Abstract
Background
Matrix metalloproteinase 1 (MMP-1) may play a role in cardiovascular disease (CVD) susceptibility by influencing plaque rupture via its ability to degrade extracellular collagens.
Methods and Results
We performed a genome-wide association analysis of circulating MMP-1 levels using 500K SNPs to identify genes influencing variation in serum MMP-1 levels in 778 healthy Amish adults. Serum MMP-1 levels, logarithm-transformed and adjusted for age and sex, were screened for association with SNPs using mixed model variance components to account for familial relatedness. Median MMP-1 level was 3.05 ng/mL (inter-quartile range: 1.82 to 5.04 ng/mL) with an estimated heritability of 81% (P < 0.0001). Serum MMP-1 levels were strongly associated with a cluster of 179 SNPs extending over an 11.5 megabase region on chromosome 11q. The peak association was with rs495366 (P = 5.73 × 10−34), located within the region between MMP-1 and MMP-3 and having a minor allele frequency of 0.36. Two other SNPs within the 11q region, rs12289128 and rs11226373, were strongly associated with MMP-1 levels after accounting for rs495366 (P ≤ 10−7). These three SNPs explained 31% of the variance in MMP-1 levels after adjusting for age and sex.
Conclusions
The study provides strong evidence that serum MMP-1 level is highly heritable and that SNPs near MMPs on chromosome 11q explain a significant portion of the variation in MMP-1 levels. Identification of the genetic variants that influence MMP-1 levels may provide insights into genetic mechanisms of CVD.
Keywords: Epidemiology, Genetics, Metalloproteinases, Population
Background
The rupture of atherosclerotic plaques is the major cause of coronary thrombosis and subsequent coronary heart disease (CHD). Matrix metalloproteinases (MMPs) are Zn2+-dependent enzymes capable of degrading many components of extracellular matrix 1–4. Several lines of evidence have demonstrated that the expression and activity of MMPs are increased in vulnerable atherosclerotic plaques 5–7. Increased release of MMPs by macrophage foam cells may induce breakdown of extracellular components and weaken the fibrous cap, predisposing the atherosclerotic plaques to disruption and embolic events 3, 8.
MMP-1, one of the key MMPs, plays a critical role in extracellular matrix remodeling via its ability to degrade several components in the interstitial matrix, including collagen I, II, and III 3, 4. The expression of MMP-1 is elevated in human atherosclerotic plaques compared to normal arteries 7, suggesting a role of MMP-1 in the matrix degradation that predisposes plaque. MMP-1 is also implicated in the regulation of the platelet aggregation that follows plaque disruption 9. Because aggregation of platelets and formation of thrombosis may result in subsequent occurrence of acute coronary events, regulators of this process are of great clinical significance. The relevance of circulating MMP-1 levels, however, is unclear. There is limited data, if any, on the relation of MMP-1 expression levels in tissues with their concentrations in the circulation, and only a few small, cross-sectional epidemiologic studies have assessed the relation of serum MMP-1 levels to cardiovascular outcomes 10–19. A few genetic association studies have been carried out evaluating the relation of MMP-1 genotypes and CHD risk, and results from these have been inconsistent13, 20–24.
There is some data from in vitro studies suggesting that polymorphisms in the MMP-1 promoter region are associated with transcriptional levels of the MMP-1 gene and MMP-1 expression in atherosclerotic plaques 20, 25. However, no data have been published examining associations between genetic variants and circulating MMP-1 levels in humans. Identifying genetic variants that control MMP-1 levels may facilitate our understanding of the pathophysiology of CHD and provide novel opportunities for individualized interventions to prevent or treat CHD.
We hypothesized that variants in multiple genes may mediate the complex pathways that are involved in the regulation and activation of MMP-1. To detect these genes, we carried out a genome-wide association study of serum MMP-1 levels.
Methods
Study subjects
The Heredity and Phenotype Intervention (HAPI) Heart Study was initiated in 2002. Participants of this study comprised adults from the Old Order Amish community of Lancaster County, PA, who were recruited over a three-year period. Study participants were aged 20 years and older and relatively healthy based on a variety of exclusion criteria, including severe hypertension (blood pressure > 180/105 mm Hg), among others. The study aims, recruitment procedures, and ascertainment criteria have been described previously 26. The study included 868 participants, 792 of whom had available DNA and serum MMP-1 measurements. Clinical characteristics were similar between participants with and without serum MMP-1 measurements. Fourteen additional individuals were excluded from final analysis due to genotyping issues (see below), leaving 778 subjects. By virtue of the unique ancestral history of the Amish, these subjects comprised a large number of relative pairs, including 259 parent-offspring pairs, 519 sibling pairs, 11 grandparent-grandchild pairs, 373 avuncular pairs, and 163 first cousin pairs.
The protocol was approved by the Institutional Review Board of the University of Maryland, Baltimore and other participating institutions. Informed consent, including permission to contact relatives, was obtained before participation.
Examination of subjects and laboratory methods
All study participants underwent a physical examination during their visit to the Amish Research Clinic in Strasburg, PA. Subjects were withdrawn from all medications, vitamins and supplements for 7 days prior to their initial assessment. Anthropometry (including height, weight, and waist circumference), medical and family history, and health habits (e.g. smoking status) were obtained by trained research staff using standard methods. Data on medical history, family history and health habits were collected by staff-administered questionnaires. Blood pressure was measured in triplicate after the subject had been sitting quietly for 5 minutes by the use of a standard sphygmomanometer. Blood samples were placed at 4°C, processed within 30 to 60 minutes, and frozen at −80°C until lab assay. Lipid profile (total and high-density lipoprotein (HDL) cholesterol and triglycerides) was assayed in the Quest laboratory in Horsham, PA. MMP-1 serum levels, measured in the fasting state, were determined by an enzyme-linked immunosorbent assay (ELISA) (R&D Systems, Minneapolis, MN) in the University of Maryland Cytokine Core Laboratory in Baltimore, Maryland. MMP-1 levels were measured in duplicate, and the mean values of duplicates were used for data analyses. Because study participants were recruited over a three-year period, MMP-1 levels were measured in five different batches throughout the study period, and the effect of batch was included in the regression model using dummy variables. The detection range of MMP-1 values was between 0.16 and 10 ng/ml, and the intra-assay coefficient of variation was 7.5%. MMP-1 values that were above (n = 33) and below (n = 2) detection range were assigned the maximum and minimum values of the detection range, respectively. Excluding samples outside the detection range did not change results substantially.
