Abstract
Purpose: The purpose of this study was to develop an advanced computer-aided detection (CAD) scheme utilizing massive-training artificial neural networks (MTANNs) to allow detection of “difficult” polyps in CT colonography (CTC) and to evaluate its performance on false-negative (FN) CTC cases that radiologists “missed” in a multicenter clinical trial.
Methods: The authors developed an advanced CAD scheme consisting of an initial polyp-detection scheme for identification of polyp candidates and a mixture of expert MTANNs for substantial reduction in false positives (FPs) while maintaining sensitivity. The initial polyp-detection scheme consisted of (1) colon segmentation based on anatomy-based extraction and colon-based analysis and (2) detection of polyp candidates based on a morphologic analysis on the segmented colon. The mixture of expert MTANNs consisted of (1) supervised enhancement of polyps and suppression of various types of nonpolyps, (2) a scoring scheme for converting output voxels into a score for each polyp candidate, and (3) combining scores from multiple MTANNs by the use of a mixing artificial neural network. For testing the advanced CAD scheme, they created a database containing 24 FN cases with 23 polyps (range of 6–15 mm; average of 8 mm) and a mass (35 mm), which were “missed” by radiologists in CTC in the original trial in which 15 institutions participated.
Results: The initial polyp-detection scheme detected 63% (15∕24) of the missed polyps with 21.0 (505∕24) FPs per patient. The MTANNs removed 76% of the FPs with loss of one true positive; thus, the performance of the advanced CAD scheme was improved to a sensitivity of 58% (14∕24) with 8.6 (207∕24) FPs per patient, whereas a conventional CAD scheme yielded a sensitivity of 25% at the same FP rate (the difference was statistically significant).
Conclusions: With the advanced MTANN CAD scheme, 58% of the polyps missed by radiologists in the original trial were detected and with a reasonable number of FPs. The results suggest that the use of an advanced MTANN CAD scheme may potentially enhance the detection of “difficult” polyps.
Keywords: virtual colonoscopy, computer-aided diagnosis, missed lesions, colorectal cancer screening, false negatives
INTRODUCTION
Colorectal cancer is the second leading cause of cancer-related death in the United States. Evidence suggests that early detection and removal of polyps can reduce the incidence of colorectal cancer.1 Further, when colorectal cancer is detected at an early, localized stage, the 5-yr relative survival rate is 90%.2 CT colonography (CTC), also known as “virtual colonoscopy,” has gained considerable attention as an effective technique for detecting colorectal polyps and neoplasms by the use of a CT scan of the colon.3 CTC provides an option for a colorectal cancer examination that is less uncomfortable,4, 5 less invasive, and less costly6 than for colonoscopy. A variety of data now support CTC as a sensitive and specific method for detection of polyps.7, 8, 9, 10, 11 Accordingly, several national societies including the American Cancer Society have endorsed CTC as an option for colorectal cancer screening of average risk, asymptomatic patients.12
Importantly, skilled interpretation of CTC requires specific training and is associated with a “learning curve,” and variability in training may contribute to variability in reader expertise. The variability in the reported sensitivity as well as the training level in several large multicenter clinical trials is consistent with this concept,7, 8, 9, 13, 14 and the propensity for perceptual errors15 remains problematic. Computer-aided detection (CAD) may substantially enhance polyp detection16, 17 not only by improving radiologists’ diagnostic performance but also by reducing reader variability.
Researchers have developed CAD schemes for polyp detection in CTC. Summers et al.18 developed a CAD scheme based on the curvature of the surface of the colonic wall. Jerebko et al.19 incorporated a standard artificial neural network (ANN) to classify polyp candidates in the CAD scheme and improved the performance by utilizing a committee of ANNs (Ref. 20) and a committee of support vector machines.21 Their scheme yielded a sensitivity of 90% (35∕39) for polyps (⩾3 mm) with 31.4 FPs per patient.19 Recently, a CAD scheme from Summers et al. was tested and compared to a commercial CAD product (polyp enhanced viewing software, Siemens Medical Solutions, Forchheim, Germany).22 Their CAD scheme yielded a sensitivity of 83% (30∕36) for polyps (⩾6 mm) with 5.2 FPs per patient, whereas the commercial product yielded a sensitivity of 56% (20∕36) with 1.2 FPs per patient for the same database. Li et al.23 improved the performance of their CAD scheme for medium-size polyps (6–9 mm) by incorporating the wavelet transform, and they reported a sensitivity of 71% (32∕45) for medium-size polyps with 5.4 FPs per patient. Another commercial CAD product (ColonCAD, Phillips Medical Systems, Best, the Netherlands) was tested and yielded a sensitivity of 68% (60∕88) for polyps (⩾6 mm), but the number of FPs was not reported.24 Kiss et al.25 reported on a CAD scheme based on convexity and sphericity and used a standard ANN for the reduction in FPs. Their scheme yielded a sensitivity of 80% (12∕15) for polyps (⩾5 mm) with 8.2 FPs per patient. We developed a CAD scheme based on the shape index,26 feature analysis,27 and a mixture of expert three-dimensional (3D) MTANNs and achieved a sensitivity of 96% (27∕28) for polyps (⩾5 mm) with 1.1 FPs per patient.28 Thus, the reported performance of the existing CAD schemes ranges between by-polyp sensitivities of 56% and 96% with FP rates between 1.1 and 31.4 per patient.
Although current CAD schemes for the detection of polyps in CTC appear to be useful, some limitations remain. One of the major limitations with current CAD is a lack of evaluation in the setting of “difficult” polyps, particularly those which readers fail to detect by using standard techniques. It is notable that previously reported studies18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 of the utility of CAD routinely studied datasets including polyps that had been detected by readers. CAD benefits cannot be fully evaluated based on such true-positive (TP) polyps because these polyps are likely to be detected without CAD. Another major limitation of CAD is that it is associated with a relatively large number of false positives (FPs), which could adversely affect the clinical application of CAD to colorectal cancer screening. A large number of FPs are likely to confound the radiologist’s task of image interpretation and thus lower the efficiency. Thus, it is important to reduce the number of FPs as much as possible while maintaining a high sensitivity. Therefore, a major challenge in CAD development is the detection of difficult polyps which radiologists are likely to miss, with a reasonable number of FPs.
