SUMMARY
Nonparametric varying-coefficient models are commonly used for analysis of data measured repeatedly over time, including longitudinal and functional responses data. While many procedures have been developed for estimating the varying-coefficients, the problem of variable selection for such models has not been addressed. In this article, we present a regularized estimation procedure for variable selection that combines basis function approximations and the smoothly clipped absolute deviation (SCAD) penalty. The proposed procedure simultaneously selects significant variables with time-varying effects and estimates the nonzero smooth coefficient functions. Under suitable conditions, we have established the theoretical properties of our procedure, including consistency in variable selection and the oracle property in estimation. Here the oracle property means that the asymptotic distribution of an estimated coefficient function is the same as that when it is known a priori which variables are in the model. The method is illustrated with simulations and two real data examples, one for identifying risk factors in the study of AIDS and one using microarray time-course gene expression data to identify the transcription factors related to the yeast cell cycle process.
key words and phrases : Functional response, Longitudinal data, Nonparametric function estimation, Regularized estimation, Oracle property
1 Introduction
Varying-coefficient (VC) models (Hastie and Tibshirani, 1993) are commonly used for studying the time-dependent effects of covariates on responses measured repeatedly. Such models can be used for longitudinal data where subjects are often measured repeatedly over a given period of time, so that the measurements within each subject are possibly correlated with each other (Diggle et al., 1994; Rice, 2004). Another setting is the functional response models (Rice, 2004), where the ith response is a smooth real function yi(t), i = 1,…, n, t ∈ 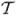 = [0, T], although in practice only yi(tij), j = 1,…, Ji are observed. For both settings, the response Y (t) is a random process and the predictor X(t) = (X(1)(t),…,X(p)(t))T is a p-dimensional random process. In applications, observations for n randomly selected subjects are obtained as (yi(tij); xi(tij)) for the ith subject at discrete time point tij, i = 1,…,n and j = 1,…,Ji. The linear varying-coefficient model can be written as
= [0, T], although in practice only yi(tij), j = 1,…, Ji are observed. For both settings, the response Y (t) is a random process and the predictor X(t) = (X(1)(t),…,X(p)(t))T is a p-dimensional random process. In applications, observations for n randomly selected subjects are obtained as (yi(tij); xi(tij)) for the ith subject at discrete time point tij, i = 1,…,n and j = 1,…,Ji. The linear varying-coefficient model can be written as
| (1) |
where β(t) = (β1(t),…,βp(t))T is a p-dimensional vector of smooth functions of t, and εi(t), i = 1,…,n are independently identically distributed random processes, independent of xi(t). This model has the simplicity of linear models but also has the flexibility of allowing time-varying covariate effects. Since its introduction to the longitudinal data setting by Hoover, Rice, Wu and Yang (1998), many methods have been developed for estimation and inference of Model (1). See, for example, Wu, Chiang and Hoover (1998) and Fan and Zhang (2000) for the local polynomial kernel method; Huang, Wu and Zhou (2002, 2004) and Qu and Li (2006) for basis expansion and the spline methods, and Chiang, Rice and Wu (2001) for the smoothing spline method. One important problem not well studied in the literature is how to select significant variables in Model (1). The goal of this paper is to estimate β(t) nonparametrically and select relevant predictors xk(t) with non-zero functional coefficient βk(t), based on observations {(yi(tij), xi(tij)); i = 1,…,n, j = 1,…,Ji}.
Traditional methods for variable selection include hypothesis testing and using information criteria such as AIC and BIC. Recently, regularized estimation has received much attention as a way of performing variable selection for parametric regression models (see Bickel and Li, 2006 for a review). Existing regularization procedures for variable selection include LASSO (Tibshirani, 1996), smoothly clipped absolute deviation (SCAD) (Fan and Li, 2001) and their extensions (Yuan and Lin, 2006; Zou and Hastie, 2005; Zou, 2006). However, these regularized estimation procedures were developed for regression models where parameters are Euclidean and cannot be applied directly to the nonparametric varying-coefficients models where parameters are nonparametric smooth functions. Moreover, these procedures are studied mainly for independent data; an exception is Fan and Li (2004) where the SCAD penalty was used for variable selection in longitudinal data analysis. On the other hand, some recent results on regularization methods with diverging number of parameters (e.g., Fan & Peng, 2004; Huang, Horowitz & Ma, 2007) are related to nonparametric settings, but the model error due to function approximation has not been of concern in that literature.
Regularized estimation for selecting relevant variables in nonparametric settings has been developed in the general context of Smoothing Spline Analysis of Variance (ANOVA) Models. Lin and Zhang (2006) developed COSSO for component selection and smoothing in Smoothing Spline ANOVA. The COSSO method was extended by Zhang and Lin (2006) to nonparametric regression in exponential families, by Zhang (2006) to Support Vector Machines and by Leng and Zhang (2007) to hazard regression. Lin and Zhang (2006) studied rates of convergence of COSSO estimators; the rest of the papers focused on computational algorithms and did not provide theoretical results. Asymptotic distribution results have not been developed for COSSO and its extensions. Moreover, these approaches have focused on independent data, not on longitudinal data that we study in this paper.
In this paper we use the method of basis expansion for estimating smooth functions in varying coefficient models because of the simplicity of the basis expansion approach as illustrated in Huang et al. (2002). We extend the application of the SCAD penalty to a nonparametric setting. In addition to developing a computational algorithm, we study the asymptotic property of the estimator. We show that our procedure is consistent in variable selection, that is, the probability that it correctly selects the true model tends to one. Furthermore, we show that our procedure has an oracle property, that is, the asymptotic distribution of an estimated coefficient function is the same as that when it is known a priori which variables are in the model. Such results are new in nonparametric settings.
The rest of the paper is organized as follows. We first describe in Section 2 the regularized estimation procedure using basis expansion and the SCAD penalty. A computational algorithm and method of selecting the tuning parameter are given in Section 3. We then present theoretical results in Section 4, including the consistency in variable selection and the oracle property. Some simulation results are shown in Section 5. Section 6 illustrates the proposed method using two real data examples, a CD4 dataset and a microarray time-course gene expression dataset. Technical proofs are gathered in the Appendix.
2 Basis Function Expansion and Regularized Estimation Using the SCAD Penalty
Huang, Wu and Zhou (2002) proposed to estimate the unknown time-varying coefficient functions using basis expansion. Suppose that the coefficient βk(t) can be approximated by a basis expansion , where Lk is the number of basis functions in approximating the function βk(t). Model (1) becomes
| (2) |
The parameters γkl in the basis expansion can be estimated by minimizing
| (3) |
where wi are weights, taking the value 1 if we treat all observations equally or 1/ni if we treat each subject equally. An estimate of βk(t) is obtained by where γ̂kl are minimizers of (3). Various basis systems such as Fourier bases, polynomial bases and B-spline bases can be used in the basis expansion. Huang, Wu and Zhou (2002) studied consistency and rates of convergence of such estimators for general basis choices and Huang, Wu and Zhou (2004) studied asymptotic normality of the estimators when the basis functions Bkl are splines.
Now suppose some variables are not relevant in the regression so that the corresponding coefficient functions are zero functions. We introduce a regularization penalty to (3) so that these zero coefficient functions will be estimated as identically zero. To this end, it is convenient to rewrite (3) using function space notation. Let 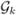 denote all functions that have the form
for a given basis system {Bkl(t)}. Then (3) can be written as (1/n)
where
. Let pλ(u), u ≥0, be a nonnegative penalty function that depends on a penalty parameter λ. It is assumed that pλ(0) = 0 and pλ(u) is nondecreasing as a function of u. Let ||gk|| denote the L2-norm of the function gk. We minimize over gk ∈
denote all functions that have the form
for a given basis system {Bkl(t)}. Then (3) can be written as (1/n)
where
. Let pλ(u), u ≥0, be a nonnegative penalty function that depends on a penalty parameter λ. It is assumed that pλ(0) = 0 and pλ(u) is nondecreasing as a function of u. Let ||gk|| denote the L2-norm of the function gk. We minimize over gk ∈ 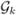 the penalized criterion
the penalized criterion
| (4) |
There are two sets of tuning parameters: the Lk’s control the smoothness of the coefficient functions and the λ governs variable selection or sparsity of the model. Selection of these parameters will be discussed in Section 4. In our implementation of the method, we use B-splines as the basis functions. Thus Lk = nk + d + 1 where nk is the number of interior knots for gk and d is the degree of the spline. The interior knots of the splines can be either equally spaced or placed on the sample quantiles of the data so that there are about the same number of observations between any two adjacent knots. We use equally spaced knots for all numerical examples in this paper.