DNA extraction and genotyping
Genomic DNA was isolated from whole blood for genetic analyses. The quality of DNA was assessed by single SNP TaqMan genotyping (ABI) as well as by agarose gel electrophoresis. Whole genome genotyping was performed using the Affymetrix Genechip® Human Mapping 500K Array Set, which included 238,304 SNPs from a StyI chip and 262,264 SNPs from an NspI chip. Two aliquots of 250 ng genomic DNA samples were each digested with NspI and StyI restriction enzymes, and ligated with each corresponding adaptor before they underwent polymerase chain reaction (PCR). The PCR products were then purified, fragmented, and labeled for hybridization to each corresponding SNP array. After hybridization and washing, each array was scanned on a GeneChip Scanner 3000 (Affymetrix). Genotypes were called using Bayesian Robust Linear Model with Mahalanobis distance classifier (BRLMM) algorithm.
Quality control
Fourteen subjects were excluded from analysis due to genotyping issues, including inadequate call rates (n = 1) and/or excessive relationship incompatibilities consistent with sample mix-ups (n = 13). We targeted for analysis only SNPs on autosomes and thus excluded 10,536 SNPs on the sex chromosome. Additional SNPs were excluded if they deviated excessively from Hardy-Weinberg Equilibrium (HWE) proportions (P ≤ 0.0001, n = 17,282), had genotype call rates <95% (n = 52,498) or had minor allele frequencies (MAF) < 2% (n = 96,052, including monomorphic SNPs). Departure from Hardy-Weinberg equilibrium was assessed by chi-square test without regard for family structure. Following these exclusions, we analyzed a total of 338,079 autosomal SNPs in 778 individuals.
Statistical analysis
Summary statistics of baseline clinical characteristics were expressed as unadjusted means ± standard deviations (SD) using the STATA statistical package (version 9.2; StataCorp, College Station, Texas) unless indicated otherwise. MMP-1 levels were natural logarithm-transformed prior to analysis to remove skewness. The correlation (r) between ln-transformed MMP-1 levels and selected cardiovascular risk factors (e.g., BMI, blood pressure, and lipid levels) was computed by estimating the proportionate reduction in the variance of the model associated with inclusion of the considered covariates, taking the square root of this quantity, and then incorporating the direction of association from the covariate beta value. The effects of age, age2, and sex were included in each model along with age (and age2) by sex interactions to allow for age effects to vary by sex. Trait heritability was defined as the proportion of the total trait variance attributable to additive effect of genes and was estimated by modeling the phenotypic covariance (conditional upon covariate effects) between any two individuals in the pedigree as a function of their degree of biological relationship. Estimations of correlations and heritability were obtained using variance component analysis as implemented in the SOLAR (Sequential Oligogenic Linkage Analysis Routines) to account for familial relatedness of the data (version 4.0.7; Southwest Foundation for Biomedical Research, San Antonio, TX) 27.
Genome-wide association analysis of serum MMP-1 levels was performed using a mixed model approach that models variation in MMP-1 levels as a function of measured environmental covariates, measured genotype, and a polygenic component to account for phenotypic correlation due to relatedness. A t-score was used to assess significance of the beta coefficient for the measured genotype under an additive model. We included age, age2, sex, age-by-sex and age2-by-sex interactions, and assay batch in the model. The polygenic component was modeled as a random effect using the relationship matrix derived from the complete 14-generation Amish pedigree to control for the relatedness of all subjects in the study. This polygenic component corresponds to the residual heritability in the trait after accounting for the fixed effects included in the model. These analyses were carried out using in-house software developed by Dr. J.R. O’Connell in our group. Genome-wide association results were plotted using Haploview 4.0 28 . A genome-wide significance level of α = 1.0 × 10−7 (~ 0.05/338,079) was defined based on Bonferroni correction. Linkage disequilibrium (LD) statistics (|D’| and r2) were calculated among SNPs using Haploview 4.0 28
Haplotypes were inferred using the EM algorithm employed by the SNPHAP program developed by David Clayton (http://www-gene.cimr.cam.ac.uk/clayton/software). Linkage analyses were carried out using variance component approach implemented in SOLAR to estimate the effects of SNPs after accounting for a linkage effect at the associated locus. The total variance of phenotype (ln-MMP-1 levels) was partitioned into components attributable to measured covariates (e.g. age, sex), measured genotypes, additive polygenic effects and an additive quantitative trait loci effect (i.e. the linkage component).
Results
Characteristics of the participants
A total of 778 Amish participants from the Heredity and Phenotype Intervention (HAPI) Heart Study were included in the analysis. The mean age was 43.2 ± 13.6 years, and 426 (54.8%) of participants were male. Baseline clinical characteristics are summarized by sex in Table 1. The median MMP-1 level in the population was 3.05 ng/mL (inter-quartile range: 1.82 to 5.04 ng/mL), with levels slightly higher in women than men, and increasing with increasing age (r = 0.11, P < 0.0001).
Table 1.
Baseline characteristics (mean (SD)) of 778 Amish participants in the study
| Characteristics | Total | Male | Female |
|---|---|---|---|
| N=778 | N=426 | N=352 | |
| Age (y) | 43.2 (13.6) | 41.6 (13.1) | 45.0 (13.9) |
| Body mass index (kg/m2) | 26.6 (4.4) | 25.6 (3.2) | 27.8 (5.3) |
| Fasting triglyceride (mg/dL) | 67.5 (40.4) | 63.1 (36.3) | 72.8 (44.3) |
| Fasting total cholesterol (mg/dL) | 209 (47) | 203 (44) | 216 (50) |
| HDL/LDL cholesterol ratio | 0.4 (0.2) | 0.4 (0.2) | 0.5 (0.2) |
| Systolic blood pressure (mmHg) | 121 (14) | 121 (12) | 121 (16) |
| Diastolic blood pressure (mmHg) | 77 (9) | 78 (9) | 76 (8) |
| Pulse pressure (mmHg) | 45 (11) | 44 (11) | 46 (12) |
| Previous myocardial infarction or stroke (%)* | 8 (1.03) | 6 (1.42) | 2 (0.57) |
| Diabetes (%)* | 5 (0.65) | 3 (0.71) | 2 (0.58) |
| Current smoking status (pipe, cigar & cigarette smokers) (%)* |
--- | 80 (19.0) | 0 (0) |
| Serum MMP-1 percentile cutoffs (ng/mL) | |||
| 25th | 1.82 | 1.78 | 1.95 |
| 50th | 3.05 | 2.93 | 3.18 |
| 75th | 5.04 | 4.68 | 5.48 |
Abbreviations: HDL/LDL: high-density lipoprotein/low-density lipoprotein
self-reported
Heritability of MMP-1 level was 81 ± 8 % after adjusting for the effects of age and sex (P < 0.0001), and remained high (h2 = 78 ± 8%, P < 0.0001) after additional adjustments for the effects of BMI, systolic blood pressure (SBP), diastolic blood pressure (DBP), fasting triglyceride, cholesterol levels, HDL/LDL cholesterol ratio and smoking status.
