Abstract
The TGF-β isoforms, TGF-β1, -β2, and -β3, share greater than 70% sequence identity and are almost structurally identical. TGF-β2 differs from the others, however, in that it binds the TGF-β type II receptor (TβR-II) with much lower affinity than either TGF-β1 or -β3. It has been previously shown that three conserved interfacial residues, Arg25, Val92, Arg94, in TGF-β1 and -β3 are responsible for their high-affinity interaction with TβR-II. In this study, the role of each of these residues was examined by creating single, double, and triple substitutions resulting in both TGF-β3 loss-of-function and TGF-β2 gain-of-function variants. One-dimensional 1H NMR spectra of the variants confirmed a lack of large structural perturbations. Affinities, kinetics, and thermodynamics for TβR-II binding were determined by surface plasmon resonance biosensor analysis. Double substitutions revealed that nearly all of the high-affinity binding is contributed by Arg25 and Arg94. Single site substitutions showed that Arg94 makes the greatest contribution. Substitution of Arg25 and Arg94 with alanine verified the requirement of the arginine guanidinium functional groups for the highly specific hydrogen-bonded ion pairs formed between Arg25 and Arg94 of TGF-β1 and -β3, and Glu119 and Asp32 of TβR-II. Further kinetic and thermodynamic analyses confirmed that Arg25 and Arg94 are primarily responsible for high-affinity binding and also revealed that noninterfacial longer range effects emanating from the TGF-β structural framework contribute slightly to TβR-II binding. Growth inhibition assays showed that binding changes generally correlate directly with changes in function; however, a role Val92 in this cellular context was uncovered.
Transforming growth factor β (TGF-β)1 isoforms are 25 kDa homodimeric polypeptide signaling ligands that control multiple cellular processes including proliferation, extracellular matrix deposition, and epithelial–mesenchymal transition (1). TGF-βs and other structurally related members of the TGF-β superfamily, such as activins, bone morphogenic proteins (BMPs), and growth and differentiation factors (GDFs), exert their biological effects by binding and bringing together two pairs of structurally similar, single-pass trans-membrane receptors, known as the type I and type II receptors, TβR-I and TβR-II (2). The ligand-mediated assembly of these two receptor types into a signaling complex initiates an intracellular phosphorylation cascade, wherein the constitutively active type II kinase transphosphorylates the type I kinase in a conserved glycine-serine repeat, known as the GS box (3). This activates the type I kinase which in turn phosphorylates receptor-regulated Smad proteins, which complex with the common mediator Smad, Smad4, and translocate to the nucleus where they interact with various coactivators and corepressors to regulate transcription of target genes (4).
There are three isoforms of TGF-β, termed TGF-β1, -β2, and -β3. They are present in most metazoans, including frogs, birds, and mammals, and they are highly conserved, sharing 70–80% sequence identity in most organisms. The isoforms exhibit somewhat overlapping spatial and temporal patterns of expression throughout development, yet in spite of this they appear to carry out distinct roles in vivo, as suggested by the severe and nonoverlapping phenotypes of the TGF-β isoform specific null mice (5-7).
All three TGF-β isoforms transduce their signals by binding TβR-I and TβR-II (8). However, TGF-β2 differs from TGF-β1 and TGF-β3 in that it binds TβR-II with an affinity that is 100–1000-fold lower (9-11). The reduced affinity of this ligand isoform for TβR-II underlies the lower biological activity that it exhibits in proliferation assays in cells that lack the coreceptor, β-glycan; i.e., in these assays the EC50 of TGF-β2 is 100–500-fold higher than that of TGF-β1 and -β3. In contrast, cells that express the coreceptor in addition to the type I and type II receptors respond roughly equally to the three isoforms. This results from the ability of β-glycan to bind TGF-β2 with a higher affinity than TGF-β1 and -β3 (12) and to promote the assembly of the TβRI–TβR-II signaling complex with TGF-β2 (13).
Important unanswered questions regarding TGF-β2 concern the underlying basis for its reduced affinity for TβR-II and the mechanism by which β-glycan promotes assembly of the signaling complex with TGF-β2. Important clues to the reduced affinity were derived from the crystal structure of the TβR-II–TGF-β3 complex which showed that the interfacial residues of TGF-β3 are identical to those in the other high-affinity isoform, TGF-β1, but are conservatively substituted at three positions in TGF-β2 (Arg25, Val92, and Arg94 in TGF-βs 1 and 3; Lys25, Ile92, and Lys94 in TGF-β2) (14). The crystal structures of the TβR-II–TGF-β3 binary complex, as well as the TβR-I–TβR-II–TGF-β3 ternary complex (15), also showed that the guanidinium groups of Arg25 and Arg94 in TGF-β3 each form doubly hydrogen bonded ion pairs with the carboxylate groups of Glu119 and Asp32, respectively, on TβR-II (Figure 1). In other ligands of the TGF-β superfamily, these two positions are substituted with residues other than arginine and lysine, which is thought to account in part for the distinct manner by which they bind their type II receptors (16, 17). In a previous report, we demonstrated that substitution of the three variable interfacial residues in TGF-β2 to those from TGF-β1 and -β3 resulted in a binding affinity comparable to that of TGF-β1 and -β3 (18). These three residues therefore underlie the reduced affinity of TGF-β2 for TβR-II.
Figure 1.
Key residues of the TβR-II–TGF-β3 interaction. The binding interface of TβR-II (red) and TGF-β3 (blue) based on the X-ray crystal structure (PDB 1KTZ) (14) is shown. Hydrogen bonds are indicated as green dashes. The two doubly hydrogen bonded ion pairs, Asp32:Arg94 and Glu119:Arg25, anchor opposite sides of the hydrophobic interface. Both acidic residues of TβR-II that directly interact with TGF-β3 are themselves stabilized by hydrogen bonds. In particular, Asp32 is stabilized by a complex hydrogen bond network involving Tyr85, Glu75, and water 207. Phe30 contacts and orients Arg94. Glu119 is stabilized by a hydrogen bond from the hydroxyl group of Thr51. Ile53 is a key residue of the hydrophobic interface and makes contacts with Val92 and Trp32.
The objective of the present study was to further characterize the importance of the three variable residues in the binding of TβR-II to the three TGF-β isoforms. To determine the contribution of each, we generated the TGF-β3 loss-of-function variants R25K, V92I, and R94K, as well as the TGF-β2 gain-of-function variants K25R, I92V, K94R, K25R/I92V, I92V/K94R, K25R/K94R, and K25R/I92V/K94R, and measured their affinities, kinetics, and thermodynamics for binding TβR-II using a surface plasmon resonance (SPR) biosensor. The results show that arginine residues 25 and 94 contribute most of the high-affinity interaction between TGF-β3 and TβR-II, with the affinity of the TGF-β2-K25R/I92V/K94R (TGF-β2-TM) and TGF-β2-K25R/K94R variants approaching that of TGF-β3. To further examine the role of these two doubly hydrogen bonded pairs, we individually substituted Arg25 and Arg94 in TGF-β3 with alanine. The affinities of these two variants for TβR-II were only marginally lower than those of the conservative lysine-substituted TGF-β3 loss-of-function variants, confirming that the doubly hydrogen bonded ion pairs formed in the TβR-II–TGF-β3 (and TβR-II–TGF-β1) interfaces are highly specific and are required for the high-affinity interaction with TβR-II. Further kinetic, thermodynamic, and functional analyses confirmed that Arg25 and Arg94 are primarily responsible for high-affinity binding but also revealed a subtle role for the ligand structural framework in TβR-II binding, as well as a role for Val92 in cellular signaling. The information presented here, in particular the relative importance of the two arginine residues, may aid in the development of novel structure-based TGF-β inhibitors.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human TGF-β2, human TGF-β3, and variants, as well as human TβR-II and variants, which include the first 136 residues of the mature extracellular domain, were expressed, refolded, and purified from Escherichia coli inclusion bodies as previously described (19, 20).
