Abstract
Background
Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy (ARVD/C) is an inherited disorder typically caused by mutations in components of the cardiac desmosome. The prevalence and significance of desmosome mutations among ARVD/C patients in North America has not previously been described. We report comprehensive desmosome genetic analysis for 100 North Americans with clinically confirmed or suspected ARVD/C.
Methods and results
In 82 individuals with ARVD/C and 18 people with suspected ARVD/C, DNA sequence analysis was performed on PKP2, DSG2, DSP, DSC2, and JUP. In those with ARVD/C, 52% harbored a desmosome mutation. A majority of these mutations occurred in PKP2. Notably, 3 of the individuals studied have a mutation in more than one gene. Patients with a desmosome mutation were more likely to have experienced ventricular tachycardia (73% versus 44%) and they presented at a younger age (33 versus 41 years) compared to those without a desmosome mutation. Males with ARVD/C were more likely to carry a desmosome mutation than females (63% versus 38%). A mutation was identified in 5/18 (28%) patients with suspected ARVD. In this smaller subgroup there were no significant phenotypic differences identified between individuals with a desmosome mutation compared to those without a mutation.
Conclusions
Our study shows that in 52% of North Americans with ARVD/C a mutation in one of the cardiac desmosome genes can be identified. Compared to those without a desmosome gene mutation, individuals with a desmosome gene mutation had earlier onset ARVD/C and were more likely to have ventricular tachycardia.
Keywords: Cardiomyopathy, ventricular tachycardia, sudden cardiac death, genetics, desmosome
Introduction
Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy (ARVD/C) is an inherited form of cardiomyopathy characterized histologically by fibrofatty replacement of the right ventricular myocardium1,2. Affected individuals typically present with palpitations, syncope, ventricular tachycardia, right heart failure, or sudden cardiac death (SCD)2–4. In one reported series of individuals with this condition, 23% experienced sudden cardiac death as their presenting symptom4. Currently the diagnosis is based on clinical characteristics assembled by an expert task force encompassing a complex series of both major and minor criteria5.
The incidence and prevalence of ARVD/C is uncertain and may vary regionally. In Northeast Italy ARVD/C was found to be responsible for 22.4% of SCD in young athletes and in 8.2% of SCD in non-athletes6. A report from Germany indicated a prevalence of 1 per 1000 individuals7. Others estimate the prevalence in Europe and North America to be 1 per 5000 individuals8,9.
In recent years research has focused on identifying the genetic basis for this condition. In 2000 McKoy et al identified a homozygous mutation in JUP in patients with Naxos disease10. This autosomal recessive form of ARVD/C is accompanied by palmoplantar keratoderma and woolly hair. JUP encodes junction plakoglobin, one of the desmosomal proteins. Carvajal syndrome is a similar recessive disorder of left ventricular cardiomyopathy with skin and hair abnormalities caused by homozygous mutations in DSP, encoding desmoplakin11. These seminal findings led to the discovery of heterozygous mutations in genes encoding these and other desmosomal proteins in patients with non-syndromic ARVD/C. In addition to mutations in DSP and JUP, nonsyndromic ARVD/C may be caused by mutations in PKP2, DSG2, or DSC212–16. Mutations in genes encoding non-desmosomal proteins may also cause ARVD/C. Alterations of the 5′ and 3′ regulatory elements of TGFB3 encoding transforming growth factor beta-3 have each been reported17. Mutations in RYR2, encoding the cardiac ryanodine receptor, result in a form of arrhythmic cardiomyopathy without significant structural abnormalities18. This phenotype is variably referred to as ARVD/C or catecholaminergic polymorphic ventricular tachycardia (CPVT)18,19. More recently, a mutation in TMEM43, encoding a transmembrane protein with ties to an adipogenic transcription factor was reported as the cause for ARVD5, a subtype of ARVD/C with prominent left ventricular involvement20.
An autosomal dominant pattern of inheritance for nonsyndromic ARVD/C is seen among approximately 30% of affected individuals4. This may reflect both reduced penetrance and variable expressivity in families segregating this condition21,22. In addition, both compound heterozygosity and autosomal recessive mutations have been reported in nonsyndromic ARVD/C, suggesting that additional genetic factors may influence phenotypic manifestations15,23,24. To date, a mutation in more than 1 desmosome gene has not been associated with this condition, suggesting that further genetic analysis may not be necessary if a single mutation is recognized.
Comprehensive desmosome genetic analysis has been reported by only one group in England22,25. They described analysis of 200 individuals in 69 families, finding a pathogenic mutation in 20 families (29%)25. As clinical use of genetic testing for ARVD/C becomes more widely used, the importance of understanding the likelihood of discovery of a pathogenic mutation becomes more prominent. Just as the rate of PKP2 mutations is different among unique cohorts, variability in other desmosome genes is also relevant14,26–29. We report the first comprehensive desmosome genetic analysis for a large North American cohort of individuals with clinically confirmed or suspected ARVD/C.
Methods
Patient recruitment and evaluation
Eighty-two unrelated ARVD/C probands and 18 unrelated individuals with suspected ARVD/C gave written informed consent to participate in a study approved by the Johns Hopkins University Institutional Review Board. History and medical records were obtained at enrollment and at yearly intervals. The age of onset of ARVD/C symptoms was defined as the age at which a patient experienced symptoms related to ARVD/C. Symptoms include palpitations, syncope, ventricular tachycardia, sudden cardiac death, edema, and chest pain. Family history was obtained by interviewing patients and family members. Patients were evaluated by physical examination. Additional testing included 12-lead ECG (n=100), 24-hour Holter monitoring (n=74), exercise testing, signal-averaged ECG (SAECG; n=87), imaging studies including echocardiography and MRI (n=100), and right ventricular endomyocardial biopsy (n=34). The presence and morphology of ventricular ectopy or ventricular tachycardia were identified using the results of standard 12-lead ECGs, Holter monitoring and stress testing. The SAECG was considered positive for late potentials if a patient, not previously known to have a right bundle branch block, showed any 2 out of the following abnormalities: (1) a filtered QRS duration ≥114 ms, (2) low-amplitude signal duration ≥38 ms, or (3) RMS ≤20mV. Biopsies of right ventricular lateral wall were assessed for fibrofatty replacement in the presence of surviving strands of cardiomyocytes.
