Abstract
Small hairpin RNAs (shRNAs) with 19-base-pair, or shorter, stems (short shRNAs [sshRNAs]) have been found to constitute a class whose mechanism of action appears to be distinct from that of small interfering RNAs (siRNAs) or longer shRNAs. These sshRNAs can be as active as canonical siRNAs or longer shRNAs. Their activity is affected by whether the antisense strand is positioned 5′ or 3′ to the loop (L or R sshRNAs, respectively). Dicer seems not to be involved in the processing of sshRNAs, although the mechanism of target gene suppression by these hairpins is through Ago2-mediated mRNA cleavage. In this study, the effects of chemical modifications on the potency, serum stability, and innate immune response of sshRNAs were investigated. Deoxynucleotide substitution and 2′-O-methyl (2′-OMe) modification in the sense strand and loop did not affect silencing activity, but, unlike with siRNAs, when placed in the antisense strand these modifications were detrimental. Conjugation with bulky groups at the 5′-end of L sshRNAs or 3′-end of R sshRNAs had a negative impact on the potency. Unmodified sshRNAs in dimer form or with blunt ends were immunostimulatory. Some modifications such as 3′-end conjugation and phosphorothioate linkages on the backbone of the sshRNAs could also induce inflammatory cytokine production. However, 2′-OMe substitution of sshRNAs abrogated the innate immune response and improved the serum stability of the hairpins.
Keywords: small hairpin RNA, shRNA, stem–loop, chemical modification, innate immune response, serum stability
INTRODUCTION
RNA interference (RNAi) is an evolutionarily conserved, highly specific pathway whereby double-stranded RNA (dsRNA) causes sequence-specific inhibition of gene expression. RNAi has been heavily used as a research tool in vitro as well as in vivo. Its potential as a therapeutic approach has also been widely pursued (Elbashir et al. 2001; Xia et al. 2002, 2004; Dorsett and Tuschl 2004; Harper et al. 2005; Wang et al. 2005; Amarzguioui et al. 2006; Behlke 2006; Bernards et al. 2006; Chang et al. 2006; Fewell and Schmitt 2006; Vlassov et al. 2007). A variety of RNA structures can be used to induce RNAi, including siRNAs, “Dicer-substrate” RNAs of ∼24–30 base pairs (bp), long dsRNAs, and small hairpin RNAs (shRNAs), either in synthetic or expressed form. Among synthetic RNAi triggers, we (and others) have found a special class of shRNA, short shRNAs (sshRNA), that have identical, or in some cases, better efficacy than siRNAs that target the same sequences (Li et al. 2007; Vlassov et al. 2007). These sshRNAs have a stem length of 19 bp or less, a connection of 0–10 nucleotides (nt) between the antisense strand and the sense strand, and optionally a 2-base 3′-overhang (Ge et al. 2010). The connection or loop is sometimes preferred at the 3′-end of the antisense strand to have better RNAi activity (L sshRNAs, as compared with R sshRNAs that contain the loop at the 5′-end of the antisense strand) (McManus et al. 2002; Harborth et al. 2003; Ge et al. 2010). Dicer seems not to be involved in the processing of sshRNAs (Siolas et al. 2005; Ge et al. 2010). 5′-RACE analysis suggests that the target gene suppression by sshRNA is via RNAi-mediated messenger RNA cleavage (Ge et al. 2010). These sshRNAs are of interest for several reasons: first, in some important model systems, they have been shown to be more potent than corresponding siRNAs (Vlassov et al. 2007); second, being shorter than standard shRNAs, they are less costly to manufacture; and finally, their high potency despite not being substrates for Dicer has implications for the mechanism of action of both siRNAs and shRNAs.
A number of studies have demonstrated the potential for synthetic siRNAs and shRNAs to activate mammalian immune responses (Kariko et al. 2004; Kim et al. 2004; Hornung et al. 2005; Judge et al. 2005; Sioud 2005; Marques et al. 2006; Schlee et al. 2006; Judge and MacLachlan 2008; Robbins et al. 2008). Toll-like receptors (TLR3, TLR7, and TLR8), protein kinase R (PKR), and the cytosolic RNA helicase retinoic acid-inducible gene-I (RIG-I) are involved in the recognition of synthetic dsRNAs and their activation of the innate immune response (Judge and MacLachlan 2008; Robbins et al. 2009). Several features of RNA, including length, sequence, and structure, could be involved in the recognition by these receptors (Hornung et al. 2005; Judge et al. 2005; Forsbach et al. 2008; Robbins et al. 2009). Although some level of inflammatory cytokine expression may be beneficial to antiviral or even antitumor therapeutics (Poeck et al. 2008), the toxicities associated with excessive cytokine release and associated inflammatory syndromes would be an undesirable side effect for most applications. Chemical modifications have been found to reduce the immune stimulatory capability of RNA duplexes (Morrissey et al. 2005; Behlke 2008; Judge and MacLachlan 2008; Watts et al. 2008; Robbins et al. 2009). Chemical modifications have also been developed to improve pharmacological properties of siRNAs, including stability against nucleases found in biological fluids and improved cellular uptake (Bumcrot et al. 2006; Debart et al. 2007).
While chemical modifications of siRNAs and Dicer-substrate RNAs have been extensively studied (Nawrot and Sipa 2006; Zhang et al. 2006; Collingwood et al. 2008; Robbins et al. 2009), synthetic sshRNAs have not been similarly studied. Because of their different structures and probable differences in how they are processed by the RNAi machinery, modifications that have proven effective for siRNAs or Dicer-substrate RNAs may not necessarily be effective with sshRNAs. We therefore undertook an investigation of the effects of different chemical modifications on the potency of sshRNAs, their ability to stimulate innate immunity, and their stability against enzymatic degradation in serum. We find that chemical modifications have different impacts on the efficacy of L and R sshRNAs. For L sshRNAs, RNAi activity was reduced when 2′-O-methyl (2′-OMe) or 2′-deoxy modifications were introduced into nucleotides of the antisense strand or when bulky chemical groups were conjugated at the 5′-end of the hairpin. On the other hand, 2′-OMe or 2′-deoxy modification of nucleotides in the sense strand and loop, 3′-end conjugation, or phosphorothioate (PS) bonds at the open ends of the hairpin or in the loop did not affect the potency of L sshRNAs. For R sshRNAs, a certain degree of 2′-OMe modification on the antisense strand was tolerated, but 3′-end conjugation decreased their efficacy. sshRNAs in dimer form or with blunt ends are immune activators. Cytokine expression was also found to be up-regulated by the addition of modifications such as 3′-end conjugation and PS linkages on the hairpins. Modification with 2′-OMe improved the serum stability of sshRNAs and minimized their activation of the innate immune response.
