Abstract
The external domains of the HIV-1 envelope glycoprotein (gp120 and the gp41 ectodomain, collectively known as gp140) contain all known viral neutralization epitopes. Various strategies have been used to create soluble trimers of the envelope to mimic the structure of the native viral protein, including mutation of the gp120-gp41 cleavage site, introduction of disulfide bonds, and fusion to heterologous trimerization motifs. We compared the effects on quaternary structure, antigenicity, and immunogenicity of three such motifs: T4 fibritin, a GCN4 variant, and the E. coli aspartate transcarbamoylase catalytic subunit. Fusion of each motif to the C-terminus of a non-cleavable JRCSF gp140(-) envelope protein led to enhanced trimerization but had limited effects on the antigenic profile and CD4 binding ability of the trimers. Immunization of rabbits provided no evidence that the trimerized gp140(-) constructs induced significantly improved neutralizing antibodies to several HIV-1 pseudoviruses, compared to gp140 lacking a trimerization motif. However, modest differences in both binding specificity and neutralizing antibody responses were observed among the various immunogens.
Keywords: Vaccine, HIV-1, Envelope, trimerization motifs, ATCase, T4 fibritin, GCN, CD4, monoclonal antibody, immunization, neutralizing antibody
Introduction
The ongoing HIV-1 epidemic has resulted in 2.7 million new infections and 2.1 million AIDS deaths in 2007 for a total of 33 million people living with HIV/AIDS (UNAIDS, 2008). Prophylactic measures against HIV-1 are clearly required, and the discovery of vaccine candidates that elicit broadly neutralizing antibody responses against HIV-1 is an important goal (Karlsson Hedestam et al., 2008; Montefiori et al., 2007; Walker and Burton, 2008). Some insights for vaccine research can be gained from a better understanding of the antibody response to HIV-1 during natural infection. Most HIV-infected individuals develop neutralizing antibodies with limited cross-reactivity (Hu and Stamatatos, 2007; Karlsson Hedestam et al., 2008; Richman et al., 2003), a result that is likely due to the predominance of autologous neutralization determinants in envelope (Env) glycoproteins. However, the existence of some patients who go on to develop broadly neutralizing immune sera suggests that such antibodies can be elicited under certain conditions (Binley et al., 2008; Carotenuto et al., 1998; Li et al., 2007; Pilgrim et al., 1997; Sather et al., 2009). Several monoclonal antibodies (mAbs) capable of neutralizing a broad range of primary isolates from various subtypes have been isolated from HIV-1 infected individuals. These demonstrate that conserved neutralization epitopes are present in Env (Binley et al., 2004; Calarese et al., 2003; Saphire et al., 2001; Sharon et al., 2003; Zwick et al., 2001). Studies with non-human primates have further demonstrated that broadly neutralizing antibodies can protect from infection (Baba et al., 2000; Hessell et al., 2009; Hofmann-Lehmann et al., 2001; Mascola, 2003; Nishimura et al., 2002; Shibata et al., 1999). Although it is not known what magnitude and breadth of neutralization will confer some measure of protection against HIV-1 infection, it is clear that current Env immunogens elicit antibodies that neutralize only a small fraction of circulating isolates (Montefiori et al., 2007).
The HIV-1 Env complex on the surface of infectious viral particles is the target of neutralizing antibodies. This complex is formed by a trimer of heterodimers of the gp120 and gp41 subunits of the Env transmembrane protein (Dey et al., 2008; Hu and Stamatatos, 2007; Karlsson Hedestam et al., 2008; Poignard et al., 2003; Poignard et al., 2001). The Env structures on the viral membrane are apparently heterogeneous (Dey et al., 2008; Moore et al., 2006; Poignard et al., 2003; Sougrat et al., 2007). Only complexes formed by intact gp120-gp41 trimers are thought to be functional and capable of mediating HIV-1 entry into target cells (Moore et al., 2006; Poignard et al., 2003; Poignard et al., 2001). Non-functional forms of Env could serve as decoys to prevent the host immune system from recognizing conserved neutralization epitopes (Dey et al., 2008; Moore et al., 2006; Poignard et al., 2003). Recent cryo-electron tomographic studies on SIV and HIV-1 particles have provided further evidence that the viral Env forms three-lobed trimeric structures containing the membrane-distal gp120, with considerable heterogeneity (Liu et al., 2008; Sougrat et al., 2007; Zanetti et al., 2006; Zhu et al., 2006).
Clinical testing of several vaccines based on gp120 has shown a lack of protective efficacy (Pitisuttithum et al., 2006). Some investigators have proposed that soluble trimers of the gp120-gp41 Env ectodomain (i.e., lacking the transmembrane and cytoplasmic domains) might better preserve or mimic the structure of functional Env complexes (Burton et al., 2004; Schulke et al., 2002; Srivastava et al., 2003b; Yang et al., 2002; Zhang et al., 2007). The inclusion of the gp41 ectodomain sequences is also likely to be a positive factor in immunogenicity (Grundner et al., 2004) since gp41 contains helical regions that often harbor T-helper epitopes,. The Env ectodomain contains all of the known neutralization epitopes. This gp140 polypeptide, if cleaved at the gp120-gp41 junction, will typically result in free gp120 subunits along with a thermodynamically favored 6-helix bundle formed by the gp41 moiety. Mutating the furin cleavage site to inhibit processing of gp140 creates a polypeptide [referred to as gp140(-)] that has a tendency to form a range of heterogeneous oligomers (Earl et al., 1994; Yang et al., 2000a; Yang et al., 2000b; Yang et al., 2002), although some modifications can stabilize oligomerization (Srivastava et al., 2003a). These modified gp140 immunogens induce high avidity antibodies in rabbits that are less focused on linear V3 epitopes, suggesting a possible benefit to using non-cleavable gp140 (Sharma et al., 2006). Another modified form of gp140, called gp140ΔCFI, lacks the gp120-gp41 cleavage site, the fusion peptide, and interhelical regions. This protein was shown to elicit an improved antibody response compared to a series of other Env forms (Chakrabarti et al., 2002). A recent study using Env immunogens from the HIV-1 R2 strain demonstrated that gp140(-) elicited broader and more potent neutralization responses than did gp120 (Zhang et al., 2007).
Several studies have shown that cleavage-defective Env proteins are antigenically different from processed Env (Binley et al., 2000; Herrera et al., 2005; Moore et al., 2006; Pancera and Wyatt, 2005). It is therefore not clear to what extent the structure of gp140(-) resembles native, fusion-competent Env complexes. To express processed and stable trimers of gp140, Binley et al. introduced an intermolecular disulfide bond between the gp120 and gp41 subunits to form a complex called SOS gp140 (Binley et al., 2000). Although still predominantly monomeric, SOS gp140 reacted more strongly with the neutralizing b12 mAb and less strongly with non-neutralizing antibodies compared to a non-cleavable gp140 that was comprised of a mixture of noncovalently associated and disulfide-linked dimers, trimers and tetramers (Schulke et al., 2002). Subsequently, an isoleucine-to-proline substitution was introduced at position 559 in the N-terminal heptad region of gp41 to increase the stability of SOS gp140 (Sanders et al., 2002). This SOSIP gp140 variant is fully cleaved, has desirable antigenic properties, and is predominantly trimeric (Sanders et al., 2002). Immunogenicity studies using rabbits showed that SOSIP gp140 trimers elicited approximately three-fold lower titers of anti-gp120 antibodies than did monomeric gp120 with similar neutralization profiles (Beddows et al., 2005). More recently, specific amino acids in the N-terminus of the gp41 ectodomain have been shown to further enhance the stability of SOSIP gp140 trimers (Dey et al., 2007). These substitutions in the Env polypeptide reduced the expression of non-trimeric envelope forms on pseudoviruses and were associated with a decreased binding of non-neutralizing antibodies (Dey et al., 2008).
Several small trimerization motifs have been fused to the C-terminus of gp140(-) to stabilize the formation of Env trimers. Among them are a 32 amino-acid form of the GCN4 transcription factor (GCN), a 27 amino-acid trimerization domain from the C-terminus of bacteriophage T4 fibritin (T4F), and a soluble trimerization domain of chicken cartilage matrix (CART) protein (Selvarajah et al., 2008; Yang et al., 2000a; Yang et al., 2002). Fusion of these motifs promotes trimer formation of gp140(-), which exhibits preferential binding to the neutralizing mAbs b12 and 2G12 with reduced binding to non-neutralizing antibodies (Selvarajah et al., 2008; Yang et al., 2000b; Yang et al., 2002). Immunization of rabbits with the gp140(-)GCN fusion protein elicited neutralizing antibodies of greater potency and breath than did either gp120 or solid-phase proteoliposomes containing a cleavage-defective Env (Grundner et al., 2005). A separate immunization study using mice showed that the gp140(-)T4F fusion was only marginally more effective than gp120 at eliciting neutralizing antibodies (Bower et al., 2006).
Another trimerization domain that has been fused to gp140 is the catalytic chain of aspartate transcarbamoylase (ATCase). The ATCase holoenzyme is composed of 12 polypeptide subunits with 6 catalytic chains (34 kDa each) and 6 regulatory chains (17 kDa each). The catalytic chains (ATC) are packed into 2 trimers and the catalytic center is composed of residues from adjacent chains within a trimer (Helmstaedt, Krappmann, and Braus, 2001). Consequently, ATCase enzyme activity serves as a convenient way to detect trimerization of an Env-ATC fusion protein. Chen et al. reported that fusion of ATC significantly stabilized SIVmac32H gp140(-) trimers and allowed the purification of a homogeneous fusion protein that retained ATCase activity (Chen et al., 2004). In the present study, we show that the ATC polypeptide has a similar stabilizing effect on an HIV-1 gp140 molecule. We then compare the quaternary structure, antigenicity, and immunogenicity of gp140(-) fusion proteins created using the ATC, T4F, and GCN trimerization motifs.
