Abstract
Background
Pulse wave velocity (PWV), a non-invasive index of central arterial stiffness, is a potent predictor of cardiovascular mortality and morbidity. Heritability and linkage studies have pointed toward a genetic component affecting PWV. We conducted a genome-wide association study to identify single nucleotide polymorphisms (SNPs) associated with PWV.
Methods and Results
The study cohort included participants from the SardiNIA study for whom PWV measures were available. Genotyping was performed in 4,221 individuals, using either the Affymetrix 500K or the Affymetrix 10K mapping array sets (with imputation of the missing genotypes). Associations with PWV were evaluated using an additive genetic model that included age, age2 and sex as covariates. The findings were tested for replication in an independent internal Sardinian cohort of 1,828 individuals, using a custom-chip designed to include the top 43 non-redundant SNPs associated with PWV. Of the loci that were tested for association with PWV, the nonsynonymous SNP rs3742207 in the COL4A1 gene on chromosome 13 and SNP rs1495448 in the MAGI1 gene on chromosome 3 were successfully replicated (p=7.08×10−7 and p=1.06×10−5 respectively for the combined analyses). The association between rs3742207 and PWV was also successfully replicated (p=0.02) in an independent population, the Old Order Amish, leading to an overall p=5.16×10−8.
Conclusions
A genome-wide association study identified a SNP in the COL4A1 gene that was significantly associated with PWV in two populations. Collagen type 4 is the major structural component of basement membranes, suggesting that previously unrecognized cell-matrix interactions may exert an important role in regulating arterial stiffness.
Keywords: Arterial Stiffness, Pulse Wave Velocity, Genome-Wide Association Study, COL4A1, MAGI1, genetics
Introduction
Central arterial stiffening is one of the hallmarks of arterial aging. Carotid-femoral pulse wave velocity (PWV) is the preferred non-invasive measure of central arterial stiffness1. PWV is increased in patients with cardiovascular conditions such as hypertension, diabetes, metabolic syndrome, and atherosclerosis2. Furthermore, PWV is an independent predictor of hypertension3 and of coronary heart disease and stroke in healthy subjects4 and an independent predictor of mortality in the general population5, in hypertensive subjects6, in older community dwelling individuals7, and in patients with end-stage renal disease8. Thus, understanding the mechanisms of central arterial stiffening may lead to effective interventions that could improve PWV and favorably impact cardiovascular morbidity and mortality.
Increasing central arterial stiffness has traditionally been thought to result from the breakdown of elastin in central arterial walls, due to the repeated cycles of distension and recoil of the central aorta, and from the deposition and cross-linking of collagen. There is increasing recognition that arterial stiffening is influenced by lifestyle (e.g. dietary salt consumption, exercise), and is regulated by several signaling pathways (e.g. nitric oxide)2. Furthermore, gene expression studies9, heritability10,11 and linkage11 analyses, and a genome wide association study (GWAS) performed in 644 subjects from the Framingham Heart Study12 are all consistent with the likely involvement of genetic factors in modulating the variability in PWV.
To look for these factors, we performed a GWAS in a large founder population (see supplementary materials) from Sardinia. Furthermore, to confirm the validity of our findings we tested the association of the detected loci with PWV both in a second Sardinian cohort and in a separate founder population, the Old Older Amish.
Methods
Study Sample
The SardiNIA study recruited 6,148 men and women aged 14–102 years (62.5% of the eligible population)10, from a cluster of four towns in the Lanusei valley on the island of Sardinia. The cohort includes 34,469 relative pairs, 4,933 sibling pairs, 180 half-sibling pairs, 4,014 first cousins, 4,256 parent–child pairs, 675 grandparent–grandchild pairs, and 6,400 avuncular pairs in addition to other more distant relatives. Most subjects (95%) had all 4 grand-parents born in Sardinia10, and environmental factors have remained relatively homogeneous. To achieve the accrual goal for the study, the project was advertised through provincial, religious, and municipal authorities; in local television, newspaper, and radio messages; through local physicians and by mailings and phone calls. All subjects underwent extensive phenotyping, which included assessment of traditional cardiovascular risk factors (e.g. blood pressure, cholesterol fractions, etc.) with standard methodologies, as well as the assessment of arterial stiffness by PWV. Blood draws yielded lymphocytes for subsequent DNA extraction. All subjects provided a written informed consent for participation in the study that was approved by both the Sardinian and the National Institute on Aging’s Institutional Review Boards.
PWV measurement
PWV was measured in triplicate using nondirectional transcutaneous Doppler probes (model 810A, 9 to 10-Mhz probes, Parks Medical Electronics, Inc., Aloha, OR) as previously described13: A minimum of 10 arterial flow waves from the right common carotid artery and the right femoral artery were simultaneously recorded and averaged. PWV was calculated as distance divided by time: The distance traveled by the flow wave was measured with an external tape measure over the body surface, and was calculated as the distance between the manubrium and the femoral sampling site, minus the distance between the manubrium and the carotid sampling site; The time traveled by the flow wave was measured as the time delay between the feet of simultaneously recorded carotid and femoral arterial waveforms. The waveforms were simultaneously collected by 2 sonographers, one recording at the carotid site and one recording at the femoral site. All data were subsequently analyzed by a single investigator (AS) who was blinded to the clinical characteristics of the subjects. Details of a reproducibility study for PWV are provided in the supplementary material. Forty four individuals did not contribute PWV data to the analysis because of poor quality waveforms (N=21) or because of atrial fibrillation (N=23).