RESULTS AND DISCUSSION
2′-O-methyl modification of the antisense strand affects the activity of sshRNAs
An O-methyl group at the 2′-position of the ribosyl ring is a modification that occurs naturally in mammalian ribosomal and transfer RNAs. It has been shown that 2′-OMe modification can bring various benefits to siRNAs and Dicer-substrate RNAs with little effect on potency, depending on the precise modification pattern employed (Behlke 2008). For instance, 2′-OMe modification of alternate nucleotides in both strands of siRNAs resulted in increased serum stability without loss of RNAi activity (Czauderna et al. 2003; Allerson et al. 2005; Choung et al. 2006). Selected uridine, guanosine, or adenosine in both strands of siRNA could be 2′-O-methylated to abrogate immune stimulation without affecting efficacy (Kariko et al. 2005; Judge et al. 2006). Even a single 2′-OMe substitution at position 2 (from the 5′-end) of the antisense strand significantly reduced antisense strand-mediated off-target effects (Jackson et al. 2006).
Here we have investigated whether the advantages of 2′-OMe modification could be also applied to sshRNAs without affecting their activity. In addition to examining effects on the sense and the antisense strands, modification of the loop was also studied. A firefly luciferase (fLuc) reporter plasmid whose expression is driven by the HCV internal ribosome entry site (IRES) was cotransfected with anti-HCV sshRNAs into human 293FT cells. Reduction of fLuc expression due to the IRES-specific sshRNAs (compared with controls without sshRNA or with a scrambled sshRNA) reflects their efficacy. No difference in fLuc expression was found between the no-sshRNA control and the scrambled control (data not shown).
Modifications of a synthetic, blunt-ended L sshRNA (SG105) were studied first, with an unmodified siRNA targeting the same HCV IRES mRNA sequence (si19-3) used for comparison. We used sshRNAs that were blunt-ended rather than with 3′ extensions because, at least for SG105, there was no potency benefit to adding the extra two nucleotides (Ge et al. 2010). Modification at every second nucleotide of the antisense strand (beginning with the 5′-terminal nucleotide) severely compromised activity (Fig. 1, SG216). In striking contrast to the situation with siRNAs, similar impairment was seen with sshRNAs whose antisense and sense strands were both modified, either at alternate nucleotides or at all uridines (SG203 and SG205). However, when alternating 2′-OMe and 2′-OH nucleotides were placed only in the sense strand (SG204), the efficacy was not affected and even appeared to be slightly better than the unmodified parent sshRNA (SG105) and the control si19-3. The dinucleotide UU that connects the 3′-end of the antisense and the 5′-end of the sense strands of these sshRNAs could be also 2′-O-methylated on both uridines without affecting activity (SG202). When the UU sequence and the sense strand were both modified, again there was no loss of efficacy (Fig. 1B, SG224). Since the base pair immediately adjacent to the UU dinucleotide that connects the antisense and sense strands may not actually be base-paired due to the potential strain from a 2-nt loop, we also examined whether 2′-OMe modification of the CUUG sequence (Fig. 1C, mCUmUG [where “m” represents 2′-OMe]) could be combined with a modified sense strand (SG217). As seen in Figure 1C, the IC50 of SG217 (IC50 = 6.5 pM) was only about twofold higher than the unmodified SG105 (3.5 pM). Similarly, with a special L sshRNA that contains a 19-nt antisense strand directly connected to a 17-nt sense strand without any linking nucleotides (SG119), alternate modifications in the sense strand (SG235) did not affect efficacy, but modification in the 3′-end of the antisense strand (which in this construct forms the loop) slightly reduced the activity (Fig. 1D, SG236).
FIGURE 1.
Comparison of the activities of L sshRNAs with 2′-OMe modifications in the stem and/or loop regions. The sshRNAs were compared for their potency of suppressing the expression of HCV IRES-fLuc reporter in 293FT cells, by cotransfecting them in triplicate with the reporter DNA. Unmodified si19-3, specific for the same target, was used as a positive control. (A–C) Comparison of SG105 and its derivatives containing 2′-OMe modifications in the stem and/or loop. (D) Comparison of SG119 and its derivatives containing 2′-OMe modifications in the stem and loop. Each construct is identified by sequence number and a short-hand notation for the region modified; for example, as-2 means 2′-O-methylated at position 2 of the antisense strand; ss-alt means 2′-O-methylated at every other nucleotide of the sense strand.
Interestingly, although siRNAs are known to fully tolerate modification at the second nucleotide from the 5′-end of the antisense strand (Jackson et al. 2006), the same strategy (intended to reduce immune stimulation) reduced the potency of L sshRNAs somewhat (Fig. 1B, SG225, SG226). In contrast to L sshRNAs but like siRNAs, when the loop was placed at the 3′-end of the sense strand (R sshRNA), the sshRNA with modification at position 2 of the antisense strand and alternate nucleotides on the sense strand (SG233) proved to be as active as its unmodified version (SG68) (Fig. 2A).
FIGURE 2.
Comparison of the activities of L and R sshRNAs with 2′-OMe modifications in the stem and/or loop regions. (A) Comparison of R sshRNAs with and without 2′-OMe modifications in the antisense and the sense strands. (B) Two L sshRNAs with target sequences different from si19-3 and from each other were compared for their target (IRES-fLuc) knockdown with and without 2′-OMe modifications in the stem and loop.
To test whether the patterns of 2′-OMe modification found in SG105 and its derivatives also hold for L sshRNAs with other sequences, dose responses for two sshRNAs targeting sequences different from SG105 (SG118 and SG108) were compared in the presence or absence of 2′-OMe modification. Modification at alternate nucleotides on the sense strand and both uridines of the loop, but no part of the antisense strand (SG237), provided similar potency compared with its unmodified parent, SG118 (Fig. 2B). In addition, single modifications at positions 15 and 17 resulted in little or no reduction in activity (data not shown). Thus, it appears to be true, independent of targeting sequence, that L sshRNAs maintain high potency when modified with 2′-OMe at the loop (if it is not part of the antisense sequence) and on alternate nucleotides or selected uridines of the sense strand.
