Abstract
GAF domains regulate the catalytic activity of certain vertebrate cyclic nucleotide phosphodiesterases (PDEs) by allosteric, non-catalytic binding of cyclic nucleotides. GAF domains arranged in tandem are found in PDE2, -5, -6, 10, and -11, all of which regulate the cellular concentrations of the second messengers cAMP and/or cGMP. Nucleotide binding to GAF domains affects the overall conformation and the catalytic activity of full-length PDEs. The cyclic nucleotide-bound GAF domains from PDE2, -5, -6, and -10 all adopt a conserved fold but show subtle differences within the binding pocket architecture that account for a large range of nucleotide affinities and selectivity. NMR data and details from the structure of full-length nucleotide-free PDE2A reveal the dynamic nature and magnitude of the conformational change that accompanies nucleotide binding. The discussed GAF domain structures further reveal differences in dimerization properties and highlight the structural diversity within GAF domain-containing PDEs.
Introduction
GAF domains form one of the largest and most widespread domain families found in all kingdoms of life (Anantharaman et al., 2001). Though rare in human proteins (among which they are found only in PDEs), there are about 2,000 GAF domain-containing proteins (Schultz et al., 1998) in which GAF domains have been shown to provide a variety of functions including binding of small molecules, protein-protein interactions (incl. dimerization), and other processes. However, the vast majority of GAF domains have not been studied in any detail so that their functions and ligand-binding potentials are, in general, poorly understood. The acronym GAF is derived from the first three protein families identified with this domain, namely mammalian cGMP-dependent phosphodiesterases (PDEs), Anabaena adenylyl cyclases, and E. Coli FhlA (Aravind and Ponting, 1997). Similarities in sequence and structure reveal a distant relationship to Per-ARNT-Sim (PAS) domains, another ligand-binding superfamily with a similar fold (Aravind and Ponting, 1997; Ho et al., 2000).
A subfamily of GAF domains has evolved as cyclic nucleotide (cNMP)-binding domains that allosterically regulate the catalytic activity of cyclic nucleotide phosphodiesterases (PDEs), in particular PDE2, -5, -6, -10, and -11. These PDEs contain two N-terminally located GAF domains, of which, according to the nomenclature, the more N-terminal domain is labeled as GAF A and the more C-terminal as GAF B. PDEs regulate the cellular concentrations of the cyclic nucleotides cAMP and cGMP, both of which function as essential second messengers and modulate a large number of cellular pathways (Beavo and Brunton, 2002). Through their central role in many disease-related pathways, PDEs are excellent drug targets and reached “blockbuster” status through the development of Viagra™, Cialis™, and Levitra™, all of which target PDE5 and are mainly used to treat male erectile dysfunction (Bender and Beavo, 2006; Omori and Kotera, 2007).
To date, only one GAF domain in each PDE monomer has been shown to bind cyclic nucleotide. The GAF A domains of PDE5, -6, and -11 and the GAF B domain of PDE2 selectively bind cGMP, whereas the GAF B domain of PDE10 selectively binds cAMP. Binding of cGMP to the GAF domains from PDE2, and -5 increases the catalytic activity of the respective PDE (Martins et al., 1982; Rybalkin et al., 2003a). In the case of PDE5, allosteric cGMP binding enhances phosphorylation through the cGMP-dependent protein kinase, which in turn increases PDE5 activity and cGMP binding affinity of GAF A (Francis et al., 2002; Rybalkin et al., 2003b). Binding of cGMP to GAF A of PDE6 increases affinity for the Pγ-subunit, an intrinsically disordered protein that inhibits the catalytic activity of PDE6 when bound (Muradov et al., 2002; Song et al., 2008), and alters the affinity for certain catalytic site inhibitors (Zhang et al., 2008). Less is known about the GAF domain-dependent regulatory mechanisms of PDE10 and PDE11. Binding of cAMP to the GAF B domain of full-length PDE10A2 and binding of cGMP to the GAF A domain of full-length PDE11A4 has recently been demonstrated (Matthiesen and Nielsen, 2009). Although direct activation by cyclic nucleotide binding of PDE10 and -11 has been suggested in a study with chimeric protein constructs comprised of the catalytic domain of the cyanobacterial adenylyl cyclase cyaB1 and the tandem GAF domains from PDE10 and -11 (Gross-Langenhoff et al., 2006), no direct activation was observed for the full-length PDE proteins when assayed with cyclic nucleotide analogues (Matthiesen and Nielsen, 2009). Further investigations are necessary to determine whether other factors (such as phosphorylation or membrane attachment) control a potential GAF-dependent regulation of the catalytic activity from PDE10 and -11.
This review focuses on the atomic-level structures and the derived functional implications of the cNMP-binding GAF domains from PDEs. Nucleotide binding determinants, cNMP-dependent conformational change, and dimerization properties of GAF domains are discussed. The review also highlights the diversity of GAF domains within the protein family of cyclic nucleotide PDEs and gives an outlook for GAF domains as potential drug targets.
Structure of the GAF Domain
The first reported atomic resolution structure of a GAF domain was the crystal structure of the dimeric YKG9, a yeast protein of unknown function (Ho et al., 2000). Though no ligand was found to be bound to the protein, the majority of the overall domain topology is preserved in the cNMP-binding GAF domains from cyclic nucleotide PDEs. The first atomic resolution structure of any PDE GAF domain was the 2.9 Å crystal structure of the tandem GAF domains from PDE2A (Martinez et al., 2002). The structure revealed a parallel homodimer in which GAF A contains no ligand but makes dimerization contact with a second GAF A, whereas GAF B does not contribute to the dimerization interface but binds cGMP in a deeply buried pocket. In contrast, the tandem GAF domains from the cyanobacterial adenylyl cyclase cyaB2 form an antiparallel dimer in which GAF A from one protomer makes dimerization contact with GAF B from a second protomer and vice versa. Further, both GAF A and B of cyaB2 contain cAMP binding sites (Martinez et al., 2005), a fact that stands in contrast to the tandem GAF domain units of PDEs in which only one GAF domain (either GAF A or B) contains the cNMP binding site.
