Abstract
Iron regulatory proteins (IRPs) are iron-regulated RNA binding proteins that, along with iron-responsive elements (IREs), control the translation of a diverse set of mRNA with 5′ IRE. Dysregulation of IRP action causes disease with etiology that may reflect differential control of IRE-containing mRNA. IREs are defined by a conserved stem–loop structure including a midstem bulge at C8 and a terminal CAGUGH sequence that forms an AGU pseudo-triloop and N19 bulge. C8 and the pseudo-triloop nucleotides make the majority of the 22 identified bonds with IRP1. We show that IRP1 binds 5′ IREs in a hierarchy extending over a ninefold range of affinities that encompasses changes in IRE binding affinity observed with human L-ferritin IRE mutants. The limits of this IRE binding hierarchy are predicted to arise due to small differences in binding energy (e.g., equivalent to one H-bond). We demonstrate that multiple regions of the IRE stem not predicted to contact IRP1 help establish the binding hierarchy with the sequence and structure of the C8 region displaying a major role. In contrast, base-pairing and stacking in the upper stem region proximal to the terminal loop had a minor role. Unexpectedly, an N20 bulge compensated for the lack of an N19 bulge, suggesting the existence of novel IREs. Taken together, we suggest that a regulatory binding hierarchy is established through the impact of the IRE stem on the strength, not the number, of bonds between C8 or pseudo-triloop nucleotides and IRP1 or through their impact on an induced fit mechanism of binding.
Keywords: iron regulatory protein, iron-responsive element, RNA binding protein, RNA–protein interactions, iron metabolism, aconitase
INTRODUCTION
Iron regulatory proteins (IRPs) function as critical components of a sensory and regulatory network that controls cellular iron homeostasis in vertebrates (Wallander et al. 2006; Leipuviene and Theil 2007; Muckenthaler et al. 2008). IRPs bind iron-responsive elements (IREs) in the untranslated region of mRNA encoding proteins involved in iron metabolism, or in the adaptive response to changes in iron availability, and regulate their translation or stability. IRP1 and IRP2 are orthologous proteins that are largely but not completely functionally redundant. Underscoring the central need for appropriate regulation of iron metabolism in growth and development is the observation that genetic ablation of both IRP1 and IRP2 causes embryonic lethality in mice, and their intestinal function alone is essential (Galy et al. 2005, 2008; Smith et al. 2006). In low iron, IRPs bind IREs, and when present in the 5′ UTR, they block 43S ribosomal recruitment and hence mRNA translation (for review, see Muckenthaler et al. 2008). To date six mammalian mRNAs encoding proteins of widely different function have been identified that possess 5′ IREs. These mRNAs encode the erythroid-specific isoform of 5-aminolevulinate synthase (eALAS), which catalyzes the first step in heme synthesis; the H and L subunits of the iron storage protein ferritin; the iron exporter ferroportin; mitochondrial aconitase (m-acon), which is the second enzyme of the tricarboxylic acid cycle; and the hypoxia-inducible transcription factor HIF 2α (also known as EPAS1) (Wallander et al. 2006; Leipuviene and Theil 2007; Muckenthaler et al. 2008). Given the varied function of proteins encoded by mRNA containing 5′ IREs, it seems likely that IRPs differentially control translation of these mRNAs.
Differences in translational regulation of 5′-IRE-containing mRNA by IRPs form a critical basis of an adaptive cellular response to changes in organismal iron status. Multiple lines of evidence indicate that IRPs differentially regulate the translation of mRNAs with 5′ IREs. First, dietary iron deficiency alone renders ferritin protein undetectable, while m-acon protein is only decreased twofold in liver (for review, see Wallander et al. 2006). Additionally, iron excess preferentially activates ferritin synthesis by an order of magnitude more than m-acon synthesis. Second, in erythroid progenitor cells the translational activity of ferritin mRNA is less sensitive to activation by iron than eALAS mRNA, which correlates to the relative need for expression of the encoded proteins to support erythroid differentiation (Schranzhofer et al. 2006). Third, genetic ablation of both IRPs in intestine dramatically increased ferritin and ferroportin protein levels yet only modestly increased m-acon protein, apparently because ferritin and ferroportin mRNA translation are more strongly repressed by IRP action (Galy et al. 2008). These findings, along with the observation that IRPs appear to differentially interact with the ferritin and m-acon IREs, suggest that IREs differ in their regulatory potential for dictating control of mRNA translation (Kim et al. 1996; Ke et al. 2000). In this study, we focused on determining if an RNA binding hierarchy exists between IRP1 and 5′ IREs and what sequences within these regulatory elements modulate this hierarchy.
IRP1 RNA binding activity is controlled by its iron-dependent formation of an Fe-S cluster in the protein, converting it to the cytosolic isoform of aconitase (c-acon), which cannot bind RNA. In order to bind the IRE, c-acon undergoes a large-scale structural rearrangement of its four domains upon loss of the Fe-S cluster to form IRP1 and then is believed to bind to the IRE in a manner consistent with the principle of induced fit (Williamson 2000; Dupuy et al. 2006; Walden et al. 2006). The ferritin IRE adopts a stem–loop structure in solution that is similar but not identical to that formed when bound to IRP1 (Sierzputowska-Gracz et al. 1995; Laing and Hall 1996; McCallum and Pardi 2003; Walden et al. 2006). The central elements of the IRE required for recognition by IRP are a midstem C bulge, referred to here as C8, separated from a terminal loop of sequence CAGUGH (H = U, C, or A) by an upper stem of 5 base pairs (bp) and sufficient base-pairing below C8 to stabilize the IRE secondary structure (Dandekar et al. 1991; Leipuviene and Theil 2007; Volz 2008). The first (C14) and the fifth (G18) nucleotides of the terminal loop base-pair, forming an AGU pseudo-triloop (Henderson et al. 1994; Sierzputowska-Gracz et al. 1995; Addess et al. 1997; Walden et al. 2006). We refer to these components of the IRE as the core IRE. Increasing the length of the upper stem or the size of the terminal loop, altering the identity of the conserved unpaired loop nucleotides, or disrupting the secondary structure of the IRE decreases IRP affinity (Barton et al. 1990; Leibold et al. 1990; Bettany et al. 1992; Kikinis et al. 1995). Additionally, a lower stem beneath the midstem C bulge is essential for the high-affinity binding by IRP1 to the ferritin IRE (Barton et al. 1990; Harrell et al. 1991). The central basis for the strength and specificity of the IRP1/IRE interaction lies in the presence of two large spatially separated RNA binding sites in IRP1 that promote a total of 22 intermolecular bonds primarily with the unpaired nucleotides of the C bulge and pseudo-triloop. These bonds give rise to this unique RNA/protein complex (Walden et al. 2006).
If an RNA binding hierarchy exists among IREs, it likely arises due to sequence and structural differences of IREs that are localized to the stem region and that modify the number or strength of protein–RNA bonds, or allow for additional IRE-specific bonds with IRP. In this regard, it is of interest that in the ferritin IRE/IRP1 complex, there are several protein/RNA bonds in the stem region at least one of which (Asn685:G26) should allow for unique bonds with this IRE (Walden et al. 2006). The ferritin IRE also has a more complex midstem bulge structure that appears to contribute to its selective recognition by IRP2 relative to other IREs (Ke et al. 1998, 2000). Additionally, IRPs can bind some IREs with a nonconsensus bulged nucleotide on the 3′ side of the stem (e.g., DMT1 and HIF 2α) or to naturally occurring or SELEX-generated RNA stem–loops that differ significantly from the canonical IRE sequence or structure (Henderson et al. 1994, 1996; Butt et al. 1996; Meehan and Connell 2001; Rogers et al. 2002). Taken together, these findings indicate that the rules of what defines the IRE remain to be fully elucidated.
To understand how mRNAs with 5′ IREs may be differentially recognized, we determined the affinity of IRP1 for all known 5′ IREs. We find that IRP1 bind these IREs with affinities arrayed in a hierarchical manner that relates to the strength of their regulation by IRP in vivo. Our examination of previously identified but uncharacterized IRE mutants that give rise to hereditary hyperferritinemia cataract syndrome further substantiates a role for modest changes in IRE binding affinity in the action of IRPs in vivo. Using a structure–function approach, we show that multiple determinants within the core IRE region function in concert to modulate IRP1 binding affinity and provide evidence suggesting that limited changes in the strength of bonds between IRP1 and the C8 or pseudo-triloop regions of the IRE could be sufficient to establish differential binding. Unexpectedly, our studies show that the consensus secondary structure of IRE does not represent the full range of structures recognized by IRP1, suggesting the existence of novel IREs. Taken together, our study supports the concept that multiple determinants within the IRE stem act in an integrated manner to dictate their varied regulatory potential.
RESULTS
The IRE and its interaction with IRP1
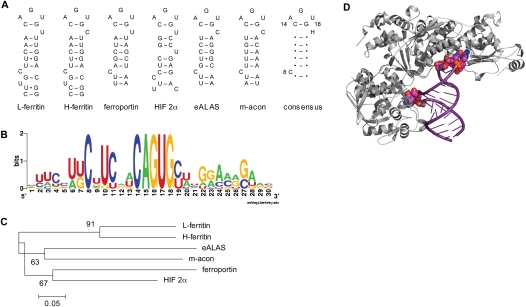
The IRE is defined by conserved sequence and secondary structure (Fig. 1A; Wallander et al. 2006; Leipuviene and Theil 2007; Muckenthaler et al. 2008; Volz 2008). As noted above, the elements of the core IRE are a stem–loop with a CAGUGH terminal loop, an unpaired/bulged cytosine (C8) 5 bp to the 5′ of the loop, and sufficient additional stem below C8 to stabilize IRE secondary structure. Analysis of the core IRE region for the six known mammalian mRNAs with 5′ IREs revealed that, other than the C8 bulge, the nucleotides of the stem are not well conserved across IRE families (Fig. 1B; Leipuviene and Theil 2007; Volz 2008). The ferritin and HIF 2α IREs differ from the core 5′ IRE structure due to the presence of additional bulge nucleotide(s) that, at least in the case of ferritin (Ke et al. 1998, 2000), affect IRP recognition. These differences in stem sequence and structure are the basis for the observed phylogenetic relationship between IRE families that is illustrated by the relatedness of ferritin IREs to themselves but less so with other IREs as noted by others (Fig. 1C; Piccinelli and Samuelsson 2007). The evidence suggests that the ferritin IRE is the ancestral element among the 10 natural IRE-containing mRNAs known to date (Piccinelli and Samuelsson 2007). The sequence and structural divergence in the IRE stem across vertebrate IRE families, relative to the ferritin IRE, argues for the functional diversity of these post-transcriptional regulatory elements.
FIGURE 1.