Deoxyribonucleotide substitutions in the antisense strand affect the activity of sshRNAs
It was reported that the seed region of the antisense strand and its complementary sequence on the sense strand could be substituted with deoxyribonucleotides without affecting the target gene knockdown activity of siRNAs. Moreover, off-target effects were significantly reduced with this pattern of DNA substitution (Ui-Tei et al. 2008). We tested whether this pattern of modification could also be applied to sshRNAs. As with 2′-OMe modification, 2′-deoxy substitution of the first six nucleotides from the 5′-end of the antisense strand of L sshRNAs (SG206 and SG208) resulted in lower efficacy than when the same substitution was introduced at the 3′-end of the sense strand (SG207) (Fig. 3).
FIGURE 3.
Comparison of the activities of L sshRNAs with DNA substitutions in the duplex region. sshRNAs against the same target were compared for their potency of suppressing the expression of HCV IRES-fLuc reporter in 293FT cells. The sshRNAs with and without modifications were cotransfected in triplicate with the reporter DNA. Unmodified si19-3, specific for the same target, was used as a positive control.
Phosphorothioate linkages at the open end of sshRNAs are well tolerated
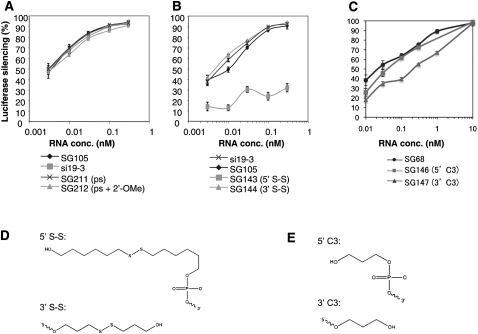
3′-Exonuclease is one of the primary classes of nucleases present in mammalian serum, and modifications at the 3′-end of DNA oligonucleotides and siRNAs have been shown to slow the degradation of these molecules in serum (Eder et al. 1991; Amarzguioui et al. 2003; Braasch et al. 2003; Chiu and Rana 2003; Harborth et al. 2003). Specifically, the phosphate backbone can be stabilized by phosphorothioate (PS) or boranophosphate modifications. 2′-Modifications of the ribose sugar also provide some degree of nuclease resistance. Hence we investigated whether the addition of PS modifications at the 5′-end of the antisense strand and the 3′-end of the sense strand would affect the efficacy of L sshRNAs (SG211). SG211 was found to be fully as active as its unmodified parent, SG105 (Fig. 4A). The further introduction of alternate 2′-OMe modifications to the sense strand (SG212) also did not affect the potency of the PS-modified sshRNA.
FIGURE 4.
Effect of PS modification and end conjugations. sshRNAs against the same target were compared for their dose responses against the reporter DNA HCV IRES-fLuc in 293FT cells. The sshRNAs with and without modifications were cotransfected in triplicate with the reporter DNA. Unmodified siRNA, si19-3, specific for the same target, was used as a positive control. The experiments were repeated at least twice. (A) Comparison of L sshRNAs with phosphorothioate modifications at the end of the stem opposite the loop. (B) Comparison of L sshRNAs with a disulfide-containing group conjugated to the 5′- or 3′-end. (C) Comparison of R sshRNAs with a C3 group conjugated to the 5′- or 3′-end. (D) disulfide-containing group. (E) C3 group.
Conjugation with multi-atom groups at free ends of the antisense strand reduces the activity of sshRNAs
End conjugation of siRNAs with cholesterol or α-tocopherol has been shown to facilitate their delivery to the liver (Soutschek et al. 2004; Nishina et al. 2008). Other delivery agents such as peptides, lipid nanoparticles, or antibodies could be also conjugated to the end of siRNAs. To evaluate whether such a strategy affects the activity of sshRNAs, a group containing a disulfide linkage (Fig. 4D) was conjugated to either the 5′- or 3′-end of the hairpin, and target knockdown was measured. As shown in Figure 4B, conjugation at the 5′-end of the antisense strand (SG143) completely abolished the activity of an L sshRNA, whereas conjugation at the 3′-end of the sense strand (SG144) resulted in retention of full efficacy. The antisense strand is at the 5′-end of L sshRNAs, and the presence of a 5′-phosphate is essential for binding to Dicer and/or Ago2 in RISC. Since 5′-conjugation presumably blocks the phosphorylation of 5′-OH ends of synthetic RNAs that normally occurs upon their transfection into the cell, the loss of RNAi activity by this modification is not surprising (Jinek and Doudna 2009). A similar phenomenon was observed with siRNAs when end conjugation occurred at the 5′-end of the antisense strand (Amarzguioui et al. 2006).
R sshRNAs were also tested for the influence of end-conjugation on their activities. Unlike with L sshRNAs, conjugation of a C3 group (Fig. 4E) at the 5′-end of the sense strand (SG146) of an R sshRNA did not significantly affect its efficacy, whereas conjugation of this group at the 3′-end of the antisense strand (SG147) was more detrimental (Fig. 4C). This activity loss might be due to alteration of the overhang at the 3′-end of the antisense strand, which interacts with the PAZ domain of Ago2 to facilitate RISC loading (Jinek and Doudna 2009).
2′-OMe modification can improve the serum stability of sshRNAs
Although dsRNAs are more stable than single-stranded RNAs (ssRNAs), sshRNAs without chemical modification are still very sensitive to nucleases. An unmodified sshRNA with a 5-nt loop (SG68) was largely degraded within 5 min upon incubation in 10% human serum at 37°C (Fig. 5). Faint bands with sizes ∼20–30 nt were detected in denaturing polyacrylamide gels, suggesting that the loop of the hairpin may be the most vulnerable region for serum nucleases. Interestingly, an sshRNA with a 2-nt UU loop and no 3′-overhang (SG105) was stable for up to 2 h of incubation in 10% human serum. 2′-OMe modification of the UU loop (SG202) did not increase its stability. Because the major degradation product seen for SG68, appearing after 5 min of incubation, corresponds to a half-molecule (21–23 nt) (Fig. 5), and in the case of SG105 it appears only after 2 h, we conclude that minimizing the loop size appears to confer nuclease resistance. With 2′-OMe modification at alternate nucleotides of the sense strand and each nucleotide in the UU loop, the molecule (SG224 and SG204) remained largely intact for up to 6 h in 10% serum (Fig. 5).
FIGURE 5.
