Abstract
Targeted inactivations of RNA-binding proteins (RNA-BPs) can lead to huge phenotypical defects. These defects are due to the deregulation of certain mRNAs. However, we generally do not know, among the hundreds of mRNAs that are normally controlled by one RNA-BP, which are responsible for the observed phenotypes. Here, we designed an antisense oligonucleotide (“target protector”) that masks the binding site of the RNA-BP CUG-binding protein 1 (CUGBP1) on the mRNA Suppressor of Hairless [Su(H)] that encodes a key player of Notch signaling. We showed that injecting this oligonucleotide into Xenopus embryos specifically inhibited the binding of CUGBP1 to the mRNA. This caused the derepression of Su(H) mRNA, the overexpression of Su(H) protein, and a phenotypic defect, loss of somitic segmentation, similar to that caused by a knockdown of CUGBP1. To demonstrate a causal relationship between Su(H) derepression and the segmentation defects, a rescue experiment was designed. Embryonic development was restored when the translation of Su(H) mRNA was re-repressed and the level of Su(H) protein was reduced to a normal level. This “target protector and rescue assay” demonstrates that the phenotypic defects associated with CUGBP1 inactivation in Xenopus are essentially due to the deregulation of Su(H) mRNA. Similar approaches may be largely used to uncover the links between the phenotype caused by the inactivation of an RNA-BP and the identity of the RNAs associated with that protein.
Keywords: RNA-binding protein, mRNA stability, somitic segmentation, CUGBP1, Suppressor of Hairless, Notch
INTRODUCTION
The nuclear and cytoplasmic fates of eukaryotic mRNAs rely on interactions with a number of molecules, such as regulatory RNAs or proteins. RNA-binding proteins (RNA-BPs) play central roles in controlling events like pre-mRNA maturation, mRNA nucleocytoplasmic export, cytoplasmic transport, translation, and degradation. Immunoprecipitations of some RNA-BPs and large-scale identification of co-immunoprecipitated mRNAs (by microarray hybridization or deep sequencing) showed that RNA-BPs are able to interact with a large number of target RNAs (Lopez de Silanes et al. 2004, 2005; Girard et al. 2006; Graindorge et al. 2008; Stoecklin et al. 2008).
Several gene inactivations or knockdown experiments of RNA-BPs have been published. These inactivations generally lead to huge phenotypic effects or even lethality. These phenotypic alterations may be caused by the deregulation of all, or at least most, of the mRNAs that are normally controlled by the inactivated RNA-BP. Alternatively, they may be caused by the deregulation of only one or a limited number of them. For example, tristetraprolin (TTP) directs bound mRNAs, including that encoding TNFα, to rapid degradation (Carballo et al. 1998). Mice in which the TTP gene was inactivated display several immunological defects, and they overexpress TNFα. A treatment of these mice by neutralizing antibodies against TNFα efficiently cures them (Taylor et al. 1996), showing that derepression of TNFα mRNA is the main cause of the mutant phenotype associated with TTP inactivation.
In contrast to the above example, we generally do not know how the inactivation of an RNA-BP leads to phenotypic alterations, even for the RNA-BPs whose mRNA targets are known. To understand the molecular reasons for altered developmental phenotypes associated with inactivation of an RNA-BP, it is necessary to individually inhibit the regulation exerted by that factor on one of its RNA targets. Ideally, both the regulations exerted by that factor on its other mRNA targets, and the regulations exerted on that RNA by other factors, should remain intact.
Here, we used the RNA-BP CUG-binding protein 1 (CUGBP1, formerly EDEN-BP in Xenopus) and the mRNA Suppressor of Hairless [Su(H), an actor of Notch signaling] to test if morpholino antisense oligonucleotides can be used to specifically inhibit the interaction between an mRNA and a protein, and to analyze the phenotypical consequences of that inhibition. Knockdown of the RNA-BP CUGBP1 in Xenopus causes defects in somitic segmentation. Somitic segmentation is the developmental process that leads to the appearance of repeated units, the somites, along the embryonic antero-posterior axis. It is easy to score by external observation of the embryos. Su(H) mRNA is normally repressed by CUGBP1 and is overexpressed in CUGBP1 knockdown embryos (Gautier-Courteille et al. 2004). However, whether Su(H) derepression and segmentation defects are causally related remains an open question, as the knockdown of CUGBP1 may deregulate several mRNA targets of CUGBP1 in addition to Su(H) (Graindorge et al. 2008). We showed here that a morpholino antisense oligonucleotide designed to prevent CUGBP1 from binding to Su(H) mRNA (“target protector”) (Choi et al. 2007) led to somitic defects similar to those caused by CUGBP1 knockdown. Proof for the specificity of that effect was brought by a phenotypic rescue experiment. Hence, this “target protector and rescue assay” (TPRA) demonstrated that derepression of Su(H) is the main cause of the phenotype associated with CUGBP1 loss-of-function in Xenopus.