There are many ways to specify the penalty function pλ(·). If pλ(u) = λu, then the penalty term in (4) is similar to that in COSSO. However, we will use the SCAD penalty function by Fan and Li (2001), defined as
| (5) |
The penalty function (5) is a quadratic spline function with two knots at λ and aλ, where a is another tuning parameter. Fan and Li (2001) suggested that a = 3.7 is a reasonable choice, which was also adopted in this paper. Use of the SCAD penalty allows us to obtain nice theoretical properties such as consistent variable selection and the oracle property for the proposed method.
For gk(t) = Σl γklBkl(t) ∈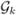 , its squared L2-norm can be written as a quadratic form in γk = (γk1,…,γkLk)T. Let Rk = (rij) Lk× Lk be a matrix with entries
. Then
. Set
. The penalized weighted least squares criterion (4) can be written as
, its squared L2-norm can be written as a quadratic form in γk = (γk1,…,γkLk)T. Let Rk = (rij) Lk× Lk be a matrix with entries
. Then
. Set
. The penalized weighted least squares criterion (4) can be written as
| (6) |
To express the criterion function (6) using vectors and matrices, we define
Ui(tij) = (xi(tij)TB(tij))T, Ui = (Ui(ti1),…,Ui(tiJi))T, and . We also define yi = (yi(ti1),…,yi(tiJi))T and . The criterion function (6) can be rewritten as
| (7) |
where W is a diagonal weight matrix with wi repeated ni times.
Remark
The penalized weighted least squares criterion here does not take into account the within-subject correlation typically present in the longitudinal data, because the correlation structure is usually unknown a priori. A popular method for dealing with within-subject correlation is to employ a working correlation structure as in the generalized estimation equations (GEE, Zeger and Liang, 1996). The idea of GEE can be easily adapted into our methodolgy. Specifically, the weight matrix W in (7) can be constructed using a working correlation structure, such as AR or compound symmetry. Actually the criteria (4) and (6) correspond to a working independence correlation structure. Using working independence correlation won’t affect the consistency of variable selection (Section 5). On the other hand, using an appropriate working correlation structure may yield more efficient function estimation if the structure is correctly specified.
3 Computational Algorithm
Because of non-differentiability of the penalized loss (7), the commonly-used gradient-based optimization method is not applicable. In this section, we develop an iterative algorithm using local quadratic approximation of the non-convex penalty pλ(||γk||2). Following Fan and Li (2001), in a neighborhood of a given positive u0 ∈ ℝ+,
| (8) |
In our algorithm, a similar quadratic approximation is used by substituting u with ||γk||k in (8), k = 1,…,p. Given an initial value of with , pλ(||γk||k) can be approximated by a quadratic form
As a consequence, removing an irrelevant constant, the penalized loss (7) becomes
| (9) |
where where Rk are defined above equation (6). This is a quadratic form whose minimizer satisfies
| (10) |
The above discussion leads to the following algorithm:
Step 1: Initialize γ = γ(1).
Step 2: Given γ(m), update γ to γ(m+1) by solving equation (10) where γ0 is set to be γ(m)
Step 3: Iterate Step 2 until convergence of γ.
The initial estimation of γ in Step 1 can be obtained using a ridge regression, which substitutes pλ(||γk||2) in the penalized loss (7) with the quadratic function . The ridge regression has a closed form solution. At any iteration of Step 2, if some is smaller than a cutoff value ε > 0, we set γ̂k = 0 and treat x(k)(t) as irrelevant. In our implementation of the algorithm ε is set to 10−3.
Remark
Due to the non-convexity of pl(γ), the algorithm sometimes may fail to converge and the search falls into an infinite loop surrounding several local minimizers. In this situation, we choose the local minimizer that gives the smallest value of pl(γ).
4 Selection of Tuning Parameters
To implement the proposed method, we need to choose the tuning parameters: Lk, k = 1,…,p and λ, where Lk controls the smoothness of β̂k(t), while λ determines the variable selection. In section 5, we will show that our method is consistent in variable selection when these tuning parameters grow or decay at a proper rate with n. In practice, however, we need a data-driven procedure to select the tuning parameters. We propose to use the “leave-one-observation-out” cross-validation (CV) when the errors are independent and use the “leave-one-subject-out” cross-validation for correlated errors.
Below we develop some computational shortcuts using suitable approximations to prevent actual computation of leave-one-out estimates. Although the derivation of model selection criteria generally holds, we focus our presentation on the situation when Lk = L for all βk(t), k = 1,…, p. To simplify our presentation, we also set W = I; extension to general W is straightforward, simply replacing y with W1/2y and U with W1/2U in the derivation below. At the convergence of our algorithm, the nonzero components are estimated as γ̂ = {UTU + (n/2)Ωλ(γ̂)}−1UTy, which can be considered as the solution of the ridge regression
| (11) |
The hat matrix of this ridge regression is denoted by H(L, λ) = U{UTU+(n/2) Ωλ(γ̂)}−1UT. The fitted y from this ridge regression satisfies ŷ = H(L, λ)y.
When the errors εij are independent for different tij, j = 1,…,Ji, leave-one-observation-out CV is a reasonable choice for selecting tuning parameters. For the ridge regression (11), computation of leave-one-out estimates for CV can be avoided (Hastie and Tibshirani, 1993). The CV criterion has a closed form expression
| (12) |
where {H(L; λ)}(ij);(ij) is the diagonal element of H(L, λ) that corresponds to the observation at time tij. The corresponding GCV criterion, which replaces the diagonal elements in the above CV formula by their average, is
| (13) |
When the errors εij are not independent and the correlation structure of εi(t) is unknown, the leave-one-observation-out CV is unsuitable, we need to use leave-one-subject-out CV (Rice and Silverman, 1991; Hoover et al., 1998; and Huang et al., 2002). Let γ̂(−i) be the solution of (6) after deleting the ith subject. The leave-one-subject-out CV can be written as
| (14) |
The CV2(L, λ) can be viewed as an estimate of the true prediction error. However, computation of CV2 is intensive, since it requires minimization of (6) n times. To overcome this difficulty, we propose to minimize a delete-one version of (11) instead of (6) in computing CV2, where γ̂ in Ωλ(γ̂) of (11) is the estimate based on the whole dataset. The approximated CV error is
| (15) |
where γ̂★(−i) is obtained by solving (11) instead of (6), deleting the ith subject. It turns out a computation shortcut is available for (15) as we discuss next.
The computation shortcut relies on the following “leave-one-subject-out” formula, the proof of which is given in Appendix A.
Lemma 1
Define , and let γ̃(i) be the minimizer of (11) with y substituted by ỹ(i). Then, Uiγ̂★(−i) = Uiγ̂(i).