Serum stability of sshRNAs. sshRNAs with and without 2′-OMe modifications were incubated with 10% human serum (Sigma-Aldrich) for various times at 37°C. At each time point, an aliquot was taken out, mixed with gel loading buffer (Ambion), and immediately stored in −80°C. The samples were analyzed by 12% denaturing PAGE (12% polyacrylamide, 20% formamide, and 8 M urea) and were stained with SYBR Gold (Invitrogen).
For in vivo application, although formulation into lipoplex or other complexes may insulate the RNA from nucleases to various degrees, it is likely that most or all formulations will benefit from additional stabilization of the RNA by chemical modification.
2′-OMe modification can reduce the innate immune response
Numerous results have demonstrated the capability of unmodified shRNAs and siRNAs to induce the undesired expression of proinflammatory cytokines such as type I interferon (IFN), IL-6, and TNF-α (Robbins et al. 2009). TLR3, TLR7/8, dsRNA-dependent PKR, and RIG-I are involved in the recognition and activation of the innate immune response (Reynolds et al. 2006; Behlke 2008; Judge and MacLachlan 2008; Robbins et al. 2009). Although sshRNAs have a duplex length of 19 bp or less, shorter than common shRNAs and 21-bp siRNAs, they may still be immune activators if they contain certain sequences or structural features. We thus evaluated the cytokine-inducing ability of sshRNAs with and without various chemical modifications.
Freshly purified human peripheral blood mononuclear cells (PBMCs) and human fetal lung fibroblast (MRC-5) cells were used in these experiments. PBMCs are a mixed immune cell population that are representative of the natural spectrum of immune receptors in vivo (Judge et al. 2006). Unlike many other cell lines, which may have defects in the IFN response pathway, MRC-5 cells are known to remain sensitive to immunostimulatory oligonucleotides (Marques et al. 2006).
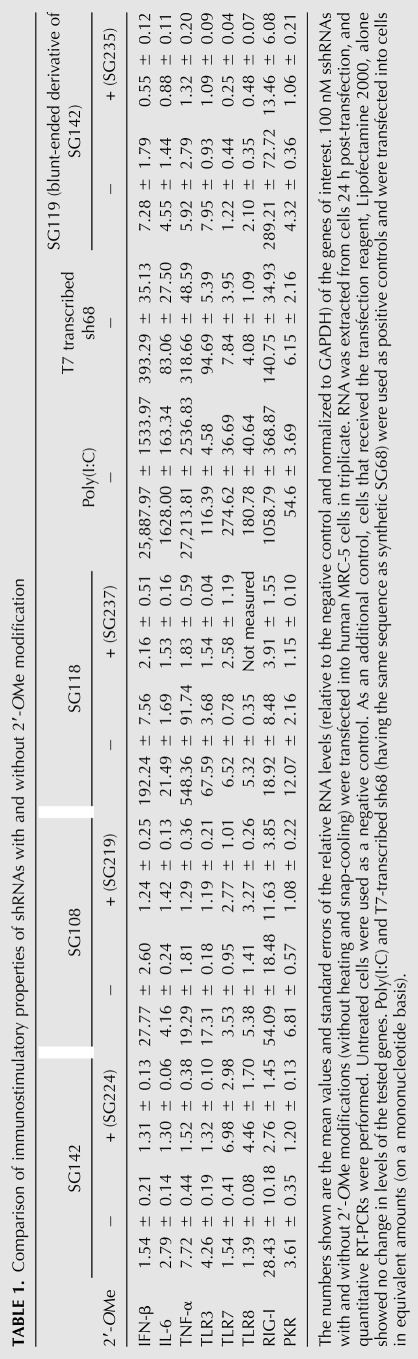
Poly(I:C) (Pharmacia) and a T7-transcribed shRNA were used as positive controls (Hornung et al. 2006), the former because it is a well-known inducer of proinflammatory cytokines, and the latter because, like all products of transcription by phage polymerases, it has an IFN-inducing 5′-triphosphate group (Robbins et al. 2009). Indeed, both controls showed induction of OAS1, IFN-β, IL-6, and TNF-α in PBMCs and MRC-5 cells (Fig. 6; Table 1; data not shown). In addition to proinflammatory cytokines, we also examined the expression of innate immune response mediators such as TLRs, RIG-I, and PKR. As shown in Table 1, Poly(I:C) strongly up-regulated all the mRNAs tested, especially the TLRs and RIG-I. The induction of TLR7 and TLR8 mRNAs in MRC-5 cells by Poly(I:C) is unexpected since both are believed to express only on immune cells. T7-transcribed shRNA specifically up-regulated RIG-I transcription (Robbins et al. 2009). Interestingly, this RNA molecule also stimulated the expression of TLR3 (Table 1).
FIGURE 6.
Type I IFN-responsive gene OAS1-induction by sshRNAs in human PBMCs. sshRNA with and without 2′-OMe modifications (20 nM) were transfected in triplicate into freshly purified human PBMCs. Equivalent amounts of T7 transcribed shRNA or poly(I:C) were transfected as positive controls. The transfection reagent DOTAP alone was used as negative control. The cells were harvested 24 h post-transfection, and the mRNAs of OAS1 and GAPDH were measured by quantitative RT-PCR. The relative level of OAS1 mRNA is shown, normalized to GAPDH.
TABLE 1.
Comparison of immunostimulatory properties of shRNAs with and without 2′-OMe modification
Since the sshRNAs used in this study have maximum target gene knockdown (HCV IRES-fLuc reporter model) at concentrations of 0.3–10 nM, we used 20 nM shRNAs in the initial PBMC experiment to examine their capabilities to up-regulate OAS1 and IFN expression. A high concentration of 100 nM of sshRNAs was later used in all MRC-5 cell transfections to make sure that we could detect even very modest immunostimulatory properties.
Synthetic L sshRNAs against three target sequences were investigated (SG142, SG108, and SG118). SG142 did not trigger an inflammatory cytokine response unless the 3′-overhang was removed (in Fig. 6 and Table 1, cf. SG142 [with overhang] and SG105, SG117, SG119 [without overhang]; additional data not shown), indicating a RIG-I-mediated IFN induction by blunt-ended hairpin (Takeuchi and Akira 2009). Indeed, the mRNA level of RIG-I, but not TLRs or PKR, was significantly up-regulated in MRC-5 cells when SG119 was transfected (Table 1). 2′-OMe modification of alternate nucleotides in the sense strand (SG204 and SG235) significantly reduced or abolished the up-regulation of cytokines and RIG-I (Fig. 6; Table 1).