Genotyping
We took advantage of the relatedness among individuals in our sample to substantially reduce study costs14. Specifically, because our sample includes many large families, we reasoned that genotyping a relatively small number of markers in all individuals would allow us to identify shared haplotype stretches within each family. We could then genotype a subset of the individuals in each family at higher density to characterize the haplotypes in each stretch and impute missing genotypes in other individuals in the family14.
Genotyping was performed with the Affymetrix 10K and 500K Mapping Arrays in 3,329 and 1,412 individuals respectively (436 subjects were genotyped with both chips). Individuals typed with the 500K array were specifically selected because they represented the largest families in our sample, not based on their phenotype. Furthermore, for the larger sibships in our cohort, both parents and one child were selected, whereas in the smaller sibships, only the two parents were selected. The lower density arrays were used to genotype everyone else. Except when parents and offsprings were genotyped in the same family, we tried to ensure that individuals genotyped with the high-density array were only distantly related to one another. In the 2,893 individuals typed with only the 10K panel, we took advantage of the overlapping dataset and of the relatedness of the population to estimate the missing genotypes based on stretches of shared haplotypes, using a modified Lander-Green algorithm.15, 16 This approach for estimating missing genotypes is implemented in MERLIN (http://www.sph.umich.edu/csg/abecasis/MERLIN/) and is described in detail elsewhere 16, 17 and in the supplementary materials.
Prior the imputation process, low quality markers that could affect the accuracy of dosage estimates were removed. In particular, markers that met any of the following criteria were discarded: call rate ≤ 90%, minor allele frequency ≤ 5%, excess of Mendelian inconsistencies or departure from Hardy– Weinberg equilibrium. From the 10K and the 500K chips we were able to analyze 7,407 and 356,359 markers respectively, resulting in a total of 362,129 markers after accounting for overlapping polymorphisms18. The genotype completeness rates exceeded 98%.
After completing this initial phase, we devised a custom chip to test for internal replication of our major findings and to eliminate possible genotyping errors. This chip consisted of 11,617 SNPs and included, for each of the quantitative traits in the SardiNIA study10, the top SNPs that were associated with these traits in GWAS. Forty-three unique SNPs associated with PWV were included on this chip. This custom chip was used to type the remaining 1,857 subjects recruited into the SardiNIA study who had not been typed with the 500K or 10K chips (denoted as SardiNIA stage 2). These individuals were not related to the individuals typed with the 500K chip (kinship coefficient =0) (see supplementary materials), and thus were a suitable cohort to serve as an internal replication sample. We considered a SNP to be internally replicated if the direction of effect was in the same direction as the initial study with p < 0.05.
Statistical analyses
To ensure adequate control of type I error rates, an inverse normal transformation was applied to PWV prior to analysis, to reduce the impact of outliers and minimize deviations from normality (Figure S1). This transformation involves ranking all available PWV values, transforming these ranks into quantiles and finally converting the resulting quantiles into normal deviates. To perform the genome wide association analysis, a simple regression model was fitted and a variance component approach that modeled background polygenic effects was used to account for correlation between different observed phenotypes within each family17. The association analysis was performed using an additive genetic model, and was adjusted for age, age2 and sex. An initial Q/Q plot suggested that the genomic control parameter was inflated (lambda = 1.14), likely reflecting residual relatedness in this founder population. Therefore, the p-values were adjusted according to the genomic control method19 (Figure S2). The p-values derived from the initial GWAS and from the association tests on the individuals genotyped with the custom chip were then combined using the z-scores meta-analysis methods, and taking into account the number of subjects analyzed in each set and the direction and magnitude of the estimated effect.
External replication
The top findings from our study were tested in 813 subjects from a genetically distant second founder population, the Old Order Amish of Lancaster, PA. These included healthy Amish subjects enrolled in two studies, the Heredity and Phenotype Intervention (HAPI) Heart Study and the Amish Longevity Study (ALS). The HAPI Heart Study was initiated in 2002 to measure the cardiovascular response to 4 short-term interventions affecting cardiovascular risk factors and to identify the genetic and environmental determinants of these responses.20 The ALS was initiated in 2000 to identify the genetic factors associated with living to an old age, and recruited Amish individuals living to age 92 years or older, their offspring, and the offspring spouses21. PWV was assessed with the Complior®SP device (Artech Medical, Pantin, France) before the interventions were administered, and genotyping was performed with an Affymetrix 500K chip. GWAS in the Old Order Amish were adjusted for family structure, and the data transformation and statistical analyses were performed in an analogous manner to those in the SardiNIA study.
Results
After excluding subjects with atrial fibrillation or poor quality PWV tracings, a total of 4,221 and 1,828 Sardinians were considered for the initial GWA analysis and for the internal replication (SardiNIA stage 2), respectively. The demographic and clinical characteristics of the study cohort are shown in Table 1. The mean age was 43.7±17.6 years (range 14–102 years); 58% were women, 20% reported a history of smoking, 29% were hypertensive, 5% were diabetics, and only 1% reported a clinical history of myocardial infarction. As expected2, PWV increased with advancing age in a quadratic fashion (Figure 1).