Let A = {UTU +(n/2)Ωλ(γ̂)}−1UT. Partition A = (A1,…,An) into blocks of columns with each block corresponding to one subject. Then
and
As a consequence of Lemma 1,
It follows that
Therefore,
| (16) |
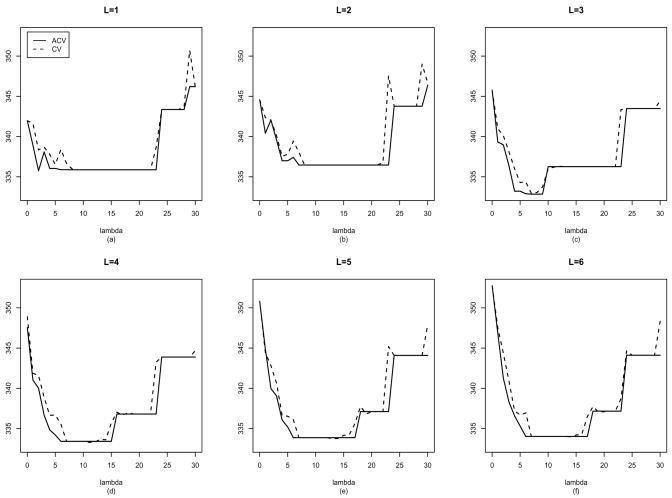
where Hii(L, λ) is the (i, i)-th diagonal block corresponding to observations for the i-th subject. Since Ji is usually not a big number, inversion of the Ji-dimensional matrices I − Hii won’t create a computation burden in the evaluation of (16). Using (16) as an approximation of the leave-one-subject-out CV helps us avoid actual computation of many delete-one estimates. The quality of the approximation is illustrated in Figure 1 using a dataset from the simulation study reported in Section 6.
Figure 1.
Illustration of the approximation of CV scores for one simulated dataset for different values of L and λ. The ACV given in(16) approximates the leave-one-subject-out CV in (14) well.
5 Large Sample Properties
For parametric regression models, Fan and Li (2001) established the large sample properties of the SCAD penalized estimates. They showed that the SCAD penalty enables consistent variable selection and has an oracle property for parameter estimation | the estimates of the nonzero regression coefficients behave as if the subset of relevant variables is already known. We show that similar large sample properties hold for our proposed method for the varying coefficients models. Note that some care needs to be taken in developing an asymptotic theory for a nonparametric setting. For example, the rate of convergence of a nonparametric estimate is not root-n. The approximation error due to use of basis expansion needs also to be carefully studied. For simplicity we shall only present our asymptotic results for working independence case. Technical proofs of all asymptotic results are given in the Appendix.
We focus our asymptotic analysis for 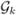 being a space of spline functions. Extension to general basis expansions can be obtained by combining the technical arguments in this paper with those in Huang et al. (2002). Define Ln = max1≤k≤p Lk and ρn = max1≤k≤p infg∈
being a space of spline functions. Extension to general basis expansions can be obtained by combining the technical arguments in this paper with those in Huang et al. (2002). Define Ln = max1≤k≤p Lk and ρn = max1≤k≤p infg∈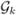 ||βk − g||L∞. Thus, ρn characterizes the approximation error due to spline approximation. Assume that only s predictors are relevant in the Model (1). Without loss of generality, let βk(t), k = 1,…,s, be the non-zero coefficient functions, and βk(t) ≡ 0, k = s + 1,…,p.
||βk − g||L∞. Thus, ρn characterizes the approximation error due to spline approximation. Assume that only s predictors are relevant in the Model (1). Without loss of generality, let βk(t), k = 1,…,s, be the non-zero coefficient functions, and βk(t) ≡ 0, k = s + 1,…,p.
We make the following technical assumptions:
(C1) The response and covariate processes (yi(t); xi(t)), i = 1,…, n, are i.i.d. as (y(t), X(t)), and the observation time points tij are i.i.d. from an unknown density f (t) on [0, T ], where f(t) are uniformly bounded away from infinity and zero.
(C2) The eigenvalues of the matrix E{X(t)XT(t)} are uniformly bound away from zero and infinity for all t.
(C3) There exists a positive constant M1 such that |Xk(t)| ≤ M1 for all t and 1 ≤ k ≤ p.
(C4) There exists a positive constant M2 such that E{ε2(t)} ≤ M2 for all t.
(C5) lim supn(maxk Lk/mink Lk) < ∞.
(C6) The process ε(t) can be decomposed as the sum of two independent stochastic processes, that is, ε(t) = ε(1)(t) + ε(2)(t), where ε(1) is an arbitrary mean zero process and ε(2) is a process of measurement errors that are independent at different time points and have mean zero and constant variance σ2.
The same set of assumptions has been used in Huang et al. (2004). Our results can be extended to cases with deterministic observation times; the assumption on independent observation times in (C1) can also be relaxed (Remarks 3.1 and 3.2 of Huang et al., 2004).
Define . When wi = 1/Ji, or wi = 1/Ji with Ji uniformly bounded, we can show that rn ≍ (Ln/n)1/2
Theorem 1
Suppose Conditions (C1)–(C5) hold, limn→∞ ρn = 0 and
| (17) |
Then, with a choice of λn such that λn → 0 and λn/max{rn, ρn} → ∞, we have
β̂k = 0, k = s + 1,…,p, with probability approaching 1.
||β̂k − βk||L2 = Op(max(rn, ρn)), k = 1,…,s.
Part (a) of Theorem 1 says that the proposed penalized least squares method is consistent in variable selection, that is, it can identify the zero coefficient functions with probability tending to 1. Part (b) of the the theorem provides the rate of convergence in estimating the non-zero coefficient functions.
Corollary 1
Suppose that Conditions (C1)–(C4) hold and that βk(t) have bounded second derivatives, k = 1,…,s, and βk(t) = 0, k = s + 1,…,p. Let 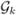 be a space of splines with degree no less than 1 and with Lk equally spaced interior knots, where Lk ≍ n1/5, k = 1,…,p. If λn → 0 and n2/5 λn → ∞, then,
be a space of splines with degree no less than 1 and with Lk equally spaced interior knots, where Lk ≍ n1/5, k = 1,…,p. If λn → 0 and n2/5 λn → ∞, then,
β̂k = 0, k = s + 1,…,p, with probability approaching 1.
||β̂k − βk||L2 = Op(n−2/5), k = 1,…, s.
Note that the rate of convergence given in part (b) of Corollary 1 is the optimal rate for nonparametric regression with independent, identically distributed data under the same smoothness assumption on the unknown function (Stone, 1982).
Now we consider the asymptotic variance of proposed estimate. Let β(1) = (β1,…, βs)T denote the vector of non-zero coefficient functions and let β̂(1) = (β̂1,…,β̂s)T its estimate obtained by minimizing (6). Let . Let U(1) denote the selected columns of U corresponding to β(1) and similarly let denote the selected diagonal blocks of Ωλ. By part (a) of Theorem 1, with probability tending to one, γ̂k, k = s + 1,…,p, are vectors of 0’s. Thus the quadratic approximation (9) yields
| (18) |
Let be the rows of U(1) corresponding to subject i. Let
| (19) |
The asymptotic variance of γ̂(1) is
| (20) |
whose diagonal blocks give Avar(γ̂k), k = 1,…,s. Since β̂(1)(t) = (B(1))T(t)γ̂(1) where B(1)(t) are the first s rows of B(t), we have that Avar {β̂(1)(t)} = (B(1))T(t)Avar(γ̂(1))B(1)(t), the diagonal elements of which are Avar{β̂k(t)}, k = 1,…,s.
Let β̃k(t) = E{β̂k(t)} be the conditional mean given all observed X’s. Let β̃(1)(t) = (β̃1(t),… β̃s(t)). For positive definite matrices A, let A 1/2 denote the unique square root of A and let A−1/2 = (A−1)1/2. Let Avar*{β̂(1)(t)} denote a modification of Avar{β̂(1)(t)} by replacing Ωλn in (19) with 0.
Theorem 2
Suppose Conditions (C1)–(C6) hold, limn → ∞ ρn = 0, limn→ ∞ Ln maxi Ji/n = 0, and (17) holds. Then, as n → ∞, {Avar* (β̂(1)(t))}−1/2{β̂(1)(t) − β̃(1)(t)} → N(0, I) in distribution, and in particular, {Avar* (β̂k(t))}−1/2{β̂k(t) − β̃k(t)} → N (0, 1), k = 1,…, s.