The diagnosis of ARVD/C was based on the criteria set by the Task Force of the Working Groups of Myocardial and Pericardial Disease of the European Society of Cardiology5. The diagnosis of suspected ARVD/C was based on meeting at least 2 minor or 1 major and 1 minor criteria from these Task Force Criteria.
Genetic Analysis
In this cohort of 100 North American Caucasians, 52 probands had not previously undergone genetic analysis for ARVD/C and they were added to 48 individuals who had previously been analyzed only for mutations in PKP2 and/or DSG215,24,26. All 100 of these patients had novel analysis of 3 genes (DSP, JUP, and DSC2) and 52 patients had initial analysis of all 5 genes, so that the entire cohort had complete sequence investigation of all 5 desmosome genes in which mutations have been reported to cause ARVD/C.
Genomic DNA was extracted from whole blood using QIAmp DNA blood maxi kits (Qiagen, Inc., Valencia, California). Polymerase chain reaction (PCR) products were generated for sequence analysis using intronic primers (primer sequences available from the authors on request) flanking each exon in PKP2, DSP, DSG2, JUP, and DSC2. PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, Inc., Valencia, California). Bidirectional sequence analysis was performed using an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, California) and chromatograms were analyzed manually and with Sequencer 4.1 software.
All novel sequence variants were classified as mutation or polymorphism. We defined a mutation as not occurring among 400 unrelated, unaffected, race-matched, control chromosomes (NIGMS Human Genetic Cell Repository, Coriell Institute for Medical Research), altering one or more conserved amino acids, and segregating with disease in the family when that information was available. All other novel variants were classified as polymorphisms. Sequence variants that were previously reported as mutations were considered mutations unless we could show otherwise. Controls were tested for the presence or absence of a novel restriction enzyme digest site if the DNA variant altered such a site, or by Taqman genotyping assays (Applied Biosystems, Foster City, California) if the sequence variant did not alter any restriction sites. Novel missense protein variants were analyzed by ClustalW (MacVector, Inc, Cary, NC), Polyphen (http://genetics.bwh.harvard.edu/pph/), and MUpro (http://www.ics.uci.edu/~baldig/mutation.html) for conservation, stability, and likelihood for pathogenic effect.
To clarify the sequence of DSG2 cDNA based on discordant reports of sequence for the first exon30,31, total RNA was extracted from human hearts, reverse transcribed, and 5′ rapid amplification of cDNA ends (RACE) was performed using nested primers in the 3rd and 4th exons. Products were sequenced to determine if an alternate start site exists in cardiac tissue.
Statistical Analysis
Statistical analysis was performed using the Statistical Package for Social Sciences (SPSS version 16.0; SPSS, Inc, Chicago, Ill). Continuous variables between 2 groups were compared by unpaired t test when comparing two groups, and ANOVA in case of multiple groups. Categorical variables were compared by chi-squared test in case of large samples and by Fisher’s exact test in case of small sample size (N<5 in two of the groups). Kaplan-Meier analysis and log-rank test was used when establishing and comparing symptom-and/or VT-free survival. Cox proportional hazards model was used to adjust for the effect of gender on VT-free survival. A p value ≤0.05 was considered statistically significant.
Results
Clinical characteristics and mutational analysis
Eighty-two patients (59% male) diagnosed with ARVD/C are included in this study. Clinical characteristics are summarized in Table 1 with more detailed information in Supplemental Table 1. Mean age at enrollment was 44±14 years. Age of diagnosis varied between 2 and 76 years (mean 37±14 years) with the first symptoms of ARVD/C arising at age 2 to 76 years (mean 33±14 years).
Table 1.
Baseline characteristics
| Characteristic | (n=82) |
|---|---|
| Gender (male), n (%) | 48 (59%) |
| Age, yrs (range) | 44±14 (15–80) |
| Age diagnosis, yrs (range) | 37±14 (2–76) |
| First ARVD related symptom, n (%) | |
| Palpitations | 20 (24%) |
| Syncope | 15 (18%) |
| VT | 33 (40%) |
| SCD | 1 (1%) |
| other | 13 (17%) |
| Family history, n (%) | |
| ARVD on autopsy (= major criterion) | 18 (22%) |
| SCD expected to be caused by ARVD/C | 8 (10%) |
| ARVD based on current criteria | 18 (22%) |
| Minor criterion (without presence of major criterion) | 11 (13%) |
| Repolarization abnormalities, n (%) | |
| T wave inversions V1-V3 (N=81) | 73 (90%) |
| Depolarization abnormalities, n (%) | |
| Epsilon wave in V1-V3 (N=78) | 11 (14%) |
| QRS prolongation in V1-V3 (N=79) | 34 (43%) |
| Late potential on SAECG (N=71) | 49 (69%) |
| Arrhythmias, n (% or range) | |
| LBBB-VT on ECG, Holter or X-ECG (N=80) | 47 (59%) |
| Age first recorded VT | 37±13 (2–80) |
| >1000 PVCs on Holter (N=57) | 37 (65%) |
| Minor dysfunction and structural alterations, n (%) (N=82) | 42 (51%) |
| Major dysfunction and structural alterations, n (%) (N=82) | 36 (44%) |
| Fibro-fatty replacement on endomyocardial biopsy, n (%) (N=33) | 13 (39%) |
Two different sequences have been reported for the first exon of DSG230,31. Analysis of DSG2 mRNA from human hearts by 5′-RACE was performed to determine if alternative splicing or an alternate first exon was present as previously reported for DSG2 sequence based on a human cancer cell line31. This demonstrated only one sequence for the DSG2 first exon30. Transcript sequence analysis was restricted to this first exon, and both cDNA and translated protein numbering was based on NCBI Reference Sequence NM_001943.