Genome-wide association results
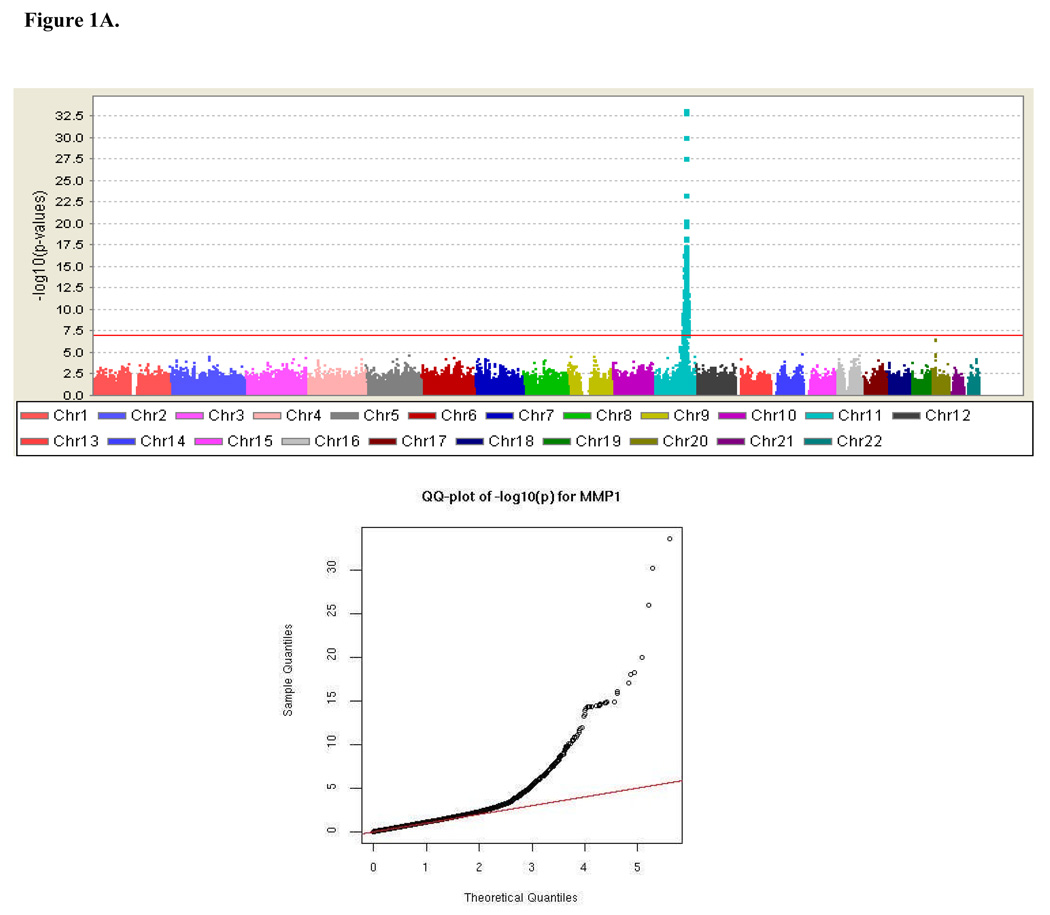
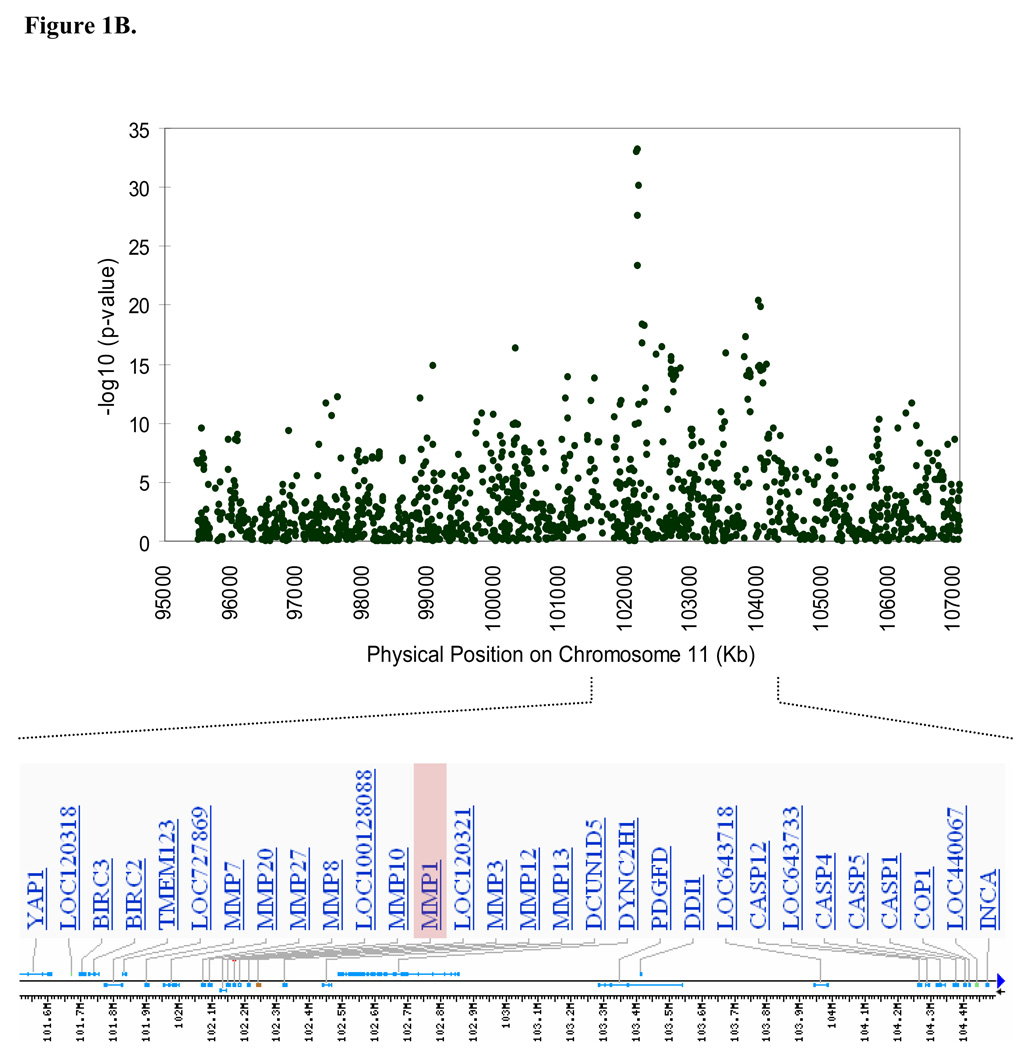
Results from the genome-wide association analysis are summarized in Figure 1A. A total of 179 SNPs, all clustered within an 11,454 kb region on chromosome 11q (95,569,593-107,024,176 bp), were significantly associated with MMP-1 levels under an additive model at the predefined genome-wide significant level of 10−7 (P ranging from 9.57 × 10−8 to 5.73 × 10−34). The most strongly associated SNPs were near a cluster of MMP genes (MMP-1, MMP-3, MMP-7, MMP-8, MMP-10, MMP-12, MMP-13, MMP-20 and MMP-27) (Figure 1B). Details of the 179 significant SNPs are provided in Data Supplement Table S1. In addition to the associated SNPs on chromosome 11, a modest association was also observed in the region of the solute carrier family 24, member 3 (SLC24A3) gene on chromosome 20p, which did not quite reach genome wide significance (rs3790268, P = 2.96 × 10−7).
Figure 1.
Genome-wide association results. (A) Negative log of p-values and a quantile-quantile (Q-Q) plot showing the association of single nucleotide polymorphisms (SNPs) with MMP-1 serum levels across the entire genome using mixed model variance components analysis, and (B) Association signals and genes present in the region of significance on chromosome 11.
To quantify the impact of the excessive number of significant results on chromosome 11 on the quantile-quantile plot in Figure 1A, we calculated genomic control lambdas (λ) for the full GWAS, excluding chromosome 11, and for chromosome 11 alone using median(χ2)/0.456. The λ for chromosome 11 was much higher than for the genome average (1.528 vs. 1.08), consistent with the inflation on chromosome 11 being due to extensive linkage disequilibrium (LD) across the associated region (Data Supplement Figure S1), reflecting a strong linkage signal (shown below). Our measured genotype model does not account for covariance between individuals at the SNP due to alleles shared identical-by-descent (IBD); thus, the P-values are susceptible to inflation in regions of strong linkage. To account for this inflation, a genomic control λ = 1.528 was applied to all analyses pertaining to the chromosome 11 region presented in this paper, except the linkage analyses shown below.
The five most strongly associated SNPs (genomic control (GC)-corrected P ≤ 10−18) fell within a 28.5 kb region near the genes encoding MMP-1 and MMP-3, with 4 of the SNPs located within the intergenic region between MMP-1 and MMP-3 (i.e. upstream of MMP-1 and downstream of MMP-3) (rs603050, P = 1.28 × 10−22; rs11225434, P = 2.07 × 10−19; rs495366, P = 8.39 × 10−23; rs7926920, P = 4.75 × 10−18; all P are GC-corrected), and the fifth SNP located in intron 8 of MMP-3 (rs650108, GC-corrected P = 1.06 × 10−20). The most strongly associated SNP, rs495366, explained 17.5% of residual MMP-1 variation after adjusting for age and sex. This SNP was highly correlated with rs603050 and rs650108 (r2 = 0.99 and 0.98, respectively). The other two SNPs, rs11225434 and rs7926920, were highly correlated with each other (D’ = 0.997, r2=0.96), but were less correlated with rs495366 (D’ = 1.0 and 0.99, r2 =0.34 and 0.36, respectively). The LD structure of the five most strongly associated SNPs, along with SNPs located in MMP-1, MMP-3 and the intergenic region between MMP-1 and MMP-3 is summarized in Figure 2.
Figure 2.
Linkage disequilibrium structure (r2) of SNPs in MMP-1, MMP-3 and their intergenic region in the Amish.
To evaluate whether the multiple-associated SNPs on chromosome 11q represented distinct signals, we employed a forward regression model building analysis within the mixed model framework using SNPs across the MMP-1 associated region of 95,569,593 - 107,024,176 bp. The initial model included the baseline covariates only. Given the current model, a chi square likelihood ratio test is computed for each SNP, then the SNP with the lowest p-value, enters the model. The process was repeated until no SNP passed a chi square with P-value threshold 10−7 (without genomic control adjustment). By this approach, rs495366 was the first to be added into the model; two other SNPs, rs12289128 and rs11226373, were sequentially identified and added into the model. While rs12289128 and rs495366 were located close to each other, rs11226373 is located about 1.65 Mb away (Table 2). The closest gene to rs11226373 is platelet-derived growth factor D (PDGFD). The P-values of SNPs from the model building are shown in Data Supplement Figure S2.
Table 2.
Pairwise linkage disequilibrium (LD) and association results of rs495366, rs12289128 and rs11226373
| Pairwise LD (|D’| / r2) |
Geometric mean of MMP-1 levels by genotypes |
Conditional P while added into the model sequentially* |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| RS ID | Position | MAF (%) |
With rs495366 |
With rs12289128 |
Geno- type |
N (%) |
MMP-1 level (ng/ml) |
Marginal P* |
|
| rs495366 | 102,200,318 | 36.1 | --- | --- | CC | 311 (40.6) | 3.96 | 8.39 × 10−23 | 8.39 × 10−23 |
| CT | 358 (46.7) | 2.81 | |||||||
| TT | 98 (12.8) | 1.51 | |||||||
| rs12289128 | 102,185,176 | 14.5 | 1.0/0.1 | --- | GG | 573 (73.7) | 3.08 | 0.026 | 6.65 × 10−10 |
| GC | 185 (23.8) | 2.76 | |||||||
| CC | 20 (2.6) | 2.22 | |||||||
| rs11226373 | 103,839,449 | 15.2 | 0.9/0.08 | 0.03/0.001 | TT | 555 (71.6) | 2.58 | 1.50 × 10−12 | 7.85 × 10−07 |
| TC | 205 (26.5) | 4.18 | |||||||
| CC | 15 (1.9) | 5.21 | |||||||
Abbreviations: MAF, minor allele frequency; N, number of individuals.
