GPD). (B) D818 cells bearing the plasmids pTCA-Hsp82, pTCA-Hsp82/G313N, and pTCA-Hsp82/E431K were sporulated and efficiency calculated as in Figure 5. (C) Wild-type diploid cells were treated with the indicated concentrations of macbecin during sporulation and efficiency calculated as in Figure 5. Error bars represent the SD from two replicates. (D) D818 cells bearing the pTCA-Hsp82-yEm-RFP, pTCA-Hsp82/G313N-yEm-RFP and pTCA-Hsp82/E431K-yEm-RFP plasmids (see Materials and Methods) were grown to quiescent phase and subcellular distribution assessed.
To investigate the localization status of the hsp82-G313N and hsp82-E431K proteins, we constructed Hsp90-fluorescent protein chimeras by using the recently described yeast-enhanced monomeric red fluorescent protein (yEm-RFP; Keppler-Ross et al., 2008). This approach was chosen to aid microscopic detection because the hsp82-G313N and hsp82-E431K mutants and their corresponding wild-type parent are expressed from low copy plasmids, and yEm-RFP is exceptionally bright. Wild type and both mutant HSP82 proteins displayed the expected cytoplasmic localization pattern during log phase growth. Strikingly, hsp82-G313N-yEmRFP, and to a much lesser extent hsp82-E431K-yEmRFP, failed to localize to the nucleus in comparison with the wild-type chimera, quantified in Figure 7D. These findings establish a compelling correlation between Hsp90 functional status and subcellular localization and suggest that these two characteristics may be functionally linked.
DISCUSSION
In this report, we uncover a new role for the Hsp90 chaperone in growth and development based on nutrient-responsive nuclear accumulation. The yeast p23 orthologue Sba1 was also found to accumulate in the nucleus during quiescence. These findings suggest that a minimal Hsp90 chaperone system is required for one or more nuclear processes during periods of relative metabolic inactivity. In support of this conjecture, both Hsp90 and p23 have recently been implicated in exclusively nuclear activities. Toogun et al. have shown a requirement for both Hsp90 and p23 to promote telomerase activity by increasing telomerase binding to DNA as well as nucleotide processivity (Toogun et al., 2007, 2008). In addition, loss of Sba1 results in defects in telomere length maintenance, a hallmark of cellular senescence (Toogun et al., 2007). In another report, a delay in transcriptional activation of the galactose transcriptional pathway was observed when components of both the Hsp90 and Hsp70 chaperone machineries were absent. On activation of the galactose pathway, inhibitory nucleosomes are rapidly removed from the GAL1 promoter under wild-type conditions, leading to gene expression, but this removal is significantly delayed in cells defective in Hsp90/Hsp70 chaperone function (Floer et al., 2008). In higher eukaryotes, the Hsp90/Hsp70 chaperone systems are also needed for proper targeting of client proteins such as p53 and steroid receptors into the nucleus, thus facilitating their DNA binding activities (Pratt and Toft, 2003). These findings highlight important roles for Hsp90 involving both nuclear-localized clients and clients that shuttle from the cytoplasm to nucleus as part of their functional cycles.
The need for specialized localization of protein chaperones has also been observed with other yeast chaperones. Stochaj and coworkers demonstrated that under normal growth conditions, the Hsp70 Ssa4 shuttles in and out of the nucleus, yet upon different stresses it accumulates in the nucleus and redistributes to the cytoplasm once the cells recover (Quan et al., 2006). Ssa4 nuclear retention in response to stress is due to relocation of the nuclear export factor Msn5, which transits to the cytoplasm upon stress (Quan et al., 2006). Nuclear accumulation is therefore not an intrinsic property of Ssa4 but rather an indirect effect of inhibition of the export machinery. Conversely, the yeast Hsp70 Ssb1 is a cytoplasmic, ribosome-associated Hsp70 molecular chaperone that is involved in the folding of newly made polypeptide chains. Its colocalization with ribosomes is central to its function, and a NES has been identified which restricts the chaperone to the cytoplasm, thus maximizing ribosome-binding opportunities (Shulga et al., 1999). We have determined that nuclear accumulation of Hsp90 is a response to gradual glucose starvation as cells progress through the diauxic shift into respiratory growth. Intriguingly, the unfoldase chaperone Hsp104 interacts with the Hsp90 cochaperones Cpr7, Sti1, and Cns1 exclusively in respiring yeast, demonstrating that metabolic conditions may influence chaperone partnering and likely function (Abbas-Terki et al., 2001). Further work will be required to define the nature of the signal and effectors required for nutrient regulation of Hsp90 localization.