Our purpose was to develop a CAD scheme utilizing 3D massive-training artificial neural networks (MTANNs)28, 29 to allow detection of difficult polyps in CTC and to evaluate its performance on false-negative (FN) cases that reporting radiologists actually “missed” during their initial reading in a large multicenter clinical trial.
MATERIALS AND METHODS
The Institutional Review Board (IRB) approved this retrospective study. Informed consent for use of cases in this study was waived by the IRB because patient data was deidentified. This study complied with the Health Insurance Portability and Accountability Act, met all standards for good clinical research according to the NIH’s and local IRB’s guidelines.
CTC databases
Our testing database consisted of CTC scans obtained from a previous multicenter clinical trial14 that included air-contrast barium enema, same-day CTC and colonoscopy, and segmental unblinding for each subject, followed by robust reconciliation of all lesions utilizing the data from all three imaging examinations (thereby assuring accuracy of the reported consensus colon findings). 614 high-risk subjects participating in the original trial were scanned in both supine and prone positions with a multidetector-row CT system with collimations of 1.0–2.5 mm and reconstruction intervals of 1.0–2.5 mm. Each CT slice had a spatial resolution of 0.5–0.7 mm∕pixel. The reference standard was a final reconciliation of the unblinded lesions identified on all of the three examinations.
In the original trial, 155 patients had 234 clinically significant polyps (6 mm or larger in size). Among them, 69 patients had FN interpretations [i.e., the by-patient sensitivity was 55% (86∕155)]. These patients had 114 “missed” polyps∕masses, which were not detected by reporting radiologists during their initial clinical reading. Causes of errors included observer errors [51% (35∕69); observer perceptual or observer measurement], technical errors [23% (16∕69); artifact, distension, fluid, excessive stool, etc.], and nonreconcilable [26% (18∕69); polyps that were not found at retrospective analysis].15 The perceptual errors15 are associated with polyps that failed to be detected by the observers. The measurement errors15 refer to the errors associated with undermeasurement of polyp size as compared to colonoscopy findings as the “reference standard.” Such polyps were counted as FNs in the study.15 In this study, we focused on FN cases with observer errors because the aim of computer-aided diagnosis is to prevent observer errors. Technical errors including miselectronic cleansing should be minimized by removal of the source of each technical error.
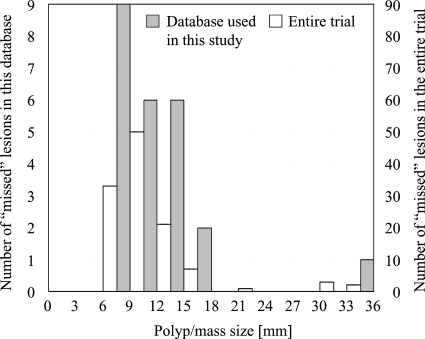
For the evaluation of our CAD scheme, we created a database with the inclusion criterion that each case had at least one polyp that was visible on both supine and prone views. As a result, we obtained the 14 FN cases with 13 polyps and a mass due to the observer errors in our database. To test our CAD scheme more critically, we added ten FN cases containing ten polyps that were visible only on either view to the visible-on-both subdatabase (i.e., the 14 FN cases) and obtained a testing database containing a total of 24 FN cases with 23 polyps and a mass (we did not include cases with DICOM header corruption or cases with missing CT data in either position; note that they were all available FN cases). A radiologist experienced in CTC (>1000 cases read) reviewed CTC cases carefully and determined the locations of polyps with reference to colonoscopy reports. Polyp sizes ranged from 6 to 15 mm, with an average of 8.3 mm. The mass size was 35 mm. The size distributions of FN polyps∕masses in the entire trial and in the total database used in this study are shown in Fig. 1. 14, 7, 2, and 1 lesion were adenoma, hyperplastic, normal (one was a hamartoma; a detailed result was not available for the other lesion), and unknown (pathology result was not available), respectively. An experienced radiologist determined the difficulty of detection for each polyp∕mass as difficult, moderate, and easy (the definitions are described in Sec. 2C). The radiologist also determined the morphology of each polyp.
Figure 1.
Distributions of sizes of FN polyps in the entire trial and in the database used in this study. Note the two different scales on the left and right vertical axes.
Our training database consisted of CTC scans obtained from 14 patients, acquired at the University of Chicago Medical Center, which were completely different from the testing database. We used the training database for training the entire CAD scheme except for a mixing ANN which was tested with a leave-one-lesion-out cross-validation test. Because of the nature of a leave-one-lesion-out cross-validation test, we used the testing database for training and testing the mixing ANN. The 14 patients had 26 polyps, 12 of which were 5–9 mm and 14 were 10–25 mm in size. All polyps were detected by radiologists in CTC, i.e., true-positive CTC cases. Each CT slice had a spatial resolution of 0.5–0.7 mm.
CAD scheme utilizing 3D massive-training ANN (3D MTANN)
Our CAD scheme was comprised of an initial polyp-detection scheme consisting of (1) colon segmentation based on anatomy-based extraction and colon-based analysis30 and (2) detection of polyp candidates based on morphologic analysis on the segmented colon,31 and a “mixture of expert” 3D MTANNs for FP reduction,28 as shown in Fig. 2.
Figure 2.
Flowchart of our CAD scheme utilizing 3D MTANNs for the detection of polyps∕masses in CTC.
Initial polyp-detection scheme
Technical details of the initial polyp-detection scheme have been described in Refs. 30, 31. To summarize, the segmentation process consisted of two major steps: (1) Anatomy-based extraction and (2) colon-based analysis. The anatomy-based segmentation consisted of the following steps. The volume outside the body was segmented based on CT values thresholding, followed by a 3D connectivity test; and the resulting volume is called an “air mask.” Bone structures that correspond to the spine, pelvis, and parts of the ribs in the original volume were segmented in the same manner. The resulting volume is called a “bone mask.” The 3D gradient of the CT value was calculated at each voxel that does not belong to the volume defined by the union of the two masks (i.e., air and bone masks); those voxels that have gradient and CT values greater than predefined threshold values were retained. Finally, the connected component that has the largest number of voxels was identified as the extracted colon.