Genomic control-corrected P value (under additive model), adjusted for baseline covariates (age, age2, sex, age-by-sex and age2-by-sex)
Table 2 shows the geometric mean values of MMP-1 levels by genotypes of rs495366, rs12289128 and rs11226373. Both the marginal effects of genotype on MMP-1 levels are shown, as well as their conditional effects estimated by adding the three SNPs sequentially into the model (in the order of rs495366, rs12289128 and rs1226373). Minor allele frequencies of these three SNPs ranged from 0.15 – 0.36. The major alleles of rs495366 and rs12289182, and the minor allele of rs11226373, were associated with increasing MMP-1 serum levels. Collectively, these 3 SNPs accounted for approximately 31% of the variance in age- and sex-adjusted levels of MMP-1 (GC-corrected P = 7.4 × 10−35 for 3 SNPs vs. 0 SNP, 3 d.f.). Additional analyses revealed no evidence for SNP*SNP interaction effects on MMP-1 levels (data not shown). After accounting for age, sex, and the effects of these three SNPs on serum MMP-1 levels, the residual heritability of this trait was 71%.
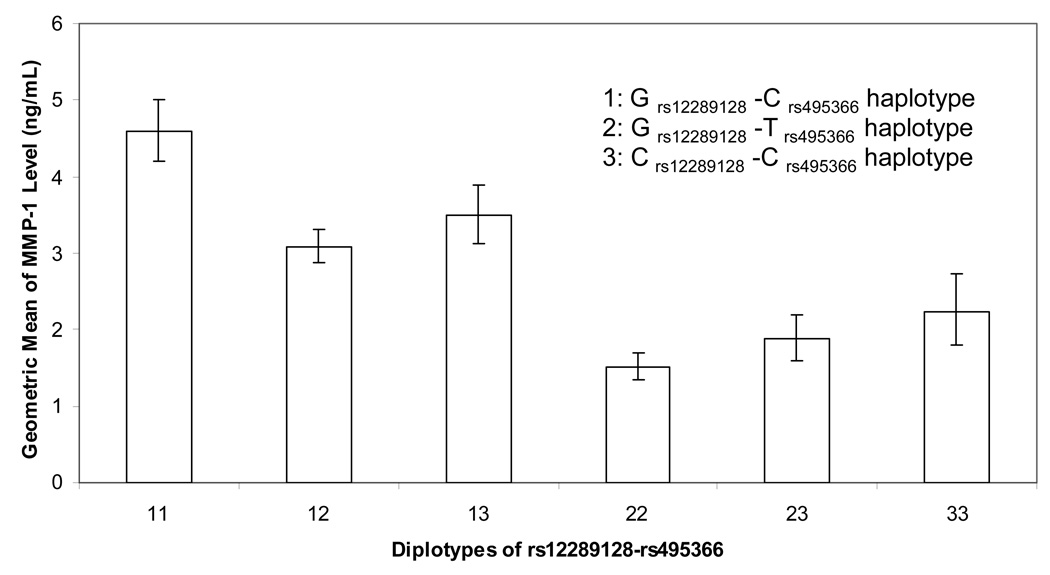
Interestingly, rs12289128 is not associated with MMP-1 levels itself, but enters the model to refine the rs495366 signal through a haplotype as both SNPs are in the same LD block and have |D’| = 1. Unambiguous haplotypes were constructed between rs12289128 (G/C polymorphism) and rs495366 (C/T polymorphism): G-C (0.493), G-T (0.362), and C-C (0.145), with frequencies indicated in parentheses. The geometric means of MMP-1 levels corresponding to the six dipolotypes are shown in Figure 3. There was a significant trend of increasing MMP-1 levels with increasing copies of G-C haplotype (GC-corrected P of trend = 1.25 × 10−29; 24 % of MMP-1 variation explained). A 3-locus haplotype analysis based on SNPs rs12289128, rs495366 and rs11226373 generated 6 different haplotypes, although no clear dose-response relationship between any of the haplotypes and MMP-1 serum levels was apparent (data not shown). Individual haplotypes could not be unambiguously assigned to all subjects due to incomplete LD.
Figure 3.
Geometric mean of MMP-1 serum levels by the diplotypes of rs12289128-rs495366.
To investigate the linkage signal to serum MMP-1 levels in the 11q region and its impact on our measured genotype analysis, we computed the two-point LOD score based on IBD constructed by rs495366-rs12289128 haplotype. Not unexpectedly, there was strong evidence for linkage to serum MMP-1 levels in the 11q region (LOD = 19.4). While taking the local linkage effect into account, the association of rs495366 with MMP-1 levels diminished substantially but remained highly significant (P = 1.40 × 10−18; 11.8% MMP-1 variation explained). Similarly, the combined association of MMP-1 levels with the 3 SNPs, rs495366, rs12289128 and rs11226373, was also diminished, but remained highly significant (P = 1.82 × 10−34 for 3 SNPs vs. 0 SNPs, 3 d.f.; 23.6% MMP-1 variation explained). The residual heritability of MMP-1 levels was 61% after accounting for the effects of the 3 SNPs and the linkage component in the age- and sex-adjusted model, suggesting that there remains a genetic influence on serum MMP-1 levels emanating from outside of the chromosome 11q region.
Associations with other cardiovascular risk factors
Having detected strong and compelling associations between these SNPs and serum MMP-1 levels, we then assessed the clinical significance of MMP-1 variation in our sample by assessing its correlation with various cardiovascular risk factors. Age and sex-adjusted serum MMP-1 levels were significantly correlated with fasting triglycerides (r = 0.14, P = 0.0006) and pulse pressure (r = 0.08, P = 0.04) and borderline correlated with HDL/LDL ratio (r = −0.06, P = 0.05) and SBP (r =0.06, P = 0.09). There was little correlation of serum MMP-1 levels with other cardiovascular risk factors (Data Supplement Table S2). Similarly, the three MMP-1-associated SNPs showed little evidence for association with any of these cardiovascular risk factors (Data Supplement Table S2).