Note that Avar*{β̂(1)(t)} is exactly the same asymptotic variance of the non-penalized weighted least squares estimate using only those covariates corresponding to nonzero coefficient functions (Theorem 3, Huang et al., 2004). Theorem 2 implies that our penalized least squares estimate has the so-called oracle property — the joint asymptotic distribution of estimates of nonzero coefficient functions are the same as that of the non-penalized least squares estimate that utilizes the information about zero coefficient functions.
Theorem 2 can be used to construct pointwise asymptotic confidence intervals for β̃k(t), k = 1,…,s. Theorem 4 and Corollary 1 of Huang et al. (2004) discuss how to bound the biases β̃k(t) − βk(t) and under what conditions the biases are negligible relative to variance. Those results are applicable to the current context. When constructing the confidence intervals, in order to improve finite sample performance, we still use (19) and the sandwich formula (20) to calculate the variance. In actual calculation, we need replace cov(yi) in (19) by its estimate where ei is the residual vector for subject i.
6 Monte Carlo Simulation
We conducted simulation studies to assess the performance of the proposed procedure. In each simulation run, we generated a simple random sample of 200 subjects according to the model used in Huang et al. (2002), which assumes
The first three variables xi(t), i = 1,…,3, are the true relevant covariates, which are simulated the same way as in Huang et al. (2002): x1(t) is sampled uniformly from [t/10, 2+ t/10] at any given time point t; x2(t), conditioning on x1(t), is Gaussian with mean zero and variance (1 + x1(t))/(2 + x1(t)); x3(t), independent of x1 and x2, is a Bernoulli random variable with success rate 0.6. In addition to xk, k = 1, 2, 3, 20 redundant variables xk(t), k = 4,…,23, are simulated to demonstrate the performance of variable selection, where each xk(t), independent of each other, is a random realization of a Gaussian process with covariance structure cov(xk(t); xk(s)) = 4 exp(− |t − s|). The random error ε(t) is given by Z(t) + E(t), where Z(t) has the same distribution as xk(t), k = 4,…,23, and E(t) are independent measurement errors from N(0, 4) distribution at each time t. The parameter τ in the model is used to control the signal-to-noise ratio of the model. We consider models with τ = 1.00, 4.00, 5.66 and 8.00. The coefficients βk(t), k = 0,…,3, corresponding to the constant term and the first three variables, are given by
(see solid lines of Figure 2) while the remaining coefficients, corresponding to the irrelevant variables, are given by βk(t) = 0, k = 4,…, 23. The observation time points tij are generated following the same scheme as in Huang et al. (2002), where each subject has a set of “scheduled” time points {1,…,30}, and each scheduled time has a probability of 60% of being skipped. Then, the actual observation time tij is obtained by adding a random perturbation from Uniform(− 0.5, 0.5) to the nonskipped scheduled time.
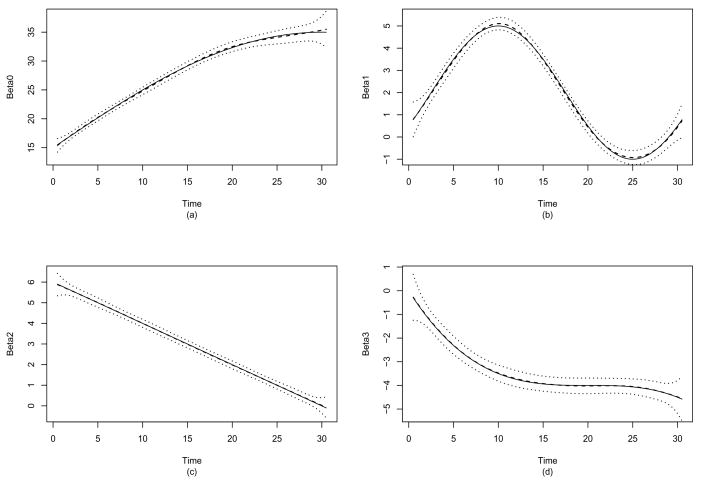
Figure 2.
Simulation study for τ = 1.00. True (solid lines) and average of estimated (dashed lines) time-varying coefficients β0(t) (a), β1(t) (b), β2(t) (c) and β3(t) (+/− 1 point-wise SD) over 500 replications.
For each simulated dataset, the penalized least squares criterion (4) was minimized over a space of cubic splines with equally spaced knots. The number of knots and the tuning parameter λ were selected by minimizing the ACV2 criterion in (15) as described in Section 4 (see Figure 1 for an example). We repeated the simulations 500 times. Table 1 shows the results of variable selection of our proposed procedure for four different models with different levels of noise variances. Clearly the level of noise had a great effect on how well the proposed method selected the exact correct models. When the noise level was low (τ = 1.00), the method selected the exact correct model in 79% of the replications. In addition, out of these 500 replications, the variables 1–3 were selected in each of the runs. In contrast, the number of times each of the 20 irrelevant variables were selected among the 500 simulation runs ranged from 3 to 12 with an average of 7.3 times (i.e., 1.46%). However, when the noise level was high (τ = 8.00), the method selected the exact true model in only 26% of the replications and the average number of times that the three relevant variables were selected was reduced to 2.24. However, it is interesting to note that the average number of times that the irrelevant variables were selected remained to be small (See Aver.I in Table 1). The simulations indicate that the proposed procedure indeed provides an effective method for selecting variables with time-varying coefficients.
Table 1.
Simulation results for models with different noise levels (τ) based on 500 replications. Perc.T: percentage of replications that the exact true model was selected; Aver.R: average of the numbers of the three relevant variables that were selected in the model; Aver.I: average of the numbers of the irrelevant variables that were selected in the model; Aver.Bias2: average of the squared biases; Aver.Var: average of the empirical variances; 95%Cov.Prob: 95% coverage probability based on the replications when the true models were selected. For Aver.Bias2, Aver.Var and 95%Cov.Prob, all values were averaged over a grid of 300 points and 500 replications. Numbers in parentheses are results of the oracle estimators that assume the true models are known.
| Noise level (τ) |
||||
|---|---|---|---|---|
| 1.00 | 4.00 | 5.66 | 8.00 | |
| Variable selection | ||||
| Perc.T | 0.79 | 0.68 | 0.48 | 0.26 |
| Aver.R | 3.00 | 2.94 | 2.73 | 2.24 |
| Aver.I | 0.29 | 0.47 | 0.55 | 0.54 |
| Aver.Bias2 | ||||
| β0(t) | 0.021 (0.023) | 0.39 (0.37) | 1.60 (0.42) | 4.55 (0.42) |
| β1(t) | 0.003 (0.004) | 0.067 (0.067) | 0.29 (0.076) | 0.97 (0.073) |
| β2(t) | 0.000089 (0.0002) | 0.0022 (0.0022) | 0.0047 (0.0043) | 0.026 (0.0089) |
| β3(t) | 0.00023 (0.0004) | 0.0079 (0.0060) | 0.061 (0.010) | 0.88 (0.02) |
| Aver.Var | ||||
| β0(t) | 0.98 (0.95) | 14.11 (12.53) | 28.29 (23.98) | 49.09 (47.25) |
| β1(t) | 0.12 (0.11) | 1.95 (1.52) | 4.08 (2.84) | 7.15 (5.52) |
| β2(t) | 0.049 (0.047) | 0.70 (0.67) | 1.37 (1.31) | 3.06 (2.63) |
| β3(t) | 0.15 (0.15) | 2.13 (2.01) | 4.70 (3.96) | 9.98 (7.88) |
| 95%Cov.Prob | ||||
| β0(t) | 0.929 | 0.935 | 0.937 | 0.926 |
| β1(t) | 0.924 | 0.929 | 0.930 | 0.929 |
| β2(t) | 0.936 | 0.940 | 0.946 | 0.942 |
| β3(t) | 0.944 | 0.949 | 0.947 | 0.949 |
To examine how well the method estimates the coefficient functions, Figure 2 shows the estimates of the time-varying coefficients of βk(t), k = 0, 1, 2, 3 for the model with τ = 1.00, indicating that the estimates fit the true function very well. Table 1 shows the averages of the squared biases and the empirical variances of the estimates of βk(t), k = 0, 1, 2, 3 averaged over a grid of 300 points. In general, the biases were small, except for the estimates of β0(t) when the noise level was high, in which case the proposed method did not select the variables well. Similarly, the variances of the estimates of β0(t) were also relatively larger due to the fact that the magnitude of true β0(t) is larger than the other three functions. As a comparison, we also calculated the biases and the empirical variances of the estimates based on the true models (i.e., the oracle estimators, see Table 1). Again, we observed that the both biases and empirical variances were very comparable to the oracle estimators when the noise level was low. When the noise level was high, due to the fact that the method tends to select fewer relevant variables, the biases and the empirical variances became large.