RESULTS AND DISCUSSION
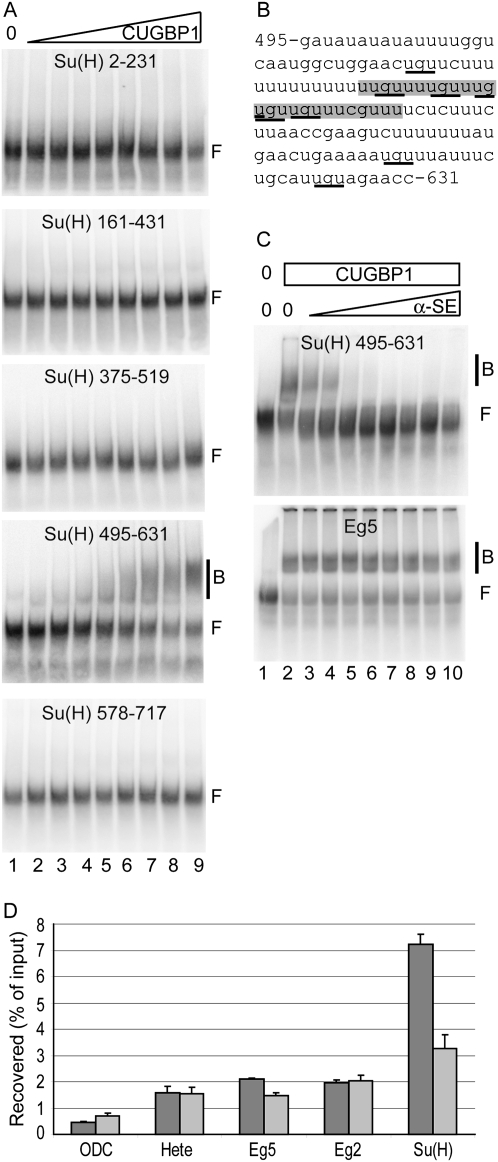
To identify the mRNA sequence element in Su(H) mRNA bound by CUGBP1, five RNAs corresponding to overlapping regions of the 3′ untranslated region (3′UTR) were tested by EMSA (Electrophoretic Mobility Shift Assay). Figure 1A shows that only the RNA corresponding to region 495–631 of the 3′UTR (numbered from the stop codon) was bound by CUGBP1. The corresponding sequence is shown in Figure 1B. A hallmark of the EDEN sequences (sequences bound by CUGBP1) is their richness in trinucleotides UGU (Marquis et al. 2006). Five such UGUs are concentrated within a small subpart (highlighted in gray in Fig. 1B) of region 495–631. No similar concentration of UGU is detected in any other region of Su(H) 3′UTR. A morpholino antisense oligonucleotide [α-SE, anti-Su(H) EDEN] was designed to hybridize to the putative EDEN of Su(H). In EMSA (Fig. 1C, upper panel), the α-SE morpholino abolished the interaction between Su(H) RNA and CUGBP1 in a dose-dependent manner. The specificity of the α-SE morpholino was verified using Eg5 3′UTR. This probe contains an EDEN whose sequence is different from Su(H) EDEN, although still UGU-rich (UAUAUAUGUGUGUCUAUCGUCACUUGUAUGUCAAAUAUU [UGUs are underlined]) (Paillard et al. 1998). In EMSA, even a large amount of α-SE morpholino had no effect on the interaction between CUGBP1 and Eg5 (Fig. 1C, lower panel). Hence, α-SE is not a general inhibitor of CUGBP1 binding.
FIGURE 1.