Discussion
To our knowledge, this study is the first to evaluate the genetic contribution to variation in serum MMP-1 levels in a large human population. Our analyses revealed that variation in MMP-1 levels is highly heritable (h2 = 81%) in the Amish, with strong and compelling evidence for association of serum MMP-1 levels with a large cluster of SNPs in an 11.4 Mb region on chromosome 11q. Of note, the strongest evidence for association was to a group of SNPs located near a cluster of MMP genes, particularly between MMP-1 and MMP-3. The estimated effect sizes of the associated SNPs were very high, with rs495366, the most strongly associated SNP, accounting by itself for 17.5% of the variance in MMP-1 levels. Two other SNPs in this region, rs12289128 and rs11226373, were also strongly associated with serum MMP-1 levels, and the three SNPs collectively accounted for 31% of the trait variance.
The location of these associated SNPs near the MMP gene cluster lends strong biological plausibility to the observed associations. Several in vitro20, 25 and epidemiologic13, 20–24 studies have shown genetic variants in the MMP-1 promoter to be associated with transcriptional level of MMP-1 and/or CVD endpoints (Data Supplement Table S3). The peak associations observed in our study (i.e., rs495366, rs603050, rs11225434 and rs7926920) were also located in the upstream region of MMP-1 gene, consistent with the existence of functional variants in the MMP-1 promoter region affecting expression. However, further studies are needed to verify this speculation given that the previously reported promoter polymorphisms were not highly correlated with our most strongly associated SNP (i.e. rs495366) based on the HAPMAP Caucasian population or with other associated SNPs in the MMP-1 upstream region. The absence of an association between intragenic MMP-1 SNPs and serum MMP-1 levels is possibly due to suboptimal SNP coverage of the MMP-1 gene by the Affymetrix 500K panel given that this panel includes only 3 SNPs within MMP-1 despite the fact that this gene spans a region of 8,243 bp.
Our analyses do not permit us to resolve whether the observed associations are attributable to a single functional SNP or to multiple functional SNPs residing in the MMP gene cluster. However, our analysis revealed that the rs12289128-rs495366 haplotype provides an even stronger association with MMP-1 serum levels than either SNP alone, suggesting that this haplotype is tagging a single (unmeasured) functional polymorphism better than either single SNP alone. Including rs11226373, which is partially correlated with the other two SNPs, in the haplotype analysis did not provide a clear dose-response relationship with MMP-1 serum levels. Therefore, it remains unclear whether rs11226373 is independently associated with MMP-1 levels or simply tags the same (unmeasured) causal polymorphism as do rs12289128 and rs495366. The latter is possible despite the >1.5 MB distance of rs11226373 from rs12289128 and rs495366 given the long-range LD present in this region in the Amish.
Previous studies suggest that MMP-1 may play an important role in atherosclerotic plaque disruption by influencing stability of the plaque fibrous cap6, 7, 29. For example, the expression of MMP-1 is elevated in human atherosclerotic plaques, particularly in shoulder regions and areas of foam cell accumulation in the plaques 7, and is higher in plaques with thin cap compared to those with a thick fibrous cap29. Foam cells isolated from rabbit aortic lesions are capable of synthesizing and releasing MMP-1 constitutively in vitro 30. Furthermore, treating patients with carotid endarterectomy with MMP inhibitor reduces the MMP-1 concentration in carotid plaques 31. These findings suggest a critical role of MMP-1 in the matrix degradation that predisposes plaque rupture and acute coronary syndromes. Furthermore, MMP-1 can induce tyrosine phosphorylation of intracellular proteins in platelets and the movement of β3 integrins to areas of cell contact and thus prime platelets for aggregation 9, indicating a potential role of MMP-1 in thrombotic events.
MMP-1 is found to be highly expressed in smooth muscle, pancreatic islets, cardiac myocytes and bronchial epithelial cells and normal vascular tissues (data sources: UCSC Genome Bioinformatics Site: http://genome.ucsc.edu; GeneHub-GEPIS database: http://www.cgl.ucsf.edu/Research/genentech/genehub-gepis/index.html). However, there has been limited data, if any, on how well MMP-1 expression levels in tissues correlate with their concentrations in the circulation, and the clinical significance of elevated MMP-1 levels in humans remains unclear. MMP-1 serum levels increase with increasing age as shown in our data and reported previously in Chinese women 32. In two relatively small studies, higher plasma MMP-1 serum levels were correlated with increasing carotid-femoral pulse-wave velocity (PWV), a marker of arterial stiffness 10 and with increasing pulse pressure and mean blood pressure 11 while in our large study, we observed significant, albeit modest, correlations of MMP-1 levels with fasting triglycerides and pulse pressure.
Only a few studies have reported correlations of circulating MMP-1 levels with CHD outcomes. Several studies have reported higher circulating MMP-1 levels to be associated with acute MI and/or unstable angina 14–16 and ruptured plaques12, suggesting that MMP-1 levels may be a marker of plaque instability, although others have failed to find such an association13. MMP-1 levels have also been associated carotid stenosis 17 and left ventricular remodeling among patients with cardiovascular disease 15, 18, 19. Given the limitations and inconsistency of existing studies, the utility of MMP-1 serum levels as a marker for screening, diagnosis or identifying individuals at risk of various disease outcomes remains unproven. Studies assessing the effects of polymorphisms in the MMP-1 on cardiovascular events have found inconsistent associations13, 20–24. The lack of association of MMP-1 SNPs with cardiovascular risk factors in our data is possibly due either subjects in our study being relatively healthy with few individuals having symptomatic CHD or an absence of a true relationship effect between MMP-1-associated genetic variants and cardiovascular risk.
We conclude that serum MMP-1 levels are highly heritable and that genetic variation at a locus on chromosome 11q that includes MMP-1 is a major determinant of MMP-1 levels. Given the relatively high frequency of the associated SNPs in this Amish population (15–36%), we expect that the strong associations detected will be present in other populations as well (especially European Caucasians). However, the large number of associated SNPs spanning across a relatively large region will make it difficult to localize the causative SNP(s) in this population based on statistical criteria alone. Conducting additional genetic association studies and functional characterization of the most likely causative variants in other populations will be needed to better define the genetic architecture of MMP-1 levels and to determine the relevance of genetic variation at this locus to CHD.