SPR Instrumentation and Supplies
CM-5 sensor chips, Tween 20, and the Biacore amine coupling kit were purchased from GE Healthcare Bio-Sciences Corp. (Piscataway, NJ). HEPES buffer, EDTA, and NaCl were purchased from Sigma-Aldrich (Oakville, Ontario, Canada). All surface plasmon resonance assays were carried out using a Biacore 3000 instrument (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) and HBS running buffer (10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA, and 0.005% Tween 20 at pH 7.4) at a flow rate of 25 μL/min and a temperature of 25 °C.
SPR Biosensor Kinetic Assays
TGF-β ligands were covalently immobilized to a Biacore CM-5 chip using standard N-hydroxysuccinimide (NHS) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) amine-coupling methodology (18). Surface TGF-β densities were used that yielded 100-200 resonance unit (RU) responses upon injection of 500 nM TβR-II. Initially, three instrument primes were followed by five 50 μL buffer blank injections with a 10 μL HCl mock regeneration. Samples (50 μL) of TβR-II were injected (300 s dissociation), and the surface was regenerated with 10 μL of 10 mM HCl followed by the default injection needle cleaning procedure. One duplicate (500 nM) was included per run to verify reproducibility, and three independent runs were performed, each consisting of a 2-fold dilution series of a 500 nM TβRII stock solution. Each set was injected in a random order and included two buffer blank injections. The resulting sensorgrams were aligned and double referenced using a mock-activated surface and blank buffer injections. Kinetic data were evaluated by globally fitting the sensorgrams to a heterogeneous ligand model using the Biacore software BiaEvaluation version 4.1 (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). The equilibrium KDs were determined from the resulting kinetic association and dissociation rates (kd/ka), and a minimum of three independent runs were used to generate the reported standard deviations.
SPR Biosensor Steady-State Assays
TGF-β ligands were covalently immobilized as described for the SPR biosensor kinetic assays. Initially, three instrument primes were followed by five 50 μL buffer blank injections with a 10 μL 10 mM HCl mock regeneration. Samples (25 μL) of TβR-II were injected (30 s dissociation), and the surface was regenerated with 10 μL of 10 mM HCl followed by the default injection needle cleaning procedure. Each run consisted of a dilution series of TβR-II with a concentration range from 10 to 0.13 μM. These TβR-II samples were randomly injected along with two buffer blanks over TGF-β variants immobilized at approximately 500 RUs. A maximum concentration of 10 μM was used to conserve sample and to minimize bulk-shift artifacts that can occur when high concentrations of TβR-II are injected over the surface. Sensorgrams were double referenced, as with the kinetic assays, and average plateau RU values for each injection curve were determined using the average fit function in BiaEvaluation. The resulting RU values were used to generate saturation plots, and the KD from each was determined using the steady-state affinity fit in BiaEvaluation (one site ligand). The KDs from a minimum of three independent runs were used to generate the reported standard deviations.
Determination of Thermodynamic Parameters Using an SPR Biosensor Assay
The same procedure as described for the SPR biosensor kinetic assay was carried out at five different temperatures (5, 11, 18, 25, and 32 °C) for TGF-β3 and the TGF-β2-TM. Each injection series at a new temperature was preceded by a glycerol normalization procedure. The equilibrium enthalpy and entropy changes, ΔH and ΔS, respectively, were determined by linear regression analysis of a van't Hoff plot (ln KD vs 1/T) using the equation ln KD = ΔH/(RT) – ΔS/R, where R is the universal gas constant and T is the temperature in kelvin. A linear plot indicates that the equilibrium enthalpy and entropy values remain constant over the temperature range studied. Since the plots were essentially linear in this study, it was assumed that these parameters were independent of temperature. Similarly, the quasi-thermodynamic parameters for the association and dissociation steps, ΔH‡ and ΔS‡, were estimated using a transition state theory-based approach, i.e., Eyring plots (ln(kh/TkB) vs 1/T) using the equation ln(k/T) = −ΔH‡/(RT) + ln(kB/h) + ΔS‡/R, where kB is the Boltzman constant, h is the Plank constant, T is degrees kelvin, and k is either the forward or reverse kinetic rate constant for the TβR-II-TGF-β interaction. The change in free energy at 298 K, ΔG values, for equilibrium binding or for the association and dissociation steps was determined from the equation ΔG = ΔH – TΔS.
Cell Growth Inhibition Assay
The assay was performed as previously described with minor modifications as noted (11). Briefly, 24-well plates were seeded with 25000 cells in 1 mL of DMEM containing 10% FBS (Wisent Inc., St-Bruno, Quebec, Canada) and 100 μg/mL endothelial mitogen (EM) (Biomedical Technologies Inc., Stoughton, MA) and allowed to grow for 24 h at 37 °C. After this time, fresh medium lacking EM was added, and TGF-β2 or variants were added to 31.3 pM final concentration in quadruplicate. The cells were incubated for 48 h total. At 40 h the cells were pulsed with 0.5 μCi of [methyl-3H]deoxythymidine (Amersham Inc., Piscataway, NJ) for 8 h to measure DNA synthesis. Cells were washed, trypsinized, and added to 5 mL of UniverSol scintillation fluid (MP Biomedicals Inc., Solon, OH), and incorporated [methyl-3H]deoxythymidine was measured using a LKB 1219 Rackbeta liquid scintillation counter (PerkinElmer Life and Analytical Sciences, Wood-bridge, Ontario, Canada).
One-Dimensional 1H NMR Spectra
One-dimensional 1H NMR spectra of TGF-β2 and TGF-β3 variants were acquired at a 1H frequency of 500 MHz and 45 °C using a Bruker (Billerica, MA) Avance500 spectrometer equipped with a 5 mm 1H–{13C,15N} triple-axis gradient probe. Samples were 0.05–0.1 mM and were prepared in 10 mM phosphate and 5% 2H2O at pH 3.80. Spectra were acquired using a watergate solvent suppression scheme (21) with 1024–4096 scans and a 1 s recycle time. Spectra were processed by apodizing the FID with a 90° shifted squared sine bell shifted, by applying a solvent convolution filter, and by baseline correcting the resulting spectra with a third-order polynomial.