Table 1. Demographic and clinical characteristics of the SardiNIA cohort (overall, and stratified according to the phase of genotyping), and of the Old Order Amish study cohort.
| SardiNIA |
Old Order Amish | |||
|---|---|---|---|---|
| Overall | Initial GWAS | Stage 2 | ||
| N=6,049 | N=4,221 | N=1,828 | N=826 | |
| Age (years) | 43.7±17.6 | 43.1±17.4 | 44.1±16.9 | 46.4±15.1 |
| Sex (male) | 2569 (42.5%) | 1841 (43.6%) | 728 (39.8%) | 443 (53.6%) |
| Smoking (ever) | 1237 (20.3%) | 850 (20.1%) | 387 (21.2%) | 211 (25.5) |
| Height (cm) | 159.9±9.1 | 159.9±9.0 | 160.1±9.1 | 167.1±8.8 |
| Weight (kg) | 64.9±13.3 | 64.8±13.3 | 65.0±13.2 | 73.8±12.7 |
| BMI (kg/m2) | 25.3±4.7 | 25.3±4.7 | 25.3±4.6 | 26.4±4.3 |
| Systolic Blood Pressure (mmHg) | 125±18 | 125±18 | 125±18 | 120±15 |
| Diastolic Blood Pressure (mmHg) | 77±11 | 77±11 | 77±11 | 75±9 |
| Heart Rate (beats/min) | 67±11 | 67±11 | 68±11 | 64±9 |
| PWV (cm/sec) | 670±217 | 669±209 | 672±207 | 547±143 |
| Total cholesterol (mg/dL) | 209±43 | 208±42 | 210±42 | 210±47 |
| Triglycerides (mg/dL) | 88±68 | 87±68 | 89±69 | 72±44 |
| LDL (mg/dL) | 127±36 | 127±35 | 127±36 | 139±42 |
| HDL (mg/dL) | 64±15 | 64±15 | 64±15 | 56±15 |
| Fasting glucose (mg/dL) | 90±23 | 90±23 | 90±25 | 87±10 |
| Creatinine (mg/dL) | 0.8±0.2 | 0.8±0.2 | 0.8±0.2 | 0.8±0.2 |
| Antihypertensive medications | 752 (12.4%) | 544 (12.2%) | 208 (11.4%) | 18 (2.2%) |
Data are expressed as Mean±SD or N (%)
Figure 1.
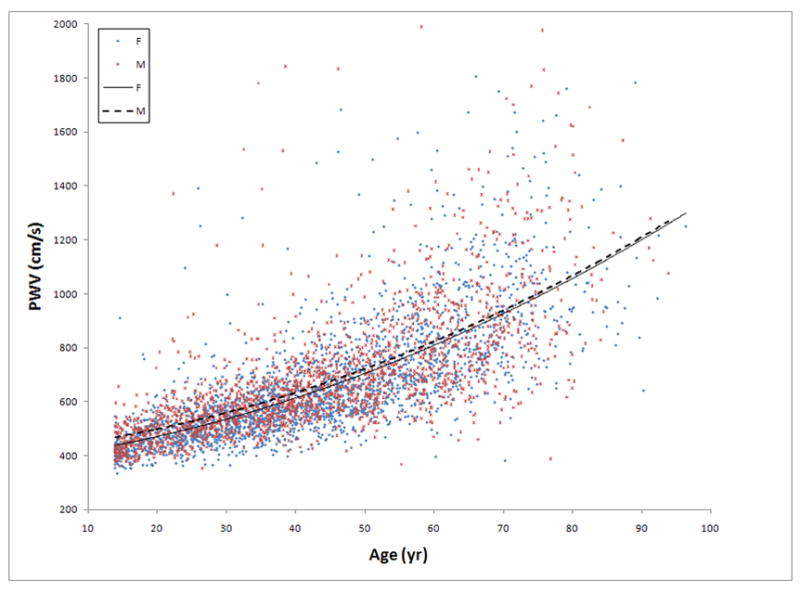
The relationship of pulse wave velocity (PWV) and age in men and women in the SardiNIA study cohort who were genotyped with the 500K/10K Gene Array Set. The best fit regression curves (quadratic) are shown.
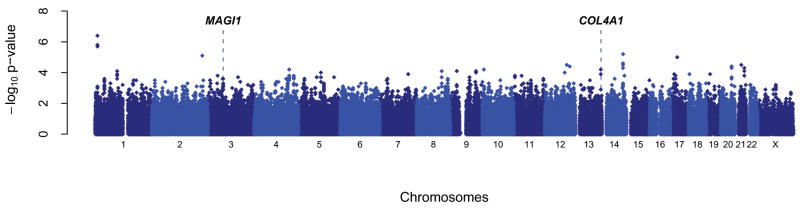
We first conducted GWAS to survey the genome for common variants associated with PWV. Figure 2 graphically summarizes the associations of PWV with the >329,129 SNPs with a minor allele frequency >5% that passed quality control checks and transmission disequilibrium testing. The top 100 hits are shown in Table S1. Promising findings were noted on chromosomes 1, 2, 4, 12–14, 17, 20, 21, encompassing 18 SNPs with p<2×10−5.