Results
Effect of trimerization motifs on expression of JRCSF gp140
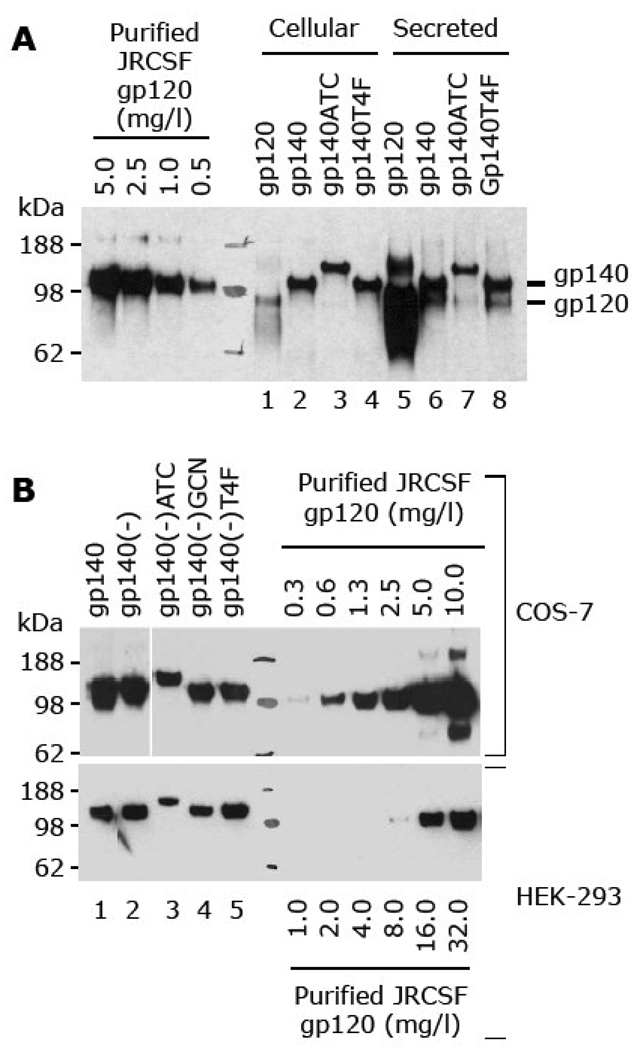
We fused the coding sequences of the T4F and ATC trimerization motifs to the 3’ end of a gene encoding gp140 from the JRCSF strain of HIV-1 as described in Materials and Methods. The expression plasmid constructs were transfected into COS-7 cells along with plasmids expressing JRCSF gp120 and gp140. Western blot analysis showed that both fusion constructs produced roughly equal amounts of intracellular and secreted protein, similar to that of unmodified gp140 (Fig. 1A). Cleavage of gp140 was incomplete in all three cases, and the proportion of gp120 varied, ranging from 10–30% of the uncleaved polypeptide (Fig. 1A, lanes 6–8).
Fig. 1.
Effect of trimerization motifs on JRCSF gp140 and gp140(-) expression. Expression plasmids were transiently transfected into the indicated cells and incubated for three days in serum-free medium. Envelope expression was analyzed by Western blot using a mouse polyclonal anti-gp120 serum (Du et al., 2008). Equal volumes of purified JRCSF gp120 at the indicated concentrations were analyzed as controls on the same gel. Lane numbers are indicated at the bottom of each blot image. (A) Transient transfection of plasmids encoding gp120 (Du et al., 2008), gp140, gp140ATC and gp140T4F in COS-7 cells. Ten µl of each supernatant and an equivalent amount of cellular extract were loaded on SDS-PAGE. (B) Transient transfection of plasmids encoding gp140, gp140(-), gp140(-)ATC, gp140(-)GCN, and gp140(-)T4F in COS-7 and HEK-293 cells. Ten µl of each supernatant were loaded on SDS-PAGE. The concentrations of the purified JRCSF gp120 controls for COS-7 and HEK-293 transfections are given above and below the Western blot images, respectively.
We next compared the effect of the ATC, GCN and T4F trimerization motifs on non-cleavable JRCSF gp140(-). The mutations introduced into the primary and secondary furin cleavage site of gp140(-) were based on those reported previously in the design of uncleaved SF162 gp140 (Srivastava et al., 2003a). Transient transfections were performed using both COS-7 and HEK-293 cells. Western blot analysis showed that the expression levels of gp140 and gp140(-) were similar (Fig. 1B, lanes 1 and 2) and that the mutations introduced into furin cleavage sites were sufficient to prevent cleavage of gp140(-) in both cell types (Fig. 1B). The presence of the T4F and GCN motifs did not affect the expression level of gp140(-), while the ATC polypeptide reduced expression by up to 50% (Fig. 1A, lane 7; Fig. 1B, lane 3).
Based on the above findings, we chose to study fusion proteins based on gp140(-) to avoid potential complications arising from a mixture of cleaved and uncleaved gp140 forms. Since the expression of codon-optimized Env genes was ~2–3-fold higher in HEK-293 cells compared to COS-7 cells (Fig. 1B), we decided to use HEK-293 cells for most of the experiments. However, COS-7 cells were used to transiently express Env genes for partial purification and blue native PAGE (BN-PAGE) analysis since this cell line is easier to handle than HEK-293 cells, particularly at large scale.
Trimerization motifs promote trimer formation of non-cleavable JRCSF gp140
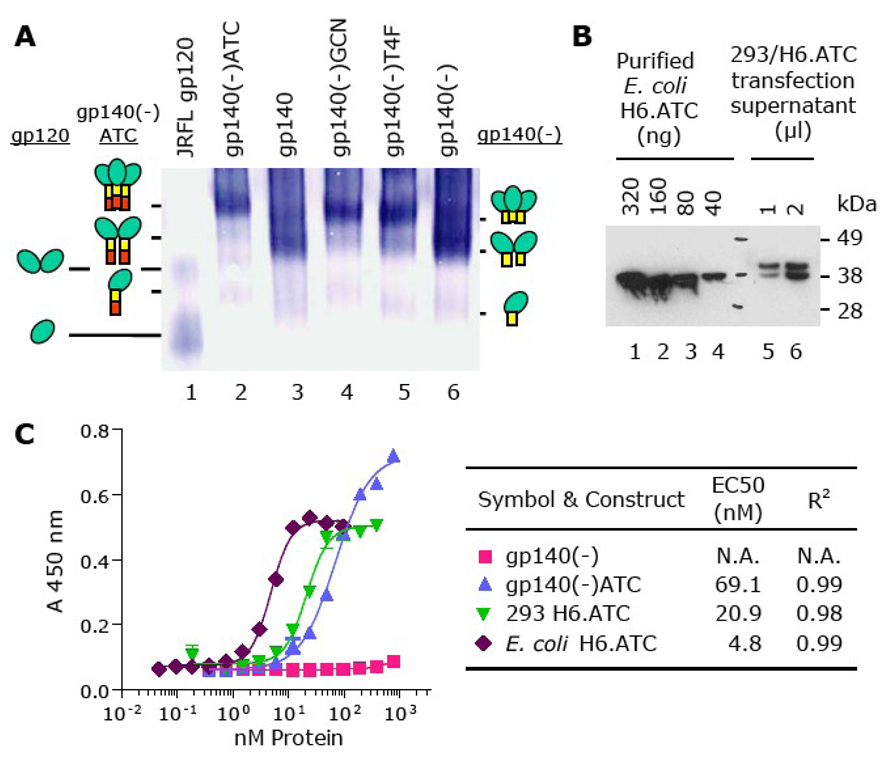
The gp140 variants were transiently expressed in COS-7 cells (Fig. 1B), purified using Galanthus nivalis agglutinin-agarose columns, and separated on BN-PAGE. Fig. 2A shows that both gp140 and gp140(-) consist of a mixture of dimer, trimer and higher oligomers, with dimers being predominant. In contrast, gp140(-)T4F, gp140(-)GCN and gp140(-)ATC were all composed of a predominant trimer population, demonstrating that addition of any of the three motifs to the C-terminus of gp140(-) stabilizes trimers (Fig. 2A, compare lanes 2, 4, and 5 to lanes 3 and 6).
Fig. 2.
Effect of trimerization motifs on oligomerization of gp140(-). (A) BN-PAGE analysis of trimer formation. Env proteins from the COS-7 transfections shown in Fig. 1B were purified by lectin affinity resin and run on BN-PAGE gels. Purified JRFL gp120 (Schulke et al., 2002) was run in parallel as a control. The expected positions of monomers, dimers, and trimers for gp120, gp140(-) and gp140(-)ATC are indicated with cartoons. (B) Expression analysis of H6.ATC. A mammalian expression vector encoding H6.ATC was transiently transfected into HEK-293 cells. The culture supernatant was harvested three days post-transfection and analyzed by Western blot using an anti-histidine tag monoclonal antibody along with H6.ATC purified from E. coli. (C) Assay of ATCase enzymatic activity. The EC50 values were calculated for the H6.ATC protein purified from E. coli, H6.ATC expressed in HEK-293 supernatants (from Fig. 2B), and gp140(-)ATC expressed in HEK-293 supernatants (from Fig. 1B). HEK-293 supernatant containing gp140(-) was used to determine background activity.