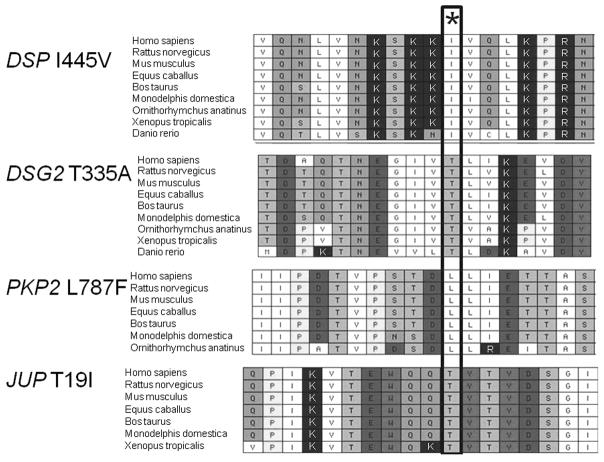
Within the desmosome genes sequenced in this cohort of 100 people, several novel and previously reported DNA sequence variants were identified. Those DNA sequence variants which are predicted not to alter the protein structure or amino acid sequence were not studied further. Some non-synonymous variants have already been determined to be polymorphisms and were also not studied further. The novel non-synonymous sequence variants were assessed in a population of 200 race-matched unaffected individuals (400 control chromosomes). Sequence variants which altered a conserved amino acid and were not present among controls were considered to be mutations. Supplemental table 2 includes 29 non-synonymous variants that were excluded as mutations due to presence among controls or lack of conservation of the encoded amino acid. We observed 31 unique non-synonymous mutations among the 100 probands; among these, 23 were previously reported and 8 are novel (Table 2). Four of the novel mutations result in frameshift and disruption of many conserved amino acids or premature termination, and 4 of the novel mutations are missense alterations of 1 conserved amino acid. Conservation of the mutated amino acid is shown in Figure 1.
Table 2.
Desmosome Mutations
| Gene | Nucleotide change | Amino acid or RNA change | Previous report | N (n=55) |
|---|---|---|---|---|
| PKP2 | c.145_148delCAGA | p.Thr50SerfsX61 | + | 4 |
| c.217_218dup (c.216insG) | p.Asn74AlafsX39 | + | 1 | |
| c.235C>T | p.Arg79X | + | 2 | |
| c.419C>T | p.Ser140Phe | + | 1 | |
| c.1171-2A>G | r.spl | + | 1 | |
| c.1237C>T | p.Arg413X | + | 2 | |
| c.1271T>C | p.Phe424Ser | + | 1 | |
| c.1307_1315del-ins8 | p.Leu436HisfsX11 | − | 1 | |
| c.1368delA | p.Lys456AsnfsX3 | + | 1 | |
| c.1613G>A | p.Trp538X | + | 5 | |
| c.1643delG (c.1642delG) | p.Gly548ValfsX15 | + | 1 | |
| c.1759G>A | p.Val587Ile | + | 1 | |
| c.2013delC (c.2011delC) | p.Lys672ArgfsX12 | + | 1 | |
| c.2145+1G>C | r.spl | − | 1 | |
| c.2146-1G>C | r.spl | + | 10 | |
| c.2197_2202delinsG | p.His733AlafsX8 | + | 3 | |
| c.2359C>T | p.Leu787Phe | − | 1 | |
| c.2484C->T | r.2483_2489del | + | 2 | |
| c.2489+1G>A | r.spl | + | 3 | |
| c.2489+1G>T | r.spl | − | 1 | |
| c.2509delA | p.Ser837ValfsX94 | + | 1 | |
| DSG2 | c.137G>A | p.Arg46Gln | + | 1 |
| c.146G>A | p.Arg49His | + | 2 | |
| c.166G>A | p.Val56Met | + | 1 | |
| c.918G>A | p.Trp306X | + | 1 | |
| c.1003A>G | p.Thr335Ala | − | 1 | |
| c.1520G>A | p.Cys507Tyr | + | 1 | |
| c.2434G>T | p.Gly812Cys | + | 1 | |
| c.829-1_835del | r.spl | − | 1 | |
| DSP | c.1331 A>G | p.Ile445Val | − | 1 |
| JUP | c.56C>T | p.Thr19Ile | − | 1 |
Each desmosome gene mutation is represented by its nucleotide and amino acid alteration. Those that have previously been reported are indicated with (+) and novel mutations are represented with (−) in column 4. Mutation nomenclature follows the Human Genome Variation Society guidelines (http://www.hgvs.org/mutnomen). For PKP2 mutations that have previously been reported with different cDNA designation, the prior nomenclature is provided in parentheses. Mutations that disrupt a splice site are noted as “r.spl” to indicate abnormally spliced mRNA.
Figure 1.
Conservation of the mutated amino acid in 4 novel missense mutations. Amino acids are represented by their standard abbreviations (I for isoleucine, T for threonine, L for leucine). * indicates site of mutation. Species are shown by their standard nomenclature.
In the cohort with ARVD/C, one or more desmosome gene mutations were identified in 43 people (52%). Thirty-seven of these (86%) had a single heterozygous desmosome gene mutation, while 6 people (14%) had more than 1 mutation (5 with 2 mutations, and 1 patient with 3 mutations), resulting in a cumulative total of 50 mutations. Forty-five percent of the ARVD/C cohort have one or more mutation in PKP2 (n=37), 9% in DSG2 (n=7), 1% in DSP (n=1), and 1% in JUP (n=1). We identified no mutations in DSC2. Mutations in more than one gene (digenic heterozygosity) were present in 4%. The remaining 48% have no discernible mutation in these desmosome genes.