Figure 2.
Summary of genome-wide association studies (GWAS) for pulse wave velocity (PWV) in the SardiNIA study cohort. The −log10 of the p-values for the associations of PWV with 362,169 SNPs that passed quality control filters are plotted according to the positions of these SNPs along the p to q arms of each chromosome (1–22, X). The positions of COL4A1 and MAGI1, the two genes that were replicated in the SardiNIA stage 2 analysis, are highlighted.
To further evaluate these initial results, a secondary custom chip from Affymetrix was devised based on the top findings from the GWAS. For PWV, the top 85 SNPs were considered; of these, 43 were not in strong linkage disequilibrium and were included in the custom chip. The 1,828 individuals typed with this custom chip were genetically independent from those genotyped in the initial analysis. Thus, the custom chip helped validate the initial findings, and provided an “internal” replication within the Sardinian cohort. Of the 43 SNPs that were included in the custom designed chip, two SNPs, rs3742207 and rs1495448 in the COL4A1 and MAGI1 genes respectively, were significantly associated with PWV with the same directionality of effect as in the GWAS (Table 2). Furthermore, when the results of the initial GWAS study group and the Stage 2 study group were combined in a meta-analysis (Table 2), the association of the C allele of rs3742207 with increased PWV was strengthened (p=7.08×10−7), whereas the association of the T allele of rs1495448 with increased PWV was weakened (p=1.07×10−5).
Table 2. Summary of association results for rs3742207 (A) and rs1495448 (B).
This table summarizes the association results with PWV for the top SNPs rs3742207 (COL4A1) and rs1495448 (MAGI1). Alleles refer to the forward strand, and are ordered such that the first allele (+) is associated with increased PWV values. Effect sizes and standard errors (in cm/s) are derived from models that analyzed non-normalized PWV, whereas p-values are calculated from models that analyzed the normalized trait as described in the text.
| A) COL4A1 | |||||
|---|---|---|---|---|---|
| Study | N | Allele (+/−) | Freq | Effect (SE) | P-value |
| SardiNIA | 4,221 | C/A | 0.44 | 21.0 (4.7) | 5.94×10−5 |
| SardiNIA stage 2 | 1,828 | C/A | 0.42 | 15.2 (5.1) | 0.0035 |
| Combined SardiNIA | 7.08×10−7 | ||||
| Old Order Amish | 813 | C/A | 0.55 | 16.4 (8.2) | 0.0218 |
| Combined SardiNIA and Old Order Amish | 5.16×10−8 | ||||
| B) MAGI1 | |||||
| Study | N | Allele (+/−) | Freq | Effect (SE) | P-value |
| SardiNIA | 4,221 | T/G | 0.42 | 8.5 (4.5) | 2.76×10−4 |
| SardiNIA stage 2 | 1,828 | T/G | 0.46 | 11.0 (5.0) | 0.013 |
| Combined SardiNIA | 1.07×10−5 | ||||
| Old Order Amish | 813 | T/G | 0.50 | 2.3 (8.7) | 0.49 |
| Combined SardiNIA and Old Order Amish | 1.23×10−5 | ||||
We repeated the association analyses using models that adjusted for mean arterial pressure, creatinine and the use of blood-pressure lowering medications, which are important covariates of arterial stiffness, in addition to age, age2 and sex which were adjusted for in the base models. The association of rs3742207 with PWV was slightly strengthened, whereby the p-value decreased from 5.94×10−5 to 1.78×10−5. Next, we excluded subjects on antihypertensive medications (N=544) and subjects on dialysis (N=10), and repeated the analyses adjusting for age, age2, sex, mean arterial pressure and creatinine. The p-value for the association of rs3742207 with PWV was 2.48×10−5. In this last model, the p-value was 3.19 ×10−5 when diabetic individuals (N=50) were excluded. However, the genomic control parameters for these 3 additional models were higher than the one for the base model (lambda =1.16, 1.17, 1.17 respectively, compared to 1.14 of the initial model). Conversely, the association of PWV with rs1495448 (in the MAGI1 gene) was weakened when the analyses were repeated using these three sets of additional adjustments (p=0.0014, p=0.0038, p=0.0081 respectively).
The associations of the two loci rs3742207 and rs1495448 with PWV were further evaluated in the Amish population, a genetically distant founder population of European ancestry. The HAPI Heart Study and the ALS study participants underwent both assessment of PWV and genotyping using the Affymetrix 500K chip. We confirmed that allele C of the SNP rs3742207 (the more frequent allele in the Amish population) was associated with PWV (p=0.02) with a comparable effect size (Table 2A), whereas rs1495448 was not (p=0.49, Table 2B). Combining SardiNIA, SardiNIA stage 2, and the Old Order Amish yielded an overall p=5.16×10−8 for the association between rs3742207 and PWV.
The heritability of PWV in the SardiNIA study is 0.226 (adjusted for age, sex and age by sex interaction10. The proportion of variance in PWV (which is equivalent to the proportion of the heritable fraction) that is explained by rs3742207 is 0.87%. The overall effect size, is 18.9 cm/s (calculated as a weighted average of the effect observed in each of the 3 study cohorts, where the weights correspond to those used in the meta-analysis).