Since ATC exhibits enzymatic activity only when the catalytic subunits form a trimer (Chen et al., 2004; Helmstaedt, Krappmann, and Braus, 2001), we further investigated the efficiency of gp140(-)ATC trimer formation using an ATCase enzyme assay. We transfected HEK-293 cells with a plasmid encoding ATC fused to an N-terminal hexahistidine tag (H6.ATC) and analyzed the supernatant on Western blot (Fig. 2B). We found that a potential N-glycosylation site, predicted by the ATC polypeptide sequence, was occupied in more than 50% of HEK-293-expressed H6.ATC (Fig. 2B, compare lanes 1–4 with 5–6). When compared to a recombinant H6.ATC expressed in and purified from E. coli, the HEK-293-expressed H6.ATC showed a ~4-fold reduction in its EC50 (Fig. 2C), possibly due to the steric effects of the N-glycan on the active site. The gp140(-) and gp140(-)ATC constructs were also expressed in HEK-293. Compared to the non-specific background given by gp140(-), the gp140(-)ATC protein exhibited clearly detectable ATCase activity, although the EC50 value was 3.5-fold lower than that of the H6.ATC expressed in HEK-293 cells. The enzymatic activity of the gp140(-)ATC protein confirms that trimers are formed. Since our BN-PAGE results showed that the majority of gp140(-) ATC is trimeric, the relatively low ATCase activity (Fig. 2C) is likely due to the formation of a suboptimal active site.
Antigenicity and CD4-binding activities of various forms of JRCSF gp140
Antigenicity is one of the criteria used to judge to what extent a soluble trimeric gp140 mimics the conformation of functional viral Env complexes. It has been previously observed that functional trimers preferentially bind to broadly neutralizing antibodies such as 2G12 and b12 relative to non-neutralizing antibodies such as b6 (Moore et al., 2006; Poignard et al., 2003; Yang et al., 2002). To analyze the binding activities of the gp140(-) fusion proteins to monoclonal antibodies and CD4-Ig, we transfected HEK-293 cells with plasmids encoding JRCSF gp140, gp140(-), the three gp140(-) trimeric motif constructs, and JRCSF gp120. The expression levels were quantitated using Western blot, the concentrations of each gp120 or gp140 polypeptide were normalized to 75 nM, and equal volumes were used to coat 96-well plates for ELISA.
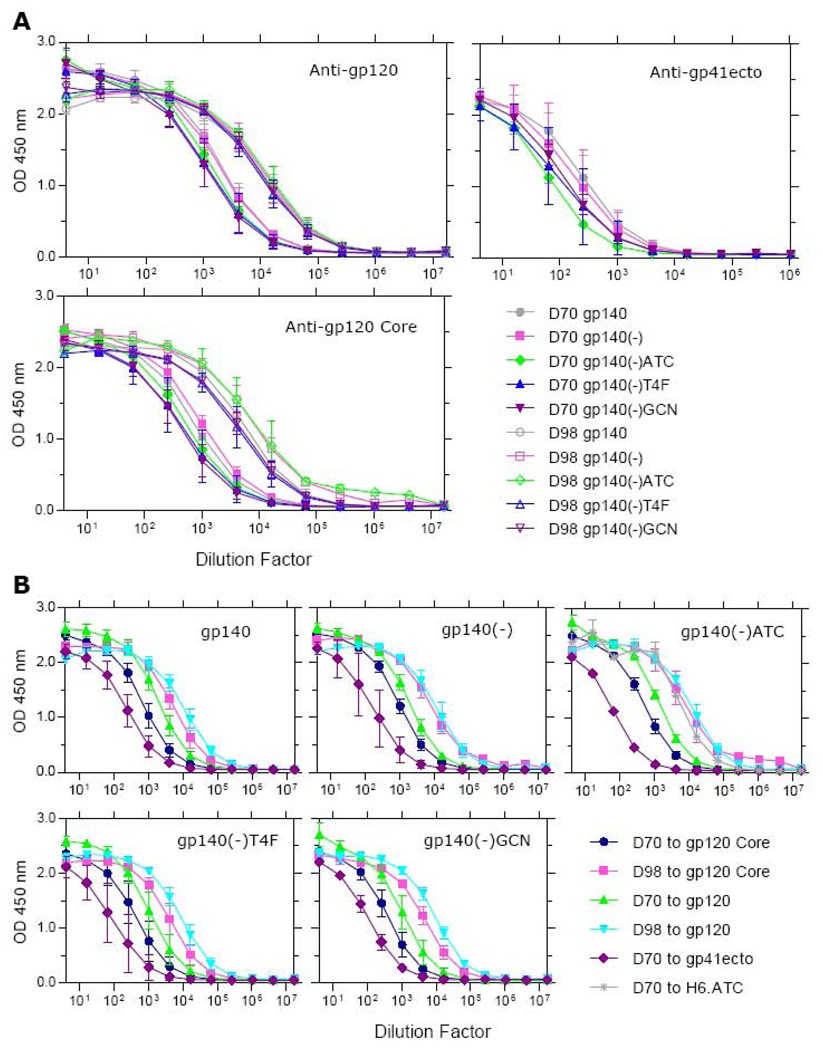
The coating efficiencies of these proteins were determined using a polyclonal antiserum to gp120 called P31JR (Fig. 3). The amounts of gp140, gp140(-), gp140(-)T4F, and gp140(-)GCN protein bound to each well were very similar, whereas that of gp140(-)ATC was somewhat less. The amounts of gp140 bound were substantially lower than that of gp120; binding signals with P31JR were half those of gp120. The effect of non-Env proteins in the culture supernatants on coating was evaluated by comparing a 75 nM sample of purified JRCSF gp120 with the HEK-293 culture supernatant containing the same concentration of gp120. We found that the P31JR binding curves were identical in two independent experiments (data not shown), suggesting that extraneous proteins do not interfere with plate coating in these experiments.
Fig. 3.
Characterization mAb- and CD4-binding activities of gp120, gp140, gp140(-), and three trimeric gp140(-) proteins. Supernatants containing the Env proteins at a concentration of 75 nM were used to coat ELISA plates. The plates were subsequently incubated with ten 1:2 dilutions of 2F5, 2G12, b6, b12, and CD4-Ig. An identical plate was incubated in parallel with ten 1:2 dilutions of the polyclonal P31JR, beginning at 1:400, to determine the Env coating efficiency. Representative data are shown from two independent experiments.
The supernatant-coated plates were probed with serial dilutions of 2F5, 2G12, b6, b12, and CD4-Ig. As shown in Fig. 3, the ligand binding activities of gp140, gp140(-), and the three gp140(-) trimeric motif fusions largely tracked their relative binding of P31JR, suggesting that mutations in the furin cleavage site as well as trimerization did not affect binding of the various monoclonal antibodies or CD4-Ig.
With the exception of gp140(-)ATC, all four gp140 variants bound 2G12 and b12 as well as or better than gp120, whereas they all bound to b6 more weakly than gp120 (Fig. 3). Although gp140(-)ATC bound b12 less strongly than did gp120, possibly due to its low coating efficiency, its binding to b6 was clearly much lower than that of gp120 (Fig. 3). Taken together, our results indicate that all JRCSF gp140 variants show a trend toward stronger binding of the neutralizing antibodies b12 and 2G12 and weaker reactivity with the non-neutralizing antibody b6 antibody.
Antibody responses induced by various forms of JRCSF gp140
We immunized 6 rabbits with the plasmid encoding gp140 and 3 rabbits each with the plasmids encoding gp140(-), gp140(-)T4F, gp140(-)ATC, and gp140(-)GCN followed by a single injection of JRCSF gp120 protein. Sera were collected on Day 70 (two weeks after the third DNA injection) and Day 98 (two weeks after the protein boost) and used to analyze antibody binding to various antigens.
Plasmids expressing the various forms of JRCSF gp140 induced similar levels of antibodies to gp120 at Day 70 (Fig. 4A, top left panel). Although not statistically significant, gp140 and gp140(-) gave slightly better anti-gp120 responses than the three trimerized forms of gp140(-). These small differences were not apparent after the protein boost, which produced a 5–10 fold increase in the level of gp120-binding antibodies.
Fig. 4.
Characterization of antibody responses induced by gp140, gp140(-), and three gp140(-) trimeric constructs. Plasmids encoding the indicated proteins were injected into rabbits at 0, 28, and 56 days followed by a single gp120 protein boost on Day 84. Sera were collected at Day 70 and Day 98 and the antibody responses to JRCSF gp120 and JRCSF gp120 Core were determined by ELISA. Day 70 sera were also used to analyze the antibody responses against the gp41 ectodomain. The Day 70 responses to ATC induced by gp140(-)ATC were also analyzed. The data shown are the average and standard deviation of all animals within each group immunized with the same immunogen. (A) Comparison of total antibody responses induced by different immunogens against each of the coating antigens. The coating antigens used for ELISA analyses are indicated in each panel. (B) Comparison of total antibody responses induced by each immunogen against different coating antigens. The immunogens are indicated in each panel.
Binding of the Day 70 sera was also tested against gp120 Core protein, in which the V1/V2, V3, and about half of the C1 and C5 regions are deleted (Fig. 4A, bottom left panel, closed symbols). We observed a larger spread in the binding activities, compared to gp120. The gp140(-) immunogen induced the highest level of binding antibodies and the gp140(-)GCN immunogen the lowest. The difference in antibody levels induced by gp140(-) and gp140(-)GCN was statistically significant for all five dilution points in linear part of the curves, (P < 0.05). The difference in antibody levels induced by gp140(-) and gp140(-)ATC was also statistically significant for three out of the same five dilutions (P < 0.05).