Features of the IRE and interaction with IRP1. (A) Secondary structure and sequence of known mammalian 5′ IRE. The core IRE is a short stem–loop structure composed of a 6-nt terminal loop, with the sequence CAGUGH, where H is usually U or C but can be A, an unpaired single nucleotide C bulge (C8) 5 bp to the 5′ of the first nucleotide of the terminal loop and sufficient base-paired stem to stabilize the secondary structure. The terminal loop forms an AGU pseudo-triloop as shown here. In addition to the core elements, IREs contain base-paired flanking sequences of varying length (see Supplemental Fig. S1). The sequence and predicted secondary structure for the known mammalian 5′ IREs include rat L-ferritin, human H-ferritin, human ferroportin, human HIF 2α, human erythroid 5′ aminolevulinate synthase (eALAS), and porcine mitochondrial aconitase (m-acon). The IRE consensus sequence and structure are displayed as dots for complementary base pairs. (B) The IRE consensus sequence derived from using the WebLogo version 2.8 (Crooks et al. 2004) is represented as information content. Sequence data for all known mammalian 5′ IRE were used. For a given position, nucleotide height represents the frequency of occurrence for a given nucleotide, and overall position height represents conservation at that position. (C) The evolutionary history of 5′ IREs was inferred using the minimum evolution method (Tamura et al. 2007). The optimal tree with the sum of branch length 1.6 is shown. The tree is drawn to scale, with the branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (5000 replicates) is given. (D) Structure of the IRP1:H-ferritin IRE complex (Walden et al. 2006). Domains 1–3 of IRP1 contact the terminal loop of the IRE, and domain 4 binds the C8 bulge. A representation of the bound IRE is presented with the unpaired nucleotides C8, A15, G16, and U17 and their phosphates forming the majority of the bonds with IRP1; these four nucleotides are shown in space-filling mode. The representations were created using PyMOL (DeLano Scientific LLC) and crystallographic file 2IPY (RCSB Protein Data Bank).
The structure of IRP1 in complex with the H-ferritin IRE provides key insights concerning how binding specificity is achieved and suggests mechanisms through which an IRE binding hierarchy might be established. In this complex, the RNA is inserted into a cleft formed between domains I–III and IV of IRP1, and the IRE binds at two large and spatially separate binding sites (Fig. 1D; Walden et al. 2006; Volz 2008). Together, these sites make 22 bonds with the RNA: 10 bonds with the unpaired nucleotides A15, G16, and U17 of the pseudo-triloop and their phosphates in a pocket formed by domains II and III, while the second site that is in domain IV makes eight bonds with the C8 bulge (Walden et al. 2006). The four remaining bonds occur between IRP1 and the IRE stem and include G26, the only known base-specific contact unique to the ferritin IRE. Any additional impacts of the IRE stem on binding affinity, as would be required in a broader regulatory hierarchy, likely arise due to stem-induced alterations in the strength or number of bonds between IRP1 and the C8 or pseudo-triloop nucleotides, or to introduction of bonds not present in the IRP1:H-ferritin complex. Our goal was to determine if an IRE binding hierarchy exists and to further define the roles of the RNA determinants of such a hierarchy.
Establishing conditions for quantification of the IRP1–IRE interaction
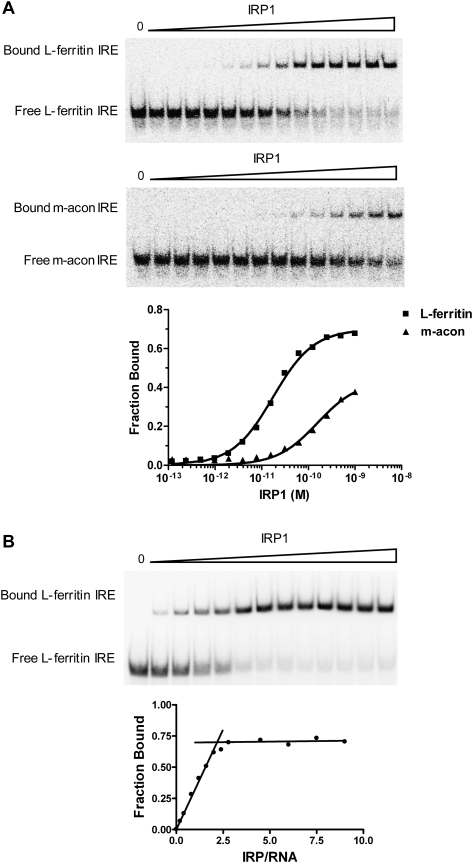
The affinity of purified IRP1 for specific IREs was determined under equilibrium conditions by direct titration of IRP1 with limiting [32P]-labeled IRE (Ryder et al. 2008). IRP1 formed a single major complex with the L-ferritin or m-acon IREs (Fig. 2A). From these data, the apparent Kd of IRP1 was determined to be 17.5 pM for L-ferritin IRE, which is in general agreement with values others have reported using cytosol (Haile et al. 1989; Barton et al. 1990) or purified protein (Alam et al. 1989; Allerson et al. 1999). The ferritin IRE binding isotherm reached saturation and was easily modeled with a Hill coefficient of 1, indicating a lack of cooperativity in binding. In contrast, use of identical conditions for the m-acon IRE was not optimal and required adjustment of the RNA concentration (cf. Fig. 2A and Fig. 3A). Although binding saturated with less than the full complement of IRE probe shifting in this and other experiments, the Kd was not dependent on the fraction shifted (data not shown). RNA that remained unbound likely represented incompletely folded RNA or is incapable of binding because of radiochemical decomposition as a consequence of the high specific radioactivity necessary for determining pM binding affinities (see Materials and Methods).
FIGURE 2.
Affinity and activity of IRP1 binding to IRE. (A) Binding curves for IRP1 with the L-ferritin and m-acon IRE as determined by EMSA. The IRP1 concentration was varied from 0.1 pM to 1.0 nM with the L-ferritin and m-acon IRE present at 0.1 pM. Results are plotted as fraction bound as a function of the concentration of active IRP1 and demonstrate that different RNA concentrations are required to establish equilibrium conditions for specific IRE. For the L-ferritin IRE (■) used, the results obtained exhibited characteristics of equilibrium binding, while the results for the m-acon IRE (▲) did not and required adjustment of the RNA concentration given its lower affinity for IRP1 (see Materials and Methods and Fig. 3A). (B) Determination of the fractional activity of purified IRP1 by stoichiometric titration. The L-ferritin IRE was kept at a constant saturating concentration (0.6 nM) while IRP1 was varied from 0.12 to 55 nM, allowing for variation of the IRP1:IRE ratio from 0.2 to ∼9. The linear increase in fraction bound saturated at an IRP1:IRE ratio of 2.3, indicating 54% active protein. The activity of the other IRP1 protein preparation used was 57% active protein (data not shown). The x-axis in panel A denotes the amount of active IRP1 protein.
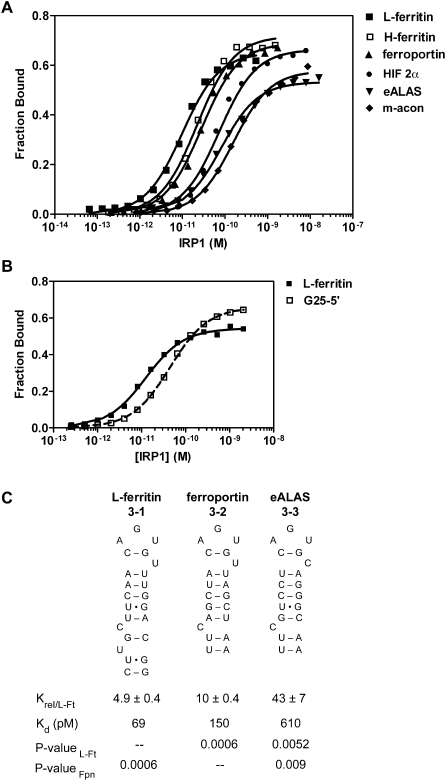
FIGURE 3.
Wild-type and mutant 5′ IRE have different affinities for IRP1. The affinity of IRP1 for 5′ IREs was determined by the direct titration of IRP1 by EMSA. (A) [32P]RNAs (63–66 nt) representing the IRE and its flanking sequence were synthesized, gel-purified, and quantified, and their affinity for IRP1 was determined. The data are representative of four to six experiments for each RNA. A nonlinear regression was performed for each experiment to obtain the Kd. Results are also reported as mean ± SEM (see Table 1). The RNAs analyzed were rat L-ferritin (■); human H-ferritin (□); human ferroportin (▲); human HIF 2α (●); human eALAS (▼); and porcine m-acon (◆). (B) Effect of mutation of the human L-ferritin IRE found in hereditary hyperferritinemia cataract syndrome (HHCS) on IRP1 binding. The wild-type (■) and an HHCS truncation mutant human L-ferritin IRE, G25-5′ (□) (Burdon et al. 2007), were synthesized, and their affinity of interaction with IRP1 was determined. (C) IRP1 affinity was determined for short L-ferritin (27 nt), Fpn (24 nt), and eALAS (24 nt) core IRE RNAs that lack flanking sequence. Equilibrium dissociation constants were normalized to that of L-ferritin and presented as Krel ± SEM (see Materials and Methods) or as the mean value for Kd. Each RNA was analyzed three times. In this panel, P-valueL-Ft and P-valueFpn refer to the comparison of the RNA of interest to the short 24–27-nt L-Ft or Fpn IREs. (N.S.) Not significant. The x-axis in panels A and B denotes the amount of active IRP1 protein.
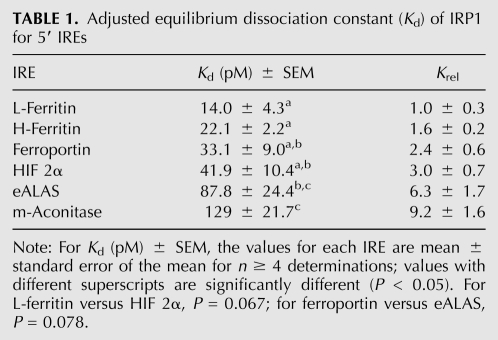
To convert apparent Kd to actual Kd values, it was necessary to determine the fraction of IRP1 protein that was active for RNA binding. Stoichiometric titration (Polach and Uhlenbeck 2002) was used to calculate the fractional activity of each IRP1 preparation using the fact that IRP1 binds the IRE with a 1:1 stoichiometry (Fig. 1D; Walden et al. 2006). The IRP1:IRE ratio was varied over nearly a 10-fold range with the RNA concentration at 35-fold higher than the apparent Kd. Binding was saturable and with breakpoint characteristic. From this analysis the activity of two IRP1 protein preparations was determined to be 54% and 57%. The mean activity of the protein preparations was used to adjust the apparent dissociation constant for the L-ferritin IRE, yielding a mean value of 14.0 ± 4.3 pM (Table 1).
TABLE 1.