Design of a morpholino antisense oligonucleotide (“α-SE”) that precludes the interaction between Su(H) and CUGBP1. (A) In vitro transcribed radioactive RNAs corresponding to the indicated regions of Su(H) 3′UTR (numbered from the stop codon) were incubated with no (lane 1) or increasing amounts (lanes 2–9) of recombinant CUGBP1. Free (F) and bound (B) RNAs were resolved by native electrophoresis and autoradiographed. (B) Sequence of Su(H) 3′UTR between nucleotides 495 and 631 (numbered from the stop codon). UGU trinucleotides are underlined, and the region complementary to α-SE morpholino is highlighted. (C) Radioactive RNAs corresponding to region 495–631 of Su(H) 3′UTR (upper panel) or Eg5 3′UTR (lower panel) were incubated with no (lane 1) or 50 nM (lanes 2–10) of recombinant CUGBP1, and increasing concentrations of α-SE morpholino (lanes 3–10). They were analyzed as in A. (D) CUGBP1 was immunoprecipitated from Xenopus egg extracts previously incubated with 5 μM of control (dark gray) or α-SE (light gray) morpholino. The indicated input and co-immunoprecipitated mRNAs were quantified by RT-qPCR. The percentages of mRNA recovered in the immunoprecipitated fractions are shown. Quantifications were in triplicate and error bars are the s.e. of the triplicates.
We next analyzed the in vivo effect of α-SE morpholino. First, we checked its effect on the interaction of CUGBP1 with Su(H) mRNA in undiluted Xenopus egg extracts. Cell-free cytoplasmic egg extracts are translationally active (Legagneux et al. 1995). CUGBP1 was immunoprecipitated from Xenopus egg extract previously incubated with control or α-SE morpholinos. The amounts of several co-immunoprecipitated mRNAs were quantified by real-time RT-PCR (Fig. 1D). ODC mRNA, that is not known to bind CUGBP1, was recovered with the lowest efficiency of all the mRNAs tested. In contrast, Hete, Eg5, Eg2, and Su(H) mRNA, which were previously shown to interact with CUGBP1 (Graindorge et al. 2008), were much more efficiently co-immunoprecipitated in the presence of the control morpholino. The amount of co-immunoprecipitated Su(H) was reduced more than twofold by the α-SE morpholino, demonstrating that this morpholino decreases the interaction between Su(H) mRNA and CUGBP1 in egg extract. Furthermore, this effect was specific, as α-SE had virtually no effect on the amount of recovered Hete, Eg5, and Eg2 mRNA (Fig. 1D). Importantly, in that experiment, we measured the capacity of α-SE morpholino to disrupt a complex between CUGBP1 and Su(H) mRNA that already exists when the morpholino is added. We expect that the morpholino would be even more efficient in preventing the formation of that complex, for example, with neo-synthesized mRNA.
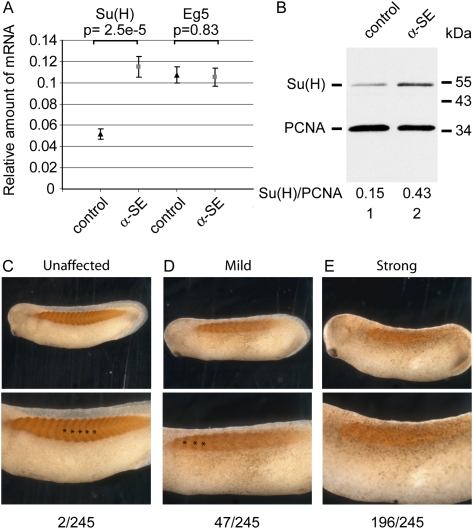
Since α-SE morpholino inhibits the interaction between CUGBP1 and Su(H) mRNA, and mRNAs that bind CUGBP1 are rapidly degraded, α-SE was expected to increase the steady-state level of Su(H) mRNA. This was verified by injecting both blastomeres of two-cell embryos with α-SE morpholinos and then allowing them to develop. Analyses by RT-qPCR showed that, relative to the level of ODC mRNA, Su(H) mRNA level in α-SE embryos was significantly higher than in controls (Fig. 2A). This was not the case for Eg5 mRNA (Fig. 2A). Consistent with Su(H) mRNA overexpression, a Western analysis showed that, as normalized to the level of PCNA, Su(H) protein level was almost threefold higher in embryos injected with α-SE morpholino than in controls (Fig. 2B).