Extracellular matrix (ECM) remodeling is an essential process in the pathogenesis of atherosclerosis and coronary heart diseases (CHD). Matrix metalloproteinases (MMPs) mediate this process by participating in the degradation of the ECM, resulting in weakening of the fibrous cap and predisposing the atherosclerotic plaques to disruption and embolic events. One of the key MMPs is MMP-1, although the relation of circulating MMP-1 levels to cardiovascular risk and its potential value as a biomarker for screening for disease is unknown. To identify genetic determinants of MMP-1 serum levels, we carried out a genome-wide association study in 778 healthy Old Order Amish individuals. We found serum MMP-1 levels to be highly heritable in this population (h2 = 81%). We identified a large cluster of SNPs spanning an 11.5 Mb region on chromosome 11q to be strongly associated with serum MMP-1 levels, with the peak association observed with rs495366 (p = 5.73 × 10−34; minor allele frequency = 0.36). This region covers a cluster of MMP genes, including MMP-1. However, there was little evidence that the MMP-1-associated SNPs were associated with any other cardiovascular risk factors in this relatively healthy population. These results suggest that genetic variation on chromosome 11q is a major determinant of circulating MMP-1 levels in the Amish; additional studies in other populations are needed to determine the relevance of genetic variation at this region to CHD.
Supplementary Material
Acknowledgments
Sources of Founding
This work was supported by NIH research grants U01 HL72515, U01 HL84756 and R01 088119, the University of Maryland General Clinical Research Center, grant M01 RR 16500, the Clinical Nutrition Research Unit of Maryland (P30 DK072488), and the Baltimore Veterans Administration Medical Center Geriatrics Research and Education Clinical Center. Dr. Cheng was supported by the Merck Foundation fellowship.
Footnotes
Disclosure
None
References
- 1.Shah PK. Role of inflammation and metalloproteinases in plaque disruption and thrombosis. Vascular Medicine. 1998;3:199–206. doi: 10.1177/1358836X9800300304. [DOI] [PubMed] [Google Scholar]
- 2.Shah PK, Galis ZS. Matrix metalloproteinase hypothesis of plaque rupture: Players keep piling up but questions remain. Circulation. 2001;104:1878–1880. [PubMed] [Google Scholar]
- 3.Jones CB, Sane DC, Herrington DM. Matrix metalloproteinases: a review of their structure and role in acute coronary syndrome. Cardiovasc Res. 2003;59:812–823. doi: 10.1016/s0008-6363(03)00516-9. [DOI] [PubMed] [Google Scholar]
- 4.Newby AC. Dual role of matrix metalloproteinases (matrixins) in intimal thickening and atherosclerotic plaque rupture. Physiol Rev. 2005;85:1–31. doi: 10.1152/physrev.00048.2003. [DOI] [PubMed] [Google Scholar]
- 5.Nikkari ST, O'Brien KD, Ferguson M, Hatsukami T, Welgus HG, Alpers CE, Clowes AW. Interstitial collagenase (MMP-1) expression in human carotid atherosclerosis. Circulation. 1995;92:1393–1398. doi: 10.1161/01.cir.92.6.1393. [DOI] [PubMed] [Google Scholar]
- 6.Sukhova GK, Schonbeck U, Rabkin E, Schoen FJ, Poole AR, Billinghurst RC, Libby P. Evidence for increased collagenolysis by interstitial collagenases-1 and -3 in vulnerable human atheromatous plaques. Circulation. 1999;99:2503–2509. doi: 10.1161/01.cir.99.19.2503. [DOI] [PubMed] [Google Scholar]
- 7.Galis ZS, Sukhova GK, Lark MW, Libby P. Increased expression of matrix metalloproteinases and matrix degrading activity in vulnerable regions of human atherosclerotic plaques. J Clin Invest. 1994;94:2493–2503. doi: 10.1172/JCI117619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galis ZS, Khatri JJ. Matrix metalloproteinases in vascular remodeling and atherogenesis: the good, the bad, and the ugly. Circ Res. 2002;90:251–262. [PubMed] [Google Scholar]
- 9.Galt SW, Lindemann S, Allen L, Medd DJ, Falk JM, McIntyre TM, Prescott SM, Kraiss LW, Zimmerman GA, Weyrich AS. Outside-in signals delivered by matrix metalloproteinase-1 regulate platelet function. Circ Res. 2002;90:1093–1099. doi: 10.1161/01.res.0000019241.12929.eb. [DOI] [PubMed] [Google Scholar]
- 10.McNulty M, Mahmud A, Spiers P, Feely J. Collagen type-I degradation is related to arterial stiffness in hypertensive and normotensive subjects. J Hum Hypertens. 2006;20:867–873. doi: 10.1038/sj.jhh.1002015. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa J, Kario K, Matsui Y, Shibasaki S, Morinari M, Kaneda R, Hoshide S, Eguchi K, Hojo Y, Shimada K. Collagen metabolism in extracellular matrix may be involved in arterial stiffness in older hypertensive patients with left ventricular hypertrophy. Hypertens Res. 2005;28:995–1001. doi: 10.1291/hypres.28.995. [DOI] [PubMed] [Google Scholar]
- 12.Zhang XW, Ge JB, Yang JM, Ge L, Wang NF, Gao Y, Li PZ, Pan H, Tong GX, Zhou L, Ye XH, Xu J. Relationship between hs-CRP, proMMP-1, TIMP-1 and coronary plaque morphology: intravascular ultrasound study. Chin Med J (Engl) 2006;119:1689–1694. [PubMed] [Google Scholar]
- 13.Hlatky MA, Ashley E, Quertermous T, Boothroyd DB, Ridker P, Southwick A, Myers RM, Iribarren C, Fortmann SP, Go AS. Matrix metalloproteinase circulating levels, genetic polymorphisms, and susceptibility to acute myocardial infarction among patients with coronary artery disease. Am Heart J. 2007;154:1043–1051. doi: 10.1016/j.ahj.2007.06.042. [DOI] [PubMed] [Google Scholar]
- 14.Inoue T, Kato T, Takayanagi K, Uchida T, Yaguchi I, Kamishirado H, Morooka S, Yoshimoto N. Circulating matrix metalloproteinase-1 and -3 in patients with an acute coronary syndrome. Am J Cardiol. 2003;92:1461–1464. doi: 10.1016/j.amjcard.2003.08.061. [DOI] [PubMed] [Google Scholar]