In the past year, three new cNMP-bound PDE GAF domain structures have been reported (Table 1). Our laboratories presented a NMR solution structure of cGMP-bound PDE5A GAF A (Heikaus et al., 2008) and a crystal structure of cGMP-bound PDE6C GAF A (Martinez et al., 2008) while a structural genomics consortium reported a crystal structure of cAMP-bound PDE10A GAF B (Handa et al., 2008).
Table 1.
Reported structures of cyclic nucleotide binding GAF domains from PDEs
| Protein | PDE2A | PDE2A | PDE5A | PDE6C | PDE10A |
|---|---|---|---|---|---|
| Organism | mouse | human | mouse | chicken | human |
| PDB-code | 1mc0 | 3ibj | 2k31 | 3dba | 2zmf |
| Method | X-ray | X-ray | NMR | X-ray | X-ray |
| Resolution | 2.9 Å | 3.0 Å | n/a | 2.6 Å | 2.1 Å |
| Nucleotide | cGMP (GAF B) | apo | cGMP | cGMP | cAMP |
| Domain | GAF A/B | GAF A/B + cat. domain | GAF A | GAF A | GAF B |
| Oligomer | Dimer (GAF A) Monomer (GAF B)* | Dimer* | Monomer | Monomer | Dimer |
| Reference | (Martinez et al., 2002) | (Pandit et al., 2009) | (Heikaus et al., 2008) | (Martinez et al., 2008) | (Handa et al., 2008) |
apo-GAF B is dimeric in the crystal structure of the full-length human PDE2A but monomeric in the crystal structure of the cGMP-bound tandem GAF domains from mouse PDE2A.
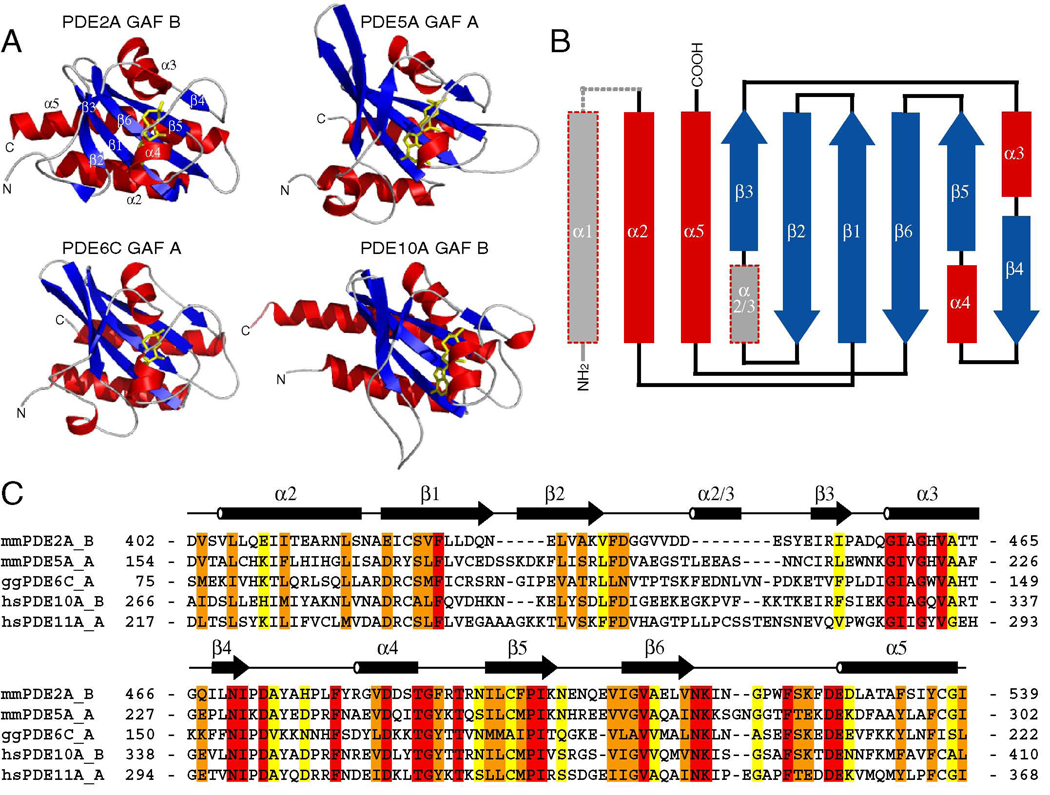
All reported structures from cyclic nucleotide-binding GAF domains reveal that GAF domains consist of a core of a six-stranded antiparallel β-sheet with the strand order 3-2-1-6-5-4 and four α-helices (Fig. 1A, B). A fifth α-helix (α1) is often packed against the domain and may take part in domain dimerization (Fig. 2A, B) but is not essential for the minimal and stably folded GAF domain entity as it is not part of the PDE5A GAF A structure. The central β-sheet is a dividing plane with helices α2 and α5 (and the N- and C-termini, respectively) on one face and helices α3 and α4 on the other face. The latter face and helices α3 and α4 form the cNMP binding site. The structures of the GAF A domains from PDE5A and PDE6C also contain a short α-helical turn that lies between β2 and β3, termed α2/3 (Fig. 1B) (Heikaus et al., 2008; Martinez et al., 2008).
Figure 1. Structures of the cNMP-bound PDE GAF Domains.
A) Structures of the cNMP-bound GAF domains from PDE2A, PDE5A, PDE6C, and PDE10A. α-helices are shown in red, β-strands in blue, and loops in gray. cNMP is shown in yellow sticks.
B) Topology of GAF domain. Elements not universally present in the structures are shown in gray. Helix α1 is not present in the solution structure of PDE5A GAF A or the crystal structure of PDE2A GAF B whereas helix α2/3 is not present in the structures from PDE2A GAF B and PDE10A GAF B.