RESULTS
Comparison of TGF-β2-TM to TGF-β2 and TGF-β3
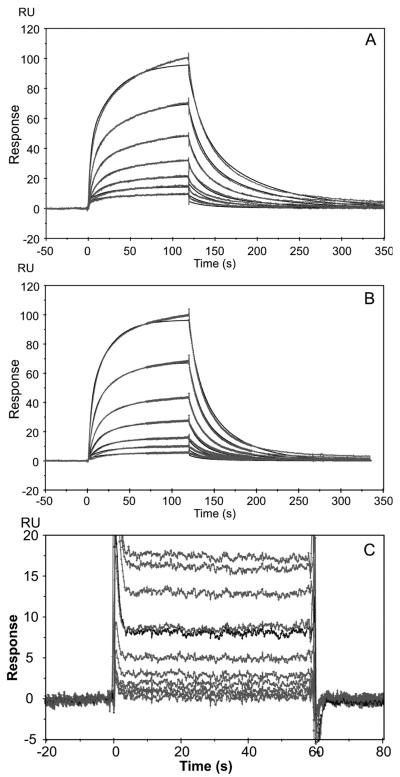
Previous analysis of the gain-of-function triple mutant TGF-β2-TM revealed that the three conserved TGF-β1 and -β3 residues, Arg25, Val92, and Arg94, when substituted into TGF-β2, imparted it with TGF-β3-like affinity for binding TβR-II (18). In that study, the binding affinity information for TGF-β2-TM was derived from steady-state SPR biosensor measurements, which yielded the equilibrium dissociation constant, KD, but not the kinetic association (ka) and dissociation (kd) rate constants. Here, we reassessed the binding characteristics of the TGF-β2-TM variant using kinetic analysis with global fitting of the data to a heterogeneous ligand model. The requirement for this type of model was previously demonstrated for TGF-β1 and -β3 and was attributed to the nonoriented immobilization of TGF-β onto the biosensor surface (22). This results in the generation of high- and low-affinity binding sites, with the high-affinity site (low KD) representing the unimpeded association of TβR-II with TGF-β. Accordingly, the values corresponding to the high-affinity site are presented as apparent KDs throughout the rest of this study. Global fit analysis of the TGF-β2-TM sensorgrams yielded a KD of approximately 100 nM for the high-affinity site (Figure 2 and Table 1), consistent with the previous steady-state measurements (18). The KD determined by global fitting of the data for TGF-β3 (73 nM, Figure 2 and Table 1) was very similar to that of TGF-β2-TM. However, there were slight differences in the kinetics of the interaction of TβR-II with TGF-β3 versus TGF-β2-TM, with TGF-β3 exhibiting a slightly slower dissociation rate (5.4 × 10−2 versus 1.1 × 10−1) and a slightly slower association rate (7.4 × 105 versus 1.1 × 106), which compensate each other resulting in similar affinities for TGF-β3 and TGF-β2-TM.
FIGURE 2.
Kinetic and steady-state analysis of TGF-β3, TGF-β2, and TGF-β2-TM. Representative sensorgrams of TβR-II interacting with immobilized TGF-β3 (A), immobilized TGF-β2-TM (B), or immobilized TGF-β2 (C). The global fit of the heterogeneous ligand model to the sensorgrams is shown in (A) and (B) as a thin solid line.
Table 1.
Summary of Kinetic and Thermodynamic Constants for TGF-β2 and TGF-β3 Variants Interacting with TβRIIa
| protein variant | KD (nM) | ka (M−1 s−1) | kd (s−1) | ΔΔGbind(β2 variants) (kcal/mol) |
|---|---|---|---|---|
| TGF-β2 | (1.7 ± 0.041) × 104 | 0.0 | ||
| TGFβ2-I92V | (1.7 ± 0.31) × 104 | 0.0 | ||
| TGFβ2-K25R | (1.1 ± 0.14) × 104 | −0.25 | ||
| TGFβ2-K94R | (6.0 ± 0.49) × 103 | −0.60 | ||
| TGFβ2-K25R/I92V | (1.1 ± 0.078) × 104 | −0.26 | ||
| TGFβ2-K25R/K94R | (1.0 ± 0.56) × 102 (χ2 ≤ 0.9) | (1.5 ± 0.61) × 106 | (1.3 ± 0.76) × 10−1 | −3.0 |
| TGFβ2-I92V/K94R | (4.6 ± 0.34) × 103 | −0.76 | ||
| TGFβ2-K25R/I92V/K94R | (1.0 ± 0.070) × 102 (χ2 ≤ 0.7) | (1.1 ± 0.14) × 106 | (1.1 ± 0.076) × 10−1 | −3.0 |
| protein variant | KD (nM) | ka (M−1 s−1) | kd (s−1) | ΔΔGbind(β3 variants) (kcal/mol) |
| TGF-β3 | 73 ± 1.9 (χ2 ≤ 2.3) | (7.4 ± 0.53) × 105 | (5.4 ± 0.35) × 10−2 | 0.0 |
| TGFβ3-V92I | (1.1 ± 0.32) × 102 (χ2 ≤ 0.3) | (7.0 ± 2.4) × 105 | (7.2 ± 1.0) × 10−2 | 0.34 |
| TGFβ3-R25K | (5.1 ± 2.4) × 102 (χ2 ≤ 0.4) | (4.4 ± 1.5) × 105 | (2.0 ± 0.22) × 10−1 | 1.2 |
| TGFβ3-R94K | (3.0 ± 0.62) × 103 | 2.3 | ||
| TGFβ3-R25A | (8.9 ± 0.90) × 102 | 1.6 | ||
| TGFβ3-R94A | (9.5 ± 1.7) × 103 | 3.0 | ||
Kinetic binding results are shown where determined. Each value is the mean of three independent runs determined from kinetic analysis or steady-state analysis (see Experimatal Procedures) with the standard deviation shown in parentheses. Where applicable, the maximum χ2 value resulting from the global fit analysis of kinetic data is shown. ΔΔGbind values were determined from ΔGwt – ΔGvariant, where ΔG = −RT ln(KD).
The affinity of TβR-II for the low-affinity isoform, TGF-β2, was determined using steady-state analysis. Global data fitting was not used since the association and dissociation rates were too fast to be accurately determined using this method (Figure 2C). The KD was determined to be approximately 16 μM, demonstrating that there is an overall 160-fold enhancement in the affinity of TGF-β2-TM, as compared to TGF-β2, for TβR-II.
Gain-of-Function Substitutions in TGF-β2
The contributions of each of the three substituted residues in TGF-β2-TM were dissected from one another by generating single and double TGF-β3 substitutions in the TGF-β2 background. Kinetic analysis with global fitting of the data was used for determining the affinities of the variants with KDs in the 1 μM range or lower, while steady-state analyses were used for variants with fast association and dissociation kinetics (typically KDs > 1 μM). Among the gain-of-function variants studied, the double arginine mutant, TGF-β2-K25R/K94R, was amenable to kinetic analysis while the remainder were studied using steady-state methods.
The measured affinities of TGF-β2 and its gain-of-function variants for TβR-II are shown in Figure 3. The KDs and the difference in the free energy change relative to that of TGF-β2 (ΔΔGbind) are presented in Table 1. There was no enhancement in affinity observed with the TGF-β2 I92V variant, while the K25R and K94R variants each led to either small (KD = 11 μM, corresponding to ΔΔGbind = −0.25 kcal/mol for K25R) or intermediate (KD = 6.0 μM, corresponding to ΔΔGbind = −0.60 kcal/mol for K94R) enhancements. Consistent with the lack of change in affinity observed for the TGF-β2 I92V variant, combining the I92V substitution with single arginine residue substitutions at positions 25 or 94 offered little, if any, improvement to the binding affinity over the single arginine variants. Significant improvement was however observed with the double arginine mutant, TGF-β2-K25R/K94R, which bound TβR-II with an affinity (KD ≅ 100 nM) which is indistinguishable from that of TGF-β2-TM (see Table 1 for standard deviations). These results show that Arg25 and Arg94 play a critical role in the bimolecular interaction between TGF-β and TβR-II, while that of Val92 is minimal.