- 15.Soejima H, Ogawa H, Sakamoto T, Miyamoto S, Kajiwara I, Kojima S, Hokamaki J, Sugiyama S, Yoshimura M, Suefuji H, Miyao Y, Fujimoto K, Miyagi H, Kishikawa H. Increased serum matrix metalloproteinase-1 concentration predicts advanced left ventricular remodeling in patients with acute myocardial infarction. Circ J. 2003;67:301–304. doi: 10.1253/circj.67.301. [DOI] [PubMed] [Google Scholar]
- 16.Tziakas DN, Chalikias GK, Parissis JT, Hatzinikolaou EI, Papadopoulos ED, Tripsiannis GA, Papadopoulou EG, Tentes IK, Karas SM, Chatseras DI. Serum profiles of matrix metalloproteinases and their tissue inhibitor in patients with acute coronary syndromes. The effects of short-term atorvastatin administration. Int J Cardiol. 2004;94:269–277. doi: 10.1016/j.ijcard.2003.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Wu YW, Yang WS, Chen MF, Lee BC, Hung CS, Liu YC, Jeng JS, Huang PJ, Kao HL. High serum level of matrix metalloproteinase-1 and its rapid surge after intervention in patients with significant carotid atherosclerosis. J Formos Med Assoc. 2008;107:93–98. doi: 10.1016/S0929-6646(08)60015-7. [DOI] [PubMed] [Google Scholar]
- 18.Hojo Y, Ikeda U, Ueno S, Arakawa H, Shimada K. Expression of matrix metalloproteinases in patients with acute myocardial infarction. Jpn Circ J. 2001;65:71–75. doi: 10.1253/jcj.65.71. [DOI] [PubMed] [Google Scholar]
- 19.Lopez B, Gonzalez A, Querejeta R, Larman M, Diez J. Alterations in the pattern of collagen deposition may contribute to the deterioration of systolic function in hypertensive patients with heart failure. J Am Coll Cardiol. 2006;48:89–96. doi: 10.1016/j.jacc.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 20.Pearce E, Tregouet DA, Samnegard A, Morgan AR, Cox C, Hamsten A, Eriksson P, Ye S. Haplotype effect of the matrix metalloproteinase-1 gene on risk of myocardial infarction. Circ Res. 2005;97:1070–1076. doi: 10.1161/01.RES.0000189302.03303.11. [DOI] [PubMed] [Google Scholar]
- 21.Ye S, Gale CR, Martyn CN. Variation in the matrix metalloproteinase-1 gene and risk of coronary heart disease. Eur Heart J. 2003;24:1668–1671. doi: 10.1016/s0195-668x(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 22.Nojiri T, Morita H, Imai Y, Maemura K, Ohno M, Ogasawara K, Aizawa T, Saito A, Hayashi D, Hirata Y, Sugiyama T, Yamazaki T, Nagai R. Genetic variations of matrix metalloproteinase-1 and -3 promoter regions and their associations with susceptibility to myocardial infarction in Japanese. International Journal of Cardiology. 2003;92:181–186. doi: 10.1016/s0167-5273(03)00100-1. [DOI] [PubMed] [Google Scholar]
- 23.Horne BD, Camp NJ, Carlquist JF, Muhlestein JB, Kolek MJ, Nicholas ZP, Anderson JL. Multiple-polymorphism associations of 7 matrix metalloproteinase and tissue inhibitor metalloproteinase genes with myocardial infarction and angiographic coronary artery disease. Am Heart J. 2007;154:751–758. doi: 10.1016/j.ahj.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han Y, Wu Z, Zhang X, Yan C, Xi S, Yang Y, Pei F, Kang J. Impact of matrix metalloproteinase-1 gene variations on risk of acute coronary syndrome. Coron Artery Dis. 2008;19:227–230. doi: 10.1097/MCA.0b013e3282f9d3d8. [DOI] [PubMed] [Google Scholar]
- 25.Rutter JL, Mitchell TI, Buttice G, Meyers J, Gusella JF, Ozelius LJ, Brinckerhoff CE. A single nucleotide polymorphism in the matrix metalloproteinase-1 promoter creates an Ets binding site and augments transcription. Cancer Res. 1998;58:5321–5325. [PubMed] [Google Scholar]
- 26.Mitchell BD, McArdle PF, Shen H, Rampersaud E, Pollin TI, Bielak LF, Jaquish C, Douglas JA, Roy-Gagnon MH, Sack P, Naglieri R, Hines S, Horenstein RB, Chang YP, Post W, Ryan KA, Brereton NH, Pakyz RE, Sorkin J, Damcott CM, O'Connell JR, Mangano C, Corretti M, Vogel R, Herzog W, Weir MR, Peyser PA, Shuldiner AR. The genetic response to short-term interventions affecting cardiovascular function: Rationale and design of the Heredity and Phenotype Intervention (HAPI) Heart Study. Am Heart J. 2008;155:823–828. doi: 10.1016/j.ahj.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Morgan AR, Rerkasem K, Gallagher PJ, Zhang B, Morris GE, Calder PC, Grimble RF, Eriksson P, McPheat WL, Shearman CP, Ye S. Differences in matrix metalloproteinase-1 and matrix metalloproteinase-12 transcript levels among carotid atherosclerotic plaques with different histopathological characteristics. Stroke. 2004;35:1310–1315. doi: 10.1161/01.STR.0000126822.01756.99. [DOI] [PubMed] [Google Scholar]
- 30.Galis ZS, Sukhova GK, Kranzhofer R, Clark S, Libby P. Macrophage foam cells from experimental atheroma constitutively produce matrix-degrading proteinases. Proc Natl Acad Sci U S A. 1995;92:402–406. doi: 10.1073/pnas.92.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Axisa B, Loftus IM, Naylor AR, Goodall S, Jones L, Bell PR, Thompson MM. Prospective, randomized, double-blind trial investigating the effect of doxycycline on matrix metalloproteinase expression within atherosclerotic carotid plaques. Stroke. 2002;33:2858–2864. doi: 10.1161/01.str.0000038098.04291.f6. [DOI] [PubMed] [Google Scholar]
- 32.Guo LJ, Luo XH, Wu XP, Shan PF, Zhang H, Cao XZ, Xie H, Liao EY. Serum concentrations of MMP-1, MMP-2, and TIMP-1 in Chinese women: Age-related changes, and the relationships with bone biochemical markers, bone mineral density. Clinica Chimica Acta. 2006;371:137–142. doi: 10.1016/j.cca.2006.02.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.