Adjusted equilibrium dissociation constant (Kd) of IRP1 for 5′ IREs
IRP1 binds 5′ IREs in a hierarchical manner
While it has been suggested that differences in IRP RNA binding affinity could be a basis for differential regulation of mRNA with 5′ IREs, no studies to date have directly compared the binding affinities of IRP1 for various IREs. Using the EMSA assay described above, we determined the equilibrium RNA binding affinity of IRP1 for all mammalian 5′ IREs. The RNAs used were ∼65 nt long to allow for the inclusion of the extensive base-paired flanking sequence found in some IREs that can affect their function (Harrell et al. 1991; Dix et al. 1993). IRP1 bound these IREs with pM affinities that varied over a ninefold range. The L-ferritin IRE had the highest affinity (14.0 ± 4.3 pM), and the m-acon IRE (129 ± 21.7 pM) had the lowest affinity (Fig. 3; Table 1). In this and subsequent figures, the results are presented as Krel; that is, the ratio of RNA binding affinities for the experimental RNA relative to a control RNA, which was usually L-ferritin. It is of interest that the mRNAs with IREs exhibiting the lowest binding affinity for IRP1 (i.e., m-acon and eALAS) are less stringently regulated at the translational level as compared to ferritin mRNAs (Schalinske et al. 1998; Schranzhofer et al. 2006). Using the affinities determined here, the difference in Gibbs standard free energy of binding between the IRP1 complex with L-ferritin versus the m-acon IRE is calculated to be 1.4 kcal/mol (delCardayre and Raines 1995; Ryder et al. 2008). Thus, the sum of contributions of the various bond types and interactions giving rise to different IRE:IRP1 complexes is similar in magnitude to the thermodynamic contribution of a single H-bond.
Mutations in the L-ferritin IRE that decrease the affinity for IRPs lead to dysregulation of ferritin synthesis. These mutations cause the disease hereditary hyperferritinemia cataract syndrome (HHCS), and the effect on IRP binding affinity correlates with the clinical severity of the disease (Girelli et al. 1995; Allerson et al. 1999). Therefore, we analyzed IRP1 affinity for L-ferritin IREs containing mutations associated with HHCS in order to evaluate the relationship between IRE binding affinity and regulation in the context of the binding hierarchy for natural IREs. We chose two HHCS L-ferritin IRE mutants that would be predicted to not disrupt IRE secondary structure in the upper stem or C8 bulge region essential for high IRP1 affinity and whose binding characteristics have not been previously examined. We first examined the A37G mutant (A12G in the numbering system used herein) (Cremonesi et al. 2003) that substitutes a wobble base pair for the A12/U21 base pair and decreased affinity 1.4-fold, although this was not statistically significant (Table 2). Next, we examined a deletion mutant that alters the start site of transcription, resulting in a truncation of the 5′ side of a distal region of the lower stem (Burdon et al. 2007). This mutation decreased affinity nearly twofold (Fig. 3B; Table 2) (P = 0.045). These results suggest that subtle alterations in IRP1 affinity that are within the ninefold range observed for natural 5′ IREs are sufficient for altered regulation of mRNA translation in vivo.
TABLE 2.
Adjusted equilibrium dissociation constant (Kd) of IRP1 for wild-type and hereditary hyperferritinemia cataract mutant human L-ferritin IREs
To determine the extent to which the RNA binding hierarchy for natural IREs was dependent on the core IRE region, we eliminated the flanking sequences from L-ferritin, ferroportin, and eALAS IREs (see Supplemental Fig. S1 for predicted secondary structures of IREs with flanking sequences). Short 24- to 27-nt RNAs were synthesized for each of these three IRE, and their affinities of interaction with IRP1 were determined (Fig. 3C). Truncation of flanking sequence resulted in a five- to sevenfold loss of affinity of the L-ferritin, ferroportin, and eALAS IREs for IRP1 (RNAs 3-1 to 3-3). Importantly, although elimination of the flanking sequences reduced the affinity of interaction of these IREs with IRP1, the relative difference in binding was not greatly altered. This finding supports the concept that the core IRE region is sufficient to establish the RNA binding hierarchy with IRP1.
Multiple regions of the IRE contribute to the selective interactions with IRP1
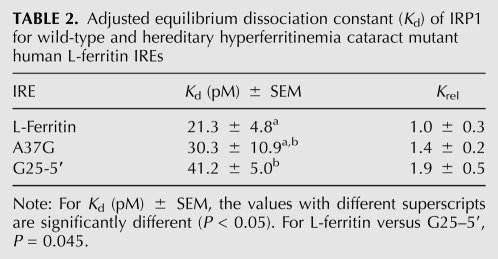
To further investigate the role of specific IRE regions in the binding to IRP1, we created chimeric IREs by swapping the flanking sequences between lower- and higher-affinity IREs. First, the L-ferritin and eALAS IREs were examined (Fig. 4A). Joining the eALAS flanking sequence with the C8 region and upper stem of L-ferritin IRE, lowered affinity relative to wild-type L-ferritin IRE by nearly sixfold and to an affinity that was not different from eALAS IRE (RNA 4-1). The fact that elimination of the L-ferritin flanking sequence had produced a similar effect (RNA 3-1) indicates that this region of eALAS IRE is not a negative or positive determinant for IRP1 recognition in the context of this chimera. For the reciprocal construct, the L-ferritin flanking region was joined to the C8 region and upper stem of eALAS IRE (RNA 4-2), and although the mean affinity for this RNA increased by 40% relative to wild-type eALAS IRE, the difference was not statistically significant. However, this chimera (RNA 4-2) bound significantly less well to IRP1 than did the wild-type L-ferritin, further demonstrating that sequence differences in the upper stem region between IREs dictate differential recognition by IRP1.
FIGURE 4.
Effect of sequence divergence in the IRE stem in modulating IRP1 affinity for 5′ IREs. Chimeric IREs were constructed by swapping the core IRE (upper stem and C-bulge) region with the flanking sequence between L-ferritin and eALAS IREs or ferroportin and eALAS IREs. The core IRE consensus structures are represented. (A) eALAS flanking sequence was joined with L-ferritin IRE, or L-ferritin flanking sequence was joined with eALAS IRE. (B) eALAS flanking sequence was joined with Fpn IRE, or Fpn flanking sequence was joined with eALAS IRE. Equilibrium dissociation constants were normalized to that of the long (66 nt) L-ferritin and presented as Krel ± SEM (see Materials and Methods) or as the mean-value for Kd. Each RNA was analyzed at least three times. P-valueL-Ft refers to the comparison of the RNA of interest to the long (66 nt) L-ferritin IRE. (N.S.) Not significant.
Second, chimeras of the ferroportin and eALAS IREs were examined (Fig. 4B). Unlike ferritin, these IREs do not have the additional U6 bulged nucleotide in the C8 region that can affect IRP binding (Bettany et al. 1992; Ke et al. 1998). When joined with the ferroportin IRE upper stem and C8 region, the eALAS IRE flanking sequence (RNA 4-3) had no impact on binding affinity relative to that of wild-type ferroportin IRE. This indicated that the ferroportin and eALAS IRE flanking sequences are functionally equivalent for binding IRP1. However, a chimera of the eALAS C8 region and upper stem joined to the ferroportin flanking regions (RNA 4-4) exhibited twofold lower affinity for IRP1 relative to the wild-type ferroportin IRE. That this chimera differs from ferroportin IRE only in the upper stem again demonstrates that sequence differences in the upper stem region of the core IRE are sufficient to establish differences in IRP1 binding. Consequently, our subsequent structure/function studies focused on the C8 and upper stem elements of the core IRE.
Divergent sequences in the C8 region optimizes IRP1 affinity for L-ferritin and eALAS IREs
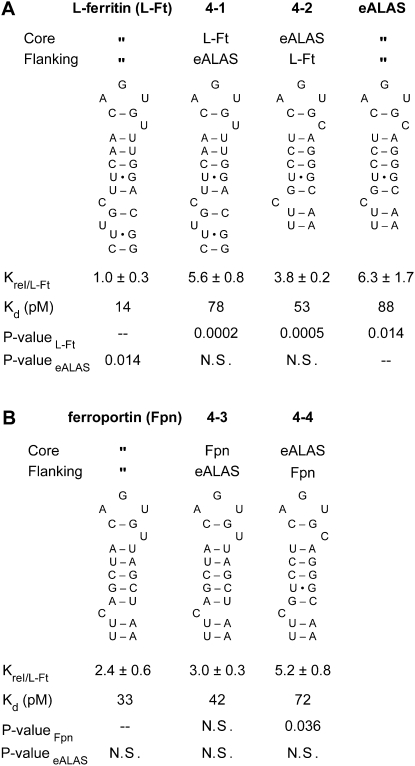
Previous studies have demonstrated that the C8 bulge is a key determinant for recognition of IRE by IRPs and that eight of 22 bonds between IRP1 and the ferritin IRE are made there. Four additional bonds occur on the 3′ side of the IRE helix below the C bulge region including a base-specific bond between Asn685 of IRP1 and G26. G26, present on the 3′ side of the stem just below and opposite C8, is a nucleotide uniquely conserved among ferritin IREs (Fig. 5A; Walden et al. 2006). Furthermore, it has been shown that the presence of a unique unpaired U (U6) 2 nucleotides (nt) to the 5′ of the C8 region enhances IRP binding to the L-ferritin IRE (Ke et al. 1998; Leipuviene and Theil 2007). Importantly, unpaired nucleotides can act as structural hinges or sites of protein contact. The flipping of unpaired nucleotides between an intrahelical versus extrahelical orientation can be a basis for regulation (Reiter et al. 2004). In this regard, it is of interest that the orientation of C8 differs depending on whether the IRE is free or bound to IRP1 (McCallum and Pardi 2003; Walden et al. 2006). Since the kinetics and extent of base-flipping can be affected by the nucleotide sequence surrounding the unpaired nucleotide (Morden et al. 1983; Hart et al. 2005; Barthel and Zacharias 2006), we investigated the role of the sequence and structure of the C8 region on IRP1 binding.
FIGURE 5.
Effect of sequence and structure divergence in the C8 region of IREs as a basis for differential recognition by IRP1. (A) Single and multiple base substitutions or a nucleotide deletion were made in the L-ferritin IRE, and their affect on affinity was determined. Base-pair substitutions were made at 10/23 and 9/24 for Fpn IRE (B) and eALAS IRE (C). Equilibrium dissociation constants were normalized to that of L-ferritin, presented as Krel ± SEM for at least three determinations for each IRE. (A) P-valueL-Ft, (B) P-valueFpn, and (C) P-valueeALAS refer to the comparison of the RNA of interest to the long wild-type IREs (L-ferritin and ferroportin, 66 nt; eALAS, 65 nt). (N.S.) Not significant. Nucleotide changes relative to wild type are circled.