Among the 55 separate mutations that occurred in the cohort of 100 (Table 2), consequences consisted of disruption of a critically conserved nucleotide at the exon-intron junction in 29%, insertions and/or deletions in 25%, missense substitution of a conserved amino acid in 24%, single nucleotide substitution resulting in premature termination codon in 18%, and cryptic splicing in 4% (Table 2).
Genotype – Phenotype relationship
Differences between patients with and without a desmosome mutation are shown in Table 3. Among males with ARVD/C, the likelihood of a desmosome gene mutation was higher (63%) than among females (38%). Patients with a desmosome mutation were more likely to have experienced VT (73% versus 44%), and experience their first episode of VT at a younger age (33±12 years versus 41±14 years). Figure 2 shows gender adjusted cumulative VT-free survival in patients with ARVD/C with and without a desmosome mutation, with a median VT-free survival of 32 years in patients with a mutation and 46 years in patients without a mutation. When stratified by gender, this difference in VT-free survival persists among women with regard to the presence or absence of a desmosome gene mutation (p=0.001) but not for men (p=0.072).
Table 3.
Comparison mutation carriers versus non-mutation carriers
| Mut + (n=43) | No Mut (n=39) | P-value | |
|---|---|---|---|
| Gender: male, n (%) | 30 (63%) | 18 (37%) | 0.03 |
| Gender: female, n (%) | 13 (38%) | 21 (62%) | |
| Age diagnosis, yrs | 33±12 | 40±15 | 0.04 |
| Age first ARVD/C related symptoms, yrs | 30±13 | 37±14 | 0.015 |
| Family history, n (%) | 16 (37%) | 15 (39%) | 0.91 |
| Family history proven on autopsy | 7 (16%) | 11 (28%) | 0.19 |
| Repolarization abnormalities, n (%) | 41 (95%) | 32 (84%) | 0.054 |
| Depolarization abnormalities, n (%) | 37 (88%) | 27 (73%) | 0.08 |
| LBBB-VT on ECG, Holter or stress-ECG | 30 (73%) | 17 (44%) | 0.007 |
| Age of first recorded VT | 33±12 | 41±14 | 0.007 |
| Minor dysfunction and structural alterations, n (%) | 20 (47%) | 22 (56%) | 0.37 |
| Major dysfunction and structural alterations, n (%) | 21 (49%) | 15 (39%) | 0.34 |
| Fibrofatty replacement on endomyocardial biopsy, n (%) | 5 (33%) | 8 (44%) | 0.51 |
Differences between patients with and without a desmosome mutation are shown here.
LBBB-VT refers to ventricular tachycardia with left bundle branch block morphology. P-values that are below 0.05 are indicated with bold-faced text.
Figure 2.
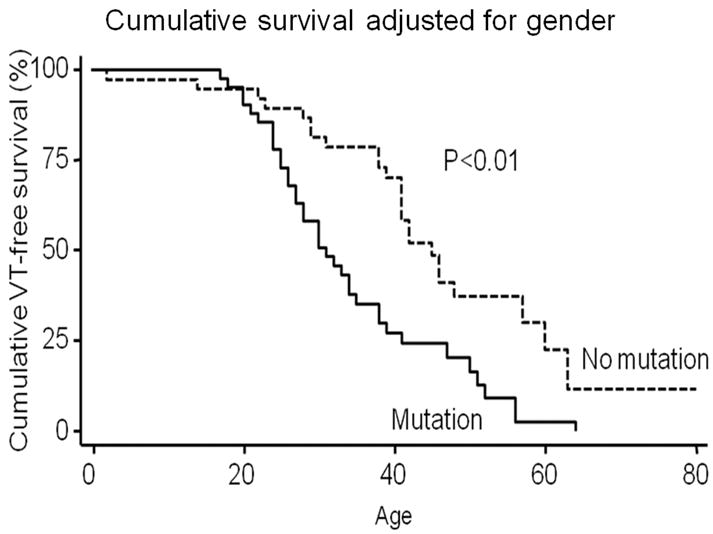
Kaplan-Meier survival analysis demonstrating cumulative VT-free survival in the study population stratified by presence of a desmosome gene mutation and adjusted for gender
When comparing the consequences of mutations, there was no significant difference found in symptom-free survival or VT survival among those with missense mutations compared to those with truncating gene mutations or splice altering mutations. There was also no significant difference recognized in VT-free survival between patients with mutations in the different desmosomal genes and between patients with one versus multiple mutations. In contrast with a prior report, we found no difference with regard to family history of ARVD/C among those with and without a desmosome gene mutation27.
Gender
In this study population there is a small male preponderance (59%). There are a few notable differences in comparison of affected men and women (Table 4). A mutation was identified in 30 out of 48 men (63%) and in 13 out of 34 women (38%) (p<0.05). In addition, men show more depolarization abnormalities and even though there is no significant difference in the prevalence of structural abnormalities, there is a difference in severity of structural abnormalities. Males in this cohort tend to have major structural abnormalities (54%), while women tend to have minor structural abnormalities (65%).
Table 4.