The gp120 protein boost enhanced the Day 98 antibody responses to gp120 Core by at least 10-fold for all five immunogens (Fig. 4A, bottom left panel, open symbols). In contrast to the responses to gp120, the differences among the anti-gp120 Core responses seen on Day 70 were still observed on Day 98. The strongest anti-gp120 Core response was given by the gp140(-)ATC immunogen, which was boosted by more than 10-fold. These results suggest that modifications of gp140 can affect the regions of gp120 that are immunogenic. Both gp140(-) and gp140(-)ATC might be better immunogens for V1/V2 and V3-independent responses than the other three gp140 variants tested.
We also analyzed the Day 70 sera for antibodies directed to the gp41 ectodomain (Fig. 4A, top right panel, closed symbols). Based on the levels of antibodies induced, the ranking of immunogens is gp140 > gp140(-) > gp140(-)GCN > gp140(-)T4F > gp140(-)ATC. Although most of these differences are rather small, the difference between gp140 (the strongest) and gp140(-)ATC (the weakest) was statistically significant for four out of the five dilution points in the linear part of the curves (P < 0.03, respectively).
To illustrate the differences of the immune responses induced by the several forms of gp140, we plotted the same set of data for each immunogen (Fig. 4B). All five constructs induced clearly detectable immune response to gp120, gp120 Core, and gp41 by Day 70. The responses to gp120 were more than 10-fold stronger than those to gp41 (Fig. 4B); the responses to gp120 Core were several fold weaker than those to gp120 (Fig. 4B). The anti-gp41 responses induced by gp140(-)ATC were very reproducible among the three animals immunized (Fig. 4B, compare top right panel with others). The Day 70 sera from immunization with gp140(-)ATC exhibited a strong antibody response to the ATC moiety (Fig. 4B, top right panel).
After the gp120 protein boost, gp140-primed rabbits continued to have relatively strong responses to gp120 compared to gp120 Core, whereas gp140(-) primed rabbits showed a similar antibody response to gp120 and gp120 Core, suggesting that there is a qualitative difference in the specificity of the antibodies induced between the two types of immunogens (Fig. 4B, compare top left and top middle panels). Among the three trimeric constructs, the gp140(-)T4F and gp140(-)GCN immunogens induced higher antibody levels to gp120 compared to gp120 Core after the gp120 boost (Fig. 4B, compare top middle and top right panels). However, the levels of antibody to gp120 and gp120 Core induced by the gp140(-)ATC immunogen were comparable, and in that respect gp140(-)ATC was similar to gp140(-) (Fig. 4B, compare top middle and top right panels).
Neutralizing antibody responses induced by various forms of JRCSF gp140
We next measured the neutralization activities induced by the various gp140 immunogens against a panel of five pseudoviruses comprising 6535, BaL, JRCSF, NL4-3, and SF162. Protein boosting clearly enhanced neutralizing titers, but the differences among the groups suggested that DNA priming defined the specificities (data not shown). Consequently, only the Day 98 results are shown here because of their higher titers (Fig. 5).
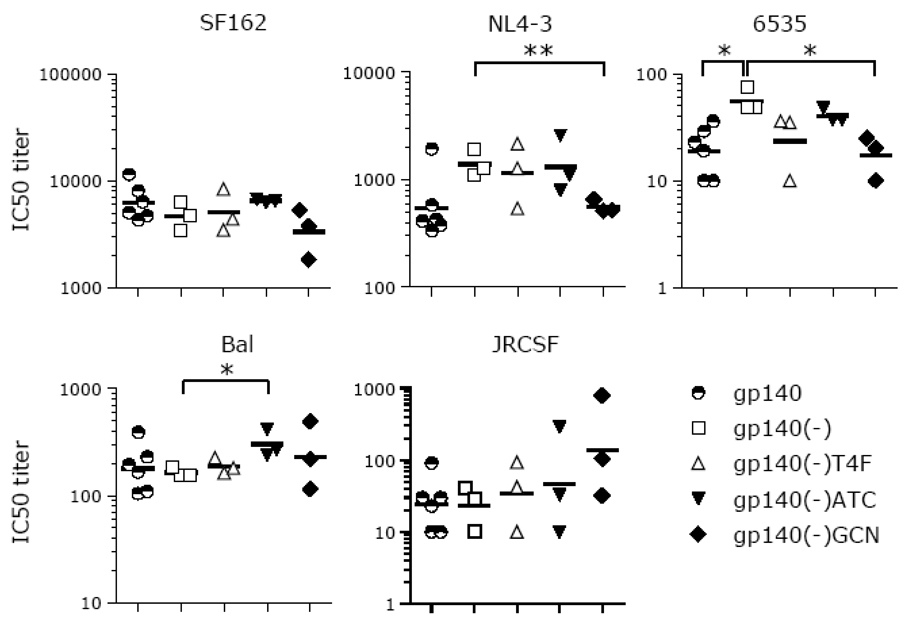
Fig. 5.
Comparison of the neutralization responses induced by gp140, gp140(-), and the three trimeric gp140(-) fusion proteins. The Day 98 sera were tested in neutralization assays against the five indicated pseudoviruses. IC50 neutralization titers were calculated and plotted for each serum-virus pair. The aMLV controls for all sera were negative (IC50 ≤10; not shown). The geometric mean titer for each group of sera is shown as a horizontal bar. For statistical analysis, a two-tailed, two-sample equal variance Student’s t-Test was performed using the log10 of the IC50 values. ** P < 0.01; * 0.01 < P < 0.05.
The cleavable and non-cleavable forms of gp140 induced similar neutralizing responses against three of the five pseudoviruses tested (BaL, JRCSF, and SF162) but gp140(-) gave a significantly stronger neutralization response against 6535 (2.9-fold; P = 0.015) and a stronger trend against NL4-3 (2.5-fold; P = 0.052). The neutralization activities induced by gp140(-)T4F were indistinguishable from those induced by gp140(-) on all five pseudoviruses tested (Fig. 5). The gp140(-)GCN immunogen induced a significantly lower neutralization titer compared to gp140(-) against two out of five pseudoviruses [6535 (3.2-fold; P = 0.019) and NL4-3 (2.4-fold; P = 0.008)] although it gave a strong autologous neutralization response to JRCSF (Fig. 5). Both gp140(-) and gp140(-)ATC elicited equally strong neutralizing responses against 3 out of 5 pseudoviruses tested (6535, NL4-3, and SF162) and gp140(-)ATC gave significantly better neutralization activity against BaL (1.8 fold; P = 0.024) and a stronger trend against JRCSF (4.2-fold; P = 0.402). Overall, there was no significant improvement for any of the trimeric gp140(-) fusion constructs compared to gp140(-).
We performed a second immunization study using the same protocol as above but with eight rabbits per group to evaluate the reproducibility of the results obtained with the constructs expressing gp140, gp140(-), and gp140(-)ATC. The Day 98 sera were assayed against three pseudoviruses, 6535, JRCSF, and NL4-3. Once again, gp140(-) gave a significantly stronger neutralization response against 6535 (1.6-fold; P = 0.018) and a stronger trend against NL4-3 (1.5-fold; P = 0.052) compared to gp140, although both immunogens induced similar neutralization responses against JRCSF. The gp140(-)ATC construct induced neutralizing activity against all three viruses that was equivalent to or greater than that induced by either the cleavable or uncleaved forms of gp140 (Fig. 6). Although not statistically significant, gp140(-)ATC once again induced higher neutralizing titers against JRCSF (3.6-fold; P = 0.173).
Fig. 6.
Comparison of the neutralization responses induced by gp140, gp140(-), and the trimeric gp140(-)ATC fusion protein. Plasmids expressing the indicated proteins were used to immunize groups of eight rabbits three times followed by a single gp120 protein boost. Sera were tested in neutralization assays against the three indicated pseudoviruses. The aMLV controls for all sera were negative (IC50 ≤10; not shown). The geometric mean titer for each group of sera is shown as a horizontal bar. * 0.01 < P < 0.05.
Discussion
An antibody-based preventative vaccine to HIV-1 should induce broad and potent virus neutralizing responses (Burton et al., 2004; Karlsson Hedestam et al., 2008; Montefiori et al., 2007; Walker and Burton, 2008). It has been suggested that the inability of current Env immunogens to elicit broadly neutralizing antibodies is related in part to structural differences between the subunit vaccine component and the functional trimeric Env complexes of the virus (Karlsson Hedestam et al., 2008; Moore et al., 2006; Poignard et al., 2003; Schulke et al., 2002; Yang et al., 2002). It is generally thought that a true mimic of the viral Env trimer should expose neutralizing epitopes including conserved sites such as the one defined by the b12 mAb (Liu et al., 2008; Moore et al., 2006; Poignard et al., 2003; Zanetti et al., 2006). Largely driven by such a hypothesis, considerable effort has been devoted to improving the trimerization of the soluble gp140 ectodomain of Env (Dey et al., 2007; Dey et al., 2008; Iyer et al., 2007; Schulke et al., 2002; Yang et al., 2000b; Yang et al., 2002). Four trimerization motifs (ATC, CART, GCN, and T4F) have been reported to stabilize gp140(-) when fused to the C-terminus of either JRFL, YU2 or SIVmac32H gp140 (Chen et al., 2004; Selvarajah et al., 2008; Yang et al., 2000a; Yang et al., 2002). We have found that GCN, T4F and ATC effectively enhance the trimerization of JRCSF gp140(-). The trimerization of gp140(-)ATC was also supported by the detection of ATCase enzymatic activity (Fig. 2).