Our examination reveals that during logarithmic growth, Hsp90 probably shuttles between the cytoplasm and the nucleus; yet, only upon quiescence does this chaperone accumulate in the nucleus. The yeast SRP1/KAP95 gene encodes the essential orthologue of the established α/β importin system required for the transport of nondiffusible proteins into the nucleus (Enenkel et al., 1995). Using a temperature-sensitive allele of KAP95, we identified this importin as being responsible for the nuclear trafficking of Hsp90. In contrast, Hsp90 nuclear accumulation was unaffected in a survey of nonessential karyopherin deletion strains (see Results). Previous work demonstrated that avian Hsp90 is a predominantly cytoplasmic protein that can be partially relocated to the nucleus with overexpression of a heterologous client receptor protein (Kang et al., 1994). However, given the extent of accumulation that we observe during quiescence it seems unlikely that Hsp90 is “piggybacking” with a sole client protein into the nucleus. In mammalian cells, specific cochaperones such as FKBP52 can alter the subcellular distribution of Hsp90, suggesting that distinct Hsp90 chaperone machines may form to regulate client protein trafficking (Harrell et al., 2004). No ubiquitous consensus sequence has been defined for nuclear localization beyond the inclusion of basic or hydrophobic stretches of amino acids. Examination of the amino acid sequences of both HSP82 and HSC82 does not reveal a close match to known NLS or NES sequences. The single amino acid change we uncovered that blocks nuclear accumulation (I578F) is obscured within the C-terminal dimerization domain and the surrounding region is not indicative of an NLS or NES. It is conceivable that this mutation might lead to a perturbation in Hsp90 tertiary or quaternary structure, preventing nuclear accumulation. This possibility is strongly supported by the finding that the G313N mutation, and to a lesser extent E431K, likewise result in cytoplasmic localization during quiescence, linking localization to Hsp90 functional status. However, inhibition of Hsp90 function per se is probably not sufficient to block nuclear transport as treatment of Hsp82-yEmRFP cells with macbecin did not result in cytoplasmic retention (data not shown). At this time, therefore, we cannot divorce Hsp90 function from subcellular localization or definitively conclude that is the causal deficiency. However, it is interesting to note the convergence of the function and localization phenotypes: in a search for mislocalizing HSP82 mutants, we uncovered a functionally impaired allele, and at least two mutants isolated on the basis of loss of function were found by us to mislocalize. Attempts to force cytoplasmic retention of wild-type Hsp82 with tandem nuclear export sequences failed due to extensive proteolysis of Hsp82-NES-GFP chimeras, suggesting that a formal test of the role of nuclear accumulation during quiescence will require further in-depth study (Edgington and Futcher, 2001).
The dramatic defects observed in spore formation and maturation in cells genetically or pharmacologically depleted of Hsp90 activity strongly implicate the chaperone in the process of yeast sporulation and suggest the existence of one or more previously unappreciated client proteins. On initiation of sporulation, three temporal categories of genes have been recognized: early, middle, and late. Early genes are generally required for meiotic chromosome dynamics, including recombination; middle genes include those involved in meiotic divisions as well as genes involved in spore biogenesis; and late genes are required for spore wall assembly (Neiman, 2005). Tightly regulated gene expression during the sporulation program is required to orchestrate these events. Importantly, we did not detect defects in meiotic nuclear division, suggesting that Hsp90 plays a critical role downstream in spore maturation. In contrast to a vegetatively growing yeast cell wall that is comprised of β-glucan and mannan, the spore wall contains four layers (inside to outside): mannan, β-glucan, chitosan, and dityrosine. It is the outer layers that confer much of the observed stress resistance characteristics to spores. Smk1 is a mitogen activated protein kinase required for spore wall assembly in S. cerevisiae (Krisak et al., 1994). Mutations in Smk1 result in improper spore wall formation in a manner very similar to what we observe in our I578F mutant (Krisak et al., 1994). Despite a report of an interaction between Hsp90 and Smk1 in a proteome-wide yeast two-hybrid analysis (Zhao et al., 2005), we did not observe an interaction in vivo via coimmunoprecipitation nor did we detect deficiency in Smk1 signaling through analysis of the Smk1-dependent, late meiotic gene reporter SPS100-lacZ (data not shown). Further work will be required to more precisely determine the identity of the Hsp90 client responsible for the severe spore assembly defects observed when the chaperone is prevented from localizing to both the premeiotic and spore nuclei.