FIGURE 3.
Summary of the dissociation constants (KD) of TGF-β2 and TGF-β2 gain-of-function variants as determined by the Biacore assays; the mean and standard deviation from triplicate runs are shown.
These results clearly rank the contribution of the three conserved interfacial residues in the TGF-β2 background as Arg94 > Arg25 ⪢ Val25. To determine whether these residues play a similar stabilizing role in TGF-β3, we generated single TGF-β2-residue substitutions in TGF-β3 and measured the extent to which they diminished TβR-II binding.
Loss-of-Function Substitutions in TGF-β3
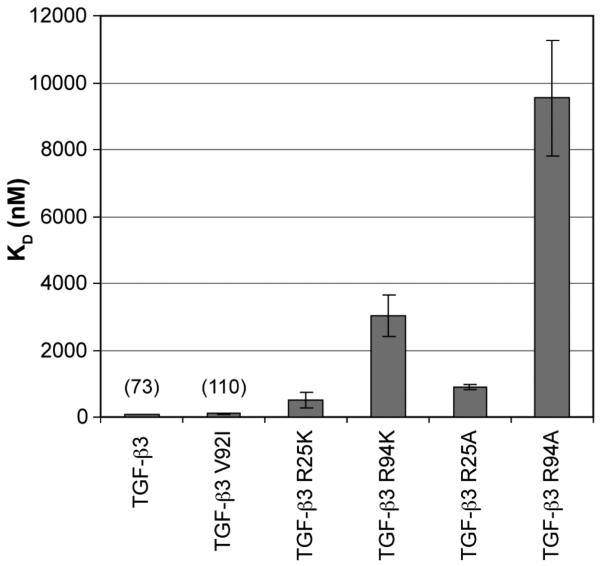
Similar to the TGF-β2 gain-of-function variants, kinetic analysis was used for determining the affinities of the TGF-β3 variants with KDs in the 1 μM range or lower, while steady-state analysis was used for variants with KDs > 1 μM. Among the loss-of-function TGF-β3 variants with TGF-β2-residue substitutions, only the TGF-β3-R94K variant was not amenable to kinetic analysis. The measured affinities of TGF-β3 and its variants for TβR-II are shown in Figure 4. The KDs and the difference in the free energy change relative to that of TGF-β3 are presented in Table 1. Consistent with the previous results from the gain-of-function TGF-β2 variants, substitution of Val92 with isoleucine led to little or no change in affinity, with the measured KD (approximately 110 nM) being nearly within the error limits of that of TGF-β3 (73 nM). Substitution of Arg25 and Arg94 with lysine led to larger reductions in affinity, with the R25K variant exhibiting a KD of 0.51 μM (corresponding to a ΔΔGbind of +1.2 kcal/mol) and the R94K variant exhibiting a KD of 3.0 μM (corresponding to a ΔΔGbind of +2.3 kcal/mol). These results match the ranking from the TGF-β2 gain-of-function variants, with Arg94 making the largest contribution to TβR-II binding in a TGF-β3 background, Arg25 making an intermediate contribution, and Val92 making little or none.
FIGURE 4.
Summary of the dissociation constants (KD) of TGF-β3, TGF-β3 loss-of-function variants, and neutral alanine substitutions as determined by the Biacore assays; the mean and standard deviation from triplicate runs are shown.
To examine further the role of the two doubly hydrogen bonded ion pairs that the guanidinium groups of Arg25 and Arg94 in TGF-β3 form with TβR-II (Figure 1), we substituted Arg25 and Arg94 in TGF-β3 with neutral alanine and tested the affinities of these variants for binding TβR-II. Steady-state analysis yielded KDs of 0.89 and 9.5 μM for the R25A and R94A variants, respectively. These KDs are slightly higher (lower affinities) than those measured for the corresponding lysine variants (R25K KD = 0.51 μM and R94K KD = 3.0 μM) indicating that, although the lysine side chains do make minor contributions to binding (corresponding to ΔΔGs of −0.4 and −0.7 kcal/mol, respectively), the doubly hydrogen bonded ion pairs formed in the TβR-II–TGF-β3 interfaces are highly specific.
The substitution results show in summary that the arginine residues at positions 25 and 94 are the key determinants for TGF-β3-like activity, as seen by essentially identical binding affinities for TGF-β3, TGF-β3-V92I, TGF-β2-TM, and TGF-β2-K25R/K94R. The isoaffinity plot of the kinetic data highlights this, with all of these variants clustered around the 100 nM binding isotherm (Figure 5). The importance of Arg25 is also emphasized by the fact that the TGF-β3-R25K variant is the only one that lies away from the 100 nM binding isotherm. It is interesting to note that, although four of the variants display almost identical affinities, there are differences in their kinetics of binding. It can be seen that the TGF-β2 gain-of-function variants (TGF-β2-TM and TGF-β2-K25R/K94R) have slightly faster association rates, i.e., are on the right half of the plot, while TGF-β3 and its loss-of-function variants (TGF-β3-V92I, TGF-β3-R25K) all have somewhat slower association rates, i.e., are on the left half of the plot. Since the interfacial residues are identical in TGF-β3 and TGF-β2-TM, it follows that this difference in association rates between TGF-β3 and TGF-β2 variants may be due to long-range effects coming from the TGF-β structural framework, which is known to be similar but not identical in TGF-β2 versus TGF-β3 (23–25). On the basis of these results, we decided to obtain an additional level of information on the interaction of TGF-β with TβR-II by carrying out a thermodynamic analysis of the binding of TβR-II to both TGF-β3 and TGF-β2-TM.
FIGURE 5.
Isoaffinity plot of all TGF-β2 and -β3 variants amenable to kinetic analysis. Each point represents the kinetic parameters derived from a single TβR-II dilution series over the indicated TGF-β variant surface. Key: TGF-β3(●), TGF-β2-TM (■), TGF-β2-K25R/K94R (▲), TGF-β3-R25K (○), and TGF-β3-V92I (×). All had their kinetic rate constants, ka and kd, determined by globally fitting their sensorgram data to a heterogeneous ligand model (BiaEvaluation v4.1). The rate constants associated with the higher affinity binding site (lowest KD) as determined by the heterogeneous ligand model were selected as the representative values for the interaction as previously determined (22).