Yang et al previously demonstrated that fusion of YU2 gp140(-) to GCN or T4F creates trimers that exhibit preferential binding of both 2G12 and b12 compared to gp120. This binding profile is thought to mimic the antigenicity of functional viral trimers (Yang et al., 2000b; Yang et al., 2002). Similarly, all three of the trimeric JRCSF gp140(-) fusion proteins studied here exhibit a higher ratio of b12 to b6 binding compared to gp120 (Fig. 3). However, these preferential binding profiles might not be the result of improved trimerization of gp140(-) but rather be due to the structural differences between gp120 and gp140(-). The gp140(-) protein, which contains no trimerization domain, also show an antigenic profile similar to that of the gp140(-) fused to trimerization motifs (Fig. 3). Thus, although all three trimerization motifs clearly improved trimer formation of gp140(-), trimerization did not result in a substantial change in the antigenicity of gp140(-) as determined by the binding of 2G12, b6, b12, and CD4-Ig (Fig. 3).
Previous studies using Env proteins from the YU2 and SF162 strains have shown that introduction of mutations at the furin cleavage site between gp120 and the gp41 ectodomain were sufficient to stabilize oligomers, including trimers (Srivastava et al., 2003a; Yang et al., 2000a). However, our BN-PAGE analysis shows that both JRCSF gp140 and gp140(-) have a similar proportion of dimer, trimer and higher order oligomers (Fig. 2A, lanes 3 and 6), suggesting that simply mutating the furin cleavage site is not be sufficient to create trimers. We compared the immunogenicity of gp140 and gp140(-) in two independent rabbit studies and found that in both cases gp140(-) elicited a statistically stronger neutralization response against 6535 and a stronger trend against NL4-3 (Fig. 5 and 6). Therefore, mutation of the furin-cleavage site appears to modestly affect the induction of neutralizing antibodies against certain isolates. Analysis of the Day 70 and Day 98 sera showed that while the gp140 and gp140(-) constructs induced similar binding antibody responses against gp120 and gp41, gp140(-) induced a stronger response to the V1, V2 and V3-deleted gp120 Core (Fig. 4), which could partially account for differences in neutralization activities.
Yang et al. reported that YU2 gp140(-)T4F and gp140(-)GCN are more immunogenic than YU2 gp120 (Yang et al., 2000b; Yang et al., 2002). Such improvements could be due to the improved trimerization of gp140(-) afforded by T4F or GCN but might also be simply due to the inclusion of the gp41 ectodomain (e.g., by providing additional T-helper epitopes). We compared the immunogenicity of JRCSF gp140(-) with or without trimerization motifs using a DNA prime and protein boost protocol. The neutralization profiles of antibodies induced by gp140(-) and gp140(-)T4F were essentially identical while gp140(-)GCN showed reduced neutralization activity to some viruses (Fig. 5). The results suggest that the enhancement of trimer formation is not always accompanied by a beneficial effect on immunogenicity.
The size of a monomeric ATCase catalytic subunit is 34 kDa, and use of this trimerization motif therefore adds 102 kDa to the size of the gp140(-) trimer. This increased size had only a modest effect on expression of either gp140ATC or gp140(-)ATC (Fig. 1). The ATC moiety in gp140(-)ATC elicited a strong antibody response in rabbits (Fig. 4) but this it did not mask the response to gp140(-). Compared to gp140(-)T4F and gp140(-)GCN, gp140(-)ATC induced similar levels of antibodies to gp120 and gp41 as measured by ELISA (Fig. 4). Although gp140(-)ATC induced somewhat weaker responses to gp120, gp120 Core, and gp41 than did gp140(-) on Day 70 (Fig. 4), the response to both gp120 and gp120 Core are indistinguishable; this is similar to the pattern observed with the gp140(-) immunogen (Fig. 4). Moreover, addition of ATC to gp140(-) resulted in a highly reproducible anti-gp41 responses at Day 70 (Fig. 4).
Compared to the sera induced by gp140(-) at Day 98, sera from immunization with gp140(-)ATC exhibited indistinguishable neutralization responses against 6535, NL4-3, and SF162 as well as stronger responses to BaL (1.8-fold; P = 0.024) and JRCSF (4.2-fold; P = 0.402) (Fig. 5). Although not statistically significant, the improved response to JRCSF induced by gp140(-)ATC was observed in a separate study with a larger number of animals per group (3.6-fold; P = 0.173) (Fig. 6). The stronger neutralization response induced by gp140(-)ATC cannot, however, be explained simply by the total antibody levels, which were similar to those induced by gp140(-) on Day 98 (Fig. 4). Taken together, our data suggest that there is a beneficial effect of ATC on the immunogenicity of gp140.
In summary, we conclude that trimer formation of wildtype gp140 sequences has limited value for enhancing the ability of HIV-1 Env to induce broadly neutralizing antibodies. However, we have compared immunogenicity using only uncleavable gp140(-) and we cannot rule out the possibility that the lack of significant improvement found here might not apply to the cleavable form of gp140. A logical next step is to improve immunogenicity by modifying the Env sequences in the gp140ATC trimer. One potential approach is directed molecular evolution, using in vitro DNA recombination to create sequence variants followed by high-throughput screening of the expressed chimeric proteins for desirable antigenic or structural properties (Locher et al., 2004). The ATC motif will allow use of enzymatic activity to identify gp140(-)ATC variants that form trimers. Screening of tens or hundreds of immunogens in animals can potentially identify vaccine candidates that exhibit an improved ability to induce neutralizing antibodies to HIV-1.
Materials and Methods
Expression vectors and JRCSF gp140 expression constructs
The mammalian expression vector pMAmp was used for both cell culture transfection and DNA-based immunization of animals (Du et al., 2008). This vector contains the CMV immediate-early promoter and the corresponding viral intron, a Kozak initiator sequence, a human tissue plasminogen activator signal sequence, an N-terminal hexahistidine tag, and a poly(A) signal from bovine growth hormone. Three in-frame restriction sites, Age I, BsiW I and NgoM IV, were engineered to facilitate cloning of gp140 genes and the coding sequences of trimerization motifs.
The DNA sequence encoding gp140 from the JRCSF strain of HIV-1 was synthesized using codons chosen from highly expressed human genes (Haas, Park, and Seed, 1996). The gp140 gene was cloned into pMAmp using the Age I and BsiW I restriction sites. The coding sequence for non-cleavable gp140(-) was created using an overlap PCR approach to introduce the following mutations at both the primary and secondary furin cleavage sites between gp120 and gp41: K488I, R489S, R490S, E495S, and R497S (Srivastava et al., 2003a).
DNA sequences encoding the trimerization motifs were cloned into pMAmp containing either gp140 or gp140(-) using the BsiW I and NgoM IV sites so that these motifs were fused in frame at the C-terminal end of the Env polypeptide coding sequence. For T4F and GCN (Yang et al., 2000b; Yang et al., 2002), we designed two oligonucleotides for each motif based on the T4 bacteriophage wac gene (GenBank Accession No. NC_000866) and a Saccharomyces cerevisiae GCN4 mRNA (AF416613), respectively. The two oligonucleotides were annealed to form a linker with engineered overhangs for directly cloning into the BsiW I and NgoM IV sites. For the ATC motif, we designed two primers to amplify the coding sequence from genomic DNA of the E. coli K12 strain using a ChargeSwitch® gDNA Mini Bacteria Kit (Invitrogen). The amplified fragment was then digested and cloned into pMAmp containing either gp140 or gp140(-) using the BsiW I and NgoM IV sites.
Cell culture and transient transfection
Human embryonic kidney HEK-293 cells (ATCC No. CRL-1573) and African green monkey kidney fibroblast-like COS-7 cells (ATCC No. CRL-1651) were maintained in Dulbecco's Modified Eagle Medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 100 units/ml of penicillin and 100 µg/ml of streptomycin. Chinese Hamster Ovary-K1 cells (ATCC No. CCL-61) were maintained in a 1:1 mixture of Dulbecco's Modified Eagle Medium / Nutrient Mixture F-12 (DMEM/F-12; Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 100 units/ml of penicillin and 100 µg/ml of streptomycin. All cell cultures were maintained at 37°C in a humidified incubator containing 5% CO2.
Cells were seeded 24 hours before transfection at 50% confluency in the appropriate serum-containing medium. FuGENE 6.0 (Roche Applied Sciences) was used for transient transfection of COS-7 cells and Lipofectamine 2000 (Invitrogen) was used for HEK-293 cells according to the manufacturer's instructions. For expression analysis, both COS-7 and HEK-293 cells were directly transfected in serum-free Opti-MEM I medium (Invitrogen) and incubated for 3 days at 37° C. Supernatants were harvested and clarified by centrifugation at 3,000 rpm (2,500 × g) for 10 minutes. Cellular extracts were prepared using CelLytic-M lysis buffer (Sigma).
Human monoclonal antibodies
The isolation of the b6 and b12 human mAbs has been described (Barbas et al., 1993; Burton et al., 1991; Roben et al., 1994). Both mAbs recognize discontinuous epitopes that overlap the functionally conserved CD4-binding site on gp120 (Pantophlet et al., 2003; Roben et al., 1994). In this study, the IgG1 forms of b6 and b12 were used. The human mAbs 2F5 and 2G12, which recognize the conserved tryptophan-rich MPER of gp41 (Muster et al., 1993; Ofek et al., 2004; Pejchal et al., 2009) and a mannose-dependent epitope on gp120 (Scanlan et al., 2002; Trkola et al., 1996), respectively, were obtained from Polymun Scientific (Vienna, Austria).
Expression and purification of control proteins
The expression and purification of both JRCSF gp120 and gp120 Core from CHO-K1 cells were following the methods described previously (Du et al., 2008). The JRCSF gp120 Core gene was designed based on the HXBc2 gp120 Core construct used for crystallization studies (Kwong et al., 1998; Wyatt et al., 1998).