After the colon was segmented, polyp candidates in the colonic wall were identified by extracting geometric features that characterize polyps at each point on the wall. Polyps adhering to the colonic wall tend to appear as relatively small, bulbous, caplike structures, and the colonic wall itself appears as a large, nearly flat cuplike structure. To characterize these shape and scale differences among polyps, folds, and colonic wall, two 3D geometric features called the volumetric shape index (SI) and volumetric curvedness32, 33 were used. The volumetric SI, SI(p), and the volumetric curvedness, CV(p), at a voxel p are defined26 as
| (1) |
| (2) |
where κ1≠κ2 are the principal curvatures defined as the eigenvalues of the Weingarten endomorphism matrix.34 The SI characterizes the topologic shape of the volume in the vicinity of a voxel. This index determines to which of the following five topologic shapes a voxel belongs: cup, rut, saddle, ridge, or cap. Voxels that belong to the cup shape have values around 0; rut, around 0.25; saddle, around 0.5; ridge, around 0.75; and cap, around 1.0; although the transition from one topologic shape to another occurs continuously. The curvedness CV of a voxel represents the magnitude of the effective curvature at the voxel, which is defined as the square root of the sum of the squared minimum and maximum curvatures at the voxel.33 To identify polyp candidates, thresholding on SI and CV was performed.
3D MTANNs for reduction in FPs
To remove various types of FPs produced by the initial polyp-detection scheme while maintaining a high sensitivity, we developed a mixture of expert 3D MTANNs.28 A schematic illustration of the principles of a 3D MTANN is shown in Fig. 3. To process 3D CTC volume data, a 3D MTANN (Ref. 29) was developed by extending the structure of a two-dimensional (2D) MTANN.35, 36, 37 The 3D MTANN consists of a linear-output ANN model for regression,38 which is capable of operating on voxel data directly. The input to the 3D MTANN is the voxel values I(x,y,z) in a subvolume VS extracted from an input volume. The output of the 3D MTANN is a continuous value, represented by
| (3) |
where NN{⋅} is the output of the linear-output ANN. The 3D MTANN is trained with input CTC volumes and the corresponding “teaching” volumes for enhancement of polyps and suppression of nonpolyps.
Figure 3.
Schematic illustration of the principles of a 3D MTANN for distinguishing polyps∕masses from FPs. The 3D MTANN was trained to enhance lesions and suppress nonlesions. Lesions such as a sessile polyp, a sessile polyp on a fold, and a mass are enhanced in the output images, whereas nonpolyps such as a rectal tube, stool, and the ileocecal value (ICV) are suppressed. By the use of a scoring scheme, each of the output images is converted to a single score, indicating the likelihood of being a lesion for each lesion candidate. Classification between lesions and nonlesions is made by thresholding of the likelihood scores.
The teaching volume contains a 3D Gaussian distribution with standard deviation σT for enhancement of polyps and suppression of nonpolyps in CTC volumes. This distribution represents the “likelihood of being a polyp” for a polyp and zero for a nonpolyp,
| (4) |
The 3D MTANN involves training with a large number of subvolume-voxel pairs. For enriching the training samples, a training volume, VT, extracted from the input CTC volume is divided voxel by voxel into a large number of overlapping subvolumes. Single voxels are extracted from the corresponding teaching volume as teaching values. The expert 3D MTANN is massively trained by the use of each of a massive number of the input subvolumes together with each of the corresponding teaching single voxels; hence, the term “massive-training ANN.” The error to be minimized by training of the nth expert 3D MTANN is represented by
| (5) |
where c is a training case number, On,c is the output of the nth expert MTANN for the cth case, Tn,c is the teaching value for the nth expert MTANN for the cth case, and Pn is the number of total training voxels in the training volume for the nth expert 3D MTANN, VTn. The expert 3D MTANN is trained by a linear-output backpropagation algorithm.39 After training, the expert 3D MTANN is expected to output higher values for a polyp and lower values for a nonpolyp. Thus, by training with input lesion∕nonlesion volumes together with teaching volumes that contain the “likelihood of being a lesion,” the 3D MTANN was trained to enhance polyps and suppress various types of nonpolyps including rectal tubes, stool with bubbles, colonic walls, folds, and solid stool in the training database.
For combining output voxels from the trained expert 3D MTANNs, we developed a 3D scoring method. A score for a given polyp candidate from the nth expert 3D MTANN is defined as
| (6) |
where
| (7) |
is a 3D Gaussian weighting function with standard deviation σn, VE is the volume for evaluation, and On(x,y,z) is the output volume of the nth trained expert 3D MTANN. The use of the 3D Gaussian weighting function allows us to combine the responses (outputs) of a trained expert 3D MTANN as a 3D distribution. This score represents the weighted sum of the estimates for the likelihood that the volume (polyp candidate) contains a polyp near the center, i.e., a higher score would indicate a polyp, and a lower score would indicate a nonpolyp.
The scores from the expert 3D MTANNs are combined by the use of a mixing ANN such that different types of nonpolyps can be distinguished from polyps. The mixing ANN consists of a linear-output multilayer ANN model with a linear-output backpropagation training algorithm38 for processing of continuous output∕teaching values. One unit is employed in the output layer for distinction between a polyp and a nonpolyp. The scores of each expert 3D MTANN are used for each input unit in the mixing ANN. The output of the mixing ANN for the cth polyp candidate is represented by
| (8) |
where NN(⋅) is the output of the linear-output ANN model and N is the number of input units. After training, the mixing ANN is expected to output a higher value for a polyp and a lower value for a nonpolyp. Thus, the output can be considered to be a likelihood of being a polyp. Each of the output images was converted to a single score, indicating the likelihood of being a lesion for each lesion candidate by the use of a scoring scheme consisting of a mixing ANN. The mixing ANN was trained and tested with the testing database of 24 FN cases by the use of a leave-one-lesion-out cross-validation test. Classification between polyps and nonpolyps was made by thresholding of the likelihood scores. The balance between a TP rate and an FP rate was determined by the selected threshold value.
The overall performance was evaluated by free-response receiver operating characteristic (FROC) analysis.40 We compared the performance of our CAD scheme utilizing the 3D MTANNs to that of a standard CAD scheme31 consisting of the same initial polyp-detection scheme, calculation of 3D pattern features of the polyp candidates, and linear discriminant analysis (LDA) for classification of the polyp candidates as polyps or nonpolyps based on the pattern features (this conventional CAD is referred to as LDA CAD). For fair comparisons with the mixing ANN, the LDA was trained and tested using same testing method (i.e., a leave-one-lesion-out cross-validation test) with the same testing database (i.e., 24 FN cases) as used for testing of the mixing ANN.