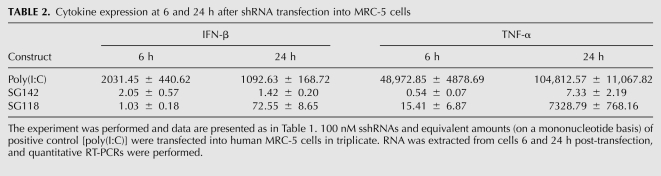
It has been reported that the selection of the time used to detect the inflammatory cytokine effect is important because the cytokine response to siRNA is transient, peaking between 2 and 8 h after administration (Robbins et al. 2008). Since the detection time we chose was 24 h after transfection of PBMCs and MRC-5 cells, to rule out the possibility that SG142 (negative in cytokine induction at 24 h) induces the innate immune response at earlier times, we compared the expression of IFN-β and TNF-α in MRC-5 cells at 6 and 24 h after the transfection of SG142. As shown in Table 2, cytokines were nearly negative at both time points when SG142 was examined. When SG118 was transfected, the expression of these cytokines was much higher at 24 h than at 6 h. The positive control, Poly(I:C), up-regulated TNF-α with similar kinetics, peaking at 24 h. However, IFN-β expression level did not increase after 6 h (Table 2). The kinetics of Poly(I:C)-induced cytokine expression in MRC-5 cells are different from what has been reported for PBMCs (Gonzalez-Gonzalez et al. 2009), probably due to differences in the cell types and their immune response mediators (Robbins et al. 2009).
TABLE 2.
Cytokine expression at 6 and 24 h after shRNA transfection into MRC-5 cells
Two other L sshRNAs, SG118 and SG108, stimulated PBMC and MRC-5 to express OAS1, IFN-β, TNF-α, and IL-6 even when the 3′-overhang was present (Fig. 6; Table 1). This implies that the sequence itself may contain a sequence motif such as GU that may be stimulatory to TLRs or other mediators of innate immune responses to RNA oligonucleotides. The RNA duplex length may also play a role in the sshRNA-induced immune response. Indeed, the synthetic sshRNAs tested in this study are present as a mixture of monomers, dimers, and even trimers (Q Ge, H Ilves, A Dallas, P Kumar, J Shorenstein, SA Kazakov, MA Behlke, and BH Johnston, in prep.). Although containing a bulge in the middle, the duplex length of a dimer sshRNA is well over 30 bp and thus a good candidate for PKR recognition (Manche et al. 1992). The dimer and trimer sshRNAs can be largely converted to monomers by heating (95°C for 4 min) and snap-cooling (ice-water bath). Upon comparing the innate immune responses of SG118 before and after this treatment, we found that the cytokine induction was high when multimers were present but nearly absent with monomers alone (before treatment, IFN-β, 33.58 ± 1.43, TNF-α, 2426.48 ± 364.31; after treatment, IFN-β, 2.23 ± 0.22, TNF-α, 2.84 ± 1.03), indicating that the presence of dimers and/or trimers, but not the monomer or the sequence of SG118, is mainly responsible for immune stimulation. This also supports the findings that the recognition and activation of some immune mediators requires a minimal length (Robbins et al. 2009). dsRNAs with a duplex length of 19 bp or less are less capable of inducing inflammatory cytokines. Modification of sshRNAs with 2′-OMe in the loop and alternate nucleotides of the sense strand (SG218 and SG219) again abrogated cytokine-inducing activity (Table 1).
2′-OMe modification has been found to be particularly effective in preventing the recognition of siRNAs by TLR7/8 and RIG-I (Hornung et al. 2006; Judge et al. 2006; Robbins et al. 2007, 2009). The modified groups interact with TLR7 without triggering signaling cascades, antagonizing TLR7-mediated activation by both RNA and small-molecule TLR7 agonists (Cekaite et al. 2007; Robbins et al. 2007). As shown in Table 3, substitution of as few as two of 42 native ribonucleotides in SG118 (without heat and snap-cool treatment) was sufficient to inhibit IFN-β and TNF-α up-regulation, suggesting that 2′-OMe modification prevents or antagonizes the recognition of dimer SG118 by PKR or perhaps some other immune response mediator. It should be noted that the majority of innate immune response experiments were performed in MRC-5 cells, a human fibroblast cell line. This cell line likely expresses several innate immune response sensors such as TLR3, RIG-I, and PKR. Thus, whether or not sshRNAs activate TLR7 and TLR8, which are exclusively expressed in immune cells, was not thoroughly studied. Nevertheless, our results show that 2′-OMe modification efficiently removes the immunostimulating properties of sshRNAs.
TABLE 3.
Immunostimulatory properties of shRNAs with various numbers of 2′-OMe modifications (24 h after transfection)
Certain modifications can induce innate immune responses in MRC-5 cells
Conjugation of a bulky group at their 3′-end (SG144) or introduction of PS linkages (SG211) in the stem did not affect the potency of L sshRNAs (Fig. 4A,B). Since the parent sshRNA SG105 induced low levels of IFN-β and TNF-α expression in MRC-5 cells even after heating and snap-cooling, we examined whether these modifications would change the cytokine expression profile. The heat and snap-cool treatment was given to all the sshRNAs before transfection to remove dimers and multimers. As shown in Table 4, both addition of 3′-end conjugation and PS linkages induced high levels of IFN-β and TNF-α expression, unlike the parent molecule, SG105. TLR3 and RIG-I were up-regulated by SG144 and SG211 more profoundly than by SG105 (data not shown). In particular, a significant amount of RIG-I mRNA was detected with SG144-transfected cells, suggesting specific activation and up-regulation of RIG-I by the presence of the 3′-end conjugation on the L sshRNA. Interestingly, the up-regulation of cytokines and RIG-I was not seen when the same group was conjugated to the 5′-end of SG105 (data not shown). It has been shown that dsRNAs with protruding termini bind RIG-I weakly and do not stimulate the activation of RIG-I efficiently in cells (Takeuchi and Akira 2009). It is not clear how the activity of RIG-I was affected differently by a disulfide group conjugated to the 3′- or 5′-ends of a blunt-ended RNA hairpin.
TABLE 4.