Thermodynamic Comparison of TGF-β3 and TGF-β2-TM
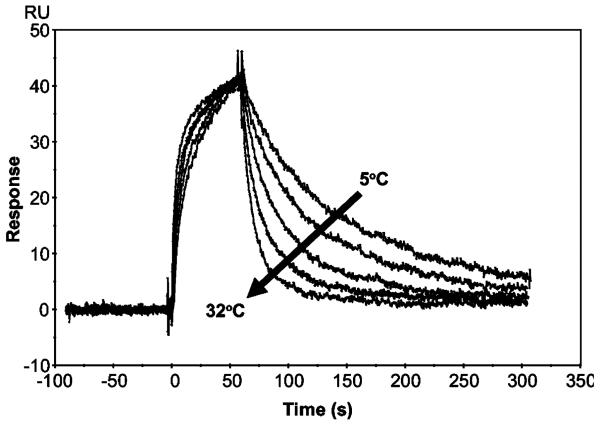
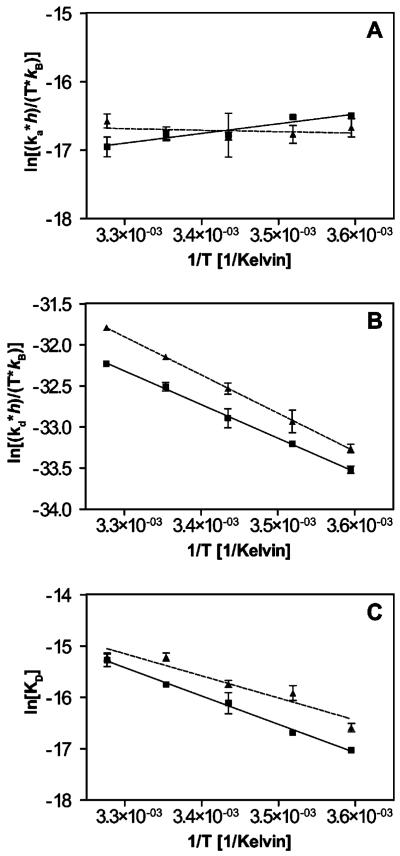
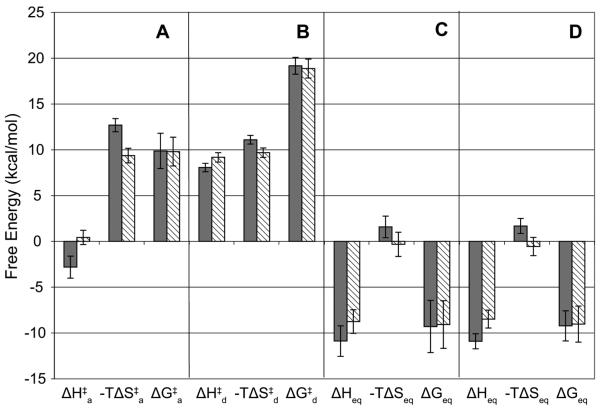
It has recently been shown that SPR biosensor analysis can be used for the determination of the thermodynamic parameters ΔG (free energy), ΔH (enthalpy), and ΔS (entropy) of interactions (26, 27). To compare the thermodynamic characteristics of the interactions of TβR-II with TGF-β2-TM versus TGF-β3, binding data were collected over a temperature range of 5–32 °C. As can be seen qualitatively by examining the overlay of sensorgrams representing a constant concentration of TβR-II flowing at different temperatures, there was a negligible effect of temperature on the association phase, but a large temperature dependence of the dissociation phase, for the TβR-II–TGF-β3 interaction (Figure 6) as well as the TβR-II–TGF-β2-TM interaction (data not shown). Thermodynamic parameters (ΔH and ΔS) of association and dissociation steps can be estimated from the temperature dependence of the kinetic constants. Accordingly, we derived kinetic constants by globally fitting the interaction data obtained at different temperatures and carried out linear regression of Eyring plots (see Experimental Procedures). The relatively temperature independent nature of the association rates is reflected in the near horizontal slopes of the association Eyring plots (Figure 7A), whereas the temperature dependence of the dissociation rates is reflected in the steeper slopes of the dissociation Eyring plots (Figure 7B). The thermodynamic parameters derived from these Eyring plots are shown in Figure 8A–C. The small difference in the slopes of the association Eyring plots for TGF-β3 versus TGF-β2-TM (Figure 7A) is reflected in somewhat different ΔH‡a values (−2.8 ± 1.2 kcal/mol for TGF-β3 versus 0.43 ± 0.78 kcal/mol for TGF-β2-TM; Figure 8A). Similarly, there is a slight difference in the estimated −TΔS‡a values for the association step for TGF-β3 and TGF-β2-TM (13 ± 0.70 and 9.4 ± 0.80 kcal/mol, respectively). When the ΔG‡a values for the association step were calculated using these ΔH‡ and −TΔS‡a values, the entropy and enthalpy differences compensated such that there was no significant difference in the ΔG‡a values for TGF-β3 and TGF-β2-TM (~10 kcal/mol for both). The thermodynamic parameters of the dissociation step (ΔH‡d, −TΔS‡d, ΔG‡d) also did not differ significantly between TGF-β3 and TGF-β2-TM (Figure 8B). Additionally, when the parameters from the association and dissociation steps were combined to derive equilibrium thermodynamic parameters (ΔHeq, −TΔSeq, ΔGeq), there was no statistical difference between the value of these parameters for TGF-β3 and TGF-β2-TM (Figure 8C). The equilibrium thermodynamic parameters can also be estimated using van't Hoff plots (Figure 7C) in which the KD values that were determined by global fitting are plotted (see Experimental Procedures). As expected, the values of ΔHeq, −TΔSeq, and ΔGeq determined by this approach (Figure 8D) are essentially the same as those determined using Eyring plots and are very similar for TGF-β3 and TGF-β2-TM.
FIGURE 6.
An overlay of sensorgrams showing the 500 nM TβR-II–TGF-β3 interaction over 5–32 °C.
FIGURE 7.
Thermodynamic plots for the interaction of TβR-II with TGF-β3 (■) and TGF-β2-TM (▲) determined from three independent SPR experiments at each temperature point. Eyring plots are shown in panels A and B for the forward (ka), and reverse (kd) interactions, respectively. Panel C shows van't Hoff plots using equilibrium KD values determined from individual rate constants derived from the SPR data (kd/ka) at each temperature indicated.
FIGURE 8.
A compendium of transition state and equilibrium entropy, enthalpy, and Gibbs' free energy values determined from the Eyring (A–C) and van't Hoff plots (D) for the TβR-II interaction with TGF-β3 (gray bars) and TGF-β2-TM (cross-hatched bars) at 298 K. The association and dissociation energies are denoted with the superscripts “a” and “d”, respectively. The standard errors generated from the plots in Figure 6 are shown.
NMR Fold Analysis
We utilized NMR spectroscopy to verify that the substitutions made in the TGF-β2 gain-of-function and the TGF-β3 loss-of-function variants altered contacts with the receptor, without perturbing the overall structure of the ligand. We prepared each of the wild-type proteins, as well as each of the variants, at a concentration of 0.05–0.1 mM in 10 mM phosphate buffer at pH 3.8 and recorded one-dimensional 1H spectra (Figure 9). This pH value was chosen as previous studies showed that it was low enough to keep the protein soluble at these concentrations but not so low as to cause unfolding (28). The spectra thus recorded exhibit numerous resolved signals, in particular methyl protons upfield of the 0.8 ppm random coil limit and backbone amide protons downfield of the 8.5 ppm random coil limit, both for the wild-type proteins and for the variants. The patterns of peaks are similar between the wild-type proteins and variants. This indicates that none of the substitutions, including those that lead to a substantial loss in the affinity of TGF-β3 for TβR-II, had any substantial effect on either the overall folding or the relative positioning of residues in the receptor binding site.