First, we reasoned that the base-specific contact that forms between G26 N2 and IRP1 could be a basis for the higher binding affinity of the ferritin IREs (Walden et al. 2006). A G26A substitution eliminated the base-specific primary amine (N2 of guanine) yet had no effect on IRP1 affinity (Fig. 5A, RNA 5-1). Likewise, a U5C substitution would H-bond N2 of G26 with O2 of C5 and would be predicted to alter the electrostatic potential in the minor groove (Xu et al. 2007). However, its affinity (RNA 5-2) was no different from wild type. We next considered whether alternative base-pairing of G26 with U6 as opposed to G26 with U5 could be a modulator of binding, perhaps during the induced fit of the RNA with IRP1. A double substitution—U6C and G26A—prevents potential base-pairing between U6 and G26 by restricting formation to the U5•G26 but not U6•G26 base pair, but had no impact on affinity (RNA 5-3). An interesting outcome of this mutant is that U6 can be replaced with C without affecting binding, which supports the concept that U6 has a structural role in the IRE, perhaps in allowing C8 to be optimally positioned for binding. Some ferritin IREs in lower organisms have a C substitution for U6 (Piccinelli and Samuelsson 2007). Others have shown that deletion of U6, found only in ferritin IREs, leads to a modest reduction in IRP1 affinity and regulates IRE selectivity among IRPs (Ke et al. 1998). IRP1 affinity was reduced threefold by deletion of U6 (RNA 5-4). Including the G26A substitution in the ΔU6 mutant did not lead to any additional change in binding (RNA 5-5). Substitution of sequence from eALAS IREs into L-ferritin IREs (RNA 5-6) in this region did not reduce affinity beyond that of U6 deletion alone (RNA 5-4). We conclude that unlike U6, G26 is not essential for optimal binding of IRP1 with the L-ferritin IREs.
We further sought to explain the role of the C8 region in the affinity differences between IRP1 and the shorter RNA of the IREs of ferroportin and eALAS, which differ only in the base pairs of the upper stem (Fig. 3C, RNA 3-2 versus RNA 3-3). A double base-pair substitution of U10•G23 and G9/C24 into ferroportin IRE (RNA 5-7) made this region conserved with eALAS IREs yet had no impact on affinity and was no different from single base-pair substitutions alone (Fig. 5B, RNAs 5-8 and 5-9). The converse base-pair substitution of G10/C23 and A9/U24 in eALAS IREs eliminated the wobble base pair at 10/23 and made these positions conserved with ferroportin IRE (Fig. 5C). Although the double base-pair substitutions (RNA 5-10) reduced Krel by nearly 70%, this difference failed to reach statistical significance (P = 0.14). Furthermore, the single base-pair substitution U9/A24 made this position conserved with ferritin and likewise was not bound differently than wild type (RNA 5-13). However, a C24U substitution made G9•U24 a wobble base pair and decreased IRP1 affinity fourfold relative to wild-type eALAS (RNA 5-14). Furthermore, compared to the double base-pair substitution (RNA 5-10), affinity was reduced by the single base-pair substitutions A9/U24 (RNA 5-12; P = 0.017), U9/A24 (RNA 5-13; P = 0.0016), and G9•U24 (RNA 5-14; P = 0.0008) into wild-type eALAS IREs. Thus, IRP1 affinity can be modulated through sequence divergence not only in the region just below C8 (e.g., the presence or absence of U6) or just above C8. Since these base pairs (10/23 and 9/24) are not predicted to bond to IRP1, they could affect binding affinity through changes in the thermodynamic stability of the C8 region that have an impact on the induced-fit mechanism. However, since these differences in affinity did not correlate with the predicted thermodynamic stability of neighboring base pairs (data not shown), it appears that the nucleotide sequence surrounding C8 alters binding through other means.
These mutations further substantiate and delineate the role of the C8 region in the selective recognition of IREs by IRP1. The unique U6 bulge 5′ of C8 that is conserved in ferritin IREs makes a significant but not predominant contribution to IRP1 recognition: about one-third of the ninefold difference in affinity between L-ferritin and m-acon can be attributed to this structural determinant. Interestingly, residue 6 does not have to be a U as insertion of a C, in the context of the G26A mutation (RNA 5-3), was without effect on binding. In the case of eALAS IREs, mutating nucleotides proximal to the C8 bulge in the upper stem so that this region matched the sequence in ferroportin increased affinity to a value not different from ferroportin. Since both of these IREs lack the unique U bulge of the ferritin elements, it suggests that multiple evolved differences between natural IREs in the C8 region impact the bonds between C8 and IRP1 and contribute to the establishment of the binding hierarchy.
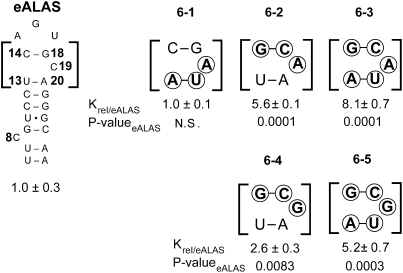
Role of base-stacking between the base pair of the terminal loop and upper stem
Because the ability to discriminate between ferritin and eALAS IREs is essential for the preferential formation of heme in red cells and the dysregulation of IRP1 therein causes anemia (Wingert et al. 2005; Camaschella et al. 2007), the eALAS IRE was further examined. Initially, the upper stem sequence adjacent to the terminal loop was investigated because we hypothesized that the variation of base-stacking and composition in this region might alter the thermodynamic stability of the pseudo-triloop, and base-pair substitutions in the upper stem can significantly affect IRP1 recognition (Leibold et al. 1990). The conserved C14/G18 base pair of the terminal loop stacks with the nonconserved nucleotides of the A13/U20 base pair of the upper stem, and the intervening nucleotide U19 is flipped/bulged out of the ferritin IRE helix (Laing and Hall 1996; Addess et al. 1997; Gdaniec et al. 1998; McCallum and Pardi 2003; Walden et al. 2006). Since our results indicate that IRP1 binds IREs with R13/Y20 (e.g., L-ferritin) with higher affinity than those with Y13/R20 (e.g., eALAS), we investigated whether IRP1 prefers the R13/Y20 base-pair order of the upper stem.
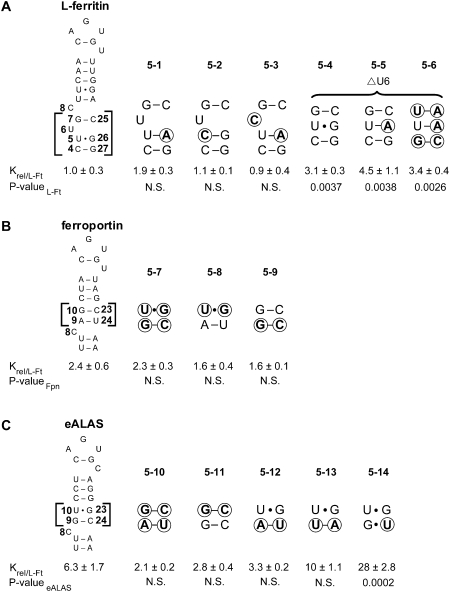
Transversions were introduced in eALAS IRE at the 13/20 base pair and at the adjacent base pair in order to test whether base-stacking effects could have a role in establishing a preference for the order of the 13/20 base pair. A cosubstitution at N19 was made that changed C to A or G to prevent terminal loop rearrangement by unwanted 14/19 base-pairing. IRP1 affinity was unaffected by transversion of the U13/A20 base pair of the upper stem with A19 (RNA 6-1). However, transversion of the terminal loop base pair alone reduced affinity sixfold with A19 (RNA 6-2) as observed by others (Henderson et al. 1996), and transversion of both base pairs further decreased affinity to eightfold less than wild type (RNA 6-3). Since base-stacking occurs across the N19 bulge (Hall and Tang 1998; Walden et al. 2006), it is of interest that IRP1’s affinity for these RNAs is weakly related to the thermodynamic stability of the base-pair substitutions (e.g., RNA 6-3 versus wild-type eALAS, ΔΔG°37 = −0.16 kcal/mol) (Xia et al. 1998). IRP1 affinity was diminished about threefold by transversion of the terminal loop base pair when N19 was G (RNA 6-4), indicating that IRP1 is sensitive to which purine base is present at position 19 (RNA 6-4 versus 6-2, P = 0.0007). Transversion of both base pairs with G19 diminished affinity fivefold, which is significantly different from in an A19 context, further supporting a limited role for thermodynamic stability in this region on IRP1 binding and also a role for purine identity at position 19 in modulating IRP1 affinity (RNA 6-5 versus 6-3, P = 0.050), which may affect binding of IRP1 to the limited number of IREs containing A at N19 (Piccinelli and Samuelsson 2007).
Role of base pairs closing the C19 bulge and terminal loop
A second mechanism through which upper stem sequence could affect IRE function relates to the impact of base pairs closing terminal loops and bulged nucleotide regions on modulating RNA structural stability (Antao and Tinoco 1992; Wu and Turner 1996). Since the base pairs 13/20 and 12/21 close the terminal loop and N19 bulge and are conserved within but not across IRE families, the impact of H-bond number and stacking stability of these base pairs on the affinity of IRP1 for 5′ IREs was investigated. To accomplish this, the 13/20 and 12/21 base pairs were systematically mutated in the eALAS IRE, and all possibilities for base-pairing and stacking involving these nucleotides were examined. While none of the mutants differed from the wild-type eALAS IRE, IRP1 affinity appeared to be sensitive to variation in Watson–Crick base-pairing or to base-stacking (e.g., RNA 7-4 versus RNAs 7-5, 7-6, or 7-7; P < 0.05) at these positions when compared among the mutants (Fig. 7, see below); however, binding affinity failed to correlate to the predicted thermodynamic stability of the 13/20 and 12/21 base pairs of these RNAs (Mathews et al. 1999). Interestingly, these results indicate that these base pairs of the natural 5′ IRE are optimized for high-affinity binding and that they are represented by RNAs 7-2 (m-acon), 7-11 (H- and L-ferritin), 7-9 (ferroportin), and 7-14 (HIF 2α), suggesting that this region of the IRE has evolved to promote maximal binding for each IRE family. Further support for this conclusion comes from the fact that insertion of the eALAS or ferroportin sequences for 13/20 and 12/21 base pairs into L-ferritin was without effect (data not shown). Taken together, we conclude that the H-bonding and base-stacking of these base pairs closing the terminal loop have a minor role in IRP1 binding in the context of the eALAS IRE.
FIGURE 7.
Effects of base-stacking and H-bond number in the upper stem on IRP1 binding. Nucleotides at the 13/20 and 12/21 positions were systematically substituted with all other 15 Watson–Crick base-pair combinations. Across a row, base pair 12/21 is varied, while down a column, 13/20 is varied. Equilibrium dissociation constants were normalized to that of long (65 nt) eALAS IRE (P-valueeALAS), presented as Krel ± SEM for n = 3 determinations per RNA. IRP1 affinity for RNA 7-4 differs from RNA 7-5 (P = 0.034), RNA 7-6 (P = 0.033), and RNA 7-7 (P = 0.015). (N.S.) Not significant. Nucleotide changes relative to wild type are circled.
Investigating the roles for nucleotide 19 in regulating IRP1 affinity
Given our observation that IRP1 affinity was sensitive to N19 identity (Fig. 6), we considered whether N19 might have an unrecognized role in contacting IRP1 or in modifying IRE structure to optimize binding. For example, in the majority of higher-affinity IREs (L-ferritin, ferroportin, and HIF 2α), N13 could base-pair with N19. Such pairing would significantly alter upper stem structure by eliminating the thermodynamically preferred N13/N20 base pair (Fig. 1A). Since this would likely alter orientation of the pseudo-triloop and reduce the affinity of interaction with IRP1, it could serve as a mechanism to limit the strength of interaction of the high-affinity IRE. Excessive repression of 5′ IRE-containing mRNA has pathological consequences (Wingert et al. 2005; Camaschella et al. 2007).