The coding sequence for ATC was subcloned into pQE-81L (Qiagen) using the BamH I and Hind III sites in frame with the initiation codon and the sequences encoding a N-terminal hexahistidine tag provided by the vector. After transformation into E. coli XL-10 Gold Ultracompetent cells (Stratagene), a single colony was picked and inoculated into 50 ml of 2X YT (Sigma) supplemented with carbenicillin (50 µg/ml) and glucose (0.5%). After incubation overnight at 37°C, 20 ml of the culture was inoculated into 100 ml of 2× YT supplemented with carbenicillin (50 µg/ml) and incubated at 37°C for 1 hour. IPTG was then added to a final concentration of 2 mM and incubation continued for 5 hours at 37°C. Cells were collected by centrifugation at 3,500 rpm (2,500 × g) at 4°C for 20 minutes and suspended in a total of 25 ml PBS (pH 7.4; Invitrogen). The cell suspension was passed three times through a French Pressure cell press (SLM Aminco Instruments) under 1000–1500 psi. Insoluble material was removed by centrifugation at 12,000 rpm (17,210 × g) at 4°C for 20 minutes using a Sorvall SS-34 rotor. The clarified cell lysate was loaded onto a 5 ml Hi-Trap Ni-column (GE Healthcare) freshly charged with Ni++ running on a BioLogic LP Chromatography System (BioRad). After washing with 150 ml of 500 mM NaCl in 20 mM NaH2PO4 (pH 7.4) containing 25 mM imidazole, H6.ATC was eluted with 0.25 M imidazole in the same buffer. The peak protein-containing fractions were buffer-exchanged and concentrated to 10 mg/ml and stored at −80°C. The purity of the final H6.ATC preparation was >95% as judged by SDS-PAGE analysis.
The H6.ATC coding sequences were also cloned into pMAmp for expression by transient expression in mammalian cells.
A CD4-Ig fusion protein was created as described (Byrn et al., 1990) with some modifications. The coding sequence for the first 26–181 amino acids of human CD4 containing the V1J1 and V2J2 domain of mature CD4 (Arthos et al., 1989) was amplified from resting CD14+ monocyte cDNA from a Human Blood Fractions MTC™ Panel (Clontech) and ligated to the 5’ end of the coding sequence of human IgG1 Fc constant region omitting the CH1 domain through an in-frame BstE II site that introduced two amino acids, VT, before proline 212 in the IgG1 hinge. The DNA fragment was cloned into pMAmp in frame with the tissue plasminogen activator leader peptide coding sequence using the Nhe I and Xba I sites. The resultant plasmid was co-transfected with pPUR (Clontech) into CHO-K1. Stable CHO cell lines expressing CD4-Ig were created and expanded to roller bottles for protein production as described previously (Du et al., 2008). For purification, one liter of culture supernatant was supplemented with 0.5 M NaCl, 1 mM of Pefabloc (Sigma), and 10 mM of benzamidine (Sigma). The supernatant was filtered on a 0.22 micron membrane and loaded onto a 5 ml HiTrap Protein A FF column (GE Healthcare) equilibrated with PBS (pH 7.4) using a BioLogic LP system. The column was washed with 80 ml of 1.5 M Glycine-NaOH buffer (pH 9.0) containing 3 M NaCl and eluted with 0.2 M Glycine-HCl buffer (pH 2.5). The eluate was immediately neutralized by 1/10 volume of 1 M Tris-HCl buffer (pH 9.0) that was pre-added to the collection tubes. The peak protein-containing fractions were buffer-exchanged and concentrated to 0.5–1.0 mg/ml and stored at −80°C. The purity of the final CD4-Ig preparation was >90% as judged by SDS-PAGE analysis.
Western blot analysis and quantitation of molar expression
Transfection supernatants were separated on 4–12% SDS-PAGE gels and the gp140 polypeptide was visualized by Western blot using either a mouse polyclonal anti-gp120 serum or an anti-histidine tag monoclonal antibody (GE Healthcare) as described previously (Du et al., 2008). For quantitation of expression, 3–4 different dilutions of each supernatant were compared to 6 serial 1:2 dilutions of purified JRCSF gp120 with a starting concentration of 200 nM on the same Western blot. Each Western blot image was scanned and saved as a non-compressed TIFF file. After the intensities of Env-specific bands were quantitated using Phoretix Array 3.0 (Nonlinear Dynamics), a standard curve was generated from the gp120 controls using non-linear regression curve fitting (GraphPad Prism 5.0) and the molar expression level in nM of gp140 variant in each supernatant was calculated.
Determination of trimer formation by BN-PAGE analysis
JRCSF gp140 variants were transiently expressed in COS-7 cells and purified on lectin columns. For each purification run, a disposable column was prepared using 200 µl of Galanthus nivalis Agglutinin-Agarose 4% resin suspension (Sigma) in a 5 ml polypropylene column with a porous polyethylene disc (Pierce). After resin storage buffer was allowed to drain by gravity, the column was washed three times with PBS (pH 7.4) supplemented with 0.1 mM CaCl2. Fifteen ml of culture supernatant containing gp140 or gp140 fusion proteins were centrifuged at 4,000 rpm for 15 minutes and loaded on the column by gravity. The flow-through was collected and passed over the column one more time. The column was then washed three times with PBS (pH 7.4) supplemented with 0.1 mM CaCl2 and eluted with 1 ml of 1 M α-methyl mannoside (Sigma) in PBS. The eluate was injected into a Dialysis Cassette (Pierce) and dialyzed in PBS at 4°C overnight. The recovered protein was further concentrated 25-fold using Microcon YM-50 centrifugal filter devices (Millipore) and stored at −70°C.
BN-PAGE analysis was performed as described in detail previously (Moore et al., 2006) except that for this study the Western blot steps following BN-PAGE separation were omitted since purified proteins were used. Briefly, protein samples were mixed with an equal volume of 2× sample buffer containing 100 mM morpholinepropanesulfonic acid, 100 mM Tris-HCl, pH 7.7, 40% glycerol, and 0.1% Coomassie Blue and loaded onto a 4–12% Bis-Tris NuPAGE gel (Invitrogen). Ferritin (GE Healthcare) was used as a size standard. Electrophoresis was carried out at 4°C for 3 hours at 100 V with 50 mM morpholinepropanesulfonic acid, 50 mM Tris-HCl, pH 7.7, containing 0.002% Coomassie Blue as cathode buffer and the same buffer without Coomassie Blue as the anode buffer. Gels were destained and dried.
ATCase assays
A 96-well ATCase assay was adapted from an original method (Prescott and Jones, 1969) modified for use with microtiter plates (Else and Herve, 1990). All chemicals used in the assay were purchased from Sigma unless specified otherwise. Since three ATCase catalytic subunits are required to form one enzymatic site (Helmstaedt, Krappmann, and Braus, 2001), the molar concentration used here is that of the trimer, although not all monomers will form trimers. The starting concentrations of purified H6.ATC from E. coli, H6.ATC expressed in HEK-293 supernatant, and gp140(-) or gp140(-)ATC expressed in HEK-293 supernatants were 93, 371 and 741 nM, respectively. A 12-point 1:2 dilution series of each sample (final volume of 50 µl) was prepared in OptiMEM I medium lacking phenol red (Invitrogen) in a 96-well U-bottom MaxiSorp plate (Nalge Nunc International). The assay plate was sealed and pre-incubated at 37°C for 30 minutes. To each well was added 100 µl of freshly prepared substrate buffer containing 15 mM carbamyl phosphate and 30 mM L-aspartate in dilution buffer (40 mM Tris-acetate, 1 mM EDTA, 376 pM BSA, 1 mM mercaptoethanol, pH 8.3). After incubation at 37°C for 30 minutes, 100 µl of freshly prepared color mix (2 parts of 0.5 % (w/v) antipyrine in 3 M H2SO4 and 1 part of 0.8 % (w/v) diacetyl monoxime in 5% (v/v) acetic acid) was added to each well. The plate was immediately sealed and color reactions were developed at 60°C in an ISOTMP forced air incubator (Fisher) for 3 hours. Colorimetric signals were acquired at 450 nm. The EC50 for each sample was calculated in GraphPad Prism 5.0 using the log(agonist) versus response non-linear regression model with variable slope based on triplicate assay results.
ELISA analysis of mAb and CD4 binding activities
Plasmid constructs expressing JRCSF gp120, gp140, and the four gp140(-) variants were transiently transfected into HEK-293 cells. The nM expression levels of Env in the harvested supernatants were quantitated and normalize to 75 nM using Opti-MEM I. Normalized supernatants were used to coat 96-well U-bottom MaxiSorp plates, 50 µl per well, at 4°C overnight. The coated plates were covered with 200 µl of Blocking Buffer (5% non-fat milk in 1× PBS plus 0.5% Tween 20) for 3 hours and then incubated at room temperature for 30 minutes with 1:2 serially diluted antibodies (2F5, 2G12, b6, or b12) or CD4-Ig in the Blocking Buffer. Binding was visualized by incubation at room temperature for 30 minutes with an anti-human IgG conjugated with horseradish peroxidase (1:5,000; Vector), followed by incubation with freshly prepared TMB substrate (Pierce) at room temperature for 10 minutes. Colorimetric signals were acquired at 450 nm. Most of liquid handling procedures were carried out by a Tecan Genesis 100 robotic station.
We first tested a wide range of concentrations for each ligand to determine a proper dilution range: 0.01 to 20.00 µg/ml for 2G12, b6, and b12 and 0.04 to 80.00 µg/ml for 2F5 and CD4-Ig. We chose starting concentration of 0.25, 0.25, 0.50, and 10.00 µg/ml for b6, b12, 2G12, and CD4-Ig, respectively, to show a full range of binding (base-line to plateau) for gp140 variants as well as the gp120 control. For 2F5, the binding activities did not reach plateau even at a concentration of 80 µg/ml, which we chose as the starting concentration.