Gender differences
| Male (n=48) | Female (n=34) | P-value | |
|---|---|---|---|
| Mutation | 30 (63%) | 13 (38%) | 0.03 |
| Age diagnosis, yrs | 38±16 | 35±10 | 0.35 |
| Age first ARVD related symptoms, yrs | 34±15 | 32±12 | 0.55 |
| Family history, n (%) | 15 (31%) | 16 (47%) | 0.15 |
| Family history proven on autopsy | 9 (19%) | 9 (27%) | 0.40 |
| Repolarization abnormalities, n (%) | 41 (87%) | 32 (94%) | 0.21 |
| Depolarization abnormalities, n (%) | 41 (89%) | 23 (70%) | 0.03 |
| Major depolarization abnormalities | 28 (61%) | 9 (28%) | 0.004 |
| LBBB-VT on ECG, Holter or stress-ECG | 31 (67%) | 16 (47%) | 0.07 |
| Age of first recorded VT | 37±15 | 37±11 | 0.93 |
| Structural alterations, n (%) | 46 (96%) | 32 (94%) | 1.00 |
| Minor dysfunction and structural alterations, n (%) | 20 (42%) | 22 (65%) | 0.04 |
| Major dysfunction and structural alterations, n (%) | 26 (54%) | 10 (29%) | 0.026 |
| Fibrofatty replacement on endomyocardial biopsy, n (%) | 9 (53%) | 4 (25%) | 0.10 |
Repolarization abnormalities are considered present if there is T-wave inversion in ECG leads V1, V2, and V3 for those aged > 14 years in the absence of right bundle branch block. Depolarization abnormalities are considered major if there are epsilon waves or localized prolongation (>110ms) of the QRS complex in right precordial leads (V1–V3), and minor if there are late potentials on signal-averaged ECG. P-values that are below 0.05 are indicated with bold-faced text.
Suspected ARVD/C
Eighteen patients (39% male) with suspected ARVD/C (2 or more criteria without meeting current Task Force diagnostic criteria) were included in this study. Clinical characteristics are summarized in Supplemental Table 3. Mean age was 41±8 years. Age of suspected diagnosis varied between 15 and 44 years (mean 35±8 years) with the first symptoms of ARVD/C arising at age 13 to 40 years (mean 31±8 years).
A desmosome gene mutation was identified in 5 patients (28%). One of them had a single heterozygous desmosome gene mutation in DSG2, and 4 had an isolated heterozygous mutation in PKP2. The mutations identified in PKP2 consisted of 2 deletions, a disruption of a critically conserved nucleotide at the intron-exon splice site, and an insertion. The isolated DSG2 mutation is a missense substitution of a conserved amino acid15. There were no significant differences between patients suspected of ARVD/C with and without mutations. Although there is a reduced prevalence of desmosome gene mutations in the cohort with suspected ARVD/C compared to those who fulfill current clinical criteria (28% versus 52%), the numbers are too small to achieve statistical significance.
Multiple mutations
One individual in this cohort was found to have three different previously reported mutations. This individual (#43, Supplemental table 1) is a 50-year old man who underwent evaluation for ARVD/C after an episode of syncope due to ventricular tachycardia. With phenotypic evaluation, he meets 2 major and 3 minor criteria. Desmosome gene sequencing identified 2 previously published mutations in PKP2 (c.2146-1G>C and c.419C>T) as well as a previously published mutation in DSG2 (p.V56M)14,30. We considered each of these mutations independently; PKP2 c.2146-1G>C results in abnormal splicing with loss of exon 11 with frameshift and alteration of many conserved amino acids, strongly supporting its pathogenicity14. The PKP2 missense mutation c.419C>T (p.S140F) alters only a single amino acid, though it has repeatedly been found in association with ARVD/C and was not seen in over 500 control chromosomes14,26,28,32. However, in the absence of a well validated functional assay for desmosome protein variants of uncertain significance, one cannot definitively ascribe a pathogenic role to this allele. Finally, the DSG2 V56M variant has also been reported in ARVD/C and this valine is highly conserved30. Although this allele was recently found among 3/617 individuals in mixed German populations (2 groups of cardiac patients without LV cardiomyopathy and 1 cohort of blood donors without phenotypic testing), it was found in 10-fold greater prevalence among people with nonischemic dilated cardiomyopathy (DCM)33. These authors proceeded to show that the presence of the DSG2 V56M allele cosegregates with disease in families with DCM, is associated with both abnormal cardiac immunostaining and abnormal electron microscopy, and is putatively functional in protein modelling studies33. We share these authors conclusion that DSG2 V56M likely plays a contributory role, though it may not be sufficient by itself to result in cardiomyopathy34.
Although compound heterozygosity (2 different mutations in the same gene) has been reported in ARVD/C, digenic heterozygosity (heterozygous mutations in 2 genes) has not previously been reported. Digenic heterozygosity was present in 2 additional individuals with ARVD/C in this cohort, emphasizing the importance of comprehensive genetic analysis even if a single gene mutation is identified.
Discussion
In an era of increasing availability and use of clinical genetic testing, it is important to consider its possibilities and limitations. Our results show diversity in the desmosome genotypes of North American probands with ARVD/C, and the genetic findings in this cohort are different than those in other reported series23,25. In addition, as previously seen with analysis restricted to PKP2, individuals with ARVD/C and any desmosome gene mutation have VT at an earlier age compared to those without a desmosome gene mutation26.
As seen in some other cohorts, the likelihood of finding a PKP2 mutation remains quite high in this group27. Though mutations occur in other desmosome genes, they are less frequently seen among this group of North Americans. Our analysis excluded individuals with left ventricular dilation and dysfunction. Lower prevalence of PKP2 mutations in other series may relate to inclusion of those with biventricular cardiomyopathy or those with predominant left ventricular involvement22,35.
Importantly, mutations in more than 1 gene were found in 3 probands (3%). This is also the first identification of three previously reported gene mutations in an individual with ARVD/C. The possibility of multiple pathogenic mutations leads to a challenge in genetic counseling. Once a disease causing mutation is identified in an individual with ARVD/C, unaffected family members may choose to be screened for the presence of this mutation to determine their risk of developing ARVD/C. Mutation carriers may modify their lifestyle to decrease the likelihood of sudden cardiac death. Family members who do not share a single desmosome mutation present in a proband may incorrectly be advised of a low risk of developing ARVD/C if more extensive testing was not performed in the proband. With the possibility of multiple mutations influencing disease expression, comprehensive cardiac desmosome genetic testing should be performed for this condition.