FIGURE 6.
Effect of altering base-stacking at the junction of the terminal loop and the upper stem of the eALAS IRE. The base pairs 14/18 and 13/20 of the eALAS IRE were systematically flipped in the presence of A19 or G19. Equilibrium dissociation constants were normalized to that of eALAS IRE, presented as Krel ± SEM for at least three determinations for each RNA. P-valueeALAS refers to the comparison of the RNA of interest to the long (65 nt) eALAS IRE. (N.S.) Not significant. Nucleotide changes relative to wild type are circled.
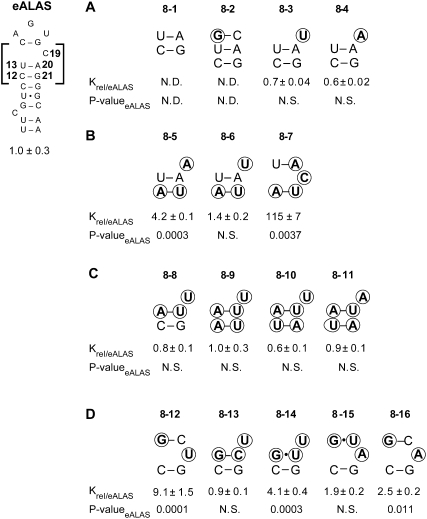
We first examined the impact of the loss of N19 through deletion or base-pairing and N19 identity on IRP1 binding. Deleting C19, as others have done (Jaffrey et al. 1993), or pairing the base lowered binding affinity to the nanomolar range, which supports the concept that N19 has an important role in positioning the terminal pseudo-triloop for optimal interaction with IRP1 (RNAs 8-1 and 8-2). However, substitutions in place of C19 in the eALAS IRE, did not alter affinity, demonstrating indifference to base identity for protein recognition in this sequence context (RNAs 8-3 and 8-4). Furthermore, U19A substitution (RNA 8-4) created the potential for the alternate base pair U13/A19 but was without effect. Because there appeared to be small impact of H-bond number and base-stacking at base pair 12/21 on IRP1 binding (Fig. 7), we tested the same hypothesis with A12/U21 substitution for C12/G21. In this background, the C19A substitution (RNA 8-5) diminished binding affinity fourfold, suggesting the formation of a nonconsensus IRE structure and further indicating that IRP1 affinity is sensitive to predicted thermodynamic changes in the upper stem (Fig. 8A,B, cf. RNAs 8-4 and 8-5). This conclusion was supported by comparison of RNA 8-6, where the C19U substitution was in the same stem as RNA 8-5, and which does not support possible N13/N19 base-pairing, was without effect.
FIGURE 8.
Investigating the role of an N19 versus N20 bulge on IRP1 recognition. The impact of the N19 bulge, its nucleotide identity, and capacity to pair with N13 and the impact of replacing the bulge at N19 with one at N20 on IRP1 binding was examined using the eALAS IRE. Equilibrium dissociation constants were normalized to that of eALAS IRE, presented as Krel ± SEM for at least n = 3 determinations for each RNA. (A) Single nucleotide substitutions were made at N19 or N19 was deleted or paired in the eALAS IRE. For RNAs 8-1 and 8-2, the affinity was not measurable by the assay and is indicated by N.D. (not determined). (B) For the A12/U21 substitution in pace of C12/G21, single nucleotide substitutions were made at C19 (RNA 8-5 and 8-6) or C19 and A20 (RNA 8-7). RNA 8-7 favors formation of an N20, not N19, bulge. (C) A transversion was introduced at U13/A20 with predicted N12/N21 base-pairing, and single nucleotide substitutions were made at C19. (D) U13G substitution was made in eALAS IRE, and N19 was either U or C; N20 was U, C, or A. P-valueeALAS refers to the comparison of the RNA of interest to the long (65 nt) eALAS IRE. (N.S.) Not significant; (N.D.) not determined, in that the EMSA results indicated that the binding affinity was in the nanomolar range. Nucleotides changes relative to wild type are circled.
In order to distinguish between RNA structures with the two alternatively bulged nucleotides A19 or A20 (RNA 8-5), we made an additional single nucleotide substitution A20C (RNA 8-7) predicted to strongly favor a U13/A19 base pair. IRP1 bound this RNA with 115-fold lower affinity than the wild-type eALAS IRE, suggesting that with C20 (RNA 8-7), the U13/A19 base pair may not form, leading to terminal loop expansion and a shortened upper stem critical to high-affinity IRP1 binding (Leibold et al. 1990; Bettany et al. 1992; Kikinis et al. 1995). We tested the potential for N13/N19 base-pairing in other sequence contexts, and surprisingly, IRP1 affinity was unaffected by predicted competition between U19 and U20 for A13, suggesting that 3′-strand base-stacking stability modifies the potential for a nonconsensus N20-bulged structure (Fig. 8C; RNAs 8-8, 8-9, 8-10, 8-11).
We further investigated whether IRP1 was capable of recognizing a predicted Y20-bulged IRE using a scenario of competitive H-bonding between Y19 and Y20 for G13 (Fig. 8D). We reasoned that the difference in H-bond number between G/C and G•U base pairs could allow preferential formation of G13/Y19 or G13/Y20 base pairs (Varani and McClain 2000). IRP1 bound the eALAS IRE double mutant (G13/C19)•U20 (RNA 8-12) with 10-fold lower affinity than the inversely competitive mutant (G13/C20)•U19 (RNA 8-13), indicating, as did RNA 8-7, that the proposed N20 IRE can impair recognition by IRP1. Interestingly, the (G13•U20)•U19 mutant (RNA 8-14) that has the capacity to display G13/Y20 or G13/Y19 base pairs bound with fivefold lower affinity than that of (G13/C20)•U19 (RNA 8-13), which should favor G13/Y20 pairing. Surprisingly, when the (G13•U19)-A20 substitution was examined, which is predicted to favor an A20 bulge (RNA 8-15), the loss of affinity observed with the (G13/Y19)•U20 competitive base-pairing (RNA 8-12 and 8-14) was rescued. Furthermore, another mutant with a predicted N20 bulge exhibited an affinity that was not greatly different from wild-type eALAS (RNA 8-16). We note that because the relative affinity of the G13•U19 and G13/C19 mutants with A20 is greater than the similarly predicted structure that would form two fewer H-bonds, differences are likely to exist between these nonnatural IRE structures (RNAs 8-15 and 8-16 versus 8-7). Nevertheless, our findings suggest that in specific sequence contexts, an N20 bulge can replace an N19 bulge. We conclude that the consensus secondary structure for IRE does not fully represent the structures recognized by IRP1.
DISCUSSION
Our study of the requirements of IRP1 for binding iron-responsive elements leads to the following major conclusions. First, IRP1 binds to IREs from 5′-IRE-containing mRNAs in an RNA binding hierarchy that suggests binding affinity is a significant determinant of differential translational control of these mRNAs. Second, multiple determinants within the IRE outside of the conserved terminal loop and C8 nucleotides determine IRP1 binding affinity. Third, while sequences flanking the IRE can substantially and selectively affect recognition by IRP1, they are not required for establishing the binding hierarchy. Fourth, IRP1 can bind with high affinity to RNA stem–loops that deviate significantly in structure from the consensus IRE, suggesting the existence of additional mRNAs in its regulon. Taken together, our findings provide new insights concerning how IRP1 differentially recognizes natural IREs, identify parameters that have been selected against during the evolution of these novel regulatory elements, and indicate that the accepted consensus IRE structure does not represent the only structures that can be recognized with high affinity by IRP1.
Establishing the IRP1 RNA binding hierarchy
Our study demonstrates that it is the summation of the impact of multiple conserved and nonconserved regions within IRE families that is integrated in determining the final strength of interaction with the binding protein. Previous studies lead to the view that, similar to tRNAs (Giege et al. 1998), the IRE is a scaffold that allows for proper presentation of critical residues, identity elements in tRNAs, required for protein binding (Walden et al. 2006). For the IRE the key elements required for recognition are the unpaired C8 in the stem and the AGU pseudo-triloop nucleotides. Although tRNA recognition by tRNA synthetases is heavily dependent on key identity elements, it is also clear that these RNAs contain minor elements that fine-tune recognition (for review, see Giege et al. 1998). However, with the exception of studies on the U6 bulge in ferritin IREs (Ke et al. 1998; Leipuviene and Theil 2007), the role of natural variation in IRE stem sequence in influencing IRP binding has been largely ignored. Our present study establishes that the sequence and structure of the nonconserved segments of the IRE are capable of significantly impacting IRP1 binding and firmly establishes that these regions of the IRE stem are not inert contributors to IRE recognition as has been previously suggested (Addess et al. 1997). Similar observations have been made regarding the selective interaction of tRNAs with their cognate synthetase (McClain et al. 1988; Frugier et al. 1998; Giege et al. 1998; Beuning and Musier-Forsyth 1999). Given that the range of binding affinity defined by the limits of the IRP1 binding spectrum can be explained by differences in the energetics of binding equivalent to one hydrogen bond, and does not involve the degree of discrimination of binding observed in other systems (Beuning and Musier-Forsyth 1999; Opperman et al. 2005), we suggest that modest effects of multiple IRE regions on the strength of a limited number of bonds or other interactions (e.g., hydrophobic, van der Waals) with the binding protein function to establish a regulatory hierarchy. Furthermore, since some bonds between IRP1 and nonconserved regions of the IRE stem are dispensable (e.g., G26 and N20), we suggest that there is plasticity in the IRP1/IRE interaction and that loss of individual bonds, and their compensation by addition of new contacts or alteration in the strength of remaining bonds, is an innate feature of the regulatory hierarchy in this system. Coupled with our demonstration that mutation of regions of the IRE stem not known to contact IRP1 can significantly alter binding affinity, as has also been observed in tRNA/tRNA synthetase interactions (Frugier et al. 1998), we suggest that a regulatory binding hierarchy is established through impacts of the stem sequence or structure on the strength of bonds between C8 and to a lesser extent the pseudo-triloop and IRP1 or through an impact on an induced-fit mechanism of IRE recognition.