Finally, we examined the coverage probabilities of the theoretical point-wise confidence intervals based on Theorem 2. For each model, the empirical coverage probabilities averaged over a grid of 300 points were presented in Table 1 for β0(t), β1(t), β2(t) and β3(t), respectively, for nominal 95% point-wise confidence intervals. The slight under-coverage is caused by bias: the GCV criterion tried to balance the bias and variance. When we used the number of knots larger than that suggested by GCV (under-smoothing), the empirical coverage probabilities got closer to the nominal level. For example, for the model with τ = 1, the average empirical coverage probabilities became 0.946, 0.949, 0.942 and 0.947 when the number of interior knots were fixed at 5.
7 Applications to Real Datasets
To demonstrate the effectiveness of the proposed methods in selecting the variables with time-varying effects and in estimating the time-varying effects, we present in this Section results from the analysis of two real datasets, including a longitudinal AIDS dataset (Kaslow et al., 1987) and a repeated measurement microarray time course gene expression dataset (Spellman et al., 1998).
7.1 Application to AIDS data
Kaslow et al. (1987) reported a Multicenter AIDS Cohort Study to obtain the repeated measurements of physical examinations, laboratory results and CD4 cell counts and percentages of the homosexual men who became HIV-positive during 1984 and 1991. All individuals were scheduled to have their measurements made at semi-annual visits, but because many individuals missed some of their scheduled visits and the HIV infections happened randomly during the study, there were unequal numbers of repeated measurements and different measurement times per individual. As a subset of the cohort, our analysis focuses on the 283 homosexual men who become HIV-positive and aims to evaluate the effects of cigarette smoking, pre-HIV infection CD4 percentage, and age at HIV infection on the mean CD4-percentage after infection. In this subset, the number of repeated measurements per subject ranged from 1 to 14, with a median of 6 and a mean of 6.57. The number of distinct measurement time points was 59. Let tij be the time in years of the jth measurement of the ith individual after HIV infection, yij be the ith individual’s CD4 percentage at time tij, and xi1 be the smoking status of the ith individual, taking a value of 1 or 0 for smoker or non-smoker. In addition, let xi2 be the centered age at HIV infection for the ith individual and xi3 be the centered pre-infection CD4 percentage. We assume the following VC model for yij,
where β0(t) is the baseline CD4 percentage, representing the mean CD4 percentage t years after the infection for a non-smoker with average pre-infection CD4 percentage and average age at HIV infection, and β1(t), β2(t), β3(t) measures the time-varying effects for cigarette smoking, age at HIV infection and pre-infection CD4 percentage, respectively, on the post-infection CD4 percentage at time t.
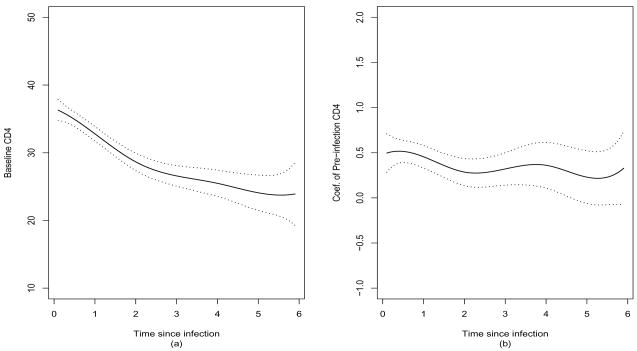
Our procedure identified two non-zero coefficients, β0(t) and β3(t), indicating that cigarette smoking and age of HIV infection do not have any effects on the poster-infection CDS4 percentage. Figure 3 shows the fitted coefficient functions (solid curves) and their 95% point-wise confidence intervals, indicating that the baseline CD4 percentage of the population decreases with time and pre-infection CD4 percentage appears to be positively associated with high post-infection CD4 percentage. These results agreed with those obtained by the local smoothing methods of Wu and Chiang (2002) and Fan and Zhang (2002) and by the basis function approximation method of Huang et al. (2002).
Figure 3.
Application to AIDS data. Estimated coefficient functions for intercept (a) and pre-infection CD4 percentages (b) (solid curves) and their point-wise 95% confidence intervals (dashed lines).
To assess our method in more challenging cases, we added 30 redundant variables, which are generated by randomly permuting the values of each predictor variable 10 times, and applied our method to the augmented dataset. We repeated the procedure 50 times. For the three observed covariates, “smoking status” was never selected, “age” was selected twice, and “pre-infection CD4 percentage” was always selected, wheras among the 30 redundant variables, two were selected twice, four were selected once, and the rest were never selected.
7.2 Application to yeast cell cycle gene expression data
The cell cycle is a tightly regulated set of processes by which cells grow, replicate their DNA, segregate their chromosomes, and divide into daughter cells. The cell cycle process is commonly divided into G1-S-G2-M stages, where the G1 stage stands for “GAP 1”, the S stage stands for “Synthesis”, during which DNA replication occurs, the G2 stage stands for “GAP 2” and the M stage stands for “mitosis”, which is when nuclear (chromosomes separate) and cytoplasmic (cytokinesis) division occur. Coordinate expression of genes whose products contribute to stage-specific functions is a key feature of the cell cycle (Simon et al., 2001). Transcription factors (TFs) have been identified that have roles in regulating transcription of a small set of yeast cell-cycle regulated genes; these include Mbp1, Swi4, Swi6, Mcm1, Fkh1, Fkh2, Ndd1, Swi5 and Ace2. Among these, Swi6, Swi4 and Mbp1 have been reported to function during the G1 stage; McM1, Ndd1 and Fkh1/2 have been reported to function during the G2 stage; and Ace2, Swi5 and Mcm1 have been reported to function during the M stage (Simon et al., 2001). However, it is not clear where these TFs regulate all cell cycle genes.
We apply our methods to identify the key transcription factors that play roles in the network of regulations from a set of gene expression measurements captured at different time points during the cell cycle. The dataset we use comes from the α-factor synchronized cultures from Spellman et al. (1998). They monitored genome-wide mRNA levels for 6178 yeast ORFs simultaneously using several different methods of synchronization including an α-factor-mediated G1 arrest, which covers approximately two cell-cycle periods with measurements at 7-min intervals for 119 mins with a total of 18 time points. Using a model-based approach, Luan and Li (2003) identified 297 cell-cycle-regulated genes based on the α-factor synchronization experiments. Let yit denote the log-expression level for gene i at time point t during the cell cycle process, for i = 1, ···, 297 and t = 1, ···, 18. We then applied the mixture model approach (Chen et al., 2007; Wang et al., 2007) using the ChIP data of Lee et al. (2002) to derive the binding probabilities xik for these 297 cell-cycle-regulated genes for a total of 96 transcriptional factors with at least one nonzero binding probability in the 297 genes. We assume the following VC model to link the binding probabilities to the gene expression levels
where βk(t) models the transcription effect of the kth TF on gene expression at time t during the cell cycle process. In this model, we assume that gene expression levels are independent conditioning on effects of the relevant transcription factors. In addition, for a given gene i, ∈its are also assumed to be independent over different time points due to the cross-sectional nature of the experiments. Our goal is to identify the transcriptional factors that might be related to the expression patterns of these 297 cell-cycle-regulated genes. Since different transcriptional factors may regulate the gene expression at different time points during the cell cycle process, their effects on gene expression are expected to be time-dependent.