Analysis of TPs, FNs, and FPs
There is no established objective metric, to our knowledge, for rating the difficulty of CTC cases. Therefore, for analyzing the computer outputs on CTC cases, a subjective decision was made by an expert unblinded to the fact that the cases were FNs in the trial.41 To analyze TPs and FNs by our MTANN CAD scheme, the unblinded radiologist subjectively graded polyps as easy, moderate, and difficult to detect by using both 2D analysis and 3D problem solving on a Vital Images workstation with VITREA 2 software (version 3.7, Vital Images, Minneapolis, MN). To analyze FPs generated by our MTANN CAD scheme, the radiologist reviewed all FPs and identified the sources of error for the FPs. The unblinded radiologist subjectively graded FPs in terms of ease of identification of the FP output as not being a polyp∕mass, into easy, moderate, and difficult by using both 2D and 3D views. An easy case is defined as “it is obviously not a polyp∕mass” when viewed on 2D and∕or 3D views. A moderate case would require interactive window∕level adjustment and paging several times. A difficult case, i.e., “pitfalls” with CAD for radiologists, would require supine∕prone comparison.
RESULTS
CAD performance
Our initial polyp-detection scheme yielded a (by-polyp=by-patient) sensitivity of 71.4% (10∕14) with 18.9 (264∕14) FPs per patient with the polyp-visible-on-both-views subdatabase including 14 cases. We applied the trained mixture of expert 3D MTANNs for reduction in the FPs. The polyps∕mass were enhanced by the 3D MTANNs in the output images, whereas nonlesions were suppressed, as illustrated in Fig. 3. The mixture of expert 3D MTANNs was able to remove 35% (93∕264) or 76% (201∕264) of the FPs with loss of 0 or 1 TP, respectively, as shown in Fig. 4. To test the generalization ability of our CAD scheme more strictly, we applied our scheme to the total database which included 10 FN polyps that were visible only on one view, in addition to the 14 cases. Our initial polyp-detection scheme yielded a sensitivity of 63% (15∕24) with 21.0 (505∕24) FPs per patient, as shown in Fig. 4. The MTANNs removed FPs substantially, and our CAD scheme achieved a sensitivity of 58% (14∕24) with 8.6 (207∕24) FPs per patient for the 24 missed lesion cases, whereas the conventional LDA CAD scheme achieved a sensitivity of 25% (6∕24) at the same FP rate. There were statistically significant differences between the sensitivity of the MTANN CAD scheme and that of the conventional LDA CAD scheme for both databases, as the 95% confidence intervals in Fig. 4 indicate, where the 95% confidence intervals were calculated under an initial-detection-and-candidate-analysis FROC model.42 The difference between the sensitivities of the MTANN CAD scheme for the polyp-visible-on-both-views cases and polyp-visible-only-on-one-view cases was not statistically significant.42 Therefore, our MTANN CAD scheme has the potential to detect 58% of missed polyp∕mass cases with a reasonable number of FPs.
Figure 4.
FROC curves for the performance of our CAD scheme utilizing 3D MTANNs and that of the conventional LDA CAD scheme for the 14-case polyp-visible-on-both-views subdatabase and the whole 24-case database. Our scheme achieved 58% sensitivity with 8.6 FPs∕patient for 24 polyps∕mass missed by reporting radiologists in the original clinical trial. The error bars indicate 95% confidence intervals.
Analysis of FN and FP sources
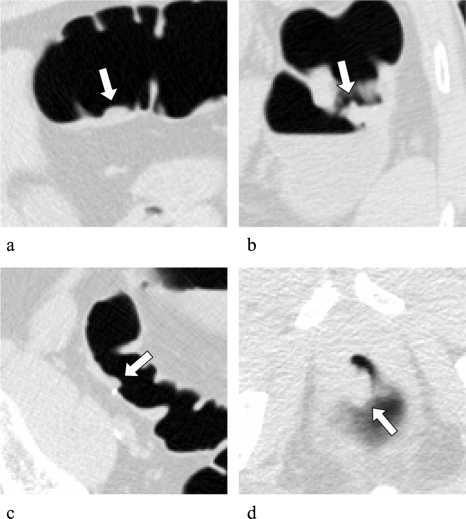
Among the 24 polyps∕mass, 17 polyps, 6 polyps, and 1 mass were classified by a radiologist into difficult, moderate, and easy, respectively. Among the 23 polyps, 12, 9, and 2 were categorized as sessile, sessile on a fold, and pedunculated, respectively. Figure 5 illustrates FN polyps detected by our MTANN CAD scheme. All three examples were graded as difficult to detect. We would expect our CAD scheme to be helpful in the detection of difficult polyps. Table 1 summarizes characteristics of polyps detected or missed by our CAD scheme. Our CAD scheme tended to miss small, sessile polyps visible only on one view, rated as difficult. There was a statistically significant difference between difficult cases and others in the detectability characteristic (chi-square test, P<0.05); there was no statistically significant difference in the other characteristics. Figure 6 illustrates FN polyps that were not detected by our CAD scheme. Polyp (a) is sessile on a fold, rated as difficult to detect, and polyp (b) is very small and sessile on a fold, rated as difficult to detect.
Figure 5.
Illustrations of polyps missed by reporting radiologists during initial reading in the original trial in 2D views (upper images) and 3D endoluminal views (lower images), which were detected by our MTANN CAD scheme. (a) A small polyp (6 mm; hyperplastic) in the sigmoid colon was detected correctly by our CAD scheme (indicated by an arrow). This polyp was missed in both CTC and reference-standard optical colonoscopy in the original trial. (b) A small polyp (6 mm; adenoma) in the sigmoid colon. (c) A sessile polyp on a fold (10 mm; adenoma) in the ascending colon.
Table 1.