Although this is the first report of developmentally regulated subcellular targeting of Hsp90 in yeast, there is precedence in higher eukaryotes. The Hsp90 homologue TRAP-1 exhibits differential localization throughout the developmental stages of the Dyctostelium amoeba (Yamaguchi et al., 2005). It is especially interesting to note that during development of prespore cells, localization of TRAP1 to the prespore-specific vacuole is essential for spore wall formation (Yamaguchi et al., 2005). Hsp90 and its cofactors also play an important role in development and tissue differentiation in Arabidopsis, as mutations in the chloroplast-specific Hsp90 homologue lead to delayed chloroplast development (Sangster and Queitsch, 2005). In the same manner, our results suggest a functional and perhaps spatial role for the sole Hsp90 in a developmental process in budding yeast.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Jill L. Johnson, Kathryn J. Ryan, Edward Winter, Avrom Caplan, and Neta Dean for providing materials and William Margolin for assistance with fluorescence microscopy. We thank Glenn Neckers for excellent technical assistance with electron microscopy. We thank Amy Trott for important initial contributions to this project. Members of the Morano laboratory provided helpful advice and discussion. This work was supported by National Institutes of Health grant GM-074696.
Abbreviation used:
- Hsp
heat-shock protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-05-0376) on November 4, 2009.
REFERENCES
- Abbas-Terki T., Donze O., Briand P. A., Picard D. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol. Cell. Biol. 2001;21:7569–7575. doi: 10.1128/MCB.21.22.7569-7575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohen S. P. Hsp90 mutants disrupt glucocorticoid receptor ligand binding and destabilize aporeceptor complexes. J. Biol. Chem. 1995;270:29433–29438. doi: 10.1074/jbc.270.49.29433. [DOI] [PubMed] [Google Scholar]
- Bohen S. P. Genetic and biochemical analysis of p23 and ansamycin antibiotics in the function of Hsp90-dependent signaling proteins. Mol. Cell. Biol. 1998;18:3330–3339. doi: 10.1128/mcb.18.6.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohen S. P., Yamamoto K. R. Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc. Natl. Acad. Sci. USA. 1993;90:11424–11428. doi: 10.1073/pnas.90.23.11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A. J., Tsai J., Casey P. J., Douglas M. G. Farnesylation of Ydj1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J. Biol. Chem. 1992;267:18890–18895. [PubMed] [Google Scholar]
- Edgington N. P., Futcher B. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J. Cell Sci. 2001;114:4599–4611. doi: 10.1242/jcs.114.24.4599. [DOI] [PubMed] [Google Scholar]
- Enenkel C., Blobel G., Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J. Biol. Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- Fang Y., Fliss A. E., Rao J., Caplan A. J. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol. Cell. Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felts S. J., Owen B. A., Nguyen P., Trepel J., Donner D. B., Toft D. O. The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J. Biol. Chem. 2000;275:3305–3312. doi: 10.1074/jbc.275.5.3305. [DOI] [PubMed] [Google Scholar]
- Floer M., Bryant G. O., Ptashne M. HSP90/70 chaperones are required for rapid nucleosome removal upon induction of the GAL genes of yeast. Proc. Natl. Acad. Sci. USA. 2008;105:2975–2980. doi: 10.1073/pnas.0800053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom G., Weekes J., Johnson J. L. Novel interaction of the Hsp90 chaperone machine with Ssl2, an essential DNA helicase in Saccharomyces cerevisiae. Curr. Genet. 2005;47:368–380. doi: 10.1007/s00294-005-0580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. V., Petsko G. A., Johnston G. C., Ringe D., Singer R. A., Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell J. M., Murphy P. J., Morishima Y., Chen H., Mansfield J. F., Galigniana M. D., Pratt W. B. Evidence for glucocorticoid receptor transport on microtubules by dynein. J. Biol. Chem. 2004;279:54647–54654. doi: 10.1074/jbc.M406863200. [DOI] [PubMed] [Google Scholar]