FIGURE 2.
α-SE morpholino derepresses endogenous Su(H) mRNA and causes segmentation defects. (A,B) Xenopus embryos at the two-cell stage were injected with 2 pmol of control or anti-Su(H) EDEN morpholino (α-SE) in each blastomere. They were allowed to develop until the tailbud stage (24–26). (A) RNAs were extracted and the amounts of Su(H) and Eg5 mRNA, relative to the amount of ODC mRNA, were measured by RT-qPCR. Shown are the medians, ±s.e., of 15 embryos for each condition. Student tests were used to compare control and α-SE morpholinos, and P-values are indicated above the bars. (B) Proteins were extracted from pools of five embryos, and one embryo equivalent was submitted to a Western analysis using antibodies directed against Su(H) and PCNA as a loading control. Positions of molecular weight markers are indicated on the right. (C–E) Two-cell embryos were injected in one blastomere with 2 pmol of anti-Su(H) EDEN morpholino (α-SE) and allowed to develop until tailbud stage. They were fixed and stained with myotome-specific 12/101 monoclonal antibody. C, D, and E are representative photographs of an unaffected embryo, an embryo with a progressive loss of segmentation toward the posterior extremity, and a nonsegmented embryo, respectively. Lower panels are higher magnifications of upper panels. In C and D, lower panels, stars show successive somites. The number of embryos that fell within each class in one representative experiment is given on bottom.
To analyze the phenotypic consequences of suppressing the down-regulation exerted by CUGBP1 on Su(H) mRNA, one blastomere of embryos at the two-cell stage was injected with α-SE morpholino. Embryos were allowed to develop to tailbud stage. They were stained with the myotome-specific 12/101 monoclonal antibody (Kintner and Brockes 1985) to make somites more visible. The side originating from the control (noninjected) blastomeres was generally similar to a noninjected control embryo (and similar to that shown in Fig. 2C), but the side originating from the blastomere injected with α-SE morpholino displayed one of three aspects. Figure 2C (unaffected) shows a normally segmented side with several chevron-shaped somites along the antero-posterior axis. In Figure 2D (mildly affected), segmentation occurred normally in the anterior part of the embryo but was abolished in the posterior-most region. Importantly, that region was still stained with the 12/101 antibody, demonstrating myotomal differentiation despite altered segmentation. Finally, in Figure 2E (strongly affected), there was no evidence for any segmentation process, despite myotomal differentiation. Respectively, 80% and 19% of the embryos were strongly and mildly affected when 2 pmol of α-SE morpholino were injected (Figs. 2C–E). We have shown previously that 90% of the embryos injected with a morpholino designed to knock down CUGBP1 display very similar segmentation defects (Gautier-Courteille et al. 2004).
These data show that α-SE morpholino derepresses Su(H) mRNA, leading to an overexpression of Su(H) protein, and causes somitic segmentation defects. A likely hypothesis is that the derepression of Su(H) is indeed responsible for the phenotypic defects, but, at this stage, off-target effects of the α-SE morpholino cannot be excluded. Translating ribosomes are able to remove morpholinos hybridized within an mRNA coding frame. Consequently, only mRNA containing a site for α-SE in their untranslated regions can be considered as possible off-targets. A Blastn search of Xenopus laevis sequences (nucleotide collection [NR/NT]) using α-SE as the query sequence was made. In addition to Su(H), it retrieved two mRNAs that contain 15 consecutive nucleotides fully complementary to α-SE in their untranslated regions, mannose-binding lectin-associated serine protease-3a (accession number BC170241) and OLT 2-1 retrotransposon-like element 10A1 gag-like protein gene (AF145965). However, since sequencing of Xenopus laevis is not complete, other off-targets might also exist.