We applied our method using the GCV defined in (13) for selecting the tuning parameter in order to identify the TFs that affect the expression changes over time for these 297 cell-cycle-regulated genes in the α-factor synchronization experiment. Our procedure identified a total of 71 TFs that are related to yeast cell-cycle processes, including 19 of the 21 known and experimentally verified cell-cycle related TFs, all showing time-dependent effects of these TFs on gene expression levels. In addition, the effects followed similar trends between the two cell cycle periods. The other two TFs, CBF1 and GCN4, were not selected by our procedure. The minimum p-values over 18 time points from simple linear regressions are 0.06 and 0.14, respectively, also indicating that CBF1 and GCN4 were not related to expression variation over time. The 52 additional TFs that were selected by our procedure almost all showed estimated periodic transcriptional effects. The identified TFs include many pairs of cooperative or synergistic pairs of TFs involved in the yeast cell cycle process reported in the literature (Banerjee and Zhang, 2003; Tsai et al., 2005). Of these 52 TFs, 34 of them belong to the cooperative pairs of the TFs identified by Banerjee and Zhang (2003). Overall, the model can explain 43% of the total variations of the gene expression levels.
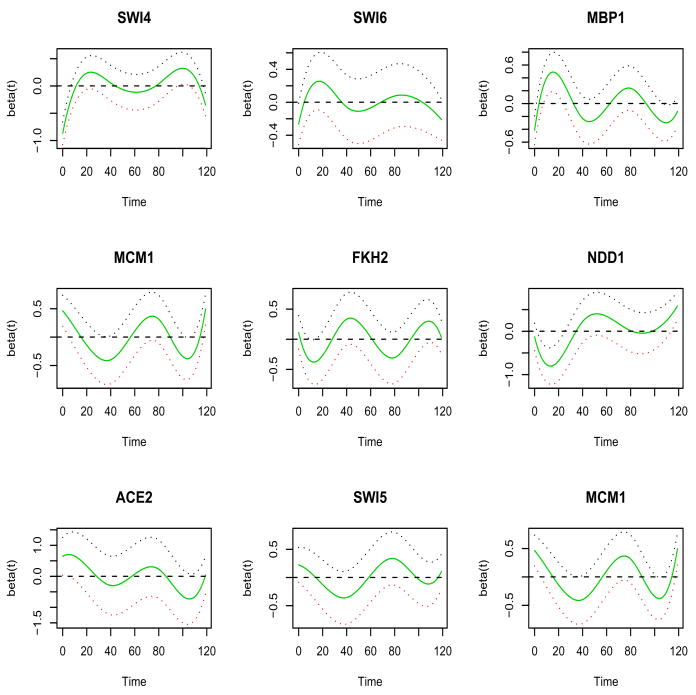
Figure 4 shows the estimated time-dependent transcriptional effects of nine of the experimentally verified TFs that are known to regulate cell-cycle-regulated genes at different stages. The top panel shows the transcriptional effects of three TFs, Swi4, Swi6 and Mbp1, which regulate gene expression at the G1 phase (Simon et al., 2001). The estimated transcriptional effects of these three TFs are quite similar with peak effects obtained at approximately the time points corresponding to the G1 phase of the cell cycle process. The middle panel shows the transcriptional effects of three TFs, Mcm1, Fkh2 and Ndd1, which regulate gene expression at the G2 phase (Simon et al., 2001). The estimated transcriptional effects of two of these three TFs, Fkh2 and Ndd1, are similar with peak effects obtained at approximately the time points corresponding to the G2 phase of the cell cycle process. The estimated effect of Mcm1 was somewhat different, however Mcm1 is also known to regulate gene expression at the M phase (Simon et al., 2001). The bottom panel of Figure 4 shows the transcriptional effects of three TFs, Swi5, Ace2 and Mcm1, which regulate gene expression at the M phase (Simon et al., 2001), indicating similar transcriptional effects of these three TFs with peak effects at approximately the time points corresponding to the M phase of the cell cycle. These plots demonstrated that our procedure can indeed identify the known cell-cycle-related TFs and point to the stages when these TFs function to regulate the expressions of cell cycle genes. As a comparison, only two of these nine TFs (SWI4 and FKH2) show certain periodic patterns in the observed gene expressions over the two cell-cycle periods, indicating that by only examining the expression profiles, we may miss many relevant transcription factors. Our method incorporates both time-course gene expression and ChIP-chip binding data and enables us to identify more transcription factors that are related to the cell-cycle process.
Figure 4.
Application to yeast cell cycle gene expression data. Estimated time-varying coefficients for selected transcription factors (TFs)(solid lines) and point-wise 90% confidence intervals (dotted lines). Top panel, TFs that regulate genes expressed at the G1 phase; middle panel, TFs that regulate genes expressed at the G2 phase; bottom panel, TFs that regulate genes expressed at the M phase.
Finally, to assess false identifications of the TFs that are related to a dynamic biological procedure, we randomly assigned the observed gene expression profiles to the 297 genes while keeping the values and the orders of the expression values for a given profile unchanged and applied our procedure again to the new datasets. The goal of such permutations was to scramble the response data and to create new expression profiles that do not depend on the motif binding probabilities in order to see to what extent the proposed procedure identifies the spurious variables. We repeated this procedure 50 times. Among the 50 runs, 5 runs selected 4 TFs, 1 run selected 3 TFs, 16 runs selected 2 TFs and the rest of the 28 runs did not select any of the TFs, indicating that our procedure indeed selects the relevant TFs with few false positives.
8 Discussion
We have proposed a regularized estimation procedure for nonparametric varying-coefficient models that can simultaneously perform variable selection and estimation of smooth coefficient functions. The procedure is applicable to both longitudinal and functional responses. Our method extends the SCAD penalty of Fan and Li (2001) from parametric settings to a nonparametric setting. We have shown that the proposed method is consistent in variable selection and has the oracle property that the asymptotic distribution of the nonparametric estimate of a smooth function with variable selection is the same as that when knowing which variables are in the model. This oracle property has been obtained for parametric settings, but our result seems to be the first of its kind in a nonparametric setting. A computation algorithm is developed based on local quadratic approximation to the criterion function. Simulation studies indicated that the proposed procedure is very effective in selecting relevant variables and in estimating the smooth regression coefficient functions. Results from application to the yeast cell cycle dataset indicates that the procedure can be effective in selecting the transcription factors that potentially play important roles in the regulation of gene expressions during the cell cycle process.
We need regularization for two purposes in our method: controlling the smoothness of the function estimates and variable selection. In our approach, the number of basis functions/the number of knots of splines is used to control the smoothness, while the SCAD parameter is used to set the threshold for variable selection. Thus, the two tasks of regularization are disentangled. To achieve finer control of smoothness, however, it is desirable to use the roughness penalty for introducing smoothness in function estimation, as in penalized splines or smoothing splines. Following the idea of COSSO, we could replace Σk pλ(||gk||) in (4) by λΣk ||gk||W where ||·||W denotes a Sobolev norm, and then minimize the modified criterion over the corresponding Sobolev space. However, such an extension of COSSO is not very attractive compared with our method, because a single tuning parameter λ serves two different purposes of regularization. It is an interesting open question to develop a unified penalization method that does both smoothing and variable selection but also has the two tasks decoupled in some manner. Recent results on regularization methods with a diverging number of parameters (Fan and Peng, 2004; Huang et al., 2008) may provide theoretical insights into such a procedure.
As we explained in Section 2, our methodology applies when a working correlation structure is used to take into account within-subject correlation for longitudinal data. We only presented asymptotic results for the working independence case, however. Extension of these results to parametrically estimated correlation structures is relatively straightforward, using the arguments in Huang, Zhang and Zhou (2007); one needs to assume that the ratio of the smallest and largest eigenvalues of the weighting matrix W is bounded. Extension to nonparametrically estimated correlation structures (Wu and Pourahmadi, 2003; Huang et al., 2006; Huang, Liu and Liu, 2007) would be more challenging. Careful study of the asymptotics of our methodology for general working correlation structures is left for future research.