C) Sequence alignment of cNMP-binding GAF domains from PDEs. Conservation was determined by ClustalW (Thompson, 1994) with identical residues highlighted in red, highly conserved residues in orange, and weakly conserved residues in yellow. The sequence of human PDE11A GAF A is shown for comparison.
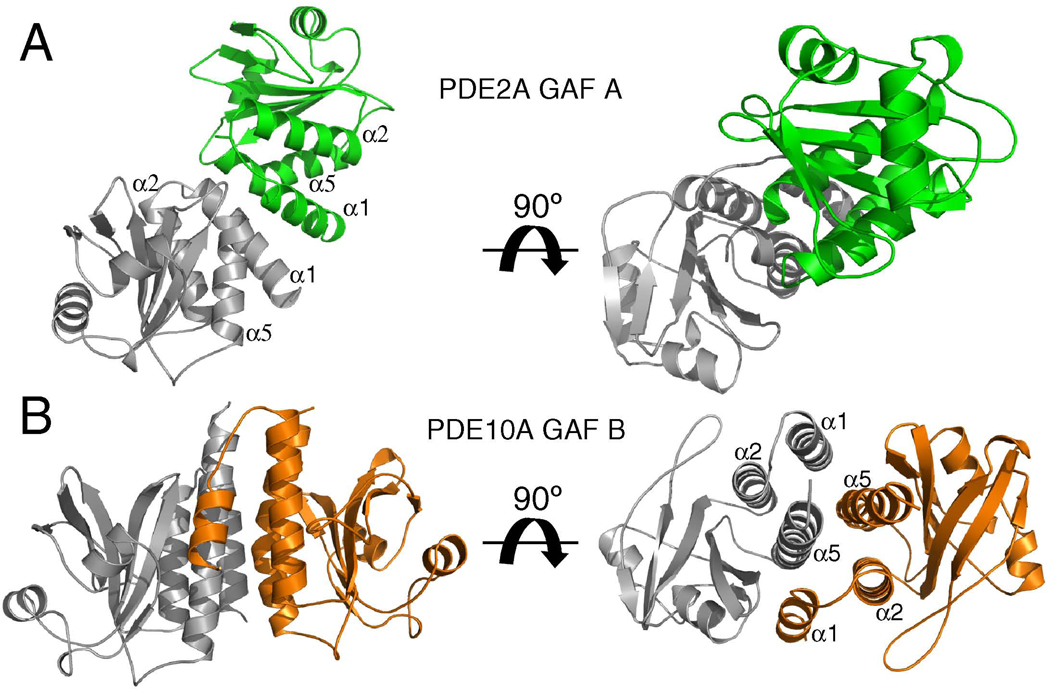
Figure 2. Dimerization interfaces of PDE2A GAF A and PDE10A GAF B.
A) Structure of the homodimer formed by PDE2A GAF A. Protomers are shown in gray and green.
B) Structure of the homodimer formed by PDE10A GAF B. Protomers are shown in gray and orange.
Two different angles are shown with right panel rotated by 90° around the x-axis towards the viewing plane. The gray PDE10A GAF B domain is structurally aligned with the gray PDE2A GAF A domain and shown in identical orientations.
GAF Domains as Dimerization interfaces
All PDEs are dimeric and contain a conserved C-terminal catalytic domain, but differ in their family-specific N-terminal regulatory domains (Bender and Beavo, 2006). Monomeric catalytic domains have been shown to be catalytically active (Fink et al., 1999) so the functional significance of the dimerization of PDEs remains unexplained. Several biochemical studies using a series of PDE constructs and the discussed GAF domain structures establish the tandem GAF domains (potentially including the N-terminal sequences) as the site of tight dimerization (Blount et al., 2006; Heikaus et al., 2008; Muradov et al., 2003; Weeks et al., 2007; Zoraghi et al., 2005).
This is supported by low resolution electron microscopy images (Kajimura et al., 2002; Kameni Tcheudji et al., 2001) in which full-length PDE5 and PDE6 both appear to be arranged as parallel dimers with contacts between GAF A and B domains, respectively. Consistent with this interpretation, the first high resolution crystal structure of any full-length PDE enzyme reveals that the cyclic nucleotide-free PDE2A enzyme forms a parallel homodimer (Pandit et al, 2009). In particular, the connecting helices between GAF A and B, and between GAF B and the catalytic domain serve as the main dimerization interfaces in the cGMP-free PDE2. This stands in contrast to the earlier reported crystal structure of the cGMP-bound tandem GAF domains (without the catalytic domain) from PDE2A in which the cGMP-bound GAF B domain makes no dimerization contact (Martinez et al., 2002), whereas GAF A forms a tight dimerization interface via its helices α1 and α5 (Fig. 2A) that is essentially identical in both PDE2A crystal structures. Though a large cGMP-dependent conformational change (see below) may influence the dimerization properties of GAF B and the GAF domain-connecting helix, it is more likely that the monomeric GAF B domain observed in the crystal structure of the tandem GAF domains is due at least in part to crystal packing and that GAF B is in fact dimeric in both states. This idea is further supported by the crystal structure of the PDE10A GAF B domain in which dimerization is also provided by helices α1 and α5, which make extensive contact with a second GAF B protomer and are both packed against the rest of the domain via helix α2. Together, the three helices from each protomer form a six-helix bundle of parallel helices (Fig. 2B). Although the equivalent helices provide the dimerization contacts in both PDE2A GAF A and PDE10A GAF B (and PDE2 GAF B) domains, the exact helical arrangement and domain orientation significantly differ (Fig. 2A, B).