FIGURE 9.
One-dimensional NMR spectra of wild-type TGF-β2, TGF-β3, and variants. Samples were prepared at a concentration of 0.05–0.1 mM in 10 mM phosphate and 5% 2H2O at pH 3.80. Spectra were recorded at 500 MHz and 45 °C using a 500 MHz spectrometer using a WATERGATE solvent suppression sequence. Spectra shown were collected using 1024–4096 scans and a recycle time of 1 s.
Growth Inhibition Assays
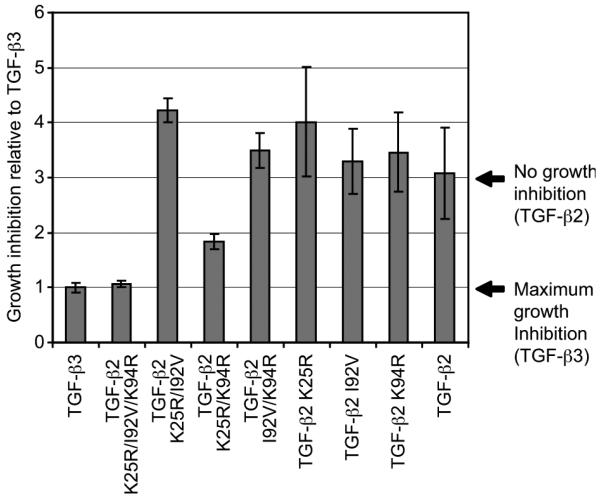
Growth inhibition assays were carried out to determine whether the gain of binding observed with the TGF-β2 variants correlated with gain of function in a cell growth inhibition assay. Fetal bovine heart endothelial (FBHE) cells were chosen for these studies since they lack detectable expression of the TGF-β coreceptor, β-glycan, and are thus relatively insensitive to growth inhibition with TGF-β2 (11). Previously, it was shown that TGF-β2-TM exhibits a growth inhibitory potency intermediate to that of TGF-β2 and TGF-β3 (18). In the present study, we used a single concentration of TGF-β ligand (31.3 pM) at which TGF-β2-TM and TGF-β3, but not TGF-β2, are expected to be growth inhibitory (11, 18). As predicted, at this concentration TGF-β2-TM exhibits a growth inhibitory response close to that of TGF-β3; however, the double arginine mutant, TGF-β2-K25R/K94R, shows an inhibition intermediate to that of TGF-β2 and TGF-β3 (Figure 10). The remaining single and double TGF-β2 variants resembled TGF-β2 in that they were not growth inhibitory at this concentration.
FIGURE 10.
Growth inhibition of FBHE cells in 10% fetal bovine serum as determined by [methyl-3H]deoxythymidine incorporation. Cell growth inhibition was normalized to TGF-β3. The bars represent mean values from triplicate or quadruplicate assays with the standard deviation indicated.
DISCUSSION
Structural analysis of the TβR-II–TGF-β3 binary and TβR-I–TβR-II–TGF-β3 ternary complexes has shown that TβR-II binds the “fingertips” of the TGF-β3 homodimer, such that the TβR-II extracellular domains bind independent of one another when forming the 2:1 binary and 2:2:1 ternary complexes (14, 15). The crystal structures further show that side chain guanidinium groups of Arg25 and Arg94 of the TGF-β3 homodimer are aligned with the side chain carboxylate groups of Glu119 and Asp32 of TβR-II to form two near ideal hydrogen-bonded ion pairs (Figure 1). Importantly, these are the only two ion pairs in the binding interface, suggesting that they likely play critical roles in determining the specificity of receptor binding. The main objective of the present study was to dissect the roles played by these two hydrogen bonded ion pairs, as well as by the third variable interfacial residue (Ile92), in TβR-II binding.
To do so, we generated TGF-β2 gain-of-function variants bearing single, double, and triple amino acid replacements from the high-affinity TGF-β isoforms, TGF-β1 and -β3. The corresponding TGF-β3 loss-of-function variants bearing single amino acid replacements from TGF-β2 were also investigated. This was done in order to ascertain the extent to which noninterfacial effects coming from the ligand structural framework, which has been shown to be very similar but not identical in the three ligand isoforms (23-25), affect binding. SPR biosensor data were used to determine the equilibrium dissociation constants for the interactions of these ligand variants with TβR-II. In appropriate cases, kinetic and thermodynamic parameters were also determined from SPR biosensor data. One-dimensional 1H NMR spectra were determined for the variants, which confirmed that the observed binding differences are not due to variations in overall fold or to large structural perturbations (Figure 9).
To serve as a basis of comparison, the equilibrium dissociation constant (KD) of TβRII binding to TGF-β2 was determined to be 17 μM using steady-state affinity analysis (Figure 2C and Table 1). This weak interaction, which has not been previously characterized, has a 230-fold higher KD than that of the interaction of TβR-II with TGF-β3, which was determined to be 73 nM. The three residue change in TGF-β2-TM brings about a 170-fold increase in affinity to yield a KD (100 nM) that is only slightly higher (lower affinity) than that of TGF-β3. There is also a remarkable difference in the kinetics between TGF-β2-TM and TGF-β2 (Figure 2).
The contributions of Arg25, Val92, and Arg94 to binding were dissected by generating single and double variants in both the TGF-β2 and TGF-β3 background. The results showed that Val92 plays only a minor role in binding, as evidenced by a lack of enhancement in affinity for the TGF-β2 gain-of-function variant TGF-β2 I92V and only a slight decrease in affinity for the TGF-β3 loss-of-function variant TGF-β3 V92I (Table 1). No detectable, or only marginal, enhancements in affinity were recorded when the I92V substitution was studied in the context of other TGF-β2 variants (compare TGF-β2-TM with TGF-β2-K25R/K94R; TGF-β2-I92V/K94R with TGF-β2-K94R; TGF-β2-K25R/I92V with TGF-β2-K25R), further supporting the minor role played by residue 92 in binding. In the crystal structure of the TβR-II–TGF-β3 complex, Val92 is located on the periphery of the binding interface and has limited (16 Å2) contact with TβR-II, thus explaining the relatively minor effect this residue has on affinity (Figure 1).
Arg25 and Arg94 alone contributed moderately to affinity, with the TGF-β2-K25R and TGF-β2-K94R gain-of-function variants exhibiting KDs decreased relative to TGF-β2 by a factor of 1.5- and 2.8-fold, respectively. As expected, an opposite pattern was observed with the TGF-β3-R25K and TGF-β3-R94K loss-of-function variants; however, the changes were somewhat more pronounced with KD increases relative to TGF-β3 of 7.0- and 41-fold, respectively. Although the magnitude of the effects is somewhat larger in the context of TGF-β3, it is nevertheless clear that both Arg25 and Arg94 contribute significantly to high-affinity binding, with the contribution of Arg94 exceeding that of Arg25.