No desmosome gene mutation was identified in nearly half of the patients. The genetic basis for their condition may be due to mutations in non-desmosome ARVD/C genes such as RYR2 or TMEM43, though phenotypes of individuals with mutations in these genes are typically different than the probands in our cohort18,20. Also, traditional sequence analysis may not identify large deletions or gene rearrangements. Additional genes with mutations resulting in ARVD/C are likely to be identified in the future. Finally, it is possible that people with ARVD/C and no discernible desmosome gene mutation may have non-genetic causes for their cardiomyopathy, such as viral infection or autoimmunity36,37.
The likelihood of finding a desmosome gene mutation was lower for individuals with suspected ARVD/C compared to those who meet current Task Force Criteria for this condition. This is important to note in the context of clinical use of desmosome gene sequencing for individuals with suspected ARVD/C. According to the current Task Force Criteria for ARVD/C, the presence or absence of a desmosome gene mutation does not influence the diagnosis of this condition.
The utility of clinical genetic testing can be interpreted in many different contexts. Priori and Napolitano have reported a scoring system for applicability of such testing38. Using their scoring system, the use of comprehensive desmosome genetic testing receives a score of 4.0, thus supporting genetic testing in ARVD/C. However, when using their scoring system for each of the desmosome genes separately, PKP2 is the only gene with a score higher than 3 for both men and women.
Because affected individuals may experience SCD as their initial manifestation, presymptomatic recognition and lifestyle modifications are easily justified. However, the low penetrance and possibility of additional genetic factors influencing the phenotype emphasizes the importance of proper genetic counseling for affected probands and at-risk family members.
Study Limitations
There are a few limitations to the study performed. First of all there is a selection bias. These people came to medical attention because they had symptoms of ARVD/C themselves or because a family member was diagnosed with ARVD/C. The people with the most severe first manifestation of disease, sudden cardiac death, are not represented in this group. Family members of these individuals are represented in this group, but they may have less severe disease. Another limitation is that there is currently no functional test available to study the consequences of mutations in desmosome genes. Despite rigorous criteria to designate novel sequence alterations as mutations or polymorphisms, misclassification is possible in the absence of functional data.
Conclusions
In a cohort of 100 individuals with clinically confirmed or suspected ARVD/C, desmosome gene mutations are frequently identified. In rare cases, more than one gene mutation is present. People with ARVD/C and a desmosome gene mutation present at a younger age and are more likely to have VT. Clinical use of comprehensive cardiac desmosome gene testing for individuals with this condition may identify family members at increased risk of developing ARVD/C.
Supplementary Material
Acknowledgments
The authors would also like to acknowledge the Johns Hopkins ARVD Program (http://www.arvd.com).
Funding sources: This work was supported by funding from the National Institutes of Health (HL088072 to DPJ), the France-Merrick Foundation, JHU Friends in Red, and the Jeff Cooper CARE Foundation. ADDH was supported by the Stichting Dr. Hendrik Muller’s Vaderlandsch Fonds, the Van Wijck-Stam-Caspers fund for cardiovascular research, and the Trajectum Scholarship. HC received funding from the Bogle Foundation, the Campanella family, the Wilmerding Endowment, the Healing Hearts Foundation, Boston Scientific, Medtronic, and St Jude Medical, Inc. to support this project in part.
Footnotes
Conflict of Interest Disclosures: HC receives honoraria from Boston Scientific, Medtronic, and St. Jude Medical, Inc and is a consultant for Medtronic. The authors have no other potential conflicts of interest to disclose.
Journal Subject Codes: [109]Clinical genetics, [89]Genetics of cardiovascular disease
References
- 1.Marcus FI, Fontaine GH, Guiraudon G, Frank R, Laurenceau JL, Malergue C, Grosgogeat Y. Right ventricular dysplasia: a report of 24 adult cases. Circulation. 1982;65:384–98. doi: 10.1161/01.cir.65.2.384. [DOI] [PubMed] [Google Scholar]
- 2.Thiene G, Nava A, Corrado D, Rossi L, Pennelli N. Right ventricular cardiomyopathy and sudden death in young people. N Engl J Med. 1988;318:129–33. doi: 10.1056/NEJM198801213180301. [DOI] [PubMed] [Google Scholar]
- 3.Marcus FI, Fontaine GH, Frank R, Gallagher JJ, Reiter MJ. Long-term follow-up in patients with arrhythmogenic right ventricular disease. Eur Heart J. 1989;10 (Suppl D):68–73. doi: 10.1093/eurheartj/10.suppl_d.68. [DOI] [PubMed] [Google Scholar]