In contrast to the GAF domains from PDE2A and PDE10A, the isolated cGMP-bound GAF A domain constructs of PDE5A (including helices α2, α5) and PDE6C (including helices α1, α2, α5) used for the respective structure determinations are both monomeric (Heikaus et al., 2008; Martinez et al., 2008). Therefore, other sequence elements located N- and C-terminal of the GAF A domain are likely necessary for tight dimerization of the PDE5 and -6 enzymes as indicated by several studies (Heikaus et al., 2008; Muradov et al., 2003; Zoraghi et al., 2005).
The structures of PDE GAF domains determined to date highlight that there are several modes of GAF domain dimerization as exemplified by PDE10 GAF B and PDE2 GAF A. Based on the available structural and biochemical data, the consensus is that all tandem GAF domains from PDEs form parallel dimers with extensive dimerization interfaces between the domain-connecting helices. The antiparallel dimerization of the tandem GAF domains from the Anabaena adenylyl cyclase may therefore be limited to evolutionarily lower organisms (Martinez et al., 2005). The unique properties and orientations of the various PDE dimerization interfaces may provide a mechanism to favor homodimerization in cells expressing multiple GAF-containing PDEs. Though the structure of PDE2A tandem GAF plus catalytic domains suggests that GAF domain dimerization is an essential regulator of catalytic activity (Pandit et al, 2009), it remains to be shown whether other regulatory processes alter the dimerization properties of the full-length enzymes. To answer this question, structures of identical protein constructs (ideally full-length) in activated and non-activated form are necessary (cNMP-bound vs. cNMP-free and/or phosphorylated vs. unphosphorylated).
Cyclic nucleotide recognition and binding mode
The tandem GAF domains of PDE2, -5, and -6 have low nanomolar affinities for cGMP (KD ≤ 10 nM) but vary in their nucleotide selectivity. GAF A from PDE6 has the highest cGMP-selectivity with a preference for cGMP over cAMP of at least 10,000-fold (Hebert et al., 1998; Huang et al., 2004), whereas PDE5A GAF A has a cGMP-selectivity of several thousand-fold (Heikaus et al., 2008). PDE2A GAF B is only moderately selective with a 10–30-fold preference for cGMP (Wu et al., 2004). No affinities for nucleotide binding to PDE10A GAF B or PDE11A GAF A have been reported to date. The affinity of PDE10A GAF B is likely in the nanomolar range (based on its slow off-rate) as it binds cAMP from the Escherichia Coli cells during expression and crystallizes bound to cAMP without the addition of cAMP (Handa et al., 2008). In all cGMP-binding GAF domains, the cGMP affinities are similarly high (in the low nanomolar range) but the cAMP affinities differ significantly, indicating that high-selectivity GAF domains contain certain negative determinants that select against cAMP. From studies using constructs of various length, it has become evident that affinities for isolated (or tandem) GAF domains are often higher than for the full-length enzymes (e.g. full-length PDE5 has cGMP affinity of ~200 nM, whereas the tandem GAF domains have an affinity of ~2–10 nM (Heikaus et al., 2008; Zoraghi et al., 2005)). This indicates that additional elements of the full-length enzymes (such as the N-terminal extension) can lower the cNMP affinity of the GAF domains, presumably by stabilizing the apo-form of the domain (Bruder et al., 2006).
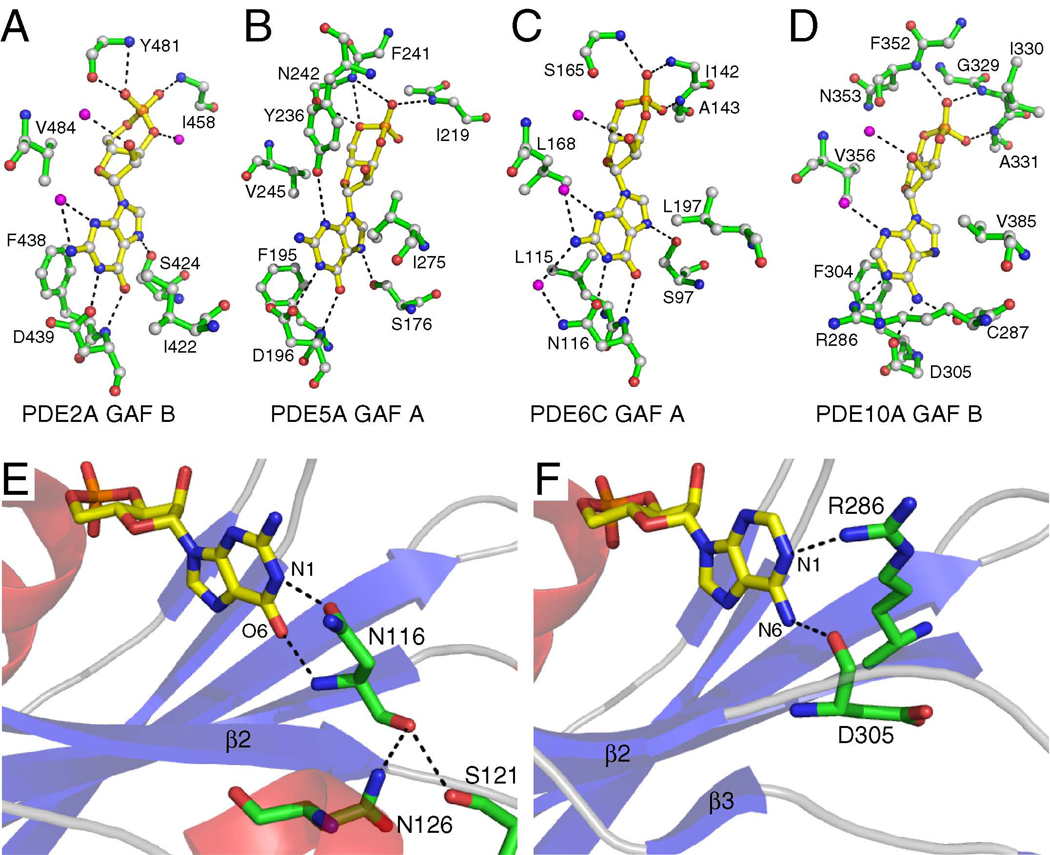
In all reported GAF domain structures, the cyclic nucleotide is deeply buried. The binding pocket is comprised of the six-stranded β-sheet, which provides the floor of the binding pocket, and α-helices α3 and α4, which provide the roof of the binding pocket (Fig. 1A). The helical dipole at the N-terminus of α3 binds the cyclic phosphate group. There are also several hydrogen bonds between protein backbone atoms and the phosphate oxygens (Fig. 3A–D). Hydrogen bond networks and hydrophobic interactions between side chains and the invariant part of the nucleotides provide important binding energy yielding the observed nanomolar binding affinities for the respective nucleotide.