Arg25 and Arg94 together contributed very significantly to affinity, with the TGF-β2-K25R/K94R variant binding TβR-II with a 170-fold lower KD than that of TGF-β2. This decrease is substantially more than the product of the reductions in KD attained in the TGF-β2-K25R and TGF-β2-K94R variants, 1.5- and 2.8-fold, respectively, indicating that the high-affinity binding is a consequence of cooperative interactions between these and other residues in the binding interface. A comparison of the structures of free (24) and TβR-II-bound TGF-β3 (14) reveals that small changes in the TβR-II binding interface occur upon binding, as previously noted (18). Hence it is possible that this cooperativity arises from subtle reorientation and rigidification of residues surrounding the first arginine once it forms the highly specific doubly hydrogen bonded ion pair with its corresponding carboxylate group on TβR-II. This cooperativity likely accounts for the much more pronounced changes in KD that occurred as the same arginine residue was removed from TGF-β3 (as opposed to being substituted into TGF-β2) since the changes in TGF-β3 occur in the context of another arginine residue, whereas in TGF-β2 they do not.
The data discussed thus far clearly indicate that the interactions between Arg25 and Arg94 of TGF-β3 and their corresponding carboxylate acceptors on TβR-II, Glu119, and Asp32, respectively, are important and likely to be highly specific. To explore this specificity further, we generated nonconservative neutral alanine variants of these two arginine residues in TGF-β3 and measured their affinities. The results show that the alanine substitutions decrease affinity as expected but that the affinities drop to levels only 2–3-fold lower than those of the conservative lysine variants. This indicates that the substituted lysine residues retain only minor binding functionality, perhaps due to interactions between their positively charged ∈-NH2 groups and the negatively charged TβR-II side chain carboxylates and/or due to the formation of weak hydrogen bonds. The fact that the differences between conservative substitution with lysine and nonconservative substitution with alanine are quite small (amounting to free energy differences of just 0.4–0.7 kcal/mol) is consistent with previous mutational analysis of the carboxylate acceptors on TβR-II, which showed that conservative Asp to Asn and Glu to Gln substitutions perturbed binding to a similar extent as nonconservative substitutions with Ala (18). As additional support for the importance of the two carboxylate acceptors on TβR-II, we have now analyzed the double alanine substituted TβR-II receptor (E119A and D32A) and have found that no binding could be detected on a TGF-β3 surface (data not shown). Thus, the data presented here, together with the previous published results, underscore the highly specific nature of the interaction between arginines 25 and 94 of TGF-β3 and their corresponding carboxylate acceptors on TβR-II.
Although TGF-β2-K25R/K94R, TGF-β2-TM, TGF-β3-V92I, and TGF-β3 all have nearly identical affinities, they exhibit slightly different kinetics with the TGF-β3 variants having slower dissociation and association rates relative to those of the TGF-β2 variants (Figure 5). These small differences in kinetics may be due to noninterfacial long-range effects arising from the TGF-β structural framework, which is similar but not identical in TGF-β2 versus TGF-β3. To investigate further the possibility that long-range effects may subtly influence the interaction of TGF-β ligands with TβR-II, we carried out a thermodynamic analysis of the binding of TGF-β3 and TGF-β2-TM. Since the interfacial residues are identical in TGF-β3 and TGF-β2-TM, a comparison of these two variants enables the evaluation of noninterfacial effects coming from the TGF-β structural framework. We observed that the thermodynamic parameters of the association step differed slightly between TGF-β3 and TGF-β2-TM (Figure 8A), supporting the idea that the noninterfacial ligand structural framework influences binding to TβR-II to a small degree. It is also interesting to note that the value of ΔHeq that we observed for the interaction of TβR-II with TGF-β3 and TGF-β2-TM (~–10 kcal/mol) is close to the median value of ΔHeq (–9.4 kcal/mol) calculated based on a data set of 43 protein–protein and 26 protein–peptide interactions reported in the literature (29). Although it is tempting to conclude that the near zero value of −TΔSeq and the negative magnitude of ΔHeq indicate that the interactions of these two ligand variants with TβR-II are enthalpically driven, it has been pointed out recently that this type of interpretation should be made with caution in the case of protein interactions (30, 31). This is due to the fact that such interactions generally involve more than one step in their pathway and that the enthalpy value represents a composite of values for the individual steps.
Growth inhibition assays were carried out on the TGF-β2 gain-of-function variants using fetal bovine heart endothelial cells to determine if binding changes correlate directly with changes in function. The results show that TGF-β3-like potency is only attained with TGF-β2-TM. The lack of activity exhibited by all the variants bearing a single arginine residue at position 25 or 94 suggests that the cells have a minimum KD requirement to achieve growth inhibition under these conditions, roughly 3 μM, which is exceeded for all of these variants. The double arginine mutant, TGF-β2-K25R/K94R exhibits a growth inhibitory potential intermediate between that of TGF-β2 and TGF-β3 or TGF-β2-TM. This is somewhat unexpected since the affinity and kinetic measurements clearly showed that Val92 only makes a minor contribution to binding TβR-II. This apparent contradiction suggests that residue Val92, in this cellular context, may interact with another protein that stabilizes the ligand–TβR-II complex on the cell surface. However, it is unlikely that this protein is TβR-I due to the absence of any contacts between Val92 and TβR-I in the recently reported TβR-I–TβR-II–TGF-β3 crystal structure (15) or β-glycan due to its lack of expression in this cell line (11). An alternative explanation for the somewhat unexpected activity of TGF-β2-K25R/K94R is that this variant may be slightly less stable during the time of incubation in cell culture.
In summary, the data presented here provide a deeper level of understanding of the critical residues required for the high-affinity interaction between TGF-β and TβR-II and of the manner by which TβR-II discriminates between the high-affinity TGF-β isoforms, TGF-β1 and -β3, and the lowaffinity isoform, TGF-β2. It is tempting to speculate based on these findings that the evolution of TGF-β isoforms with differing type II receptor binding potentials has occurred since it enables the expansion and diversification of function through TGF-β2's reliance on the coreceptor β-glycan for function. It is also worth noting that TGF-β has been identified as an important therapeutic target for treatment of a number of diseases, including liver, lung, and kidney fibrosis (32), as well as several forms of cancer, including those of the breast, pancreas, prostate, lung, and brain (33). In light of this, the information presented here, in particular the relative importance of the two arginine residues, may aid in the development of novel structure-based TGF-β inhibitors, such as those proposed by Shimanuki and coworkers, that target the TβR-II binding site on TGF-β (34).
ACKNOWLEDGMENT
We thank Andre Migneault for assistance in preparing the figures and Anne Lenferink for technical assistance.
Footnotes
This work was supported by the Genomics and Health Initiative, National Research Council, Canada, the National Institutes of Health (GM58670), and the Robert A. Welch Foundation (AQ1431).
Abbreviations: DMEM, Dulbecco/Vogt-modified Eagle's minimal essential medium; EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride; EDTA, ethylenediaminetetraacetic acid; FBS, fetal bovine serum; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; TβR-I, transforming growth factor β type I receptor; TβR-II, transforming growth factor β type II receptor; TGF-βII, transforming growth factor β; TGF-β2-TM, TGF-β2-K25R/I92V/K94R variant.