- Herman P. K. Stationary phase in yeast. Curr. Opin. Microbiol. 2002;5:602–607. doi: 10.1016/s1369-5274(02)00377-6. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Kang K. I., Devin J., Cadepond F., Jibard N., Guiochon-Mantel A., Baulieu E. E., Catelli M. G. In vivo functional protein-protein interaction: nuclear targeted hsp90 shifts cytoplasmic steroid receptor mutants into the nucleus. Proc. Natl. Acad. Sci. USA. 1994;91:340–344. doi: 10.1073/pnas.91.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler-Ross S., Noffz C., Dean N. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics. 2008;179:705–710. doi: 10.1534/genetics.108.087080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisak L., Strich R., Winters R. S., Hall J. P., Mallory M. J., Kreitzer D., Tuan R. S., Winter E. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 1994;8:2151–2161. doi: 10.1101/gad.8.18.2151. [DOI] [PubMed] [Google Scholar]
- Lillie S. H., Pringle J. R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol. 1980;143:1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. D., Morano K. A., Thiele D. J. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 1999;274:26654–26660. doi: 10.1074/jbc.274.38.26654. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N., Pemberton L. F. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Neiman A. M. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi C. E., Rabinovich E., Dancis A., Bonifacino J. S., Klausner R. D. Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctr1p in the apparent absence of endocytosis. EMBO J. 1996;15:3515–3523. [PMC free article] [PubMed] [Google Scholar]
- Pearl L. H., Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesset J., Ludwig J. R., Cox B. S., McLaughlin C. S. Effect of cell cycle position on thermotolerance in Saccharomyces cerevisiae. J. Bacteriol. 1987;169:779–784. doi: 10.1128/jb.169.2.779-784.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt W. B., Silverstein A. M., Galigniana M. D. A model for the cytoplasmic trafficking of signalling proteins involving the hsp90-binding immunophilins and p50cdc37. Cell Signal. 1999;11:839–851. doi: 10.1016/s0898-6568(99)00064-9. [DOI] [PubMed] [Google Scholar]
- Pratt W. B., Toft D. O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Quan X., Tsoulos P., Kuritzky A., Zhang R., Stochaj U. The carrier Msn5p/Kap142p promotes nuclear export of the Hsp70 Ssa4p and relocates in response to stress. Mol. Microbiol. 2006;62:592–609. doi: 10.1111/j.1365-2958.2006.05395.x. [DOI] [PubMed] [Google Scholar]
- Rexach M., Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Richter K., Reinstein J., Buchner J. A Grp on the Hsp90 mechanism. Mol. Cell. 2007;28:177–179. doi: 10.1016/j.molcel.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Richter K., Walter S., Buchner J. The Co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle. J. Mol. Biol. 2004;342:1403–1413. doi: 10.1016/j.jmb.2004.07.064. [DOI] [PubMed] [Google Scholar]
- Ryan K. J., Zhou Y., Wente S. R. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol. Biol. Cell. 2007;18:886–898. doi: 10.1091/mbc.E06-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster T. A., Queitsch C. The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr. Opin. Plant Biol. 2005;8:86–92. doi: 10.1016/j.pbi.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Shulga N., James P., Craig E. A., Goldfarb D. S. A nuclear export signal prevents Saccharomyces cerevisiae Hsp70 Ssb1p from stimulating nuclear localization signal-directed nuclear transport. J. Biol. Chem. 1999;274:16501–16507. doi: 10.1074/jbc.274.23.16501. [DOI] [PubMed] [Google Scholar]
- Smits G. J., van den Ende H., Klis F. M. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology. 2001;147:781–794. doi: 10.1099/00221287-147-4-781. [DOI] [PubMed] [Google Scholar]
- Toogun O. A., Dezwaan D. C., Freeman B. C. The hsp90 molecular chaperone modulates multiple telomerase activities. Mol. Cell. Biol. 2008;28:457–467. doi: 10.1128/MCB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toogun O. A., Zeiger W., Freeman B. C. The p23 molecular chaperone promotes functional telomerase complexes through DNA dissociation. Proc. Natl. Acad. Sci. USA. 2007;104:5765–5770. doi: 10.1073/pnas.0701442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L., Mimnaugh E. G., De Costa B., Myers C. E., Neckers L. M. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. Transmission electron microscopy of yeast. Microsc. Res. Tech. 2000;51:496–510. doi: 10.1002/1097-0029(20001215)51:6<496::AID-JEMT2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Xu Y., Lindquist S. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc. Natl. Acad. Sci. USA. 1993;90:7074–7078. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Morita T., Amagai A., Maeda Y. Changes in spatial and temporal localization of Dictyostelium homologues of TRAP1 and GRP94 revealed by immunoelectron microscopy. Exp. Cell Res. 2005;303:415–424. doi: 10.1016/j.yexcr.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Zhao R., et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.