Figure 3. Cyclic Nucleotide Recognition.
A) Binding pocket interactions between cGMP and PDE2A GAF B.
B) Binding pocket interactions between cGMP and PDE5A GAF A.
C) Binding pocket interactions between cGMP and PDE6C GAF A.
D) Binding pocket interactions between cAMP and PDE10A GAF B.
E) Interaction between α2/3 and Asn116 in PDE6C GAF A. Ser121 and Asn126 make hydrogen bond contacts to Asn116, which in turn makes nucleotide-specific contact with cGMP.
F) PDE10A GAF B does not contain α2/3. The motion range of Asp305 appears less restricted and it is rotated out to accommodate Arg286, which in turn makes nucleotide-specific contact with cAMP.
Selectivity for cGMP and cAMP is provided by intermolecular hydrogen bonds to the variant nucleotide moieties, i.e. the substitutions at C6 (carbonyl in cGMP, amino-group in cAMP), N1 (NH in cAMP), and C2 (amino-group in cGMP) of the nucleotide base (Fig. 3A–D). A single residue provides two cGMP-specific hydrogen bond contacts (one from the main chain to O6, one from the side chain to N1) in the cGMP-specific GAF domains from PDE2A, PDE5A, and, PDE6C (Fig. 3A–C, E). This residue is a conserved Asp in PDE2A and PDE5A, and an Asn in PDE6C. In contrast, the Asp in the analogous position in PDE10A is rotated away from the binding pocket in the cAMP-bound PDE10A GAF B structure allowing an Arg to make hydrogen bond contact with the N1-atom of cAMP instead (Fig. 3D, F). In PDE2A, mutation of Asp439 causes loss of nucleotide selectivity so that cAMP and cGMP both bind with essentially the same affinity (Wu et al., 2004). Mutation of Asp196 to an Ala in PDE5A leads to a complete switch of selectivity from cGMP to cAMP in GAF A constructs (Heikaus et al., 2008), whereas mutations of other residues within the binding pocket (e.g. F195A) decrease or abolish cGMP binding (Sopory et al., 2003). Based on the presence of helix α2/3 within the β2-β3-connecting loop in both high-selectivity GAF A domains from PDE5A and PDE6C, it appears that this short helix restricts the flexibility of the Asp/Asn by packing and hydrogen bonding to the backbone of the Asp/Asn, causing the side chain to be locked into position to make cGMP-specific contact (Fig. 3E). In contrast, the β2–β3-connecting loop in the low-specificity PDE2A is shorter and does not contain secondary structure elements, whereas the loop in PDE10A is longer but contains short stretches of β-strands that do not make contact to the main chain of Asp, allowing it to rotate away from the ligand, making room for an Arg that makes cAMP-specific contact (Fig. 3F). Mutational studies of the β2–β3-connecting loop (and the α2/3-helix) support such a potential role in providing nucleotide selectivity (Linder et al., 2007). In all structures, a hydrophobic residue immediate preceding the Asp/Asn (Phe438 in PDE2A, Phe195 in PDE5A, Leu115 in PDE6C, and Phe304 in PDE10A) makes hydrophobic contact with the purine ring of the cNMP molecule thereby stabilizing the binding pocket (Fig. 3A–D).
Before the first structure of a cNMP-binding GAF domain was reported, the so-called NKFDE-motif comprised of five strictly conserved residues found in cNMP-binding GAF domains from PDEs (Charbonneau et al., 1990; McAllister-Lucas et al., 1995) was proposed to be essential for nucleotide binding and the formation of the binding pocket (Ho et al., 2000; Turko et al., 1996). However, instead of being directly involved in nucleotide binding, the residues of this motif turned out to be important for the folding stability of the GAF domain through a network of hydrogen bonds and salt bridges that are located away from the binding pocket (Martinez et al., 2002). Furthermore, the motif is not involved in inter-domain interactions in any of the published crystal structures. All five residues are oriented in practically identical orientations in the cGMP-bound and -free form of PDE2A GAF B (Pandit et al, 2009) indicating that they do not play an important role in the nucleotide-dependent conformational change (see below).
Nucleotide-Dependent Conformational Changes
In all cNMP-bound GAF domain structures, the cyclic nucleotide is bound in a deeply buried pocket with almost no solvent accessibility. Large rearrangements of secondary structure elements are necessary to allow cNMP to enter and exit the binding pocket. This change from “closed” to “open” state was first demonstrated for the GAF A domains from PDE5A and PDE6C by NMR studies. In both cases, an increase of domain stability and decrease of overall flexibility upon cGMP binding was observed (Heikaus et al., 2008; Martinez et al., 2008). Practically no backbone amide NMR resonances from cGMP-free GAF A of PDE5A can be observed suggesting a conformational exchange that is intermediate on the NMR timescale. Similarly, the NMR spectra of the cGMP-bound and -free GAF A domain of PDE6C significantly differ from each other and reveal that the core of GAF A adopts two distinctly structured states with more dynamic elements.