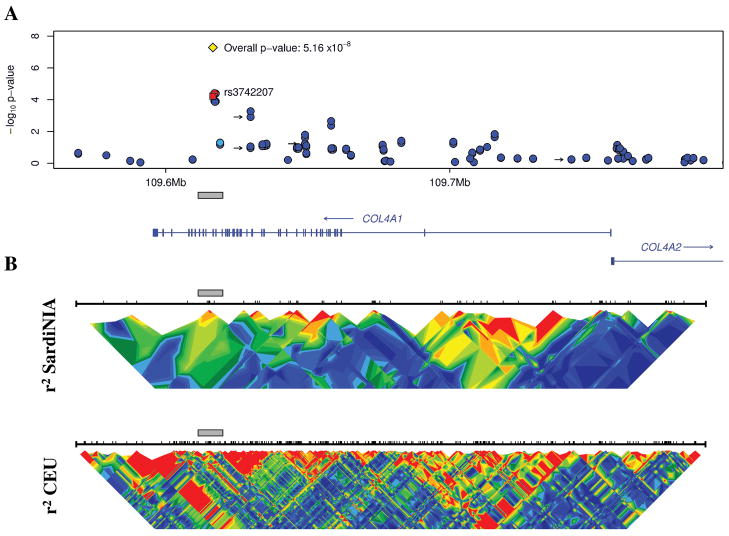
The replicated SNP rs3742207 is a common nonsynonymous coding polymorphism located in exon 45 of the Col4A1 gene. This polymorphism involves a substitution of adenine by cytosine resulting in an amino acid change from Glycine to Histidine at position 1334, which is located in a central region of the protein that consists of multiple triple-helix repeat domains. Nonetheless, further studies are necessary to determine whether the true causal variant is this SNP or another one, which, according to linkage disequilibrium structure (Figure 3)12, 22, 23, appears to lie in exon 45 or nearby.
Figure 3.
A) Summary of the association with PWV in the COL4A1 region in the SardiNIA study. Dots represent all the markers in this region that were analyzed, colored according to their linkage disequilibrium (r2) with rs3742207, which is represented by a red square. LD coloring ranges from red (high LD), to green (moderate LD), to blue (low LD). The yellow diamond indicates the overall p-value for rs3742207 obtained from combining the results of SardiNIA, SardiNIA Stage 2 and HAPI Heart. Black arrows denote markers that were also analyzed by the Framingham Heart Study12, and they are from left to right: rs2131939, rs10492497, rs496916 and rs2391823. B) A summary of the patterns of disequilibrium in the SardiNIA study cohort and in the CEU (Utah residents with ancestry from Northern and Western Europe) HapMap population, 22 where r2 values are calculated as described23 and colored as in panel A. The gray bar marks the region of association and facilitates comparisons among the panels.
Discussion
We conducted a GWAS in the SardiNIA cohort, and found that SNP rs3742207 in the COL4A1 gene was significantly associated with PWV. Furthermore, this locus was successfully replicated both in an independent sample within the SardiNIA cohort, and in the Old Order Amish population, an external genetically distant founder population of European ancestry, suggesting that this COL4A1 variant may have an effect in other populations.
Collagen Type 4
There are 6 different types of type 4 Collagen alpha chains (α1–6). Each one is encoded by a different gene and comprises repeating triple-helical domains with a characteristic G-X-Y motif interposed between the amino terminus and a globular C-terminus. These alpha chains assemble into triple helices to form type 4 Collagen. The Collagen type 4 α1 molecule consists of twenty triple helical repeats G-X-Y(X), where the first position is always a Glycine residue. Mutations in this Glycine residue cause Mendelian disorders characterized by small vessel24, or small and large vessels25 angiopathy.
Type 4 Collagen had not previously been considered to be involved in regulating arterial stiffness. Unlike Collagen types 1 and 3, which are constituents of the extracellular matrix that are found in the medial layer of the arterial walls where they impart the tensile strength to the arteries, type 4 Collagen is a structural component of basement membranes. At the present time there is no functional evidence implicating COL4A1 as a determinant of PWV. Nonetheless a speculative discussion of putative mechanisms though which COL4A1 may influence arterial stiffness is provided in the supplementary materials.
Studies of the Genetics of Arterial Stiffness
Heritability studies have consistently concluded that a genetic component likely underlies the variance in arterial stiffness26–28. Similarly, results of linkage studies for non-invasive indices of arterial stiffness11, 27, 28 suggested an underlying genetic component, even though the findings among these studies did not necessarily overlap.
Previous association studies that examined the genetic underpinnings of arterial stiffness focused mostly on polymorphisms in candidate genes that are believed to be involved in regulating arterial structure and/or function, such as nitric oxide synthase,29 angiotensin II type 1 receptor,30 collagen 1,26 G-protein β-3 subunit,31 β-adrenergic receptors,32 fibrillin 1,33 and C-reactive protein.34 These candidate gene studies, which were conducted in single populations, did not attempt replication in other populations, and often yielded discrepant results27, 29.