Our studies suggest that the heterogeneity of stem sequence and structure and its effect on the binding hierarchy act largely through modulation of contacts between IRP1 and C8 given the larger impact caused by mutations of the stem in this region as opposed to base pairs proximal to the pseudo-triloop. Previous studies demonstrated that C8 is essential for IRP1 binding and serves both as a site for multiple contacts with the binding protein and also as a hinge between the upper and lower stem regions, allowing essential rearrangement of the IRE during the induced-fit binding process (Laing and Hall 1996; Addess et al. 1997; Ke et al. 1998; Walden et al. 2006). In addition, the base pairs surrounding C8 and this cytosine itself are among the most structurally dynamic regions within the IRE (Addess et al. 1997; Gdaniec et al. 1998; McCallum and Pardi 2003). Importantly, C8 can be oriented toward the major groove of the IRE stem in the free RNA but flipped out allowing for multiple protein contacts when in complex with IRP1 (McCallum and Pardi 2003; Walden et al. 2006). Given that mutations in the stem above C8 produced as much as a 14-fold difference in binding affinity (Fig. 5C, cf. RNAs 5-10 and 5-14) and likely contribute to the difference in affinity of eALAS and ferroportin IREs for IRP1, yet these nucleotides are not predicted to contact IRP1, we suggest an impact of base pairs proximal to C8 on the structural dynamics of this critical binding residue. Support for this proposal comes from studies demonstrating that sequence proximal to a bulge can affect the kinetics and energetics of flipping of such unpaired nucleotides (Morden et al. 1983; Hart et al. 2005; Barthel and Zacharias 2006). For instance, base-flipping of U80 underlies the central role of U6 RNA in pre-mRNA splicing (Reiter et al. 2004). Alternatively, the presence of noncanonical (e.g., G•U) base pairs in the helix between C8 and the pseudo-triloop could alter the relative positioning of these critical sites of contact for IRP1 and in this manner impact bond strength and binding affinity. Noncanonical base pairs impact helix geometry in the tRNA acceptor loop and can make key contributions to tRNA identity (McClain et al. 1988; Musier-Forsyth and Schimmel 1992). However, the presence of conserved G•U base pair(s) in high- and low-affinity IREs suggests any role of non-Watson–Crick pairs in IRP binding is likely influenced by other IRE-specific determinants. Taken together, the conserved differences in nucleotide sequence and structure of the C8 region between IRE families and its possible impact on C8 conformational dynamics may represent a unique mechanism for fine-tuning the action of regulatory RNA binding proteins in controlling gene expression.
We also demonstrated that the RNA sequence flanking the IRE is a positive determinant for IRP1 recognition not just for ferritin, as had been previously observed (Harrell et al. 1991; Dix et al. 1993), but also for the ferroportin and the eALAS IRE. Addition of flanking sequence improved the binding affinity of each of the short IREs examined (Fig. 3, cf. panels A and C). The similar impact of the addition of the homologous flanking sequence for the three IREs examined suggests a general effect perhaps in stabilizing the stem–loop structure or in enhancing binding through an electrostatic mechanism. However, our results with chimeric IREs also support the concept of a unique structural and functional integration of the flanking and core regions of the ferritin IRE (Harrell et al. 1991; Dix et al. 1993). Although the extended region of base-pairing of the ferritin IRE provided by the flanking region appears to be a recent event evolutionarily (Piccinelli and Samuelsson 2007), the concept that the flanking and core regions of IREs coevolved to facilitate their optimal function merits further attention.
Biological importance of an IRE hierarchy and its implications
One central conclusion of our findings is that IREs are not recognized identically by IRP1, thereby providing a mechanistic basis for a hierarchy of translational control for 5′-IRE-containing mRNAs. Several studies have demonstrated that 5′-IRE-containing mRNAs including eALAS, m-acon, and ferritin in mammals and succinate dehydrogenase in flies are differentially regulated at the translational level (Schranzhofer et al. 2006; Surdej et al. 2008; for review, see Wallander et al. 2006). In reference to these previous studies, two questions arise concerning our present findings. First, is the difference in RNA binding affinity that we observed for natural 5′ IRE sufficient, on its own, to program differences in translational regulation? We approached this by determining the affinity of binding of IRP1 for two IRE mutants from patients suffering with HHCS, a type of cataract cause by over-accumulation of ferritin in the eye. One of the mutants studied involved a truncation of the 5′ side of the L-ferritin IRE. A plausible explanation concerning the threefold increase in serum ferritin concentration and opaqueness of the eye in the individual with this mutation (Burdon et al. 2007) is that the twofold loss of affinity for IRP1 that we observed is sufficient to cause dysregulation of L-ferritin synthesis. Along this line of reasoning, Allerson and associates have previously analyzed a range of HHCS mutants and observed a strong correlation of clinical severity of the disease with loss of RNA binding affinity (Allerson et al. 1999). Interestingly, their study demonstrated that the Pavia 2 mutant bound IRP1 with only a 1.5-fold difference in affinity relative to wild-type ferritin. A key question that arises from these comparisons made here concerns the role of IRP2 in the dysregulation of ferritin synthesis in these HHCS cases. While the exact response of specific IRE-containing mRNAs will depend on issues such as tissue-specific differences in IRP1 versus IRP2 binding activity, it is of interest to note that for several of the HHCS mutants, including Pavia 2, a similar effect on IRP1 and IRP2 binding was observed. Furthermore, these investigators observed that in the HHCS mutants analyzed, with affinity ranging from 1.5-fold to 2200-fold lower than the wild-type ferritin IRE, the serum ferritin values were highly correlated to the Krel changes for both IRP1 and IRP2, and the region of greatest responsiveness of serum ferritin occurred when Krel for the mutants varied from 1 to 10 (Allerson et al. 1999). Given these findings, we conclude that the differences in RNA binding affinity of IRP1 for natural 5′ IRE is physiologically relevant and is a prime determinant of the hierarchical regulation of translation of IRE-containing mRNA.
Second, what is the limit for lowest affinity IRE/IRP1 interaction that would still allow regulation? Although the m-acon IRE exhibits the lowest affinity within the binding hierarchy, and m-acon mRNA translation is iron regulated in cells (Schalinske et al. 1998), it is not clear if this represents the limit of IRP-dependent regulation. Factors such as the level of IRP binding activity, the concentration of mRNA ligand and the presence of ancillary factors that may enhance IRP binding (dos Santos et al. 2008) need to be considered. Clearly, cells can contain a latent pool of IRE binding activity, especially for IRP1, and the substantial activation of this pool, as can occur with NO or oxidative stress or prolonged iron deficiency, may well be sufficient to act on RNA targets containing novel IREs with weaker binding affinity than the m-acon IRE. In the case of m-acon mRNA, which is weakly repressed under normal growth conditions, translation can be strongly repressed given an appropriate degree of iron deficiency (Schalinske et al. 1998). Similar observations have been made for succinate dehydrogenase subunit b in Drosophila (Surdej et al. 2008). Hence, it seems reasonable to suggest that as-of-yet-undiscovered mRNAs containing novel 5′ IRE may extend the IRP1 regulatory hierarchy beyond the limit currently set by m-acon, possibly including the putative IRE identified by bioinformatics approaches (Sanchez et al. 2006).
Third, the implications of our findings for the regulatory fate of IRE-containing mRNAs clearly also depends on IRP2. Previous studies have established differential impacts of the C8 bulge region and the terminal loop in the recognition of IREs by IRP1 versus IRP2. IRP1 is less affected by changes in the base-pair combinations at N14/N18 but more sensitive to alterations at N15 and N17 than IRP2 (Henderson et al. 1994, 1996; Butt et al. 1996). Furthermore, the fact that IRP2 binding is more sensitive to deletion of the ferritin-specific U6 bulge than IRP1 argues that the altered structure of the C8 region in ferritin versus nonferritin IRE provides one basis for selective recognition of ferritin versus other IREs (Ke et al. 1998). It remains to be determined if IRP2 is more strongly affected by alterations in stem sequence around C8 in nonferritin IRE than is IRP1. Thus, a full appreciation for the role of nonconserved regions of the IRE stem in the regulatory hierarchy of IRE-containing mRNAs awaits a detailed analysis of their impact on IRP2 binding.
Summary
Our study of the interaction of IRP1 with 5′ IREs demonstrates that IRP1 binds these IREs in an affinity spectrum, suggesting a basis for their selective regulation and predicting a differential impact of dysregulation for a given IRE. Two scenarios for IRP dysregulation are possible, each giving rise to a different outcome. For example, when IRP activity is suppressed through the genetic ablation of an IRP (Meyron-Holz et al. 2004; Galy et al. 2005, 2008) or inactivation of the IRE through mutation (e.g., in HHCS), this would lead to constitutive activation of the translation of mRNA with 5′ IREs with a preferential response for mRNAs that are highly repressed under normal conditions such as the ferritins. On the other hand, over-activation of IRP1, as is predicted to occur in diseases decreasing Fe-S cluster biogenesis or enhancing oxidative stress (Clarke et al. 2006), could be expected to shift the equilibrium of the system causing lower-affinity IREs (e.g., eALAS) to be repressed to a level similar to that observed for higher-affinity IREs under normal conditions. Indeed, over-repression of eALAS by IRP1 is the basis for the defect in the zebrafish shiraz mutant (Wingert et al. 2005). Taken together, our work indicates that multiple aspects of IRE sequence and structure contribute to the generation of a regulatory hierarchy orchestrated by IRP, whose mRNA targets may not be all identified, and that allows for mRNA fate to be controlled over a broad range, thereby contributing to the combinatorial control of cellular function by these uniquely regulated RNA binding proteins.
MATERIALS AND METHODS
IRP1 preparation
Rabbit IRP1 protein expressed in Saccharomyces cerevisiae was purified as described (Brown et al. 1998) and stored in single-use aliquots at −80°C in 50 mM HEPES (pH 7.4), 50 mM NaCl, 5% glycerol, and 1 mM DTT in concentrations of 12 to 81 μM.
RNA production
All RNAs used in this study are listed in Supplemental Table S1. RNAs were transcribed using plasmid or oligonucleotide templates. Plasmids were constructed for synthesis of 63- to 66-nt RNAs representing the IRE regions of H- and L-ferritin, m-acon, ferroportin, eALAS, and HIF 2α. Briefly, the pSP64 poly(A) plasmid (Promega) was engineered using oligonucleotides and PCR to contain the T7 RNA polymerase promoter juxtaposed just 5′ of each IRE region. Plasmid sequences were verified by DNA sequencing. Mutant IREs were made using the oligonucleotide-based method (Milligan et al. 1987). Oligonucleotides were purified by denaturing polyacrylamide gel electrophoresis (PAGE) with their mass confirmed by mass spectrometry (Integrated DNA technologies). Transcription was performed using the Riboprobe system-T7 kit (Promega). The specific radioactivity of RNA was typically 50,000 dpm/fmol RNA, labeled using [α-32P]UTP (3000 Ci/mmol or 6000 Ci/mmol; Perkin Elmer) or [α-32P]CTP (3000 Ci/mmol; Perkin Elmer) for H-ferritin and ferroportin RNA. The corresponding nonlabeled nucleotide was added at a concentration 1.5-fold the number of moles of [32P]nucleotide. RNA was purified by 10% or 12% PAGE in 8 M urea and eluted by rocking in RNase-free 0.3 M sodium acetate (pH 5.2), followed by precipitation with ethanol and quantification by scintillation counting.