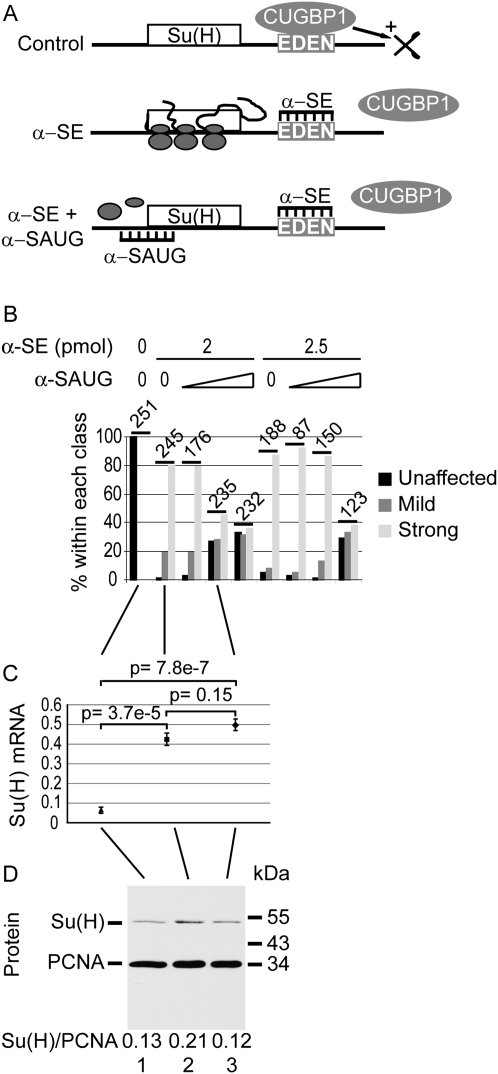
A gold standard to rule out potential off-target effects is to design a rescue experiment. Classical rescue experiments consist of adding back a molecule that was previously depleted. Here, since a down-regulation is relieved, a rescue experiment consists of re-repressing the derepressed molecule. This was done by co-injecting, with the α-SE morpholino, a second morpholino (“α-SAUG”) designed to inhibit the translation of Su(H) mRNA (Fig. 3A). A Blastn search of Xenopus laevis sequences (nr-PDB) retrieved only one mRNA containing 15 consecutive nucleotides fully complementary to either of the two α-SAUG sequences (see Materials and Methods) in its untranslated regions, BRCA1 (NM_001090779), that is not a potential α-SE target. Figure 3B shows the percentages of unaffected, mildly, and strongly affected embryos after injection of different amounts of α-SE and α-SAUG. The percentage of normally segmented embryos decreased from 100% in control embryos to <10% upon injection of either amount of α-SE morpholino. However, in both cases, at least one amount of α-SAUG could be found that partly restored segmentation (60 and 100 fmol for 2 pmol of α-SE; 100 fmol for 2.5 pmol of α-SE; P < 0.001; χ2 test). We measured the levels of Su(H) mRNA and protein in these rescue conditions. First, whereas α-SE caused a significant increase of Su(H) mRNA level as previously described, α-SAUG had no effect on that overexpression (Fig. 3C). This was expected, since α-SAUG should block the translation of Su(H) mRNA without affecting its stability. Second, as anticipated, Su(H) protein level was decreased to a control level by the injection of α-SAUG in addition to α-SE (Fig. 3D). Together, these data show that the effect of α-SE morpholino on somitic segmentation is due to a specific derepression of the down-regulation that is normally exerted by CUGBP1 on Su(H) mRNA. Furthermore, these data show that the phenotype associated with a knockdown of CUGBP1 in Xenopus embryos (Gautier-Courteille et al. 2004) is essentially due to a deregulation of Su(H) mRNA.
FIGURE 3.
Phenotypic rescue of α-SE morpholino provided by repression of Su(H) mRNA. (A) Design of a “rescue” experiment. In control embryos (upper panel), Su(H) mRNA is bound by CUGBP1, leading to a rapid degradation of the mRNA. The α-SE morpholino (middle panel) prevents CUGBP1 binding to Su(H) mRNA, hence stabilizes it, leading to protein overexpression. It also leads to phenotypic defects. If these phenotypic defects are specifically due to the derepression of Su(H) mRNA, then development should be restored by injecting an adequate amount of a second morpholino directed against the translation initiation region (α-SAUG, lower panel), which, by reducing translation, will reduce the amount of Su(H) protein to a normal level. (B) Quantification of segmentation defects. Embryos were co-injected with the indicated amounts of α-SE morpholino and 0, 10, 60, or 100 fmol of α-SAUG. Embryos were sorted at stage 24–26 according to their phenotype: unaffected (like in Fig. 2C, black), mild (like in Fig. 2D, dark gray), or strong (like in Fig. 2E, light gray). The total number of sorted embryos for each combination of morpholinos is indicated. (C,D) Xenopus embryos at the two-cell stage were injected in each blastomere with 2 pmol of control morpholino (lane 1) or 2 pmol of α-SE morpholino in the absence (lane 2) or the presence (lane 3) of 60 fmol of α-SAUG morpholino. They were allowed to develop until the tailbud stage (24–26). RNA (C) and protein (D) levels were analyzed as in Figure 2, A and B.