The first GWAS evaluating SNPs associated with arterial stiffness was performed in approximately 644 participants in the Framingham Heart Study, using a 100K Affymetrix GeneChip array11. None of the associations with the various markers of arterial stiffness reached genome-wide significance in that study. The 100K chip that was used did not include rs3742207 or any neighboring SNP in the same LD block (Supplementary Figure 3). It did include 7 other SNPs in the COL4A1 gene region, 3 of which were associated (0.01<p<0.05) with PWV in age- and sex-adjusted analyses (available at http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=search&db=gap&term=carotid-femoral%20pulse%20wave%20velocity&doptcmdl=SAnalyses). Of note, some of these markers were also associated with PWV in SardiNIA (Figure 3A) but with higher p-values than SNP rs3742207.
The GWAS in the SardiNIA study was performed in a larger cohort and with a much denser SNP map, and showed, for the first time, a link between a polymorphism in the Collagen Type 4 α1 gene and arterial stiffness. Each copy of the minor allele (C) of rs3742207 is associated with a 21 cm/s higher PWV, such that homozygotes for this minor allele have an approximately 42 cm/s (6.3%) higher PWV than homozygotes for the major allele, an effect size comparable to those reported in GWAS of other traits, including ones recently studied in the Sardinian cohort.18, 35, 36 Importantly, in a cohort of 1,678 Danes aged 40–70 years, a PWV increase of 34 cm/s was associated with a 2.9% increase in age- and sex-adjusted cardiovascular mortality5. Interestingly, rs3742207 was recently found to be associated with the prevalence of myocardial infarction in Japanese individuals37.
Limitations
This study was conducted primarily in a founder population in SardiNIA, so caution should be exercised in extending these results to other populations. However, the values and distributions of PWV in our study did not significantly differ from those obtained in other out-bred populations. Furthermore, and in contrast to the micro-isolates where the gene pool is restricted to a few variants, the number of founders of the current day Sardinian population is large enough to encompass most of the existing alleles in the European population, although with some differences in frequency. Importantly, recent studies from the SardiNIA project18, 35, 36 have demonstrated that findings in SardiNIA are reproducible in other populations; and specifically in this study, we were able to replicate the association of our top finding in an independent and genetically distant founder population of European ancestry. Nonetheless, additional studies in populations of different ethnic origin are needed to further replicate and extend our finding.
Conclusions
Using GWAS, we found that a SNP in the COL4A1 gene is strongly associated with PWV, an established independent predictor of adverse cardiovascular outcomes. Collagen type 4 is the major structural component of basement membranes, suggesting that previously unrecognized cell-matrix interactions may exert an important role in regulating arterial stiffness. Further work is needed to elucidate these mechanisms, and this could potentially lead to the development of novel interventions aimed at delaying or preventing the risks associated with accelerated arterial stiffening. This would help fulfill, in part, the high expectations for breakthroughs in basic science and in clinical medicine that are engendered by modern era genetics.
Supplementary Material
Acknowledgments
We thank Monsignore Piseddu, Bishop of Ogliastra; the Mayors of Lanusei, Ilbono, Arzana, and Elini; the head of the local Public Health Unit ASL4; and the residents of the towns for volunteering and cooperation. In addition, we are grateful to the Mayor and the administration in Lanusei for providing and furnishing the clinic site. We thank the team of physicians and nurses, who carried out the physical examinations and the recruitment personnel who enrolled the volunteers.
Funding Sources
The SardiNIA (“ProgeNIA”) team was supported by Contract NO1-AG-1-2109 from the National Institute on Aging.
This work was supported, in part, by the Intramural Research Program of the National Institute on Aging, National Institutes of Health
The Amish studies were supported by the NIH Institutional Training Grant in Cardiac and Vascular Cell Biology (T32HL072751) and by grant U01 HL72515, the University of Maryland General Clinical Research Center (GCRC) (M01 RR 16500), National Center for Research Resources, the Clinical Nutrition Research Unit of Maryland (P30DK072488), and the Paul Beeson Physician Faculty Scholars in Aging Program of the American Federation of Aging Research.