An additional plate was also prepared for each ELISA run to establish the coating efficiencies of the Env proteins in the supernatants. We used 1:2 dilutions of a polyclonal immune serum to JRCSF gp120, called P31JR, that was made by pooling sera from five rabbits immunized with JRCSF gp120 using the standard DNA electroporation and protein boost protocol. The gp120-binding signals were visualized using the same procedures as described above except that an anti-rabbit IgG conjugated with horseradish peroxidase (1:5,000; GE Healthcare) was used as the secondary antibody.
All data analysis was performed using Microsoft Excel and GraphPad Prism 5.0.
Rabbit immunizations
Selected plasmid constructs expressing gp140 variants were used for mg-scale endotoxin-free DNA preparation and subsequent immunization at Aldevron LLC (Fargo, ND) using protocols approved by the local Animal Care and Use Committee. Certified parasite-free female New Zealand White rabbits were obtained at 8 weeks of age with an average weight of 1.8 – 2.3 kg. After a one-week acclimation period, each rabbit was immunized with a total 400 µg of DNA using electroporation on Days 0, 28, and 56, (200 µg DNA injected in each of the triceps brachii). On Day 84, each rabbit received a total of 100 µg of recombinant JRCSF gp120 in 200 µl of PBS (pH 6.8) mixed with 200 µl of AS02A 2× (Voss et al., 2003) injected intramuscularly (50 µg in each of the hind muscles). Bleeds were obtained at Day 0, at 2 weeks after the third DNA injection (Day 70), and at 2 weeks after protein boosting (Day 98).
ELISA analysis of antibody responses in rabbit sera
Purified JRCSF gp120, JRCSF gp120 Core, HXB2 gp41 ectodomain (Fitzgerald Industries International), and H6.ATC were used to coat 96-well U-bottom MaxiSorp plates, 100 ng in 50 µl per well, at 4°C overnight. The coated plates were then covered with the Blocking Buffer and incubated for 3 hours at room temperature. The plates were washed and incubated at room temperature for 30 minutes with twelve 1:4 serial dilutions of the Day 70 and Day 98 sera in Blocking Buffer at an initial concentration of 1:4. The antibody binding was visualized by incubation at room temperature for 30 minutes with the anti-rabbit IgG conjugated with horseradish peroxidase (1:5,000) followed by incubation with the freshly prepared TMB substrate at room temperature for 10 minutes. Colorimetric signals were acquired at 450 nm.
Virus neutralization assays
The neutralizing activities of rabbit sera were analyzed at Monogram Biosciences, Inc. using a pseudovirus-based neutralization assay as previously described (Frost et al., 2005; Richman et al., 2003). Briefly, serial dilutions of rabbit sera are incubated with individual pseudoviruses for 18 hours at 37°C. The mixtures are used to infect U87 cells expressing CD4 plus the CCR5 and CXCR4 co-receptors. Virus infectivity is determined 72 hours post-inoculation by measuring the amount of luciferase activity expressed in infected cells. Neutralization activity is calculated by plotting the percent inhibition of luciferase activity versus serum or antibody concentration (1/dilution). The percent inhibition is derived as follows: [1 - (luciferase activity in the presence of serum or antibody/luciferase activity in the absence of serum or antibody)] × 100. Inhibition curves defined by the four-parametric sigmoidal function: f(x) = a − [b/(1 + (x/c)d)] are fitted to the data by nonlinear least squares and bootstrapping and used to calculate the serum or antibody concentration required to inhibit virus infectivity by 50%. Neutralization titers are expressed as the reciprocal of the serum dilution conferring 50% inhibition (IC50). A pseudovirus containing a non-HIV retroviral envelope from the amphotropic murine leukemia virus (aMLV) is included in the assay of each serum as a negative control. The IC50 value is considered positive if the percentage of neutralization is 3-fold greater than the aMLV control. Negative sera and sera that failed to reach 50% neutralization are assigned a value of 10, which is the lowest dilution tested for each serum. Two-tailed homoscedastic t-test analyses were performed in Excel.
Acknowledgements
This work was supported by NIH grants P01 AI056375 and R43 AI070032 to RGW. Additional support was provided by NIH grant AI58763 (JMB) and the AIDS and Infectious Disease Science Center at the Torrey Pines Institute for Molecular Studies (JMB). We are grateful to Marni England-Hill, Mariya Rzaszutak, and the Aldevron GIA™ Team for managing and performing the rabbit immunization studies. We thank Michael Zwick and Dennis Burton of The Scripps Research Institute for providing b6 and b12 human monoclonal antibodies and for helpful discussions, Gerald Voss of GSK Biologicals (Rixensart, Belgium) for supplying AS02A adjuvant, and Hermann Katinger from POLYMUN Scientific and the Institute of Applied Microbiology, University of Agricultural Sciences, Vienna, Austria for the 2F5 and 2G12 antibodies. We also thank Madan Paidhungat for providing the coding sequence for human IgG1 Fc polypeptide and Peggy Powers for critical reading of the manuscript.
Abbreviation
- aMLV
amphotropic murine leukemia virus
- ATC
E. coli aspartate transcarbamoylase catalytic subunit
- ATCase
E. coli aspartate transcarbamoylase
- BN-PAGE
blue native polyacrylamide gel electrophoresis
- ELISA
enzyme-linked immunosorbent assay
- Env
HIV-1 envelope glycoprotein
- GCN
a variant form of GCN4 trimeric helices
- gp140(-)
a noncleavable form of gp140 in which the cleavage site between gp120 and the gp41 ectodomain is mutated
- H6.ATC
ATC with an N-terminal hexahistidine tag
- kDa
kilodalton
- mAb
monoclonal antibody
- T4F
the trimerization motif of T4 fibritin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arthos J, Deen KC, Chaikin MA, Fornwald JA, Sathe G, Sattentau QJ, Clapham PR, Weiss RA, McDougal JS, Pietropaolo C, et al. Identification of the residues in human CD4 critical for the binding of HIV. Cell. 1989;57(3):469–481. doi: 10.1016/0092-8674(89)90922-7. [DOI] [PubMed] [Google Scholar]
- Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini LA, Posner MR, Katinger H, Stiegler G, Bernacky BJ, Rizvi TA, Schmidt R, Hill LR, Keeling ME, Lu Y, Wright JE, Chou TC, Ruprecht RM. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6(2):200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- Barbas CF, 3rd, Collet TA, Amberg W, Roben P, Binley JM, Hoekstra D, Cababa D, Jones TM, Williamson RA, Pilkington GR, et al. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230(3):812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- Beddows S, Schulke N, Kirschner M, Barnes K, Franti M, Michael E, Ketas T, Sanders RW, Maddon PJ, Olson WC, Moore JP. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2005;79(14):8812–8827. doi: 10.1128/JVI.79.14.8812-8827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, Wood B, Nathe C, Richman D, Tomaras GD, Bibollet-Ruche F, Robinson JE, Morris L, Shaw GM, Montefiori DC, Mascola JR. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82(23):11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of antihuman immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JF, Li Y, Wyatt R, Ross TM. HIV-1 Envgp140 trimers elicit neutralizing antibodies without efficient induction of conformational antibodies. Vaccine. 2006;24(26):5442–5451. doi: 10.1016/j.vaccine.2006.03.063. [DOI] [PubMed] [Google Scholar]
- Burton DR, Barbas CF, 3rd, Persson MA, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci U S A. 1991;88(22):10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- Byrn RA, Mordenti J, Lucas C, Smith D, Marsters SA, Johnson JS, Cossum P, Chamow SM, Wurm FM, Gregory T, et al. Biological properties of a CD4 immunoadhesin. Nature. 1990;344(6267):667–670. doi: 10.1038/344667a0. [DOI] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300(5628):2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- Carotenuto P, Looij D, Keldermans L, de Wolf F, Goudsmit J. Neutralizing antibodies are positively associated with CD4+ T-cell counts and T-cell function in long-term AIDS-free infection. Aids. 1998;12(13):1591–1600. doi: 10.1097/00002030-199813000-00005. [DOI] [PubMed] [Google Scholar]
- Chakrabarti BK, Kong WP, Wu BY, Yang ZY, Friborg J, Ling X, King SR, Montefiori DC, Nabel GJ. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol. 2002;76(11):5357–5368. doi: 10.1128/JVI.76.11.5357-5368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Cheng Y, Calder L, Harrison SC, Reinherz EL, Skehel JJ, Wiley DC. A chimeric protein of simian immunodeficiency virus envelope glycoprotein gp140 and Escherichia coli aspartate transcarbamoylase. J Virol. 2004;78(9):4508–4516. doi: 10.1128/JVI.78.9.4508-4516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey AK, David KB, Klasse PJ, Moore JP. Specific amino acids in the N-terminus of the gp41 ectodomain contribute to the stabilization of a soluble, cleaved gp140 envelope glycoprotein from human immunodeficiency virus type 1. Virology. 2007;360(1):199–208. doi: 10.1016/j.virol.2006.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey AK, David KB, Ray N, Ketas TJ, Klasse PJ, Doms RW, Moore JP. N-terminal substitutions in HIV-1 gp41 reduce the expression of non-trimeric envelope glycoproteins on the virus. Virology. 2008;372(1):187–200. doi: 10.1016/j.virol.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SX, Xu L, Viswanathan S, Whalen RG. Inhibition of V3-specific cleavage of recombinant HIV-1 gp120 produced in Chinese hamster ovary cells. Protein Expr Purif. 2008;59(2):223–231. doi: 10.1016/j.pep.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Earl PL, Broder CC, Long D, Lee SA, Peterson J, Chakrabarti S, Doms RW, Moss B. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol. 1994;68(5):3015–3026. doi: 10.1128/jvi.68.5.3015-3026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Else AJ, Herve G. A microtiter plate assay for aspartate transcarbamylase. Anal Biochem. 1990;186(2):219–221. doi: 10.1016/0003-2697(90)90069-l. [DOI] [PubMed] [Google Scholar]