The concept of “target protector” was initially developed in Zebrafish where it refers to an inhibition of the interaction between an mRNA and a miRNA (Choi et al. 2007; Li et al. 2008). In these articles, an antisense oligonucleotide was injected that hybridized to the binding site of a miRNA on an mRNA, hence precluding interaction and leading to phenotypical consequences. The injected oligonucleotide should not activate RNase H that would destroy the targeted mRNA. For that reason, morpholino oligonucleotides were used in the above studies (Choi et al. 2007; Li et al. 2008), as well as in the present one. However, any modified oligonucleotide that does not activate RNase H would probably be suitable as a target protector. Irrespective of the modified oligonucleotide, we consider it important to design rescue experiments to exclude off-target effects of the target protector. In the present study, a normal phenotype was recovered by re-repressing the translation of the mRNA that was derepressed due to target protection, but the design of rescue experiments depends on each particular case. Potentially, TPRAs could be used in any model system in which a target protector and a rescuing molecule can be introduced, such as Xenopus oocytes or embryos, Zebrafish, syncitial Drosophila embryos (by injection), or chick embryo (by electroporation). It should also allow analyzing miRNA/mRNA interactions in addition to protein/mRNA interactions. Hence, TPRA could be widely used to understand the molecular reasons for altered developmental phenotypes associated with the inactivation of factors controlling mRNA fate.
MATERIALS AND METHODS
Primer and morpholino sequences
Morpholinos
Control CCTCTTACCTAGTTACAATTTATA; α-SE AAACGAAACAACACAAACAAAACAA; α-SAUG CCAGGTTGCATAGAACAATATGATG and GCCTCTCCCCAAACTTCATTCCGCT [sequence complementary to AUG initiation codon is underlined. As a consequence of Xenopus laevis genome tetraploidization, two highly related genes exist for Su(H). Two different morpholinos were therefore required to inhibit the translation of Su(H)].
Primers used to generate transcription matrices
The T7 sequence for in vitro transcription is underlined:
Su2_fwd_T7, TAATACGACTCACTATAGGGAGACCATAAACTTTGGACTTTTACATGG;
Su161_fwd_T7, TAATACGACTCACTATAGGGAGACGTAGAAGCCACAATAATAAAATGGA;
Su375_fwd_T7, TAATACGACTCACTATAGGGAGAGTACAACTTTTGTTACCAAATAGAT;
Su495_fwd_T7, TAATACGACTCACTATAGGGAGAGATATATATATTTTGTGCAATGGCTGGC;
Su578_fwd_T7, TAATACGACTCACTATAGGGAGACCGAATGCTTTTTTTATGAACTG;
Su231_rev, TGCCCATTTTTGTAAAAATGCAGC;
Su431_rev, CTACACATACTTTAGAGCGC;
Su519_rev, GCCATTGCACAAAATATATATATC;
Su631_rev, GGTTCTCAAATGCAGAAATAAAC; and
Su717_rev, GTAGACATCAATAGTTTGTATTGC.
Primers for qPCR
Eg5_fwd, GATCGGCACAGAGTTTTCGGT;
Eg5_rev, TTGTAGCCTTTCCCCGACATC;
ODC_fwd, AAAATGGATGACTGCGAGATGGG;
ODC_rev, AATGAAGATGCTGACTGGCAAAAC;
Su(H)_fwd, GCCAGAGCCCGGACCTA;
Su(H)_rev, TTGGATGCCAGGAGACTTGAA;
Eg2_fwd, GGAGGTGGAAATTCAGTCCCA;
Eg2_rev, GGCGCATAATCGAGGATTAGG;
Hete_fwd, GGTGCTGAATGTGGCATTCTCT; and
Hete_rev, GCACACACGTTCTGGCAGATAC.