Footnotes
Subject Codes:
Genetics of cardiovascular disease
Other Vascular Research
Disclosure
None
References
- 1.Laurent S, Cockcroft J, Van Bortel L, Boutouyrie P, Giannattasio C, Hayoz D, Pannier B, Vlachopoulos C, Wilkinson I, Struijker-Boudier H. Expert consensus document on arterial stiffness: methodological issues and clinical applications. Eur Heart J. 2006;27:2588–2605. doi: 10.1093/eurheartj/ehl254. [DOI] [PubMed] [Google Scholar]
- 2.Najjar SS, Scuteri A, Lakatta EG. Arterial aging: is it an immutable cardiovascular risk factor? Hypertension. 2005;46:454–462. doi: 10.1161/01.HYP.0000177474.06749.98. [DOI] [PubMed] [Google Scholar]
- 3.Najjar SS, Scuteri A, Shetty V, Wright JG, Muller DC, Fleg JL, Spurgeon HP, Ferrucci L, Lakatta EG. Pulse Wave Velocity Is an Independent Predictor of the Longitudinal Increase in Systolic Blood Pressure and of Incident Hypertension in the Baltimore Longitudinal Study of Aging. J Am Coll Cardiol. 2008;51:1377–1383. doi: 10.1016/j.jacc.2007.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattace-Raso FU, van der Cammen TJ, Hofman A, van Popele NM, Bos ML, Schalekamp MA, Asmar R, Reneman RS, Hoeks AP, Breteler MM, Witteman JC. Arterial stiffness and risk of coronary heart disease and stroke: the Rotterdam Study. Circulation. 2006;113:657–663. doi: 10.1161/CIRCULATIONAHA.105.555235. [DOI] [PubMed] [Google Scholar]
- 5.Willum-Hansen T, Staessen JA, Torp-Pedersen C, Rasmussen S, Thijs L, Ibsen H, Jeppesen J. Prognostic value of aortic pulse wave velocity as index of arterial stiffness in the general population. Circulation. 2006;113:664–670. doi: 10.1161/CIRCULATIONAHA.105.579342. [DOI] [PubMed] [Google Scholar]
- 6.Laurent S, Boutouyrie P, Asmar R, Gautier I, Laloux B, Guize L, Ducimetiere P, Benetos A. Aortic stiffness is an independent predictor of all-cause and cardiovascular mortality in hypertensive patients. Hypertension. 2001;37:1236–1241. doi: 10.1161/01.hyp.37.5.1236. [DOI] [PubMed] [Google Scholar]
- 7.Sutton-Tyrrell K, Najjar SS, Boudreau RM, Venkitachalam L, Kupelian V, Simonsick EM, Havlik R, Lakatta EG, Spurgeon H, Kritchevsky S, Pahor M, Bauer D, Newman A. Elevated aortic pulse wave velocity, a marker of arterial stiffness, predicts cardiovascular events in well-functioning older adults. Circulation. 2005;111:3384–3390. doi: 10.1161/CIRCULATIONAHA.104.483628. [DOI] [PubMed] [Google Scholar]
- 8.Blacher J, Guerin AP, Pannier B, Marchais SJ, Safar ME, London GM. Impact of aortic stiffness on survival in end-stage renal disease. Circulation. 1999;99:2434–2439. doi: 10.1161/01.cir.99.18.2434. [DOI] [PubMed] [Google Scholar]
- 9.Durier S, Fassot C, Laurent S, Boutouyrie P, Couetil JP, Fine E, Lacolley P, Dzau VJ, Pratt RE. Physiological genomics of human arteries: quantitative relationship between gene expression and arterial stiffness. Circulation. 2003;108:1845–1851. doi: 10.1161/01.CIR.0000091407.86925.7A. [DOI] [PubMed] [Google Scholar]
- 10.Pilia G, Chen WM, Scuteri A, Orru M, Albai G, Dei M, Lai S, Usala G, Lai M, Loi P, Mameli C, Vacca L, Deiana M, Olla N, Masala M, Cao A, Najjar SS, Terracciano A, Nedorezov T, Sharov A, Zonderman AB, Abecasis GR, Costa P, Lakatta E, Schlessinger D. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell GF, DeStefano AL, Larson MG, Benjamin EJ, Chen MH, Vasan RS, Vita JA, Levy D. Heritability and a genome-wide linkage scan for arterial stiffness, wave reflection, and mean arterial pressure: the Framingham Heart Study. Circulation. 2005;112:194–199. doi: 10.1161/CIRCULATIONAHA.104.530675. [DOI] [PubMed] [Google Scholar]
- 12.Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, Vasan RS, Mitchell GF. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8 Suppl 1:S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaitkevicius PV, Fleg JL, Engel JH, O’Connor FC, Wright JG, Lakatta LE, Yin FC, Lakatta EG. Effects of age and aerobic capacity on arterial stiffness in healthy adults. Circulation. 1993;88:1456–1462. doi: 10.1161/01.cir.88.4.1456. [DOI] [PubMed] [Google Scholar]
- 14.Burdick JT, Chen WM, Abecasis GR, Cheung VG. In silico method for inferring genotypes in pedigrees. Nat Genet. 2006;38:1002–1004. doi: 10.1038/ng1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci U S A. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 17.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR. Genome-Wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell BD, McArdle PF, Shen H, Rampersaud E, Pollin TI, Bielak LF, Jaquish C, Douglas JA, Roy-Gagnon MH, Sack P, Naglieri R, Hines S, Horenstein RB, Chang YP, Post W, Ryan KA, Brereton NH, Pakyz RE, Sorkin J, Damcott CM, O’Connell JR, Mangano C, Corretti M, Vogel R, Herzog W, Weir MR, Peyser PA, Shuldiner AR. The genetic response to short-term interventions affecting cardiovascular function: rationale and design of the Heredity and Phenotype Intervention (HAPI) Heart Study. Am Heart J. 2008;155:823–828. doi: 10.1016/j.ahj.