- 4.Dalal D, Nasir K, Bomma C, Prakasa K, Tandri H, Piccini J, Roguin A, Tichnell C, James C, Russell SD, Judge DP, Abraham T, Spevak PJ, Bluemke DA, Calkins H. Arrhythmogenic right ventricular dysplasia: a United States experience. Circulation. 2005;112:3823–32. doi: 10.1161/CIRCULATIONAHA.105.542266. [DOI] [PubMed] [Google Scholar]
- 5.McKenna WJ, Thiene G, Nava A, Fontaliran F, Blomstrom-Lundqvist C, Fontaine G, Camerini F. Diagnosis of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Br Heart J. 1994;71:215–8. doi: 10.1136/hrt.71.3.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corrado D, Basso C, Schiavon M, Thiene G. Screening for Hypertrophic Cardiomyopathy in Young Athletes. N Engl J Med. 1998;339:364–369. doi: 10.1056/NEJM199808063390602. [DOI] [PubMed] [Google Scholar]
- 7.Peters S, Trummel M, Meyners W. Prevalence of right ventricular dysplasia-cardiomyopathy in a non-referral hospital. International Journal of Cardiology. 2004;97:499–501. doi: 10.1016/j.ijcard.2003.10.037. [DOI] [PubMed] [Google Scholar]
- 8.Corrado D, Thiene G. Arrhythmogenic right ventricular cardiomyopathy/dysplasia: clinical impact of molecular genetic studies. Circulation. 2006;113:1634–7. doi: 10.1161/CIRCULATIONAHA.105.616490. [DOI] [PubMed] [Google Scholar]
- 9.Marcus F, Towbin JA. The mystery of arrhythmogenic right ventricular dysplasia/cardiomyopathy: from observation to mechanistic explanation. Circulation. 2006;114:1794–5. doi: 10.1161/CIRCULATIONAHA.106.653493. [DOI] [PubMed] [Google Scholar]
- 10.McKoy G, Protonotarios N, Crosby A, Tsatsopoulou A, Anastasakis A, Coonar A, Norman M, Baboonian C, Jeffery S, McKenna WJ. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease) Lancet. 2000;355:2119–2124. doi: 10.1016/S0140-6736(00)02379-5. [DOI] [PubMed] [Google Scholar]
- 11.Norgett EE, Hatsell SJ, Carvajal-Huerta L, Ruiz Cabezas J-C, Common J, Purkis PE, Whittock N, Leigh IM, Stevens HP, Kelsell DP. Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum Mol Genet. 2000;9:2761–2766. doi: 10.1093/hmg/9.18.2761. [DOI] [PubMed] [Google Scholar]
- 12.Asimaki A, Syrris P, Wichter T, Matthias P, Saffitz JE, McKenna WJ. A novel dominant mutation in plakoglobin causes arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2007;81:964–73. doi: 10.1086/521633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rampazzo A, Nava A, Malacrida S, Beffagna G, Bauce B, Rossi V, Zimbello R, Simionati B, Basso C, Thiene G, Towbin JA, Danieli GA. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2002;71:1200–6. doi: 10.1086/344208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermott DA, Lerman BB, Markowitz SM, Ellinor PT, MacRae CA, Peters S, Grossmann KS, Drenckhahn J, Michely B, Sasse-Klaassen S, Birchmeier W, Dietz R, Breithardt G, Schulze-Bahr E, Thierfelder L. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet. 2004;36:1162–4. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- 15.Awad MM, Dalal D, Cho E, Amat-Alarcon N, James C, Tichnell C, Tucker A, Russell SD, Bluemke DA, Dietz HC, Calkins H, Judge DP. DSG2 mutations contribute to arrhythmogenic right ventricular dysplasia/cardiomyopathy. Am J Hum Genet. 2006;79:136–42. doi: 10.1086/504393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Syrris P, Ward D, Evans A, Asimaki A, Gandjbakhch E, Sen-Chowdhry S, McKenna WJ. Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Associated with Mutations in the Desmosomal Gene Desmocollin-2. Am J Hum Genet. 2006;79:978–984. doi: 10.1086/509122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beffagna G, Occhi G, Nava A, Vitiello L, Ditadi A, Basso C, Bauce B, Carraro G, Thiene G, Towbin JA, Danieli GA, Rampazzo A. Regulatory mutations in transforming growth factor-beta3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc Res. 2005;65:366–73. doi: 10.1016/j.cardiores.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Tiso N, Stephan DA, Nava A, Bagattin A, Devaney JM, Stanchi F, Larderet G, Brahmbhatt B, Brown K, Bauce B, Muriago M, Basso C, Thiene G, Danieli GA, Rampazzo A. Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2) Hum Mol Genet. 2001;10:189–94. doi: 10.1093/hmg/10.3.189. [DOI] [PubMed] [Google Scholar]
- 19.Priori SG, Napolitano C, Tiso N, Memmi M, Vignati G, Bloise R, Sorrentino V, Danieli GA. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation. 2001;103:196–200. doi: 10.1161/01.cir.103.2.196. [DOI] [PubMed] [Google Scholar]
- 20.Merner ND, Hodgkinson KA, Haywood AF, Connors S, French VM, Drenckhahn JD, Kupprion C, Ramadanova K, Thierfelder L, McKenna W, Gallagher B, Morris-Larkin L, Bassett AS, Parfrey PS, Young TL. Arrhythmogenic Right Ventricular Cardiomyopathy Type 5 Is a Fully Penetrant, Lethal Arrhythmic Disorder Caused by a Missense Mutation in the TMEM43 Gene. Am J Hum Genet. 2008 doi: 10.1016/j.ajhg.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalal D, James C, Devanagondi R, Tichnell C, Tucker A, Prakasa K, Spevak PJ, Bluemke DA, Abraham T, Russell SD, Calkins H, Judge DP. Penetrance of Mutations in Plakophilin-2 Among Families With Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy. J Am Coll Cardiol. 2006;48:1416–1424. doi: 10.1016/j.jacc.2006.06.045. [DOI] [PubMed] [Google Scholar]