Electrophoretic mobility-shift assays
Gel-shift assays were performed as described previously with the following modifications (Barton et al. 1990). RNA was folded by heating for 2 min in RNase-free 10 mM HEPES, 0.1 mM EDTA (pH 8.0), 1 mM MgCl2, and 20 mM NaCl at 90°C, followed by cooling on ice for at least 3 min. Binding reactions were performed in 20 mM HEPES (pH 7.4), 1 mM magnesium acetate, 75 mM KCl, 5% ultrapure glycerol, 20 μg/mL acetylated BSA, and 2 mM DTT. Reactions were assembled at room temperature and incubated for 10 min at 37°C followed by addition of heparin (Sigma) to 0.5 mg/mL. Bound and free were separated as described (Barton et al. 1990; Batey and Williamson 1996) except that the samples were immediately loaded onto a 4% polyacrylamide gel (60:1; acrylamide: bis-acrylamide) that had been pre-run for 30 min at 14 V/cm in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA at pH 8.0) at 4°C. Samples were loaded at 8 V/cm, after which the voltage was raised to 22 V/cm for 25 min. Gels were dried essentially as described and exposed to a phosphor screen and quantified using OptiQuant software (Perkin Elmer). Bound RNA was quantified as a percentage of the total of bound and free RNA species.
Quantification of protein activity assay
To determine the RNA binding activity of each preparation of recombinant IRP1, a stoichiometric titration experiment (Batey and Williamson 1996; Polach and Uhlenbeck 2002) was performed using lower specific activity (8000 dpm/fmol) L-ferritin IRE. The [32P]RNA concentration was 0.6 nM, and the IRP1/IRE ratio varied over nearly a 10-fold range. RNA and IRP1 were incubated for 30 min at 37°C followed by addition of heparin to 0.5 mg/mL. The two IRP1 protein preparations exhibited protein activities that were 54% ± 8% and 57% ± 11% (n = 4–5 determinations) (data not shown).
Equilibrium dissociation constant determination assay
RNA binding affinity was measured by the titration of IRP1 100-fold above and below the Kd. The RNA was kept at a low concentration, ∼150-fold lower than the affinity of IRP1 for that IRE. For the wild-type IREs, two preparations of IRP1 and at least two preparations of RNA were used to determine the absolute Kd. Because the affinity of IRP1 for H-ferritin IRE was not affected by protein preparation, a single protein preparation was used for the mutant RNAs. In a time-course experiment, 99% of maximal binding of the L-ferritin IRE to IRP1 occurred with a 5:1 molar ratio (IRP1:RNA) within 10 min (data not shown). Consequently, the use of an IRP1:RNA ratio that was 150:1 was deemed sufficient to achieve equilibrium conditions for 10-min binding assays.
Binding calculations and statistical analysis
The stoichiometric titration data were fit using dual linear regression (Polach and Uhlenbeck 2002), and the saturation breakpoint was used as a direct estimate of the fractional activity of IRP1. The binding data from individual experiments were fit to a single site-binding model, where Y = (Ymax * [IRP1])/(Kd + [IRP1]) using GraphPad Prism 4.02 (Graphpad Software, Inc.). The residuals resulting from the best-fit curves of the nonlinear regression were used to create a Q-Q plot to check for normal distribution using R 2.4.1. Visual assessment of the Q-Q plot indicated that the residuals were normally distributed. The relative binding affinity (Krel) for each RNA was calculated by dividing the Kd for each RNA by the Kd of either L-ferritin or the eALAS IRE. The standard error was calculated by transformation using the same process. Between three and six independent experiments were performed to determine Kd for each RNA. Statistical analysis of the data employed SAS 9.1.3 Service Pack 3 (SAS Institute, Inc.) on an HP Model 9000/800 on the HP-UX B.11.23 platform. Data sets were analyzed for equal or unequal variability by a modified Levene's test. For data with equal variances in the two groups (P > 0.05 on the modified Levene's test), a Pooled T-test was used to determine statistical significance. If variances differed significantly (P ≤ 0.05 on the modified Levene's test), a Cochran T-test was used. Results of the modified Levene's test are not reported. Exact P-values are reported when P < 0.05. For P > 0.05, an N.S. indicates “not significant.” Unless noted otherwise, comparison of mutant Krels are to the longer ∼65-nt wild-type RNAs.
Phylogenetic analysis
5′-IRE sequences from mammalian species was heuristically aligned based on terminal loop and C8 bulge sequence conservation. The phylogenetic tree was constructed using the minimum evolution (ME) method and MEGA version 4 software (Tamura et al. 2007). Out of 63–66 nt aligned, 39 nt positions were without gaps representing the final data set. The ME tree was searched using the close-neighbor-interchange algorithm at a search level of 1. The neighbor-joining algorithm was used to generate the initial tree. The bootstrap test was used to estimate the total number of trees where the taxa in the final tree clustered together in 5000 replicates.
Free energy calculations
The change in free energy (ΔΔG°) as a function of the difference between the dissociation constants of two RNA species was calculated by ΔΔG° = RT ln(Kd2/Kd1) (delCardayre and Raines 1995; Ryder et al. 2008). T was 37°C, the temperature of the assay.
SUPPLEMENTAL MATERIAL
The sequence of all the RNAs used in this study is provided in Supplemental Table S1 and the predicted secondary structure of the L-ferritin, ferroportin, and eALAS ∼65-nt IRE-containing RNAs is provided in Supplemental Figure S1. Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank D.A. Brow, S. Butcher, W. Walden, K. Volz, and M. Thomas Record for helpful discussions; N. Keuler and M. Clayton of the College of Agricultural and Life Sciences Statistical Consulting Service for expert advice regarding statistical analyses; and Judith Kozminski for assistance with manuscript preparation. This work was supported by NIH RO1 DK 66600 and UW-Madison Hatch Project 4885 granted to R.S.E. J.B.G. was a predoctoral trainee on NIH T32 DK 007665. While this paper was under review, a study by Khan et al. (2009) reported on the binding characteristics of L-ferritin and m-acon IRE to IRP1 and the role of iron therein.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1857210.
REFERENCES
- Addess KJ, Basilion JP, Klausner RD, Rouault TA, Pardi A. Structure and dynamics of the iron responsive element RNA: Implications for binding of the RNA by iron regulatory binding proteins. J Mol Biol. 1997;274:72–83. doi: 10.1006/jmbi.1997.1377. [DOI] [PubMed] [Google Scholar]
- Alam J, Shibahara S, Smith A. Transcriptional activation of the heme oxygenase gene by heme and cadmium in mouse hepatoma cells. J Biol Chem. 1989;264:6371–6375. [PubMed] [Google Scholar]
- Allerson CR, Cazzola M, Rouault TA. Clinical severity and thermodynamic effects of iron responsive element mutations in hereditary hyperferritinemia-cataract syndrome. J Biol Chem. 1999;274:26439–26447. doi: 10.1074/jbc.274.37.26439. [DOI] [PubMed] [Google Scholar]
- Antao VP, Tinoco JI. Thermodynamic parameters for loop formation in RNA and DNA hairpin loops. Nucleic Acids Res. 1992;20:819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel A, Zacharias M. Conformational transitions in RNA single uridine and adenosine bulge structures: A molecular dynamics free energy simulation study. Biophys J. 2006;90:2450–2462. doi: 10.1529/biophysj.105.076158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton HA, Eisenstein RS, Bomford AB, Munro HN. Determinants of the interaction of the iron responsive element binding protein with its binding site in rat L-ferritin mRNA. J Biol Chem. 1990;265:7000–7008. [PubMed] [Google Scholar]
- Batey RT, Williamson JR. Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16S rRNA: II. Specificity determinants of RNA–protein recognition. J Mol Biol. 1996;261:550–567. doi: 10.1006/jmbi.1996.0482. [DOI] [PubMed] [Google Scholar]
- Bettany AJE, Eisenstein RS, Munro HN. Mutagenesis of the IRE further defines a role for RNA secondary structure in the regulation of ferritin and TfR expression. J Biol Chem. 1992;267:16531–16537. [PubMed] [Google Scholar]
- Beuning PJ, Musier-Forsyth K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 1999;52:1–28. doi: 10.1002/(SICI)1097-0282(1999)52:1<1::AID-BIP1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Brown NM, Anderson SA, Steffen DW, Carpenter TB, Kennedy MC, Walden WE, Eisenstein RS. Novel role of phosphorylation in Fe-S cluster stability revealed by phosphomimetic mutations at Ser-138 of IRP1. Proc Natl Acad Sci. 1998;95:15235–15240. doi: 10.1073/pnas.95.26.15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon KP, Sharma S, Chen CS, Dimasi DP, Mackey DA, Craig JE. A novel deletion in the FtL gene causes HHCS by alteration of the transcription start site. Hum Mutat. 2007;28:742. doi: 10.1002/humu.9501. [DOI] [PubMed] [Google Scholar]
- Butt J, Kim H-Y, Basilion JP, Cohen S, Iwai K, Philpott CC, Altschul S, Klausner RD, Rouault TA. Differences in the RNA binding sites of IRP and potential target diversity. Proc Natl Acad Sci. 1996;93:4345–4349. doi: 10.1073/pnas.93.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camaschella C, Campanella A, De Falco L, Boschetto L, Merlini R, Silvestri L, Levi S, Iolascon A. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood. 2007;110:1353–1358. doi: 10.1182/blood-2007-02-072520. [DOI] [PubMed] [Google Scholar]
- Clarke SL, Vasanthakumar A, Anderson SA, Pondarre C, Koh CM, Deck KM, Pitula JS, Epstein CJ, Fleming MD, Eisenstein RS. Iron-responsive degradation of IRP 1 does not require the Fe-S cluster. EMBO J. 2006;25:544–553. doi: 10.1038/sj.emboj.7600954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremonesi L, Paroni R, Foglieni B, Galbiati S, Fermo I, Soriani N, Belloli S, Ruggeri G, Biasiotto G, Cazzola M, et al. Scanning mutations of the 5′UTR regulatory sequence of L-ferritin by denaturing HPLC: Identification of new mutations. Br J Haematol. 2003;121:173–179. doi: 10.1046/j.1365-2141.2003.04253.x. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar T, Stripecke R, Gray NK, Goossen B, Constable A, Johannson HE, Hentze MW. Identification of a novel iron-responsive element in murine and human erythroid delta-aminolevulinic acid synthase mRNA. EMBO J. 1991;10:1903–1909. doi: 10.1002/j.1460-2075.1991.tb07716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- delCardayre SB, Raines RT. A residue to residue hydrogen bond mediates the nucleotide specificity of ribonuclease A. J Mol Biol. 1995;252:328–336. doi: 10.1006/jmbi.1995.0500. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Lin P-N, McKenzie AR, Walden WE, Theil EC. The influence of the base-paired flanking region on structure and function of the ferritin mRNA IRE. J Mol Biol. 1993;231:230–240. doi: 10.1006/jmbi.1993.1278. [DOI] [PubMed] [Google Scholar]