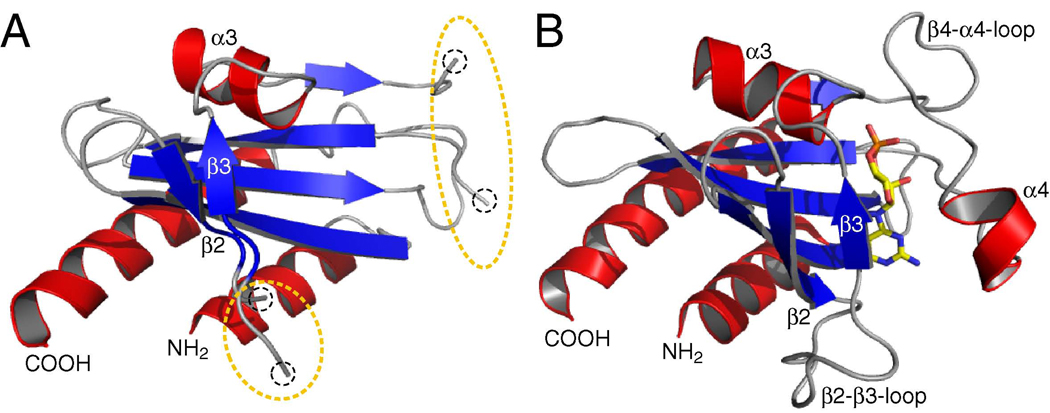
Comparison of the cGMP-bound and -free GAF B domain structures of PDE2A reveals that the six-stranded β-sheet and helices α2, α3, α5 are well-defined in both forms, whereas no density could be traced for helix α4, the β4-α4-connecting loop, and the β2–β3-connecting loop, presumably due to intrinsic disorder in the absence of cGMP (Fig. 4A, B) (Pandit et al, 2009). The β-sheet is flatter in the absence of cGMP, thereby allowing the nucleotide to enter the open binding pocket. Of the two helices that provide the roof of the binding pocket (helices α3 and α4), α3 is significantly less dynamic in the free structure than α4 (Fig. 4A, B), a fact that is supported by H-D exchange data for PDE5A GAF A, which revealed that α3 is less accessible to solvent exchange than is α4 (Heikaus et al., 2008).
Figure 4. cNMP-dependent Conformational Change.
A) cGMP-free PDE2A GAF B. Electron density could not be traced for the β4-α4-loop, α4, and the β2-β3-loop indicating structural disorder as highlighted by the dotted circles.
B) cGMP-bound PDE2A GAF B. cGMP is shown in sticks with carbon atoms in yellow.
Together these data suggest that GAF domains undergo a large-scale ligand-dependent induced fit during which entire secondary structure elements are stabilized and fixed into position. Helix α4 and the β2–β3-connecting loop (which in PDE5 and -6 encompasses the short helix α2/3, see above) are dynamic and sample multiple states. Together, they are responsible for the closure of the GAF domain upon nucleotide binding. It is likely that the charged cyclic phosphate group is attracted by the positive helix dipole created by the N-terminus from the less dynamic helix α3. Upon binding of the cyclic phosphate group, the binding pocket closes partway over the nucleotide and the cGMP- and cAMP-specific contacts (see above) lock the specific nucleotide into the binding pocket and bury it in the center of the domain. The flexibility and the large structural rearrangements within the GAF domains may explain why it has been so difficult to obtain high-resolution structures of apo GAF domains. Presumably, the presence of additional sequence elements such as the catalytic domains and the extensive dimerization interface along the tandem GAF domains stabilize the apo-GAF B domain of PDE2A sufficiently for the growth of diffraction-quality crystals that ultimately led to the full-length PDE2A crystal structure (Pandit et al, 2009).
Perspectives and Outlook
Recent advances in the structural characterization of cyclic nucleotide-binding GAF domains from PDEs have answered several questions. Four of the five cNMP-bound GAF domains from PDEs have now been described; three of them within the last year. Furthermore, the crystal structure of full-length PDE2A represents the first view of a PDE enzyme at high resolution and puts the regulatory GAF domains in relation to the catalytic domain, something that has long been regarded as the ‘Holy Grail’ in the PDE field. The recent GAF domain structures represent a major advance as they provide information about the molecular determinants of cyclic nucleotide binding and cNMP-dependent conformational change. Together, they significantly further our understanding of the GAF domain-dependent mechanism of PDE regulation.
Nevertheless, several questions about GAF domains and their functional diversity remain unanswered. The structures reveal that all GAF domains bind cNMP through a conserved pattern of interactions with helix dipole-phosphate and hydrophobic interactions. However, no absolute consensus within the protein-ligand hydrogen bond network is evident and the structures reveal that there are various ways to bind cNMP molecules with high affinity. Though GAF domains are one of the largest small-molecule-binding domain families, ligand binding has only been demonstrated for a few GAF domains and the functional significance of tandem GAF domains in PDEs with a second non-binding GAF domain also remains to be elucidated. Among PDEs, the photoreceptor phosphodiesterase 6 is structurally distinct. Whereas the catalytic subunits of cone PDE6 forms homodimer of α’-subunits, rod PDE6 forms heterodimers of α- and β-subunits. More structural studies are necessary to understand the functional significance of this cell-specific difference and the specific determinants of dimerization. The visualization of the cNMP-dependent conformational change by NMR and the comparison of cGMP-free and - bound GAF B domain from PDE2A demonstrate the magnitude of the conformational change that occurs within the GAF domain. How this large-scale induced-fit affects the overall conformation of the full-length enzyme and ultimately causes activation is still unknown. Additional crystal structures of full-length PDEs, ideally in several states of activation, are needed to answer this question.
Cyclic nucleotides play a key role in many important signaling pathways, and PDEs are the only enzymes that regulate their concentrations through hydrolysis. Agonists and antagonists that can modulate catalytic PDE-activity by binding to and stabilizing either the 'closed' or 'open' form of GAF domains have the potential to be as important therapeutically as the commercially successful catalytic-site inhibitors of PDEs. Among all PDEs, the sequence conservation and similarities in the cNMP pocket architecture are lower in GAF domains than in catalytic domains, suggesting that GAF domain-binding PDE drugs may have higher PDE-selectivity and fewer side effects than catalytic-site inhibitors.