Acknowledgments
The work of the first two authors was supported by NIH grants ES009911, CA127334 and AG025532. Jian-hua Huang’s work was partly supported by grants from the National Science Foundation (DMS-0606580) and the National Cancer Institute (CA57030). We thank two referees for helpful comments and Mr. Edmund Weisberg, MS at Penn CCEB for editorial assistance.
Appendix
A.1 Proof of Lemma 1
Use proof by contradiction. Suppose Uiγ̂★(−i) ≠ Uiγ̃(i). Denote (11) as plridge(γ, y). Then,
This contradicts the fact that γ̃(i) minimizes plridge(γ̃(i), ỹ(i)).
A.2 Useful lemmas
This subsection provides some lemmas that facilitate the proof of Theorems 1 and 2. Define ỹi = E(yi|xi), , and (β̃(t) = B(t)γ̃. Here β̃(t) can be regarded as the conditional mean of β̂(t). We have a bias-variance decomposition β̂−β = (β̂ − β̃) + (β̃ − β). Lemmas 3 and 5 quantify the rate of the convergence of ||β̃ − β||L2 and ||β̂−β̃||L2, respectively. The rate given in Lemma 5 on the variance term will be refined in part (b) of Theorem 1.
Lemma 2
Suppose that (17) holds. There exists an interval [M3, M4], 0 < M3 < M4 < ∞, such that all the eigenvalues of fall in [M3, M4] with probability approaching 1 as n → ∞.
Lemma 3
Suppose that (17) holds. Then, ||β̃ − β||L2 = OP (ρn).
Lemmas 2 and 3 are from Lemmas A.3 and A.7 in Huang et al. (2004), respectively. The proofs are omitted.
Define . When wi = 1/Ji, or wi = 1/Ji with Ji uniformly bounded, we can show that rn ≍ (Ln/n)1/2.
Lemma 4
, where |v| = ||v|| l2
Proof of Lemma 4
By the Cauchy-Schwarz inequality,
| (21) |
The expected value of the right hand side equals
| (22) |
Note that
On the other hand,
and . Therefore,
| (23) |
Combining (22) and (23) and using the Markov inequality, we obtain
which together with (21) yields the desired result.
Lemma 5
Suppose that (17) holds and that λn, rn, ρn → 0 and λn/ρn → ∞ as n → ∞. Then, ||β̂ − β̃||L2 =OP {rn + (λnρn)1/2}.
Proof of Lemma 5
Using properties of B-spline basis functions (see Section A.2 of Huang et al., 2004), we have
Sum over k to obtain
Let with δn a scaler and v a vector satisfying ||v||l2= 1. It follows that ||β̂−β̃||L2 ≍ δn. We first show that δn = Op(rn + λn) and then refine this rate of convergence in the second step of the proof.
Let εi = (εi(ti1), …, εi(tiJi))T, i = 1, …, n, and . By the definition of γ̃, UTW(y − ε − Uγ̃) = 0. Since ,
Hence,
| (24) |
where vk’s are components of v when the latter is partitioned as γ̃.
By the definition of γ̂, pl(γ̂) − pl(γ̃) ≤ 0. According to Lemma 4,
| (25) |
By Lemma 2,
| (26) |
with probability approaching 1. Using the inequality |pλ(a) − pλ(b)| ≤ λ|a − b| we obtain
| (27) |
Therefore, with probability approaching 1, which implies δn = Op(rn + λn).
Now we proceed to improve the obtained rate and show that δn = OP {rn + (λnρn)1/2}. For k = 1, …, p, we have |||γ̂k||k − ||γ̃k||k| ≤ ||γ̂k − γ̃k||k = op(1) and
| (28) |
It follows that ||γ̂k||k → ||βk||L2 and ||γ̃k||k → ||βk||L2with probability. Since ||βk||L2 > 0 for k = 1, …, s and λn → 0, we have that, with probability approaching 1, ||γ̂k||k > aλn, ||γ̃k||k > aλn, k = 1, …, s, On the other hand, ||βk||L2 = 0 for k = s + 1, …, p, so (28) implies that ||γ̃k||k = OP (ρn). Since λn/ρn → ∞, we have that with probability approaching 1, ||γ̃k||k < λn, k = s + 1, …, p. Consequently, by the definition of pλ(·), P {pλn(||γ̃k||Hk) =pλn(||γ̂||Hk)} → 1, k = 1, … s, and P{pλn(||γ̃k||Hk) = λn(||γ̃k|| Hk} → 1, k = s + 1, …, p.
Therefore,
This result combined with (24), (25) and (26) imply that, with probability approaching 1,
which in turn implies δn = OP {rn + (λnρn)1/2}. The proof is complete.
A.3 Proof of Theorem 1
To prove part (a) of Theorem 1, we use proof by contradiction. Suppose that for n large enough, there exists a constant η > 0 such that with probability at least η, there exists a k0 > s such that β̂ k0(t) ≠ 0. Then ||γ̂k0||k0 = ||β̂ k0 (·)||L2 > 0. Let γ̂* be a vector constructed by replacing γ̂k0 with 0 in γ̂. Then,
| (29) |
By Lemma 5 and the fact that βk0 (t) = 0, ||γ̂k0||k0 = ||β̂k0|| = Op{rn + (λnρn)1/2}. Since λn/max(rn, ρn) → ∞, we have that λn > ||γ̂k0||k0 and thus pλn (||γ̂k0||k0) = λn||γ̂k0||k0 with probability approaching 1. To analyze the first term on the right hand side of (29), note that
| (30) |
By the Cauchy-Schwarz inequality and Lemma 2,
It follows from the triangle inequality and Lemmas 3 and 5 that,
Hence,
| (31) |
Lemma 4 implies that
| (32) |
| (33) |
Note that on the right hand side of (33), the third term dominates both the first term and the second term, since λn/max(rn, ρn) → ∞. This contradicts the fact that pl(γ̂) − pl(γ*) ≤ 0. We have thus proved part (a).
To prove part (b), write β = ((β(1))T, (β(2))T)T, where β(1) = (β1, …, βs)T and β(2) = (βs+1, …, βp)T, and write γ = ((γ(1))T, (γ(2))T)T, where and . Similarly, write and U = (U(1); U(2)). Define the oracle version of γ̃,
| (34) |
which is obtained as if the information of the nonzero components were given; the corresponding vector of coefficient functions is denoted as β̃oracle. Note that the true vector of coefficient functions is β = ((β (1))T, 0T)T. By Lemma 3, ||β̃oracle − β||L2 = OP (ρn) = oP (1). By Lemmas 3 and 5, ||β̂ − β||L2 = oP (1). Thus, with probabiltiy approaching one, ||β̃k,oracle||L2 → ||βk||L2 > 0 and ||β̂k||L2 → ||βk||L2 > 0, for k = 1, …, s. On the other hand, by the definition of β̃oracle, ||β̃k;oracle||L2 = 0 and by part (a) of the theorem, with probability approaching one, ||β̂k||L2 = 0, for k = s + 1, …, p. Consequently,
| (35) |
with probability approaching 1. ¿From part (a) of the theorem, with probability approaching 1, γ̂ = ((γ̂(1))T, 0T)T. Let with v = ((v(1))T, 0T)T and ||v(1)||l2 = 1. Then, . By (24) and (35), and the argument following (24),
Hence, ||β̂ − β̃||L2 ≍ δn = Op(rn), which together with ||βoracle − −β||L2 = oP (ρn) implies that ||β̂ − β||L2 = Op(ρn + rn). The desired result follows.
A.4 Proof of Corollary 1
By Theorem 6.27 in Schumaker (1981), , and thus . Moreover, rn ≍ (Ln/n)1/2. Hence, the convergence rate is . When Ln ≍ n1/5, . The proof is complete.