- dos Santos CO, Dore LC, Valentine E, Shelat SG, Hardison RC, Ghosh M, Wang W, Eisenstein RS, Costa FF, Weiss MJ. An IRE-like stem–loop regulates α-hemoglobin-stabilizing protein mRNA. J Biol Chem. 2008;283:26956–26964. doi: 10.1074/jbc.M802421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy J, Volbeda A, Carpentier P, Darnault C, Moulis JM, Fontecilla-Camps JC. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure. 2006;14:129–139. doi: 10.1016/j.str.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Frugier M, Helm M, Felden B, Giege R, Florentz C. Sequences outside recognition sets are not neutral for tRNA aminoacylation. Evidence for nonpermissive combinations of nucleotides in the acceptor stem of yeast tRNAPhe. J Biol Chem. 1998;273:11605–11610. doi: 10.1074/jbc.273.19.11605. [DOI] [PubMed] [Google Scholar]
- Galy B, Ferring D, Minana B, Bell O, Janser HG, Muckenthaler M, Schumann K, Hentze MW. Altered body iron distribution and microcytosis in mice deficient in IRP2. Blood. 2005;106:2580–2589. doi: 10.1182/blood-2005-04-1365. [DOI] [PubMed] [Google Scholar]
- Galy B, Ferring-Appel D, Kaden S, Grone HJ, Hentze MW. IRP are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab. 2008;7:79–85. doi: 10.1016/j.cmet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Gdaniec Z, Sierzputowska-Gracz H, Theil EC. IRE and internal loop/bulge structure for ferritin mRNA studied by cobalt(III) hexamine binding, molecular modeling, and NMR spectroscopy. Biochem. 1998;37:1505–1512. doi: 10.1021/bi9719814. [DOI] [PubMed] [Google Scholar]
- Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girelli D, Corrocher R, Bisceglia L, Olivieri O, Franceschi LD, Zelante L, Gasparini P. Molecular basis for the recently described HHCS: A mutation in the IRE of ferritin L-subunit gene (the ‘Verona mutation’) Blood. 1995;86:4050–4053. [PubMed] [Google Scholar]
- Haile DJ, Hentze MW, Rouault TA, Harford JB, Klausner RD. Regulation of interaction of the IRE binding protein with iron-responsive RNA elements. Mol Cell Biol. 1989;9:5055–5061. doi: 10.1128/mcb.9.11.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall KB, Tang C. 13C relaxation and dynamics of the purine bases in the IRE RNA hairpin. Biochem. 1998;37:9323–9332. doi: 10.1021/bi9805285. [DOI] [PubMed] [Google Scholar]
- Harrell CM, McKenzie AR, Patino MM, Walden WE, Theil EC. Ferritin mRNA: Interactions of IRE with translational regulatory protein P-90 and the effect on base-paired flanking regions. Proc Natl Acad Sci. 1991;88:4166–4170. doi: 10.1073/pnas.88.10.4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart K, Nystrom B, Ohman M, Nilsson L. Molecular dynamics simulations and free energy calculations of base flipping in dsRNA. RNA. 2005;11:609–618. doi: 10.1261/rna.7147805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson BR, Menotti E, Bonnard C, Kuhn LC. Optimal sequence and structure of IREs. J Biol Chem. 1994;269:17481–17489. [PubMed] [Google Scholar]
- Henderson BR, Menotti E, Kuhn LC. IRP1 and IRP2 bind distinct sets of RNA target sequences. J Biol Chem. 1996;271:4900–4908. doi: 10.1074/jbc.271.9.4900. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Haile DJ, Klausner RD. The interaction between the IRE binding protein and its cognate RNA is highly dependent upon both RNA sequence and structure. Nucleic Acids Res. 1993;21:4627–4631. doi: 10.1093/nar/21.19.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Wu J, Leibold EA, Walden WE, Theil EC. Loops and bulge/loops in IRE isoforms influence IRP binding. J Biol Chem. 1998;273:23637–23640. doi: 10.1074/jbc.273.37.23637. [DOI] [PubMed] [Google Scholar]
- Ke YH, Sierzputowska-Gracz H, Gdaniec Z, Theil EC. Internal loop/bulge and hairpin loop of the IRE of ferritin mRNA contribute to maximal IRP2 binding and translational regulation in the iso-IRE/iso-IRP family. Biochemistry. 2000;39:6235–6242. doi: 10.1021/bi9924765. [DOI] [PubMed] [Google Scholar]
- Khan MA, Walden WE, Goss DJ, Theil EC. Direct Fe2+ sensing by iron-responsive messenger RNA·repressor complexes weakens binding. J Biol Chem. 2009;284:30122–30128. doi: 10.1074/jbc.M109.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikinis Z, Eisenstein RS, Bettany AJE, Munro HN. Role of RNA secondary structure of the IRE in translational regulation of ferritin synthesis. Nucleic Acids Res. 1995;23:4190–4195. doi: 10.1093/nar/23.20.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-Y, LaVaute T, Iwai K, Klausner RD, Rouault TA. Identification of a conserved and functional iron-responsive element in the 5′-untranslated region of mammalian mitochondrial aconitase. J Biol Chem. 1996;271:24226–24230. doi: 10.1074/jbc.271.39.24226. [DOI] [PubMed] [Google Scholar]
- Leibold EA, Laudano A, Yu Y. Structural requirements of iron-responsive elements for binding of the protein involved in both transferrin receptor and ferritin mRNA post-transcriptional regulation. Nucleic Acids Res. 1990;18:1819–1824. doi: 10.1093/nar/18.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipuviene R, Theil EC. The family of iron responsive RNA structures regulated by changes in cellular iron and oxygen. Cell Mol Life Sci. 2007;64:2945–2955. doi: 10.1007/s00018-007-7198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing LG, Hall KB. A model of the iron responsive element RNA hairpin loop structure determined from NMR and thermodynamic data. Biochemistry. 1996;35:13586–13596. doi: 10.1021/bi961310q. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- McCallum SA, Pardi A. Refined solution structure of the iron-responsive element RNA using residual dipolar couplings. J Mol Biol. 2003;326:1037–1050. doi: 10.1016/s0022-2836(02)01431-6. [DOI] [PubMed] [Google Scholar]
- McClain WH, Chen YM, Foss K, Schneider J. Association of tRNA acceptor identity with a helical irregularity. Science. 1988;242:1681–1684. doi: 10.1126/science.2462282. [DOI] [PubMed] [Google Scholar]
- Meehan HA, Connell GJ. The hairpin loop but not the bulged C of the iron responsive element is essential for high affinity binding to iron regulatory protein-1. J Biol Chem. 2001;276:14791–14796. doi: 10.1074/jbc.M010295200. [DOI] [PubMed] [Google Scholar]
- Meyron-Holz EG, Ghosh MC, Iwai K, LaVaute T, Brazzolotto X, Berger UV, Land W, Olivarerre-Wilson H, Grinberg A, Love P, et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using t7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morden KM, Chu YG, Martin FH, Tinoco I., Jr Unpaired cytosine in the deoxyoligonucleotide duplex dCA3CA3G•dCT6G is outside the helix. Biochemistry. 1983;22:5557–5563. [Google Scholar]
- Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K, Schimmel P. Functional contacts of a tRNA synthetase with 2′-hydroxyl groups in the RNA minor groove. Nature. 1992;357:513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- Piccinelli P, Samuelsson T. Evolution of the iron-responsive element. RNA. 2007;13:952–966. doi: 10.1261/rna.464807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DpbA. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- Reiter NJ, Blad H, Abildgaard F, Butcher SE. Dynamics in the U6 RNA intramolecular stem–loop: A base flipping conformational change. Biochemistry. 2004;43:13739–13747. doi: 10.1021/bi048815y. [DOI] [PubMed] [Google Scholar]
- Rogers JT, Randall JD, Cahill CM, Eder PS, Huang X, Gunshin H, Leiter L, McPhee J, Sarang SS, Utsuki T, et al. An iron-responsive element type II in the 5′-untranslated region of the Alzheimer's amyloid precursor protein transcript. J Biol Chem. 2002;277:45518–45528. doi: 10.1074/jbc.M207435200. [DOI] [PubMed] [Google Scholar]
- Ryder SP, Recht MI, Williamson JR. Quantitative analysis of protein–RNA interactions by gel mobility shift. Methods Mol Biol. 2008;488:99–115. doi: 10.1007/978-1-60327-475-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez M, Galy B, Dandekar T, Bengert P, Vainshtein Y, Stolte J, Muckenthaler MU, Hentze MW. Iron regulation and the cell cycle: Identification of an iron-responsive element in the 3′-untranslated region of human cell division cycle 14A mRNA by a refined microarray-based screening strategy. J Biol Chem. 2006;281:22865–22874. doi: 10.1074/jbc.M603876200. [DOI] [PubMed] [Google Scholar]
- Schalinske KL, Chen OS, Eisenstein RS. Iron differentially stimulates translation of mitochondrial aconitase and ferritin mRNAs in mammalian cells. Implications for iron regulatory proteins as regulators of mitochondrial citrate utilization. J Biol Chem. 1998;273:3740–3746. doi: 10.1074/jbc.273.6.3740. [DOI] [PubMed] [Google Scholar]
- Schranzhofer M, Schifrer M, Cabrera JA, Kopp S, Chiba P, Beug H, Mullner EW. Remodeling the regulation of iron metabolism during erythroid differentiation to ensure efficient heme biosynthesis. Blood. 2006;107:4159–4167. doi: 10.1182/blood-2005-05-1809. [DOI] [PubMed] [Google Scholar]
- Sierzputowska-Gracz H, McKenzie RA, Theil EC. The importance of a single G in the hairpin loop of the iron responsive element (IRE) in ferritin mRNA for structure: An NMR spectroscopy study. Nucleic Acids Res. 1995;23:146–153. doi: 10.1093/nar/23.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SR, Ghosh MC, Ollivierre-Wilson H, Hang Tong W, Rouault TA. Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol Dis. 2006;36:283–287. doi: 10.1016/j.bcmd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Surdej P, Richman L, Kuhn LC. Differential translational regulation of IRE-containing mRNAs in Drosophila melanogaster by endogenous IRP and a constitutive human IRP1 mutant. Insect Biochem Mol Biol. 2008;38:891–894. doi: 10.1016/j.ibmb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Varani G, McClain WH. The G•U wobble base pair: A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz K. The functional duality of iron regulatory protein 1. Curr Opin Struct Biol. 2008;18:106–111. doi: 10.1016/j.sbi.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden WE, Selezneva AI, Dupuy J, Volbeda A, Fontecilla-Camps JC, Theil EC, Volz K. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 2006;314:1903–1908. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- Wallander ML, Leibold EA, Eisenstein RS. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim Biophys Acta. 2006;1763:668–689. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JR. Induced-fit in RNA–protein recognition. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- Wu M, Turner DH. Solution structure of (rGCGGACGC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry. 1996;35:9677–9689. doi: 10.1021/bi960133q. [DOI] [PubMed] [Google Scholar]
- Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- Xu D, Landon T, Greenbaum NL, Fenley MO. The electrostatic characteristics of G•U wobble base pairs. Nucleic Acids Res. 2007;35:3836–3847. doi: 10.1093/nar/gkm274. [DOI] [PMC free article] [PubMed] [Google Scholar]