Immunostimulatory properties of L sshRNAs with 3′-end conjugation or PS bonds
The presence of many PS bonds in oligonucleotides can result in cytotoxicity, and the ability of PS oligonucleotides to nonspecifically bind to proteins may explain some if not all of their toxicity (Levin 1999; Kurreck 2003). The addition of as few as three PS bonds in sshRNAs caused significant induction of cytokines, particularly of TNF-α (Table 4). Since the hairpin has blunt ends, one possibility is that PS linkages may enhance the interaction between RIG-I and blunt-ended sshRNA. However, some single-stranded oligonucleotides with PS backbone have been shown to inhibit the signaling of TLR3 and RIG-I induced by Poly(I:C) (Ranjith-Kumar et al. 2009), suggesting that the immune activation by PS-modified dsRNAs may reflect different recognition mechanisms of RIG-I for single- and double-stranded oligonucleotides (Ranjith-Kumar et al. 2009). The up-regulation of TLR3 and RIG-I may be either an indirect effect of the release of inflammatory cytokines or the direct recognition of the modification.
2′-OMe ribosyl substitution in the sense strand (at alternate nucleotides) largely removed the innate immune responses induced by SG144 and SG211 (data not shown). Phosphorothioate modification is commonly used to stabilize oligonucleotides, and further addition of 2′-OMe allows the use of P-S without triggering an innate immune response.
dsRNA-induced innate immune response in 293FT cells
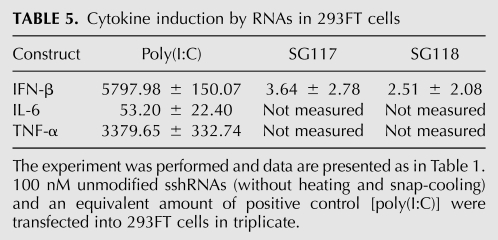
The induction of type I IFN can down-regulate the expression of many genes in a nonspecific manner. To test whether this affected the target gene knockdown experiments that were performed in 293FT cells, sshRNAs were transfected into 293FT cells without prior heating and snap-cooling. Twenty-four hours later, total RNA was extracted to measure cytokine mRNA levels. Unlike with MRC-5 cells, where IFN-β was highly up-regulated after transfection of SG117 and SG118 (Table 1; data not shown), IFN-β was not detected in 293FT cells that had received the same sshRNAs (Table 5). The control [poly(I:C)] induced IFN-β up-regulation in 293FT cells. Interestingly, RIG-I, but not TLRs or PKR, was significantly up-regulated in 293FT cells after transfection of Poly(I:C) [the relative induction by Poly(I:C) treated versus untreated was RIG-I: 322.98 ± 7.52; TLR3: 6.50 ± 1.74; TLR7: 3.75 ± 2.15; TLR8: 7.39 ± 5.18; PKR: 7.50 ± 1.48]. This contrasts drastically with the results obtained from MRC-5 cells (Table 1), suggesting that 293FT cells might be deficient in certain innate immune response capabilities.
TABLE 5.
Cytokine induction by RNAs in 293FT cells
In addition, if the target gene suppression was nonspecifically mediated by type I IFN, modifications to abrogate the inflammatory cytokine induction by sshRNAs would result in less target down-regulation. However, we found nearly identical dose response curves (Fig. 1B) between modified (SG204 and SG224, negative in cytokine induction as shown in Fig. 6 and Table 1) and unmodified sshRNAs (SG105, positive in OAS1 induction as shown in Fig. 6). This indicates that the suppression of target gene expression in 293FT cells is a specific, on-target effect.
Summary
Chemical modification affects the potency of L and R sshRNAs in different ways. L sshRNAs can tolerate deoxyribonucleotide substitution or 2′-OMe modification at the sense strand and loop, 3′-end conjugation, or PS bonds at the open ends of the hairpin, but not modifications in the antisense strand or conjugation of a bulky group at the 5′-end of the hairpin. The activity of R sshRNA is affected by 3′-end conjugation but can accommodate some 2′-OMe modifications in the antisense strand. L-sshRNAs, like siRNAs, have antisense strands with free 5′-ends. They are thus available to 5′-phosphorylation, whereupon they can presumably bind in a pocket between the MID and PIWI domains of Ago2 (Jinek and Doudna 2009). This process cannot occur if the 5′-OH is blocked, for example, through conjugation. In R-type sshRNAs, on the other hand, the 5′-end of the antisense strand is linked to the loop and cannot play the same role unless it is freed by cleavage of the backbone at precisely the right position. The greater tolerance of R than L sshRNAs for 2′-O-methylation at nucleotide 2 of the antisense strand might reflect such a different role. It remains to be determined why siRNAs but not sshRNAs can tolerate 2′-O-methylation on alternating nucleotides of the antisense strand. Further studies of the mechanism of action of sshRNAs will be reported separately (Q Ge, H Ilves, A Dallas, P Kumar, J Shorenstein, SA Kazakov, MA Behlke, and BH Johnston, in prep.).
Unmodified shRNAs can be immune activators, especially when in dimeric form or if they are blunt-ended. Certain modifications such as 3′-end-conjugation and PS linkages can exacerbate this effect. Monomer sshRNAs are less capable of activating the innate immune response than their dimeric forms, probably reflecting the influence of mediators such as PKR that are sensitive to duplex length. Importantly, as few as two 2′-OMe substitutions can reduce the innate immune response to background levels. As an added benefit, 2′-OMe modifications confer improved serum stability on sshRNAs.