Summary of the characteristics of polyps∕mass detected or missed by our MTANN CAD scheme.
| CAD TPs (n=14) | CAD FNs (n=10) | ||
|---|---|---|---|
| Lesion size | 6 mm–9 mm | 7 (29%) | 8 (33%) |
| ⩾10 mm | 7 (29%) | 2 (8%) | |
| Detectability rating | Difficult | 7 (29%) | 10 (42%) |
| Moderate | 6 (25%) | 0 (0%) | |
| Easy | 1 (4%) | 0 (0%) | |
| Visible on spine∕prone | Both views | 9 (38%) | 5 (21%) |
| Either view | 5 (21%) | 5 (21%) | |
| Morphology | Sessile | 7 (29%) | 5 (21%) |
| Sessile on fold | 6 (25%) | 3 (13%) | |
| Pedunculated | 0 (0%) | 2 (8%) | |
| Mass | 1 (4%) | 0 (0%) | |
| Pathology | Adenoma | 8 (33%) | 6 (25%) |
| Hyperplastic | 4 (17%) | 3 (13%) | |
| Normal | 2 (8%) | 0 (0%) | |
| Unknown | 0 (0%) | 1 (4%) |
Figure 6.
Illustrations of polyps missed by reporting radiologists during initial reading in the original trial in 2D views (upper images) and 3D endoluminal views (lower images), which were not detected by our CAD scheme. (a) A sessile polyp on a fold (12 mm; adenoma) in the descending colon. (b) A small sessile polyp on a fold (6 mm; hyperplastic) in the cecum.
We reviewed 63 FPs at a specific operating point (i.e., a sensitivity of 64% with 4.5 FPs per patient) for the 14-cases subdatabase and identified the sources of error, as summarized in Table 2. Twenty-five FPs were related to flexural pseudotumors or folds comprised of converging folds, haustral folds, and tenea coli. Twenty FPs were considered to be related to stool artifact. Four FPs were located in the small bowel and were therefore attributed to segmentation error and were not analyzed further. Collapsed colon segments and rectal tubes accounted for three FPs each. Two ileocecal valves (ICVs) were incorrectly marked by the CAD scheme as polyps. The remaining six FPs were grouped in the miscellaneous category, which included respiratory motion, extrinsic compression, streak artifact, and compression by or interface with the rectal catheter retention balloon.
Table 2.
FP sources of our MTANN CAD scheme. Folds, flexural pseudotumors, and stool are major sources of FPs.
| FP source | No. of FPs |
|---|---|
| Folds or flexural pseudotumors | 25 (40%) |
| Stool | 20 (32%) |
| Small bowel | 4 (6%) |
| Collapsed colon | 3 (5%) |
| Rectal tubes | 3 (5%) |
| Ileocecal valves | 2 (3%) |
| Miscellaneous sources | 6 (10%) |
We determined the subjective grading of ease of identification of the FP output as not being a polyp. Easy, moderate, and difficult cases accounted for 69%, 18%, and 13% of all FPs, respectively. Figure 7 illustrates examples of moderate and difficult cases. Easy, moderate, and difficult cases included tenea coli, respiratory motion, and a haustral fold; a collapsed colon segment, blunted folds, and stool; and stool and a hemorrhoid, respectively.
Figure 7.
Illustrations of FPs by our CAD scheme, which were categorized by subjective grading of ease. Moderate cases: (a) Stool and (b) collapsed colon segment and a fold. Difficult cases: (c) Stool and (d) a hemorrhoid.
DISCUSSION
The overarching clinical utility of a CAD scheme is dependent on both its sensitivity and specificity (i.e., its FP rate). A higher CAD sensitivity is desirable; however, it is associated with a high FP rate. Reduction in FPs is a major challenge for current CAD schemes. Some CAD studies23, 31, 18, 43, 44 have used polyps detected by radiologists in CTC, i.e., “human TP polyps.” One drawback to such an approach is that the benefit of the increased potential sensitivity of CAD will not be fully realized because these polyps are likely to be detected by radiologists without CAD. We have developed a novel MTANN technique, specifically intended to reduce FPs while maintaining a high sensitivity. In this study, we tested this technique on a population of known “FN” lesions, and we demonstrated an increase in sensitivity, yet at the same time maintaining a reasonable number of FPs. Indeed, our MTANN CAD scheme detected 58% of previously missed polyps. We believe that the sensitivity of 58% of our MTANN CAD scheme for the missed polyps would be useful in assisting radiologists in their detection of polyps in CTC.
We calculated a hypothetical performance of CTC with our MTANN CAD scheme. If our MTANN CAD was available in the trial, and if radiologists agreed on all CAD TP detections, the hypothetical sensitivity of CTC with our MTANN CAD could be 81% (the 55% by-patient sensitivity of CTC alone+the CTC FN rate+the 58% by-patient sensitivity of our MTANN CAD). It should be noted that this sensitivity calculation of CTC plus CAD is only hypothetical. We will need an observer performance study to prove this because radiologists may not agree on some CAD TP detections.
69% of the FPs classified as easy to identify as a nonpolyp would be obvious to a trained reader or radiologist as folds, stool, rectal tubes, ICVs, segmentation errors (e.g., small bowel), or technical errors. A fold might look like a polyp on 2D, but on 3D, it would readily be recognized as a fold. Technical errors due to respiratory motion or streak artifacts are also easily recognized on 2D images. CAD outputs in this category are not likely to decrease the radiologist’s efficiency because they are easily dismissed. 18% of the FPs were of moderate difficulty. In this category, there were nodular folds or stool that resembles polyps in terms of size, shape, and consistency in one window or level. Suboptimal distention also contributed to FPs in this group; thus, these FPs could be minimized by optimal attention to distension when CTC is performed.45 The nonpolyps in the “moderate” category are easily identified as nonpolyps by adjusting the window or level and paging through the area. 13% of the FPs graded as difficult can be separated conceptually into two groups. “Desirable” CAD outputs are those that an expert reader would maintain to be a polyp even after careful problem solving [e.g., Fig. 7c], and if it is sufficiently large, it would be referred for optical colonoscopy. Such marks are favorable because they help radiologists locate potential polyps. “Undesirable” CAD outputs are ones that require some time for problem solving and identification, but an expert reader would not confuse them as polyps [e.g., Fig. 7d]. Six out of eight FPs categorized as difficult were very polyplike and would be considered desirable outputs.