Biochemical methods
For EMSAs (Marquis et al. 2006), 32P-labeled RNAs were obtained by T7 in vitro transcription from PCR products using a pGbSu(H) plasmid (Gautier-Courteille et al. 2004) with the appropriate primers, or a pGbsEg5 plasmid (Paillard et al. 1998), previously linearized with BamH1. Two-fold serial dilutions of recombinant CUGBP1 starting from 400 nM, or threefold serial dilutions of morpholino starting from 500 nM were used.
For co-immunoprecipitation experiments, Xenopus egg extracts (Legagneux et al. 1995) were incubated for 1 h at 22°C in the presence of 5 μM of control or α-SE morpholino and CUGBP1 was immunoprecipitated as described (Graindorge et al. 2008). After SDS elution, RNA was extracted with Tri-reagent (Euromedex) and reverse-transcribed. The amount of RNA in each reverse-transcription reaction corresponded to the eluate of one immunoprecipitation experiment (from 40 μL of extract) or to 4 μL of total extract for the input. Real-time PCR was done in a final volume of 10 μL from 1/400 of these cDNAs using ABI Prism 7900 device (Applied), with the SybrGreen mastermix and 300 nM of each primer. For each RNA sample, quantifications were made in triplicate.
The amounts of Su(H) and Eg5 mRNA in embryos were similarly quantified by RT-qPCR, after reverse transcription of total embryo RNA. Each real-time PCR reaction was made from an amount of cDNA equivalent to 0.5 ng of total RNA. The amounts of Su(H) and Eg5 mRNA in embryos were calculated as 2ΔCt relative to the amount of ODC mRNA.
Western blots were made and revealed by ECL as previously described (Gautier-Courteille et al. 2004). They were quantified by densitometry. The primary antibodies were anti-mammalian RBP-Jκ [mammalian ortholog of Su(H); Chemicon AB5790] and anti-PCNA (Sigma P8825).
Xenopus laevis embryos procedures
One or two blastomeres of embryos at the two-cell stage were injected with 0–5 pmol of morpholino. They were allowed to develop at 16°C overnight then at 22°C for 1 d (to stages 24–26), fixed with MEMFA and immunostained with the 12/101 monoclonal antibody (Kintner and Brockes 1985) as described (Gautier-Courteille et al. 2004). Stained embryos were blind-sorted (the person sorting embryos did not know what had been injected) according to their phenotypes (unaffected by strongly affected embryos).
ACKNOWLEDGMENTS
We thank H. Beverley Osborne, Serge Hardy, and members of the EGD team for discussions. This work was supported by grants from the Association pour la Recherche Contre le Cancer (ARC4003, to L.P.) and Agence Nationale de la Recherche (ANR-07-JCJC-0097-01, to L.P.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1742610.
REFERENCES
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Gautier-Courteille C, Le Clainche C, Barreau C, Audic Y, Graindorge A, Maniey D, Osborne HB, Paillard L. EDEN-BP-dependent post-transcriptional regulation of gene expression in Xenopus somitic segmentation. Development. 2004;131:6107–6117. doi: 10.1242/dev.01528. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Graindorge A, Le Tonqueze O, Thuret R, Pollet N, Osborne HB, Audic Y. Identification of CUG-BP1/EDEN-BP target mRNAs in Xenopus tropicalis. Nucleic Acids Res. 2008;36:1861–1870. doi: 10.1093/nar/gkn031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintner CR, Brockes JP. Monoclonal antibodies to the cells of a regenerating limb. J Embryol Exp Morphol. 1985;89:37–55. [PubMed] [Google Scholar]
- Legagneux V, Omilli F, Osborne HB. Substrate-specific regulation of RNA deadenylation in Xenopus embryo and activated egg extracts. RNA. 1995;1:1001–1008. [PMC free article] [PubMed] [Google Scholar]
- Li N, Flynt AS, Kim HR, Solnica-Krezel L, Patton JG. Dispatched Homolog 2 is targeted by miR-214 through a combination of three weak microRNA recognition sites. Nucleic Acids Res. 2008;36:4277–4285. doi: 10.1093/nar/gkn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediates sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]