MATERIALS AND METHODS
Preparation of shRNAs
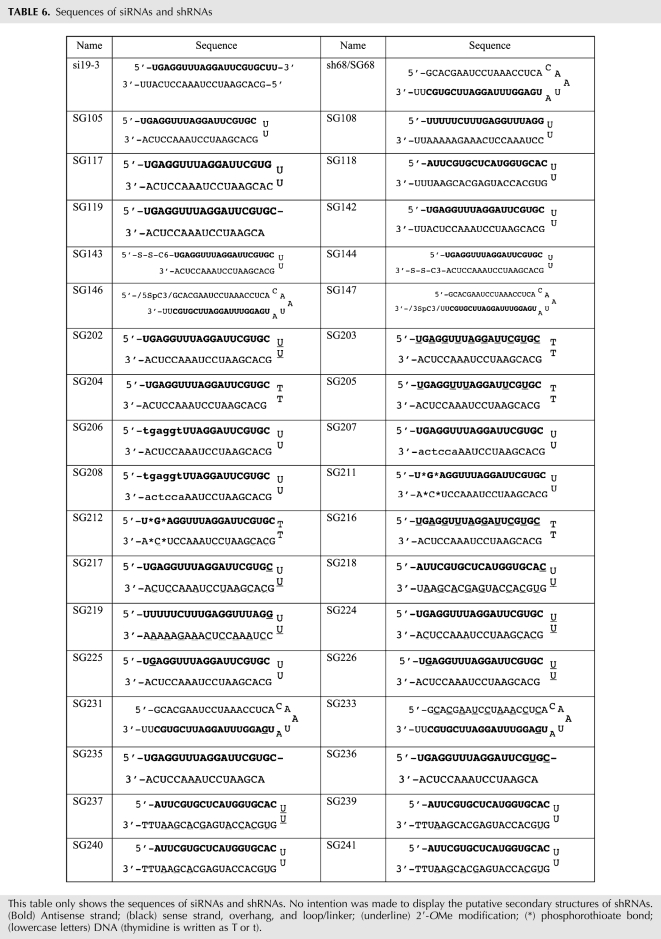
shRNAs with and without chemical modifications were chemically synthesized and HPLC-purified by Integrated DNA Technologies (IDT) and resuspended in RNase- and pyrogen-free buffer containing 20 mM KCl, 6 mM HEPES-KOH (pH 7.5), and 0.2 mM MgCl2 (Thermo Fisher Scientific, Dharmacon Products). The sequences of all the shRNAs used in this study are summarized in Table 6. The sshRNAs used for Figures 1–5 and Table 4 were heated for 4 min at 95°C and snap-cooled in an ice-water bath for 20 min before transfection. The sshRNAs used for Figure 6 and Tables 1, 2, 3, and 5 did not receive the heat and snap-cool treatment.
TABLE 6.
Sequences of siRNAs and shRNAs
Transfection and reporter gene assays
The human cell line 293FT (Invitrogen) was maintained in DMEM (Cambrex) with 10% fetal bovine serum (Hyclone), supplemented with 2 mM L-glutamine and 1 mM sodium pyruvate. One day prior to transfection, cells were seeded at 23,000 cells/well in a 96-well plate, resulting in ∼80% cell confluency at the time of transfection. Transfections were performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Thirteen nanograms of the IRES-linked reporter plasmid pSG154m (in which firefly luciferase [fLuc] expression is dependent on the IRES) (Wang et al. 2005), 20 ng of pSEAP2 control plasmid (BD Biosciences Clontech) as a transfection control, and the indicated amounts of shRNAs were cotransfected into 293FT or Huh7 cells. Forty-eight hours post-transfection, the cells were lysed, and luciferase activity was measured in a MicroLumat LB 96P luminometer (Berthold Technologies). Unless otherwise indicated, all the siRNA and shRNA samples were tested in triplicate, and two or more independent experiments were performed. The IC50s of the dose response curves were calculated using GraFit data analysis software.
Serum stability assay and electrophoresis
sshRNAs (3.35 μg) were incubated with 10% human serum (Sigma) in PBS at 37°C. An aliquot was taken out at each time point and immediately mixed with 2× gel loading buffer (Ambion) and stored at −80°C. Subsequent gel electrophoresis of the samples was performed under denaturing conditions (12% polyacrylamide, 20% formamide, and 8 M urea). The gel was stained with SYBR Gold (Invitrogen).
Proinflammatory cytokine detection
Human PBMCs were prepared from buffy coats (Stanford Blood Center) by density gradient centrifugation, washed, and then seeded in 24-well plates at 5 × 105 cells per well in RPMI 1640 containing 10% heat-inactivated fetal calf serum. Transfections were performed using DOTAP (Roche) following the manufacturer's instructions. Similarly, MRC-5 cells (human fetal lung fibroblast, CCL-171; ATCC) were seeded in 24-well plates at 6 × 104 cells per well with MEM containing 10% heat-inactivated fetal calf serum. Transfections were done using Lipofectamine 2000 following the manufacturer's instruction; 20 nM or 100 nM shRNAs were transfected in each well in triplicate. Six or 24 h later, the cells were lysed in TRIzol (Invitrogen), and total RNA was extracted according to the manufacturer's instructions. Quantitative RT-PCR was done using High-Capacity cDNA Reverse Transcription Kits, TaqMan Universal PCR Master Mix, SYBR power Master Mix, Taqman probes OAS1 (Hs00242943_m1), IFN-β (Hs01077958_s1), IL-6 (Hs99999032_m1), TNFα (Hs99999043_m1), and GAPDH (Hs99999905_m1), and a Fast 7500 real-time PCR machine (Applied Biosystems) following the manufacturer's instructions. The following primers were used:
TLR3 forward: 5′-TCCCAAGCCTTCAACGACTG-3′;
TLR3 reverse: 5′-TGGTGAAGGAGAGCTATCCACA-3′;
TLR7 forward: 5′-TTACCTGGATGGAAACCAGCTAC-3′;
TLR7 reverse: 5′-TCAAGGCTGAGAAGCTGTAAGCTA-3′;
TLR8 forward: 5′-GAGAGCCGAGACAAAAACGTTC-3′;
TLR8 reverse: 5′-TGTCGATGATGGCCAATCC-3′;
RIG-I forward: 5′-CAGTATATTCAGGCTGAG-3′;
RIG-I reverse: 5′-GGCCAGTTTTCCTTGTC-3′;
PKR forward: 5′-TCTGACTACCTGTCCTCTGGTTCTT-3′; and
PKR reverse: 5′-GCGAGTGTGCTGGTCACTAAAG-3′ (Hayashi et al. 2003; Terhorst et al. 2007).
ACKNOWLEDGMENTS
This project was partially supported by NIH grants R44AI056611 and 1R43AI074256 (to B.H.J.). We thank Dr. Peter Sarnow (Stanford University) for the dual luciferase reporter from which pSG154m was derived and Dr. Sergei Kazakov (SomaGenics) for helpful discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1901810.