One potential limitation of this study is that we evaluated what might be considered to be a relatively small number of lesions. However, the cases were selected in an entirely unbiased manner; moreover, we intentionally focused on definitive lesions that had been previously undetected (yet pathologically confirmed) in primary reads—making recruitment of cases difficult. We did not obtain a statistically significant difference between sensitivities of our CAD for polyps visible on both views and only on either view. This indicates that the reported performance would be generalizable. Additionally, it should be noted that because our CAD scheme, including the 3D MTANNs, was developed with an independent training database, any bias due to the training∕testing separation issue would be expected to be minimal.
Because the mixing ANN in the mixture of expert 3D MTANNs is a modified version of a standard multilayer ANN model (i.e., multilayer perceptron),38 it has the same property of the standard ANN model, including the “overfitting” (or overtraining) issue. If the standard ANN is trained with a very small number of cases, it will face the overfitting issue, where the ANN overfits the training cases, and it is not likely to work for nontraining cases. We did not use the training database for training the mixing ANN because the 14 cases would be too small in number for determining the free parameters of the ANN adequately. With a leave-one-out cross-validation test, however, we can use the maximally available cases for training the ANN, and we can test the trained ANN with the maximally available testing cases. Therefore, we performed a leave-one-lesion-out cross-validation test for evaluating the mixing ANN.
The mixing ANN was tested by the use of a leave-one-lesion-out cross-validation test which is generally pessimistically biased.46, 47, 48 In other words, the performance estimate obtained from the leave-one-out cross-validation test is generally lower than the “true” performance. Studies47, 48 showed that when the number of samples is small, the pessimistic bias is large, i.e., the smaller the number of samples used, the lower the performance estimated from the leave-one-out cross-validation test will be, compared to the true performance. Therefore, the performance of our CAD scheme for a different database would be comparable to (or potentially better than) that reported in this study. Although the 3D MTANNs were trained with only 10 polyps in a training database, the performance for the 24 missed polyps in an independent database was high, which reflects the robustness of the technique. This observation on the generalizability of our approach is consistent with that in our previous studies,29, 35, 36, 49, 52 which involved 109 lung nodules in thoracic CT,35 and in another, 76 malignant nodules and 413 benign nodules in thoracic CT.36
Another limitation of this study is that the subjective grading of ease of detection of polyps and ease of identification of computer FPs was done by a single radiologist. Because another radiologist would grade them differently and the grading would depend on the experience of the radiologist, it would be better to have the grading done by multiple radiologists. We believe that our analysis results of computer TPs, FNs, and FPs by a single radiologist’s grading would still be useful as a reference for other researchers to understand the characteristics of computer TPs, FNs, and FPs.
Some of the polyps missed by radiologists were very small and∕or of the sessile type (these are major causes of human misses). Some sessile-type polyps such as flat lesions are known to be histologically aggressive;50 therefore, detection of such polyps is clinically important, but they are difficult to detect because of their uncommon morphology. Our MTANN CAD scheme detected these difficult polyps correctly. It should be noted that one polyp correctly detected by our CAD scheme had been missed in both CTC and reference-standard optical colonoscopy in the trial (i.e., it was detected only on air-contrast barium enema); thus, detection of this polyp may be considered “very difficult.”
An important subcategory of sessile polyps is flat lesions (also known as nonpolypoid lesions, superficial elevated lesions, or depressed lesions).50 The definition of flat lesions has not been established yet: some experts use a “height” criterion (height <3 mm); some use a “ratio” criterion (height <1∕2 the long axis). Moreover, it is still controversial which window∕level setting (i.e., lung, soft tissue, or flat) should be used in the measurement of flat lesions, or which view (i.e., 2D or 3D) should be used. Therefore, we do not use the term, “flat lesion,” but use a “sessile polyp.”
Our study focused on FN lesions in CTC in a clinical trial. Recent CTC studies used a fecal tagging technique to improve the specificity. The effect of the fecal tagging technique on the performance of a CAD scheme would be removal of FPs due to stool. The FPs due to stool produced by our MTANN CAD scheme were 32%. If we use fecal-tagging CTC data, we may be able to reduce the FP rate of our scheme by up to 32%.
Polyps difficult to be detected by radiologists are also difficult for our CAD scheme, i.e., our scheme tended to miss small, sessile polyps visible only on one view, rated as difficult by a radiologist. Development of a technique for sessile polyps including flat lesions would be a key to the further improvement of the sensitivity. On the other hand, most CAD FPs were not difficult to be dismissed by a radiologist. This implies that a high sensitivity for difficult polyps is more important for a CAD scheme than a low FP rate. However, a lower number of FPs are desirable because a study showed that a larger number of FPs affected the reading time adversely.51
In our previous studies,28, 29 we evaluated the performance of our CAD scheme with 73 CTC cases including 14 patients with 28 polyps that had been detected by radiologists, i.e., true-positive CTC cases. Our CAD scheme yielded 96.4% (27∕28) by-polyp sensitivity for polyps 5 mm or larger, with an average of 1.1 (82∕73) FPs per patient. The results in our previous and present studies also supported that polyps difficult to detect by radiologists are also difficult for a CAD scheme.
CONCLUSION
We have shown that a CAD scheme utilizing MTANNs detected 58% of polyps missed by CTC readers and that the number of FPs remained relatively low, whereas a standard CAD scheme yielded a sensitivity of 25% (6∕24) at the same FP rate. These data imply that such a CAD scheme would be useful for detecting difficult polyps which radiologists are likely to miss, thus potentially improving readers’ sensitivity in the detection of polyps in CTC.
ACKNOWLEDGMENTS
The authors are grateful to Ms. E. F. Lanzl for improving the manuscript and Lena Gong and Joel Verceles for their assistance with experiments. This work was supported by Grant No. R01CA120549 from the National Cancer Institute∕National Institutes of Health and partially by the NIH Grant Nos. S10 RR021039 and P30 CA14599.