- Frost SD, Liu Y, Pond SL, Chappey C, Wrin T, Petropoulos CJ, Little SJ, Richman DD. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J Virol. 2005;79(10):6523–6527. doi: 10.1128/JVI.79.10.6523-6527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundner C, Li Y, Louder M, Mascola J, Yang X, Sodroski J, Wyatt R. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology. 2005;331(1):33–46. doi: 10.1016/j.virol.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Grundner C, Pancera M, Kang JM, Koch M, Sodroski J, Wyatt R. Factors limiting the immunogenicity of HIV-1 gp120 envelope glycoproteins. Virology. 2004;330(1):233–248. doi: 10.1016/j.virol.2004.08.037. [DOI] [PubMed] [Google Scholar]
- Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6(3):315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- Helmstaedt K, Krappmann S, Braus GH. Allosteric regulation of catalytic activity: Escherichia coli aspartate transcarbamoylase versus yeast chorismate mutase. Microbiol Mol Biol Rev. 2001;65(3):404–421. doi: 10.1128/MMBR.65.3.404-421.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera C, Klasse PJ, Michael E, Kake S, Barnes K, Kibler CW, Campbell-Gardener L, Si Z, Sodroski J, Moore JP, Beddows S. The impact of envelope glycoprotein cleavage on the antigenicity, infectivity, and neutralization sensitivity of Env-pseudotyped human immunodeficiency virus type 1 particles. Virology. 2005;338(1):154–172. doi: 10.1016/j.virol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, Burton DR. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009 doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann-Lehmann R, Vlasak J, Rasmussen RA, Smith BA, Baba TW, Liska V, Ferrantelli F, Montefiori DC, McClure HM, Anderson DC, Bernacky BJ, Rizvi TA, Schmidt R, Hill LR, Keeling ME, Katinger H, Stiegler G, Cavacini LA, Posner MR, Chou TC, Andersen J, Ruprecht RM. Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian-human immunodeficiency virus challenge. J Virol. 2001;75(16):7470–7480. doi: 10.1128/JVI.75.16.7470-7480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu SL, Stamatatos L. Prospects of HIV Env modification as an approach to HIV vaccine design. Curr HIV Res. 2007;5(6):507–513. doi: 10.2174/157016207782418542. [DOI] [PubMed] [Google Scholar]
- Iyer SP, Franti M, Krauchuk AA, Fisch DN, Ouattara AA, Roux KH, Krawiec L, Dey AK, Beddows S, Maddon PJ, Moore JP, Olson WC. Purified, proteolytically mature HIV type 1 SOSIP gp140 envelope trimers. AIDS Res Hum Retroviruses. 2007;23(6):817–828. doi: 10.1089/aid.2006.0261. [DOI] [PubMed] [Google Scholar]
- Karlsson Hedestam GB, Fouchier RA, Phogat S, Burton DR, Sodroski J, Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Microbiol. 2008;6(2):143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, Mascola JR. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13(9):1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locher CP, Heinrichs V, Apt D, Whalen RG. Overcoming antigenic diversity and improving vaccines using DNA shuffling and screening technologies. Expert Opin Biol Ther. 2004;4(4):589–597. doi: 10.1517/14712598.4.4.589. [DOI] [PubMed] [Google Scholar]
- Mascola JR. Defining the protective antibody response for HIV-1. Curr Mol Med. 2003;3(3):209–216. doi: 10.2174/1566524033479799. [DOI] [PubMed] [Google Scholar]
- Montefiori D, Sattentau Q, Flores J, Esparza J, Mascola J. Antibody-based HIV-1 vaccines: recent developments and future directions. PLoS Med. 2007;4(12):e348. doi: 10.1371/journal.pmed.0040348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006;80(5):2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67(11):6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Igarashi T, Haigwood N, Sadjadpour R, Plishka RJ, Buckler-White A, Shibata R, Martin MA. Determination of a statistically valid neutralization titer in plasma that confers protection against simian-human immunodeficiency virus challenge following passive transfer of high-titered neutralizing antibodies. J Virol. 2002;76(5):2123–2130. doi: 10.1128/jvi.76.5.2123-2130.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78(19):10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332(1):145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77(1):642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Gach JS, Brunel FM, Cardoso RM, Stanfield RL, Dawson PE, Burton DR, Zwick MB, Wilson IA. A conformational switch in HIV gp41 revealed by the structures of overlapping epitopes recognized by neutralizing antibodies. J Virol. 2009 doi: 10.1128/JVI.00685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim AK, Pantaleo G, Cohen OJ, Fink LM, Zhou JY, Zhou JT, Bolognesi DP, Fauci AS, Montefiori DC. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J Infect Dis. 1997;176(4):924–932. doi: 10.1086/516508. [DOI] [PubMed] [Google Scholar]
- Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, Hu D, Tappero JW, Choopanya K. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194(12):1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- Poignard P, Moulard M, Golez E, Vivona V, Franti M, Venturini S, Wang M, Parren PW, Burton DR. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J Virol. 2003;77(1):353–365. doi: 10.1128/JVI.77.1.353-365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poignard P, Saphire EO, Parren PW, Burton DR. gp120: Biologic aspects of structural features. Annu Rev Immunol. 2001;19:253–274. doi: 10.1146/annurev.immunol.19.1.253. [DOI] [PubMed] [Google Scholar]
- Prescott LM, Jones ME. Modified methods for the determination of carbamyl aspartate. Anal Biochem. 1969;32(3):408–419. doi: 10.1016/s0003-2697(69)80008-4. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100(7):4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roben P, Moore JP, Thali M, Sodroski J, Barbas CF, 3rd, Burton DR. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68(8):4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76(17):8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293(5532):1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83(2):757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol. 2002;76(14):7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulke N, Vesanen MS, Sanders RW, Zhu P, Lu M, Anselma DJ, Villa AR, Parren PW, Binley JM, Roux KH, Maddon PJ, Moore JP, Olson WC. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol. 2002;76(15):7760–7776. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarajah S, Puffer BA, Lee FH, Zhu P, Li Y, Wyatt R, Roux KH, Doms RW, Burton DR. Focused dampening of antibody response to the immunodominant variable loops by engineered soluble gp140. AIDS Res Hum Retroviruses. 2008;24(2):301–314. doi: 10.1089/aid.2007.0158. [DOI] [PubMed] [Google Scholar]
- Sharma VA, Kan E, Sun Y, Lian Y, Cisto J, Frasca V, Hilt S, Stamatatos L, Donnelly JJ, Ulmer JB, Barnett SW, Srivastava IK. Structural characteristics correlate with immune responses induced by HIV envelope glycoprotein vaccines. Virology. 2006 doi: 10.1016/j.virol.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Sharon M, Kessler N, Levy R, Zolla-Pazner S, Gorlach M, Anglister J. Alternative conformations of HIV-1 V3 loops mimic beta hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure (Camb) 2003;11(2):225–236. doi: 10.1016/s0969-2126(03)00011-x. [DOI] [PubMed] [Google Scholar]
- Shibata R, Igarashi T, Haigwood N, Buckler-White A, Ogert R, Ross W, Willey R, Cho MW, Martin MA. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5(2):204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- Sougrat R, Bartesaghi A, Lifson JD, Bennett AE, Bess JW, Zabransky DJ, Subramaniam S. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog. 2007;3(5):e63. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava IK, Stamatatos L, Kan E, Vajdy M, Lian Y, Hilt S, Martin L, Vita C, Zhu P, Roux KH, Vojtech L, D CM, Donnelly J, Ulmer JB, Barnett SW. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J Virol. 2003a;77(20):11244–11259. doi: 10.1128/JVI.77.20.11244-11259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava IK, VanDorsten K, Vojtech L, Barnett SW, Stamatatos L. Changes in the immunogenic properties of soluble gp140 human immunodeficiency virus envelope constructs upon partial deletion of the second hypervariable region. J Virol. 2003b;77(4):2310–2320. doi: 10.1128/JVI.77.4.2310-2320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS. 2008 Report on the global AIDS epidemic Executive summary. 2008 July; 2008. [Google Scholar]
- Voss G, Manson K, Montefiori D, Watkins DI, Heeney J, Wyand M, Cohen J. Prevention of disease induced by a partially heterologous AIDS virus in rhesus monkeys by using an adjuvanted multicomponent protein vaccine. J Virol. 2003;77(2):1049–1058. doi: 10.1128/JVI.77.2.1049-1058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BD, Burton DR. Toward an AIDS vaccine. Science. 2008;320(5877):760–764. doi: 10.1126/science.1152622. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Yang X, Farzan M, Wyatt R, Sodroski J. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2000a;74(12):5716–5725. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Florin L, Farzan M, Kolchinsky P, Kwong PD, Sodroski J, Wyatt R. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J Virol. 2000b;74(10):4746–4754. doi: 10.1128/jvi.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76(9):4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2006;2(8):e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PF, Cham F, Dong M, Choudhary A, Bouma P, Zhang Z, Shao Y, Feng YR, Wang L, Mathy N, Voss G, Broder CC, Quinnan GV., Jr Extensively cross-reactive anti-HIV-1 neutralizing antibodies induced by gp140 immunization. Proc Natl Acad Sci U S A. 2007;104(24):10193–10198. doi: 10.1073/pnas.0608635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441(7095):847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75(22):10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]