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorkin J, Post W, Pollin TI, O’Connell JR, Mitchell BD, Shuldiner AR. Exploring the genetics of longevity in the Old Order Amish. Mech Ageing Dev. 2005;126:347–350. doi: 10.1016/j.mad.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 22.A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis GR, Cookson WO. GOLD--graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- 24.Gould DB, Phalan FC, Breedveld GJ, van Mil SE, Smith RS, Schimenti JC, Aguglia U, van der Knaap MS, Heutink P, John SW. Mutations in Col4a1 cause perinatal cerebral hemorrhage and porencephaly. Science. 2005;308:1167–1171. doi: 10.1126/science.1109418. [DOI] [PubMed] [Google Scholar]
- 25.Plaisier E, Gribouval O, Alamowitch S, Mougenot B, Prost C, Verpont MC, Marro B, Desmettre T, Cohen SY, Roullet E, Dracon M, Fardeau M, Van Agtmael T, Kerjaschki D, Antignac C, Ronco P. COL4A1 mutations and hereditary angiopathy, nephropathy, aneurysms, and muscle cramps. N Engl J Med. 2007;357:2687–2695. doi: 10.1056/NEJMoa071906. [DOI] [PubMed] [Google Scholar]
- 26.Brull DJ, Murray LJ, Boreham CA, Ralston SH, Montgomery HE, Gallagher AM, McGuigan FE, Davey Smith G, Savage M, Humphries SE, Young IS. Effect of a COL1A1 Sp1 binding site polymorphism on arterial pulse wave velocity: an index of compliance. Hypertension. 2001;38:444–448. doi: 10.1161/01.hyp.38.3.444. [DOI] [PubMed] [Google Scholar]
- 27.Lacolley P, Gautier S, Poirier O, Pannier B, Cambien F, Benetos A. Nitric oxide synthase gene polymorphisms, blood pressure and aortic stiffness in normotensive and hypertensive subjects. J Hypertens. 1998;16:31–35. doi: 10.1097/00004872-199816010-00006. [DOI] [PubMed] [Google Scholar]
- 28.North KE, MacCluer JW, Devereux RB, Howard BV, Welty TK, Best LG, Lee ET, Fabsitz RR, Roman MJ. Heritability of carotid artery structure and function: the Strong Heart Family Study. Arterioscler Thromb Vasc Biol. 2002;22:1698–1703. doi: 10.1161/01.atv.0000032656.91352.5e. [DOI] [PubMed] [Google Scholar]
- 29.Czarnecka D, Kawecka-Jaszcz K, Stolarz K, Olszanecka A, Dembinska-Kiec A, Kiec-Wilk B. Ambulatory blood pressure, left ventricular mass and vascular phenotypes in relation to the endothelial nitric oxide synthase gene Glu298Asp and intron 4 polymorphisms in a population-based family study. J Hum Hypertens. 2005;19:413–420. doi: 10.1038/sj.jhh.1001837. [DOI] [PubMed] [Google Scholar]
- 30.Benetos A, Gautier S, Ricard S, Topouchian J, Asmar R, Poirier O, Larosa E, Guize L, Safar M, Soubrier F, Cambien F. Influence of angiotensin-converting enzyme and angiotensin II type 1 receptor gene polymorphisms on aortic stiffness in normotensive and hypertensive patients. Circulation. 1996;94:698–703. doi: 10.1161/01.cir.94.4.698. [DOI] [PubMed] [Google Scholar]
- 31.Nurnberger J, Opazo Saez A, Mitchell A, Buhrmann S, Wenzel RR, Siffert W, Philipp T, Schafers RF. The T-allele of the C825T polymorphism is associated with higher arterial stiffness in young healthy males. J Hum Hypertens. 2004;18:267–271. doi: 10.1038/sj.jhh.1001665. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, Srinivasan SR, Boerwinkle E, Berenson GS. Beta-adrenergic receptor genes are associated with arterial stiffness in black and white adults: the Bogalusa Heart Study. Am J Hypertens. 2007;20:1251–1257. doi: 10.1016/j.amjhyper.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Medley TL, Cole TJ, Gatzka CD, Wang WY, Dart AM, Kingwell BA. Fibrillin-1 genotype is associated with aortic stiffness and disease severity in patients with coronary artery disease. Circulation. 2002;105:810–815. doi: 10.1161/hc0702.104129. [DOI] [PubMed] [Google Scholar]
- 34.Morita A, Nakayama T, Doba N, Hinohara S, Soma M. Polymorphism of the C-reactive protein (CRP) gene is related to serum CRP Level and arterial pulse wave velocity in healthy elderly Japanese. Hypertens Res. 2006;29:323–331. doi: 10.1291/hypres.29.323. [DOI] [PubMed] [Google Scholar]
- 35.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, Sanna S, Maschio A, Busonero F, Usala G, Mulas A, Lai S, Dei M, Orru M, Albai G, Bandinelli S, Schlessinger D, Lakatta E, Scuteri A, Najjar SS, Guralnik J, Naitza S, Crisponi L, Cao A, Abecasis G, Ferrucci L, Uda M, Chen WM, Nagaraja R. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. PLoS Genet. 2007;3:e194. doi: 10.1371/journal.pgen.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada Y, Kato K, Oguri M, Fujimaki T, Yokoi K, Matsuo H, Watanabe S, Metoki N, Yoshida H, Satoh K, Ichihara S, Aoyagi Y, Yasunaga A, Park H, Tanaka M, Nozawa Y. Genetic risk for myocardial infarction determined by polymorphisms of candidate genes in a Japanese population. J Med Genet. 2008;45:216–221. doi: 10.1136/jmg.2007.054387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.