The recently presented GAF domain structures advance the field significantly and open the door for future investigations. Determination of complex structures (i.e. PDE6 plus Pγ), the characterization of the mechanistic details of the GAF-dependent regulation of PDEs, and design of GAF-specific drugs will all be guided by the structures.
ACKNOWLEDGEMENTS
Work on the GAF domains in our laboratories was supported by funding from National Institutes of Health Grant P01 HL44949 (to R.E.K.) and a Boehringer Ingelheim Fonds Ph.D. Scholarship (to C.C.H.). We thank Joseph A. Beavo for helpful comments.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
REFERENCES
- Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- Beavo JA, Brunton LL. Cyclic nucleotide research -- still expanding after half a century. Nat Rev Mol Cell Biol. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev. 2006;58:488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- Blount MA, Zoraghi R, Ke H, Bessay EP, Corbin JD, Francis SH. A 46-amino acid segment in phosphodiesterase-5 GAF-B domain provides for high vardenafil potency over sildenafil and tadalafil and is involved in phosphodiesterase-5 dimerization. Mol Pharmacol. 2006;70:1822–1831. doi: 10.1124/mol.106.028688. [DOI] [PubMed] [Google Scholar]
- Bruder S, Schultz A, Schultz JE. Characterization of the tandem GAF domain of human phosphodiesterase 5 using a cyanobacterial adenylyl cyclase as a reporter enzyme. J Biol Chem. 2006;281:19969–19976. doi: 10.1074/jbc.M603374200. [DOI] [PubMed] [Google Scholar]
- Charbonneau H, Prusti RK, LeTrong H, Sonnenburg WK, Mullaney PJ, Walsh KA, Beavo JA. Identification of a noncatalytic cGMP-binding domain conserved in both the cGMP-stimulated and photoreceptor cyclic nucleotide phosphodiesterases. Proc Natl Acad Sci U S A. 1990;87:288–292. doi: 10.1073/pnas.87.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink TL, Francis SH, Beasley A, Grimes KA, Corbin JD. Expression of an active, monomeric catalytic domain of the cGMP-binding cGMP-specific phosphodiesterase (PDE5) J Biol Chem. 1999;274:34613–34620. doi: 10.1074/jbc.274.49.34613. [DOI] [PubMed] [Google Scholar]
- Francis SH, Bessay EP, Kotera J, Grimes KA, Liu L, Thompson WJ, Corbin JD. Phosphorylation of isolated human phosphodiesterase-5 regulatory domain induces an apparent conformational change and increases cGMP binding affinity. J Biol Chem. 2002;277:47581–47587. doi: 10.1074/jbc.M206088200. [DOI] [PubMed] [Google Scholar]
- Gross-Langenhoff M, Hofbauer K, Weber J, Schultz A, Schultz JE. cAMP is a ligand for the tandem GAF domain of human phosphodiesterase 10 and cGMP for the tandem GAF domain of phosphodiesterase 11. J Biol Chem. 2006;281:2841–2846. doi: 10.1074/jbc.M511468200. [DOI] [PubMed] [Google Scholar]
- Handa N, Mizohata E, Kishishita S, Toyama M, Morita S, Uchikubo-Kamo T, Akasaka R, Omori K, Kotera J, Terada T. Crystal structure of the GAF-B domain from human phosphodiesterase 10A complexed with its ligand, cAMP. J Biol Chem. 2008;283:19567–19664. doi: 10.1074/jbc.M800595200. [DOI] [PubMed] [Google Scholar]
- Hebert MC, Schwede F, Jastorff B, Cote RH. Structural features of the noncatalytic cGMP binding sites of frog photoreceptor phosphodiesterase using cGMP analogs. J Biol Chem. 1998;273:5557–5565. doi: 10.1074/jbc.273.10.5557. [DOI] [PubMed] [Google Scholar]
- Heikaus CC, Stout JR, Sekharan MR, Eakin CM, Rajagopal P, Brzovic PS, Beavo JA, Klevit RE. Solution Structure of the cGMP Binding GAF Domain from Phosphodiesterase 5: INSIGHTS INTO NUCLEOTIDE SPECIFICITY, DIMERIZATION, AND cGMP-DEPENDENT CONFORMATIONAL CHANGE. J Biol Chem. 2008;283:22749–22759. doi: 10.1074/jbc.M801577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YS, Burden LM, Hurley JH. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. Embo J. 2000;19:5288–5299. doi: 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Hinds TR, Martinez SE, Doneanu C, Beavo JA. Molecular determinants of cGMP binding to chicken cone photoreceptor phosphodiesterase. J Biol Chem. 2004;279:48143–48151. doi: 10.1074/jbc.M404338200. [DOI] [PubMed] [Google Scholar]
- Kajimura N, Yamazaki M, Morikawa K, Yamazaki A, Mayanagi K. Three-dimensional structure of non-activated cGMP phosphodiesterase 6 and comparison of its image with those of activated forms. J Struct Biol. 2002;139:27–38. doi: 10.1016/s1047-8477(02)00502-6. [DOI] [PubMed] [Google Scholar]
- Kameni Tcheudji JF, Lebeau L, Virmaux N, Maftei CG, Cote RH, Lugnier C, Schultz P. Molecular organization of bovine rod cGMP-phosphodiesterase 6. J Mol Biol. 2001;310:781–791. doi: 10.1006/jmbi.2001.4813. [DOI] [PubMed] [Google Scholar]
- Linder JU, Bruder S, Schultz A, Schultz JE. Changes in purine specificity in tandem GAF chimeras from cyanobacterial cyaB1 adenylate cyclase and rat phosphodiesterase 2. Febs J. 2007;274:1514–1523. doi: 10.1111/j.1742-4658.2007.05700.x. [DOI] [PubMed] [Google Scholar]