References
- Winawer S. J. et al. , “Colorectal cancer screening: Clinical guidelines and rationale,” Gastroenterology 112(2), 594–642 (1997). 10.1053/gast.1997.v112.agast970594 [DOI] [PubMed] [Google Scholar]
- American Cancer Society, Cancer Facts & Figures 2008 (American Cancer Society, Atlanta, 2008). [Google Scholar]
- Dachman A. H., Atlas of Virtual Colonoscopy (Springer-Verlag, New York, 2003). [Google Scholar]
- Svensson M. H. et al. , “Patient acceptance of CT colonography and conventional colonoscopy: Prospective comparative study in patients with or suspected of having colorectal disease,” Radiology 222(2), 337–345 (2002). 10.1148/radiol.2222010669 [DOI] [PubMed] [Google Scholar]
- van Gelder R. E. et al. , “CT colonography and colonoscopy: Assessment of patient preference in a 5-week follow-up study,” Radiology 233(2), 328–337 (2004). 10.1148/radiol.2331031208 [DOI] [PubMed] [Google Scholar]
- Pickhardt P. J. et al. , “Cost-effectiveness of colorectal cancer screening with computed tomography colonography: The impact of not reporting diminutive lesions,” Cancer 109(11), 2213–2221 (2007). 10.1002/cncr.22668 [DOI] [PubMed] [Google Scholar]
- Pickhardt P. J. et al. , “Computed tomographic virtual colonoscopy to screen for colorectal neoplasia in asymptomatic adults,” N. Engl. J. Med. 349(23), 2191–2200 (2003). 10.1056/NEJMoa031618 [DOI] [PubMed] [Google Scholar]
- Kim D. H. et al. , “CT colonography versus colonoscopy for the detection of advanced neoplasia,” N. Engl. J. Med. 357(14), 1403–1412 (2007). 10.1056/NEJMoa070543 [DOI] [PubMed] [Google Scholar]
- Johnson C. D. et al. , “Computerized tomographic colonography: Performance evaluation in a retrospective multicenter setting,” Gastroenterology 125(3), 688–695 (2003). 10.1016/S0016-5085(03)01058-8 [DOI] [PubMed] [Google Scholar]
- Fenlon H. M. et al. , “A comparison of virtual and conventional colonoscopy for the detection of colorectal polyps,” N. Engl. J. Med. 341(20), 1496–1503 (1999). 10.1056/NEJM199911113412003 [DOI] [PubMed] [Google Scholar]
- Yee J. et al. , “Colorectal neoplasia: performance characteristics of CT colonography for detection in 300 patients,” Radiology 219(3), 685–692 (2001). [DOI] [PubMed] [Google Scholar]
- Levin B. et al. , “Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: A joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology,” Ca-Cancer J. Clin. 58(3), 130–160 (2008). 10.3322/CA.2007.0018 [DOI] [PubMed] [Google Scholar]
- Cotton P. B. et al. , “Computed tomographic colonography (virtual colonoscopy): A multicenter comparison with standard colonoscopy for detection of colorectal neoplasia,” JAMA, J. Am. Med. Assoc. 291(14), 1713–1719 (2004). 10.1001/jama.291.14.1713 [DOI] [PubMed] [Google Scholar]
- Rockey D. C. et al. , “Analysis of air contrast barium enema, computed tomographic colonography, and colonoscopy: Prospective comparison,” Lancet 365(9456), 305–311 (2005). [DOI] [PubMed] [Google Scholar]
- Doshi T. et al. , “CT colonography: False-negative interpretations,” Radiology 244(1), 165–173 (2007). 10.1148/radiol.2441061122 [DOI] [PubMed] [Google Scholar]
- Baker M. E. et al. , “Computer-aided detection of colorectal polyps: Can it improve sensitivity of less-experienced readers? Preliminary findings,” Radiology 245(1), 140–149 (2007). 10.1148/radiol.2451061116 [DOI] [PubMed] [Google Scholar]
- Petrick N. et al. , “CT colonography with computer-aided detection as a second reader: Observer performance study,” Radiology 246(1), 148–156 (2008). 10.1148/radiol.2453062161 [DOI] [PubMed] [Google Scholar]
- Summers R. M. et al. , “Automated polyp detection at CT colonography: Feasibility assessment in a human population,” Radiology 219(1), 51–59 (2001). [DOI] [PubMed] [Google Scholar]
- Jerebko A. K. et al. , “Computer-assisted detection of colonic polyps with CT colonography using neural networks and binary classification trees,” Med. Phys. 30(1), 52–60 (2003). 10.1118/1.1528178 [DOI] [PubMed] [Google Scholar]
- Jerebko A. K. et al. , “Multiple neural network classification scheme for detection of colonic polyps in CT colonography data sets,” Acad. Radiol. 10(2), 154–160 (2003). 10.1016/S1076-6332(03)80039-9 [DOI] [PubMed] [Google Scholar]
- Jerebko A. K. et al. , “Support vector machines committee classification method for computer-aided polyp detection in CT colonography,” Acad. Radiol. 12(4), 479–486 (2005). 10.1016/j.acra.2004.04.024 [DOI] [PubMed] [Google Scholar]
- Fletcher J. G. et al. , “Comparative performance of two polyp detection systems on CT colonography,” AJR, Am. J. Roentgenol. 189(2), 277–282 (2007). 10.2214/AJR.07.2289 [DOI] [PubMed] [Google Scholar]
- Li J. et al. , “Wavelet method for CT colonography computer-aided polyp detection,” Med. Phys. 35(8), 3527–3538 (2008). 10.1118/1.2938517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries A. H. et al. , “Does a computer-aided detection algorithm in a second read paradigm enhance the performance of experienced computed tomography colonography readers in a population of increased risk ?” Eur. Radiol. 19(4), 941–950 (2009). 10.1007/s00330-008-1215-3 [DOI] [PubMed] [Google Scholar]
- Kiss G. et al. , “Computer-aided diagnosis in virtual colonography via combination of surface normal and sphere fitting methods,” Eur. Radiol. 12(1), 77–81 (2002). 10.1007/s003300101040 [DOI] [PubMed] [Google Scholar]
- Yoshida H. and Nappi J., “Three-dimensional computer-aided diagnosis scheme for detection of colonic polyps,” IEEE Trans. Med. Imaging 20(12), 1261–1274 (2001). 10.1109/42.974921 [DOI] [PubMed] [Google Scholar]
- Nappi J. and Yoshida H., “Feature-guided analysis for reduction of false positives in CAD of polyps for computed tomographic colonography,” Med. Phys. 30(7), 1592–1601 (2003). 10.1118/1.1576393 [DOI] [PubMed] [Google Scholar]