- 22.Sen-Chowdhry S, Syrris P, McKenna WJ. Role of Genetic Analysis in the Management of Patients With Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy. Journal of the American College of Cardiology. 2007;50:1813–1821. doi: 10.1016/j.jacc.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Pilichou K, Nava A, Basso C, Beffagna G, Bauce B, Lorenzon A, Frigo G, Vettori A, Valente M, Towbin J, Thiene G, Danieli GA, Rampazzo A. Mutations in desmoglein-2 gene are associated with arrhythmogenic right ventricular cardiomyopathy. Circulation. 2006;113:1171–9. doi: 10.1161/CIRCULATIONAHA.105.583674. [DOI] [PubMed] [Google Scholar]
- 24.Awad MM, Dalal D, Tichnell C, James C, Tucker A, Abraham T, Spevak PJ, Calkins H, Judge DP. Recessive arrhythmogenic right ventricular dysplasia due to novel cryptic splice mutation in PKP2. Hum Mutat. 2006;27:1157. doi: 10.1002/humu.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sen-Chowdhry S, Syrris P, Ward D, Asimaki A, Sevdalis E, McKenna WJ. Clinical and genetic characterization of families with arrhythmogenic right ventricular dysplasia/cardiomyopathy provides novel insights into patterns of disease expression. Circulation. 2007;115:1710–20. doi: 10.1161/CIRCULATIONAHA.106.660241. [DOI] [PubMed] [Google Scholar]
- 26.Dalal D, Molin LH, Piccini J, Tichnell C, James C, Bomma C, Prakasa K, Towbin JA, Marcus FI, Spevak PJ, Bluemke DA, Abraham T, Russell SD, Calkins H, Judge DP. Clinical features of arrhythmogenic right ventricular dysplasia/cardiomyopathy associated with mutations in plakophilin-2. Circulation. 2006;113:1641–9. doi: 10.1161/CIRCULATIONAHA.105.568642. [DOI] [PubMed] [Google Scholar]
- 27.van Tintelen JP, Entius MM, Bhuiyan ZA, Jongbloed R, Wiesfeld ACP, Wilde AAM, van der Smagt J, Boven LG, Mannens MMAM, van Langen IM, Hofstra RMW, Otterspoor LC, Doevendans PAFM, Rodriguez L-M, van Gelder IC, Hauer RNW. Plakophilin-2 Mutations Are the Major Determinant of Familial Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy. Circulation. 2006;113:1650–1658. doi: 10.1161/CIRCULATIONAHA.105.609719. [DOI] [PubMed] [Google Scholar]
- 28.Syrris P, Ward D, Asimaki A, Sen-Chowdhry S, Ebrahim HY, Evans A, Hitomi N, Norman M, Pantazis A, Shaw AL, Elliott PM, McKenna WJ. Clinical expression of plakophilin-2 mutations in familial arrhythmogenic right ventricular cardiomyopathy. Circulation. 2006;113:356–64. doi: 10.1161/CIRCULATIONAHA.105.561654. [DOI] [PubMed] [Google Scholar]
- 29.Lahtinen AM, Lehtonen A, Kaartinen M, Toivonen L, Swan H, Widen E, Lehtonen E, Lehto VP, Kontula K. Plakophilin-2 missense mutations in arrhythmogenic right ventricular cardiomyopathy. Int J Cardiol. 2008;126:92–100. doi: 10.1016/j.ijcard.2007.03.137. [DOI] [PubMed] [Google Scholar]
- 30.Syrris P, Ward D, Asimaki A, Evans A, Sen-Chowdhry S, Hughes SE, McKenna WJ. Desmoglein-2 mutations in arrhythmogenic right ventricular cardiomyopathy: a genotype-phenotype characterization of familial disease. Eur Heart J. 2007;28:581–8. doi: 10.1093/eurheartj/ehl380. [DOI] [PubMed] [Google Scholar]
- 31.Schafer S, Koch PJ, Franke WW. Identification of the Ubiquitous Human Desmoglein, Dsg2, and the Expression Catalogue of the Desmoglein Subfamily of Desmosomal Cadherins. Exp Cell Res. 1994;211:391–399. doi: 10.1006/excr.1994.1103. [DOI] [PubMed] [Google Scholar]
- 32.Behr ER, Dalageorgou C, Christiansen M, Syrris P, Hughes S, Tome Esteban MT, Rowland E, Jeffery S, McKenna WJ. Sudden arrhythmic death syndrome: familial evaluation identifies inheritable heart disease in the majority of families. Eur Heart J. 2008;29:1670–1680. doi: 10.1093/eurheartj/ehn219. [DOI] [PubMed] [Google Scholar]
- 33.Posch MG, Posch MJ, Geier C, Erdmann B, Mueller W, Richter A, Ruppert V, Pankuweit S, Maisch B, Perrot A, Buttgereit J, Dietz R, Haverkamp W, Ozcelik C. A missense variant in desmoglein-2 predisposes to dilated cardiomyopathy. Mol Genet Metab. 2008;95:74–80. doi: 10.1016/j.ymgme.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Posch MG, Posch MJ, Perrot A, Dietz R, Ozcelik C. Variations in DSG2: V56M, V158G and V920G are not pathogenic for arrhythmogenic right ventricular dysplasia/cardiomyopathy. Nat Clin Pract Cardiovasc Med. 2008;5:E1. doi: 10.1038/ncpcardio1434. [DOI] [PubMed] [Google Scholar]
- 35.Norman M, Simpson M, Mogensen J, Shaw A, Hughes S, Syrris P, Sen-Chowdhry S, Rowland E, Crosby A, McKenna WJ. Novel mutation in desmoplakin causes arrhythmogenic left ventricular cardiomyopathy. Circulation. 2005;112:636–42. doi: 10.1161/CIRCULATIONAHA.104.532234. [DOI] [PubMed] [Google Scholar]
- 36.Calabrese F, Basso C, Carturan E, Valente M, Thiene G. Arrhythmogenic right ventricular cardiomyopathy/dysplasia: is there a role for viruses? Cardiovasc Pathol. 2006;15:11–7. doi: 10.1016/j.carpath.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Fornes P, Ratel S, Lecomte D. Pathology of arrhythmogenic right ventricular cardiomyopathy/dysplasia--an autopsy study of 20 forensic cases. J Forensic Sci. 1998;43:777–83. [PubMed] [Google Scholar]
- 38.Priori SG, Napolitano C. Role of genetic analyses in cardiology: part I: mendelian diseases: cardiac channelopathies. Circulation. 2006;113:1130–5. doi: 10.1161/CIRCULATIONAHA.105.563205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.