- Martinez SE, Bruder S, Schultz A, Zheng N, Schultz JE, Beavo JA, Linder JU. Crystal structure of the tandem GAF domains from a cyanobacterial adenylyl cyclase: modes of ligand binding and dimerization. Proc Natl Acad Sci U S A. 2005;102:3082–3087. doi: 10.1073/pnas.0409913102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez SE, Heikaus CC, Klevit RE, Beavo JA. The structure of the GAF A domain from the phosphodiesterase 6C reveals determinants of cGMP-binding, a conserved binding surface, and a large cGMP-dependent conformational change. J Biol Chem. 2008;283:25913–25919. doi: 10.1074/jbc.M802891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez SE, Wu AY, Glavas NA, Tang XB, Turley S, Hol WG, Beavo JA. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc Natl Acad Sci U S A. 2002;99:13260–13265. doi: 10.1073/pnas.192374899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins TJ, Mumby MC, Beavo JA. Purification and characterization of a cyclic GMP-stimulated cyclic nucleotide phosphodiesterase from bovine tissues. J Biol Chem. 1982;257:1973–1979. [PubMed] [Google Scholar]
- Matthiesen K, Nielsen J. Binding of cyclic nucleotides to phosphodiesterase 10A and 11A GAF domains does not stimulate catalytic activity. Biochem J. 2009 doi: 10.1042/BJ20090982. [DOI] [PubMed] [Google Scholar]
- McAllister-Lucas LM, Haik TL, Colbran JL, Sonnenburg WK, Seger D, Turko IV, Beavo JA, Francis SH, Corbin JD. An essential aspartic acid at each of two allosteric cGMP-binding sites of a cGMP-specific phosphodiesterase. J Biol Chem. 1995;270:30671–30679. doi: 10.1074/jbc.270.51.30671. [DOI] [PubMed] [Google Scholar]
- Muradov KG, Boyd KK, Martinez SE, Beavo JA, Artemyev NO. The GAFa domains of rod cGMP-phosphodiesterase 6 determine the selectivity of the enzyme dimerization. J Biol Chem. 2003;278:10594–10601. doi: 10.1074/jbc.M208456200. [DOI] [PubMed] [Google Scholar]
- Muradov KG, Granovsky AE, Schey KL, Artemyev NO. Direct interaction of the inhibitory gamma-subunit of Rod cGMP phosphodiesterase (PDE6) with the PDE6 GAFa domains. Biochemistry. 2002;41:3884–3890. doi: 10.1021/bi015935m. [DOI] [PubMed] [Google Scholar]
- Omori K, Kotera J. Overview of PDEs and their regulation. Circ Res. 2007;100:309–327. doi: 10.1161/01.RES.0000256354.95791.f1. [DOI] [PubMed] [Google Scholar]
- Pandit J, Forman MD, Fennell KF, Dillman KS, Menniti FS. Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc. Natl. Acad. Sci. USA. 2009;106:18225–18230. doi: 10.1073/pnas.0907635106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybalkin SD, Rybalkina IG, Shimizu-Albergine M, Tang XB, Beavo JA. PDE5 is converted to an activated state upon cGMP binding to the GAF A domain. Embo J. 2003a;22:469–478. doi: 10.1093/emboj/cdg051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybalkin SD, Yan C, Bornfeldt KE, Beavo JA. Cyclic GMP phosphodiesterases and regulation of smooth muscle function. Circ Res. 2003b;93:280–291. doi: 10.1161/01.RES.0000087541.15600.2B. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Guo LW, Muradov H, Artemyev NO, Ruoho AE, Markley JL. Intrinsically disordered gamma-subunit of cGMP phosphodiesterase encodes functionally relevant transient secondary and tertiary structure. Proc Natl Acad Sci U S A. 2008;105:1505–1510. doi: 10.1073/pnas.0709558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopory S, Balaji S, Srinivasan N, Visweswariah SS. Modeling and mutational analysis of the GAF domain of the cGMP-binding, cGMP-specific phosphodiesterase, PDE5. FEBS Lett. 2003;539:161–166. doi: 10.1016/s0014-5793(03)00219-9. [DOI] [PubMed] [Google Scholar]
- Thompson WJ, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turko IV, Haik TL, McAllister-Lucas LM, Burns F, Francis SH, Corbin JD. Identification of key amino acids in a conserved cGMP-binding site of cGMP-binding phosphodiesterases. A putative NKXnD motif for cGMP binding. J Biol Chem. 1996;271:22240–22244. doi: 10.1074/jbc.271.36.22240. [DOI] [PubMed] [Google Scholar]
- Weeks JL, 2nd,, Zoraghi R, Francis SH, Corbin JD. N-Terminal domain of phosphodiesterase-11A4 (PDE11A4) decreases affinity of the catalytic site for substrates and tadalafil, and is involved in oligomerization. Biochemistry. 2007;46:10353–10364. doi: 10.1021/bi7009629. [DOI] [PubMed] [Google Scholar]
- Wu AY, Tang XB, Martinez SE, Ikeda K, Beavo JA. Molecular determinants for cyclic nucleotide binding to the regulatory domains of phosphodiesterase 2A. J Biol Chem. 2004;279:37928–37938. doi: 10.1074/jbc.M404287200. [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Cahill KB, Elfenbein A, Arshavsky VY, Cote RH. Direct Allosteric Regulation between the GAF Domain and Catalytic Domain of Photoreceptor Phosphodiesterase PDE6. J Biol Chem. 2008;283:29699–29705. doi: 10.1074/jbc.M803948200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoraghi R, Bessay EP, Corbin JD, Francis SH. Structural and functional features in human PDE5A1 regulatory domain that provide for allosteric cGMP binding, dimerization, and regulation. J Biol Chem. 2005;280:12051–12063. doi: 10.1074/jbc.M413611200. [DOI] [PubMed] [Google Scholar]