A.5 Proof of Theorem 2
According to the proof of Lemma 5, with probability approaching one, ||γ̃k||k > aλn, ||γ̂ k||k > aλn, and thus pλn(||γ̃k||k) = pλn(||γ̂k||k), k = 1, …, s. By Theorem 1, with probability approaching 1, γ̂ = ((γ̂(1))T, 0T)T is a local minimizer of pl(γ). Note that pl(γ(1)) is quadratic in γ(1) = (γ1, …, γs)T when ||γk||k > aλn, k = 1, …, s. Therefore, ∂pl(γ)/∂γ(1)|γ(1)=γ̂(1),γ(2)=0 = 0, which implies
Let
Let Avar*(γ̂(1)) be a modification of Avar(γ̂(1)) given in (20) by replacing Ωλn in (19) with 0. Applying Theorem V.8 of Petrov (1975) as in the proof of Theorem 4.1 of Huang (2003) and using the arguments in the proof of Lemma A.8 of Huang et al. (2004), we obtain that, for any vector cn with dimension and whose components are not all zero,
For any s-vector an whose components are not all zero, choosing cn = (B(1)(t))Tan yields
which in turn yields the desired result.
Contributor Information
Lifeng Wang, Department of Biostatistics and Epidemiology, University of Pennsylvania School of Medicine, Philadelphia, PA 19104, lifwang@mail.med.upenn.edu.
Hongzhe Li, Department of Biostatistics and Epidemiology, University of Pennsylvania School of Medicine, Philadelphia, PA 19104, hongzhe@mail.med.upenn.edu.
Jianhua Z. Huang, Department of Statistics, Texas A&M University, College Station, TX 77843-3143, jian-hua@stat.tamu.edu.
References
- Banerjee N, Zhang MQ. Identifying cooperativity among transcription factors controlling the cell cycle in yeast. Nucleic Acids Research. 2003;31:7024–7031. doi: 10.1093/nar/gkg894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel P, Li B. Regularization in Statistics (with discussion) Test. 2006;15:271–344. [Google Scholar]
- Chen G, Jensen S, Stoeckert C. Clustering of genes into regulons using integrated modeling (CORIM) Genome Biology. 2007;8(1):R4. doi: 10.1186/gb-2007-8-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon EM, Liu XS, Lieb JD, Liu JS. Integrating regulatory motif discovery and genome-wide expression analysis; Proceedings of National Academy of Sciences; 2003. pp. 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle PJ, Liang KY, Zeger SL. Analysis of longitudinal data. Oxford: Oxford University Press; 1994. [Google Scholar]
- Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Annals of Statistics. 2004;32:407499. [Google Scholar]
- Fan J, Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. Journal of American Statistical Association. 2001;96:1348–1360. [Google Scholar]
- Fan J, Li R. New estimation and model selection procedures for semiparametric modeling in longitudinal data analysis. Journal of American Statistical Association. 2004;99:710–723. [Google Scholar]
- Fan J, Peng H. On non-concave penalized likelihood with diverging number of parameters. The Annals of Statistics. 2004;32:928–961. [Google Scholar]
- Hastie T, Tibshirani R. Varying-coefficient models. Journal of Royal Statistical Society, Ser B. 1993;55:757–796. [Google Scholar]
- Hoover DR, Rice JA, Wu CO, Yang L. Nonparametric smoothing estimates of time-varying coefficient models with longitudinal data. Biometrika. 1998;85:809–822. [Google Scholar]
- Huang J, Horowitz JL, Ma S. Asymptotic properties of bridge estimators in sparse high-dimensional regression models. The Annals of Statistics. 2008;36:587–613. [Google Scholar]
- Huang JZ, Wu CO, Zhou L. Varying-coefficient models and basis function approximation for the analysis of repeated measurements. Biometrika. 2002;89:111–128. [Google Scholar]
- Huang JZ. Local asymptotics for polynomial spline regression. The Annals of Statistics. 2003;31:1600–1635. [Google Scholar]
- Huang JZ, Wu CO, Zhou L. Polynomial spline estimation and inference for varying coefficient models with longitudinal data. Statistica Sinica. 2004;14:763–788. [Google Scholar]
- Huang JZ, Liu N, Pourahmadi M, Liu L. Covariance selection and estimation via penalised normal likelihood. Biometrika. 2006;93:85–98. [Google Scholar]
- Huang JZ, Liu L, Liu N. Estimation of large covariance matrices of longitudinal data with basis function approximations. J Comput Graph statist. 2007;16:189–209. [Google Scholar]
- Huang JZ, Zhang L, Zhou L. Efficient estimation in marginal partially linear models for longitudinal/clustered data using splines. Scandinavian Journal of Statistics. 2007;34:451–477. [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in S. cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Leng C, Zhang HH. Nonparametric model selection in hazard regression. Journal of Nonparametric Statistics. 2007;18:417–429. [Google Scholar]
- Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Lin X, Carroll RJ. Nonparametric function estimation for clustered data when the predictor is measured without/with error. Journal of American Statistical Association. 2000;95:520–534. [Google Scholar]
- Lin Y, Zhang HH. Component selection and smoothing in smoothing spline analysis of variance models – COSSO. Annals of Statistics. 2006;34:2272–2297. [Google Scholar]
- Luan Y, Li H. Clustering of time-course gene expression data using a mixed-effects model with B-splines. Bioinformatics. 2003;19:474–482. doi: 10.1093/bioinformatics/btg014. [DOI] [PubMed] [Google Scholar]
- Petrov V. Sums of Independence Random Variables. Springer-Verlag; New York: 1975. [Google Scholar]
- Rice JA. Functional and longitudinal data analysis: perspectives on smoothing. Statistica Sinica. 2004;14:631–647. [Google Scholar]
- Rice JA, Silverman BW. Estimating the mean and covariance structure non-parametrically when the data are curves. Journal of Royal Statistical Society B. 1991;53:233–243. [Google Scholar]
- Rice JA, Wu CO. Nonparametric mixed effects models for unequally sampled noisy curves. Biometrics. 2001;57:253–259. doi: 10.1111/j.0006-341x.2001.00253.x. [DOI] [PubMed] [Google Scholar]
- Schumaker LL. Spline Functions: Basic Theory. Wiley; New York: 1981. [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakola TS, Young RA. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast saccharomyces cerevisiae by microarray hybridization. Molecular Biology of Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone CJ. Optimal global rates of convergence for nonparametric regression. Annals of Statistics. 1982;10:1348–1360. [Google Scholar]
- Tibshirani RJ. Regression shrinkage and selection via the lasso. Journal of Royal Statistical Society B. 1996;58:267–288. [Google Scholar]
- Tsai HK, Lu SHH, Li WH. Statistical methods for identifying yeast cell cycle transcription factors. Proceedings of National Academy of Sciences. 2005;102(38):13532–13537. doi: 10.1073/pnas.0505874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chen G, Li H. Group SCAD regression analysis for microarray time course gene expression data. Bioinformatics. 2007;23:1486–1494. doi: 10.1093/bioinformatics/btm125. [DOI] [PubMed] [Google Scholar]
- Wu WB, Pourahmadi M. Nonparametric estimation of large covariance matrices of longitudinal data. Biometrika. 2003;90:831–44. [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. Journal of Royal Statistical Society B. 2006;68:49–67. [Google Scholar]
- Zeger SL, Diggle PJ. Semiparametric models for longitudinal data with application to CD4 cell numbers in HIV seroconverters. Biometrics. 1994;50:689–699. [PubMed] [Google Scholar]
- Zhang H. Variable selection for support vector machines via smoothing spline ANOVA. Statistica Sinica. 2006;16:659–674. [Google Scholar]
- Zhang H, Lin Y. Component Selection and Smoothing for Nonparametric Regression in Exponential Families. Statistica Sinica. 2006;16:1021–1042. [Google Scholar]
- Zou H. The adaptive Lasso and its Oracle properties. Journal of the American Statistical Association. 2006;101(476):1418–1429. [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of Royal Statistical Society B. 2005;67:301–320. [Google Scholar]