- Suzuki K. et al. , “Mixture of expert 3D massive-training ANNs for reduction of multiple types of false positives in CAD for detection of polyps in CT colonography,” Med. Phys. 35(2), 694–703 (2008). 10.1118/1.2829870 [DOI] [PubMed] [Google Scholar]
- Suzuki K. et al. , “Massive-training artificial neural network (MTANN) for reduction of false positives in computer-aided detection of polyps: Suppression of rectal tubes,” Med. Phys. 33(10), 3814–3824 (2006). 10.1118/1.2349839 [DOI] [PubMed] [Google Scholar]
- Nappi J. et al. , “Automated knowledge-guided segmentation of colonic walls for computerized detection of polyps in CT colonography,” J. Comput. Assist. Tomogr. 26(4), 493–504 (2002). 10.1097/00004728-200207000-00003 [DOI] [PubMed] [Google Scholar]
- Yoshida H. et al. , “Computerized detection of colonic polyps at CT colonography on the basis of volumetric features: Pilot study,” Radiology 222(2), 327–336 (2002). 10.1148/radiol.2222010506 [DOI] [PubMed] [Google Scholar]
- Dorai C. and Jain A., “COSMOS—A representation scheme for 3D free-form objects,” IEEE Trans. Pattern Anal. Mach. Intell. 19(10), 1115–1130 (1997). 10.1109/34.625113 [DOI] [Google Scholar]
- Koenderink J. and Vandoorn A., “Surface shape and curvature scales,” Image Vis. Comput. 10(8), 557–564 (1992). 10.1016/0262-8856(92)90076-F [DOI] [Google Scholar]
- Kobayashi S. and Nomizu K., Foundations of Differential Geometry (Interscience, New York, 1963), Vol. 1. [Google Scholar]
- Suzuki K. et al. , “Massive training artificial neural network (MTANN) for reduction of false positives in computerized detection of lung nodules in low-dose CT,” Med. Phys. 30(7), 1602–1617 (2003). 10.1118/1.1580485 [DOI] [PubMed] [Google Scholar]
- Suzuki K. et al. , “Computer-aided diagnostic scheme for distinction between benign and malignant nodules in thoracic low-dose CT by use of massive training artificial neural network,” IEEE Trans. Med. Imaging 24(9), 1138–1150 (2005). 10.1109/TMI.2005.852048 [DOI] [PubMed] [Google Scholar]
- Suzuki K. et al. , “Image-processing technique for suppressing ribs in chest radiographs by means of massive training artificial neural network (MTANN),” IEEE Trans. Med. Imaging 25(4), 406–416 (2006). 10.1109/TMI.2006.871549 [DOI] [PubMed] [Google Scholar]
- Suzuki K. et al. , “Extraction of left ventricular contours from left ventriculograms by means of a neural edge detector,” IEEE Trans. Med. Imaging 23(3), 330–339 (2004). 10.1109/TMI.2004.824238 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Horiba I., and Sugie N., “Neural edge enhancer for supervised edge enhancement from noisy images,” IEEE Trans. Pattern Anal. Mach. Intell. 25(12), 1582–1596 (2003). 10.1109/TPAMI.2003.1251151 [DOI] [Google Scholar]
- Egan J. P., Greenberg G. Z., and Schulman A. I., “Operating characteristics, signal detectability, and the method of free response,” J. Acoust. Soc. Am. 33, 993–1007 (1961). 10.1121/1.1908935 [DOI] [Google Scholar]
- Dachman A. H. et al. , “Formative evaluation of standardized training for CT colonographic image interpretation by novice readers,” Radiology 249(1), 167–177 (2008). 10.1148/radiol.2491080059 [DOI] [PubMed] [Google Scholar]
- Edwards D. C. et al. , “Maximum likelihood fitting of FROC curves under an initial-detection-and-candidate-analysis model,” Med. Phys. 29(12), 2861–2870 (2002). 10.1118/1.1524631 [DOI] [PubMed] [Google Scholar]
- Paik D. S. et al. , “Surface normal overlap: A computer-aided detection algorithm with application to colonic polyps and lung nodules in helical CT,” IEEE Trans. Med. Imaging 23(6), 661–675 (2004). 10.1109/TMI.2004.826362 [DOI] [PubMed] [Google Scholar]
- Wang Z. et al. , “Reduction of false positives by internal features for polyp detection in CT-based virtual colonoscopy,” Med. Phys. 32(12), 3602–3616 (2005). 10.1118/1.2122447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dachman A. H. and Zalis M. E., “Quality and consistency in CT colonography and research reporting,” Radiology 230(2), 319–323 (2004). 10.1148/radiol.2302031113 [DOI] [PubMed] [Google Scholar]
- Fukunaga K., Introduction to Statistical Pattern Recognition, 2nd ed. (Academic, San Diego, 1990). [Google Scholar]
- Sahiner B., Chan H. P., and Hadjiiski L., “Classifier performance estimation under the constraint of a finite sample size: Resampling schemes applied to neural network classifiers,” Neural Networks 21(2–3), 476–483 (2008). 10.1016/j.neunet.2007.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahiner B., Chan H. P., and Hadjiiski L., “Classifier performance prediction for computer-aided diagnosis using a limited dataset,” Med. Phys. 35(4), 1559–1570 (2008). 10.1118/1.2868757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. and Doi K., “How can a massive training artificial neural network (MTANN) be trained with a small number of cases in the distinction between nodules and vessels in thoracic CT ?” Acad. Radiol. 12(10), 1333–1341 (2005). 10.1016/j.acra.2005.06.017 [DOI] [PubMed] [Google Scholar]
- Soetikno R. M. et al. , “Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults,” JAMA, J. Am. Med. Assoc. 299(9), 1027–1035 (2008). 10.1001/jama.299.9.1027 [DOI] [PubMed] [Google Scholar]
- Taylor S. A. et al. , “CT colonography and computer-aided detection: Effect of false-positive results on reader specificity and reading efficiency in a low-prevalence screening population,” Radiology 247(1), 133–140 (2008). [DOI] [PubMed] [Google Scholar]
- Suzuki K., “Supervised ‘lesion-enhancement’ filter by use of a massive-training artificial neural network (MTANN) in computer-aided diagnosis (CAD),” Phys. Med. Biol. 54(18), S31–S45 (2009). 10.1088/0031-9155/54/18/S03 [DOI] [PMC free article] [PubMed] [Google Scholar]