REFERENCES
- Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared with unmodified small interfering RNA. J Med Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarzguioui M, Lundberg P, Cantin E, Hagstrom J, Behlke MA, Rossi JJ. Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat Protoc. 2006;1:508–517. doi: 10.1038/nprot.2006.72. [DOI] [PubMed] [Google Scholar]
- Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nat Methods. 2006;3:701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: A potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekaite L, Furset G, Hovig E, Sioud M. Gene expression analysis in blood cells in response to unmodified and 2′-modified siRNAs reveals TLR-dependent and independent effects. J Mol Biol. 2007;365:90–108. doi: 10.1016/j.jmb.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Chang K, Elledge SJ, Hannon GJ. Lessons from nature: MicroRNA-based shRNA libraries. Nat Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Rana TM. siRNA function in RNAi: A chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choung S, Kim YJ, Kim S, Park HO, Choi YC. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem Biophys Res Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Collingwood MA, Rose SD, Huang L, Hillier C, Amarzguioui M, Wiiger MT, Soifer HS, Rossi JJ, Behlke MA. Chemical modification patterns compatible with high potency dicer-substrate small interfering RNAs. Oligonucleotides. 2008;18:187–200. doi: 10.1089/oli.2008.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilizing modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debart F, Abes S, Deglane G, Moulton HM, Clair P, Gait MJ, Vasseur JJ, Lebleu B. Chemical modifications to improve the cellular uptake of oligonucleotides. Curr Top Med Chem. 2007;7:727–737. doi: 10.2174/156802607780487704. [DOI] [PubMed] [Google Scholar]
- Dorsett Y, Tuschl T. siRNAs: Applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- Eder PS, DeVine RJ, Dagle JM, Walder JA. Substrate specificity and kinetics of degradation of antisense oligonucleotides by a 3′ exonuclease in plasma. Antisense Res Dev. 1991;1:141–151. doi: 10.1089/ard.1991.1.141. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fewell GD, Schmitt K. Vector-based RNAi approaches for stable, inducible and genome-wide screens. Drug Discov Today. 2006;11:975–982. doi: 10.1016/j.drudis.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Forsbach A, Nemorin JG, Montino C, Muller C, Samulowitz U, Vicari AP, Jurk M, Mutwiri GK, Krieg AM, Lipford GB, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- Ge Q, Ilves H, Dallas A, Kumar P, Shorenstein J, Kazakov SA, Johnston BH. Minimal-length short hairpin RNAs: The relationship of structure and RNAi activity. RNA. 2010 doi: 10.1261/rna.1894510. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gonzalez E, Ra H, Hickerson RP, Wang Q, Piyawattanametha W, Mandella MJ, Kino GS, Leake D, Avilion AA, Solgaard O, et al. siRNA silencing of keratinocyte-specific GFP expression in a transgenic mouse skin model. Gene Ther. 2009;16:963–972. doi: 10.1038/gt.2009.62. [DOI] [PubMed] [Google Scholar]
- Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, Means TK, Luster AD. Toll-like receptors stimulate human neutrophil function. Blood. 2003;102:2660–2669. doi: 10.1182/blood-2003-04-1078. [DOI] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces ‘off-target’ transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- Judge A, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum Gene Ther. 2008;19:111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Kariko K, Bhuyan P, Capodici J, Weissman D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J Immunol. 2004;172:6545–6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Kim DH, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- Levin AA. A review of the issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim Biophys Acta. 1999;1489:69–84. doi: 10.1016/s0167-4781(99)00140-2. [DOI] [PubMed] [Google Scholar]
- Li L, Lin X, Khvorova A, Fesik SW, Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA. 2007;13:1765–1774. doi: 10.1261/rna.599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- McManus MT, Petersen CP, Haines BB, Chen J, Sharp PA. Gene silencing using micro-RNA designed hairpins. RNA. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Nawrot B, Sipa K. Chemical and structural diversity of siRNA molecules. Curr Top Med Chem. 2006;6:913–925. doi: 10.2174/156802606777303658. [DOI] [PubMed] [Google Scholar]
- Nishina K, Unno T, Uno Y, Kubodera T, Kanouchi T, Mizusawa H, Yokota T. Efficient in vivo delivery of siRNA to the liver by conjugation of α-tocopherol. Mol Ther. 2008;16:734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- Poeck H, Besch R, Maihoefer C, Renn M, Tormo D, Morskaya SS, Kirschnek S, Gaffal E, Landsberg J, Hellmuth J, et al. 5′-Triphosphate-siRNA: Turning gene silencing and Rig-I activation against melanoma. Nat Med. 2008;14:1256–1263. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- Ranjith-Kumar CT, Murali A, Dong W, Srisathiyanarayanan D, Vaughan R, Ortiz-Alacantara J, Bhardwaj K, Li X, Li P, Kao CC. Agonist and antagonist recognition by RIG-I, a cytoplasmic innate immunity receptor. J Biol Chem. 2009;284:1155–1165. doi: 10.1074/jbc.M806219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Anderson EM, Vermeulen A, Fedorov Y, Robinson K, Leake D, Karpilow J, Marshall WS, Khvorova A. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA. 2006;12:988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther. 2007;15:1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, Ambegia E, Choi C, Yaworski E, Palmer L, McClintock K, MacLachlan I. Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation. Hum Gene Ther. 2008;19:991–999. doi: 10.1089/hum.2008.131. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- Schlee M, Hornung V, Hartmann G. siRNA and isRNA: Two edges of one sword. Mol Ther. 2006;14:463–470. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J Mol Biol. 2005;348:1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhorst D, Kalali BN, Weidinger S, Illig T, Novak N, Ring J, Ollert M, Mempel M. Monocyte-derived dendritic cells from highly atopic individuals are not impaired in their pro-inflammatory response to toll-like receptor ligands. Clin Exp Allergy. 2007;37:381–390. doi: 10.1111/j.1365-2222.2006.02639.x. [DOI] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K. Functional dissection of siRNA sequence by systematic DNA substitution: Modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov AV, Korba B, Farrar K, Mukerjee S, Seyhan AA, Ilves H, Kaspar RL, Leake D, Kazakov SA, Johnston BH. shRNAs targeting hepatitis C: Effects of sequence and structural features, and comparision with siRNA. Oligonucleotides. 2007;17:223–236. doi: 10.1089/oli.2006.0069. [DOI] [PubMed] [Google Scholar]
- Wang Q, Contag CH, Ilves H, Johnston BH, Kaspar RL. Small hairpin RNAs efficiently inhibit hepatitis C IRES-mediated gene expression in human tissue culture cells and a mouse model. Mol Ther. 2005;12:562–568. doi: 10.1016/j.ymthe.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: Tools and applications. Drug Discov Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- Zhang HY, Du Q, Wahlestedt C, Liang Z. RNA Interference with chemically modified siRNA. Curr Top Med Chem. 2006;6:893–900. doi: 10.2174/156802606777303676. [DOI] [PubMed] [Google Scholar]