REFERENCES
- 1.Roberts AB, Sporn MB. The transforming growth factor-betas. In: Roberts AB, Sporn MB, editors. Peptide Growth Factors and their Receptors. Springer; Heidelberg: 1990. pp. 421–472. [Google Scholar]
- 2.Derynck R. TGF-beta-receptor-mediated signaling. Trends Biochem. Sci. 1994;19:548–553. doi: 10.1016/0968-0004(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 3.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 4.Massague J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 5.Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development (Cambridge, England) 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Proetzel G, Pawlowski SA, Wiles MV, Yin M, Boivin GP, Howles PN, Ding J, Ferguson MW, Doetschman T. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat. Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massague J. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 9.De Crescenzo G, Pham PL, Durocher Y, Chao H, O'Connor-McCourt MD. Enhancement of the antagonistic potency of transforming growth factor-beta receptor extracellular domains by coiled coil-induced homo- and heterodimerization. J. Biol. Chem. 2004;279:26013–26018. doi: 10.1074/jbc.M400655200. [DOI] [PubMed] [Google Scholar]
- 10.De Crescenzo G, Litowski JR, Hodges RS, O'Connor-McCourt MD. Real-time monitoring of the interactions of two-stranded de novo designed coiled-coils: effect of chain length on the kinetic and thermodynamic constants of binding. Biochemistry. 2003;42:1754–1763. doi: 10.1021/bi0268450. [DOI] [PubMed] [Google Scholar]
- 11.Cheifetz S, Hernandez H, Laiho M, ten Dijke P, Iwata KK, Massague J. Distinct transforming growth factor-beta (TGF-beta) receptor subsets as determinants of cellular responsiveness to three TGF-beta isoforms. J. Biol. Chem. 1990;265:20533–20538. [PubMed] [Google Scholar]
- 12.Vilchis-Landeros MM, Montiel JL, Mendoza V, Mendoza-Hernandez G, Lopez-Casillas F. Recombinant soluble betaglycan is a potent and isoform-selective transforming growth factor-beta neutralizing agent. Biochem. J. 2001;355:215–222. doi: 10.1042/0264-6021:3550215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Casillas F, Wrana JL, Massague J. Betaglycan presents ligand to the TGF beta signaling receptor. Cell. 1993;73:1435–1444. doi: 10.1016/0092-8674(93)90368-z. [DOI] [PubMed] [Google Scholar]
- 14.Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP. Crystal structure of the human TbetaR2 ectodomain-TGF-beta3 complex. Nat. Struct. Biol. 2002;9:203–208. doi: 10.1038/nsb766. [DOI] [PubMed] [Google Scholar]
- 15.Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP. Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol. Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 16.Thompson TB, Woodruff TK, Jardetzky TS. Structures of an ActRIIB:activin A complex reveal a novel binding mode for TGF-beta ligand:receptor interactions. EMBO J. 2003;22:1555–1566. doi: 10.1093/emboj/cdg156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol. Cell. 2003;11:605–617. doi: 10.1016/s1097-2765(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 18.De Crescenzo G, Hinck CS, Shu Z, Zuniga J, Yang J, Tang Y, Baardsnes J, Mendoza V, Sun L, Lopez-Casillas F, O'Connor-McCourt M, Hinck AP. Three key residues underlie the differential affinity of the TGFbeta isoforms for the TGFbeta type II receptor. J. Mol. Biol. 2006;355:47–62. doi: 10.1016/j.jmb.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 19.Hinck AP, Walker KP, III, Martin NR, Deep S, Hinck CS, Freedberg DI. Sequential resonance assignments of the extracellular ligand binding domain of the human TGF-beta type II receptor. J. Biomol. NMR. 2000;18:369–370. doi: 10.1023/a:1026775321886. [DOI] [PubMed] [Google Scholar]
- 20.Cerletti N. Process for the production biologically active dimeric protein. U.S. Patent. 2000:6057430. [Google Scholar]
- 21.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 22.De Crescenzo G, Grothe S, Zwaagstra J, Tsang M, O'Connor-McCourt MD. Real-time monitoring of the interactions of transforming growth factor-beta (TGF-beta) isoforms with latency-associated protein and the ectodomains of the TGF-beta type II and III receptors reveals different kinetic models and stoichiometries of binding. J. Biol. Chem. 2001;276:29632–29643. doi: 10.1074/jbc.M009765200. [DOI] [PubMed] [Google Scholar]
- 23.Schlunegger MP, Grutter MG. An unusual feature revealed by the crystal structure at 2.2 Å resolution of human transforming growth factor-beta 2. Nature. 1992;358:430–434. doi: 10.1038/358430a0. [DOI] [PubMed] [Google Scholar]
- 24.Mittl PR, Priestle JP, Cox DA, McMaster G, Cerletti N, Grutter MG. The crystal structure of TGF-beta 3 and comparison to TGF-beta 2: implications for receptor binding. Protein Sci. 1996;5:1261–1271. doi: 10.1002/pro.5560050705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daopin S, Piez KA, Ogawa Y, Davies DR. Crystal structure of transforming growth factor-beta 2: an unusual fold for the superfamily. Science (New York) 1992;257:369–373. doi: 10.1126/science.1631557. [DOI] [PubMed] [Google Scholar]
- 26.Myszka DG, Abdiche YN, Arisaka F, Byron O, Eisenstein E, Hensley P, Thomson JA, Lombardo CR, Schwarz F, Stafford W, Doyle ML. The ABRF-MIRG'02 study: assembly state, thermodynamic, and kinetic analysis of an enzyme/inhibitor interaction. J. Biomol. Tech. 2003;14:247–269. [PMC free article] [PubMed] [Google Scholar]
- 27.Moll D, Schweinsberg S, Hammann C, Herberg FW. Comparative thermodynamic analysis of cyclic nucleotide binding to protein kinase A. Biol. Chem. 2007;388:163–172. doi: 10.1515/BC.2007.018. [DOI] [PubMed] [Google Scholar]
- 28.Pellaud J, Schote U, Arvinte T, Seelig J. Conformation and self-association of human recombinant transforming growth factor-beta3 in aqueous solutions. J. Biol. Chem. 1999;274:7699–7704. doi: 10.1074/jbc.274.12.7699. [DOI] [PubMed] [Google Scholar]
- 29.Stites WE. Protein–protein interactions: interface structure, binding thermodynamics, and mutational analysis. Chem. Rev. 1997;97:1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- 30.Winzor DJ, Jackson CM. Interpretation of the temperature dependence of equilibrium and rate constants. J. Mol. Recognit. 2006;19:389–407. doi: 10.1002/jmr.799. [DOI] [PubMed] [Google Scholar]
- 31.Winzor DJ, Jackson CM. Interpretation of the temperature dependence of rate constants in biosensor studies. Anal. Biochem. 2005;337:289–293. doi: 10.1016/j.ab.2004.10.049. [DOI] [PubMed] [Google Scholar]
- 32.Cutroneo KR. TGF-beta-induced fibrosis and SMAD signaling: oligo decoys as natural therapeutics for inhibition of tissue fibrosis and scarring. Wound Repair Regen. 2007;15(Suppl 1):S54–S60. doi: 10.1111/j.1524-475X.2007.00226.x. [DOI] [PubMed] [Google Scholar]
- 33.Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimanuki T, Hara T, Furuya T, Imamura T, Miyazono K. Modulation of the functional binding sites for TGF-beta on the type II receptor leads to suppression of TGF-beta signaling. Oncogene. 2007;26:3311–3320. doi: 10.1038/sj.onc.1210123. [DOI] [PubMed] [Google Scholar]