Abstract
Murid herpesvirus 4 (MuHV-4) microRNAs were previously cloned from latently infected tumor cells and predicted to be processed from a series of RNA polymerase III primary transcripts. We detected maturely processed MuHV-4 miRNAs within total RNA from lytically infected cells in vitro and infected tissues ex vivo, using a highly sensitive reverse ligation meditated RT-PCR strategy. We determined that the MuHV-4 microRNAs are biologically active during infection by a luciferase reporter system. We experimentally demonstrated that transcription of the MuHV-4 microRNAs is by RNA polymerase III by α-amanitin insensitivity and by specific deletion of the RNA polymerase III type 2-like promoter elements of MuHV-4, resulting in the complete loss of miRNA detection and function. Finally, we demonstrate that these 10 viral miRNAs, each transcribed from highly conserved and novel polymerase III promoter elements, vary markedly in their relative abundance and activity.
Keywords: microRNA, herpesvirus, RNA polymerase III, reverse ligation mediated-reverse transcription-polymerase chain reaction, noncoding RNA
INTRODUCTION
MicroRNAs (miRNAs) are short, noncoding RNAs that function to regulate protein production levels of target messenger RNAs (mRNAs) in multicellular organisms (Bartel 2004). Canonical miRNA biogenesis is typically described as beginning with the transcription of a primary-miRNA (pri-miRNA) transcript via RNA polymerase II (pol II) (Lee et al. 2004). Post-transcriptional processing of the pri-miRNA is initiated in the nucleus by Drosha cleavage of the pri-miRNA stem–loop to yield an intermediately processed precursor-miRNA (pre-miRNA) (Lee et al. 2003; Kim 2005; Zeng and Cullen 2006). In the cytoplasm, the pre-miRNA is cleaved by Dicer to yield maturely processed RNA duplex molecules (Hutvágner et al. 2001), one strand of which is selectively loaded into the miRNA-containing RNA-induced silencing complex (miRISC) (Khvorova et al. 2003; Maniataki and Mourelatos 2005). In most cases, sequence-specific interactions between the maturely processed miRNA within the miRISC and the 3′ untranslated region (3′UTR) of the target mRNA leads to translational inhibition or degradation of the target mRNAs, resulting in decreased protein production from the target mRNAs (Bartel 2004; Carthew and Sontheimer 2009). Control of protein expression by endogenous cellular miRNAs has been revealed to play a key role in several important cellular processes, including embryonic development, hematopoiesis, stress resistance, fat metabolism, brain morphogenesis, and tumorigenesis (Metzler et al. 2004; Alvarez-Garcia and Miska 2005; Gregory and Shiekhattar 2005; Krützfeldt et al. 2006).
Recently it was demonstrated that viruses, including many herpesvirus examined to date, encode miRNA genes (Pfeffer et al. 2005, 2004; Gottwein and Cullen 2008; Cullen 2009). Biological functions associated with virally encoded miRNAs during infection have been linked to latency within herpesvirus and retrovirus infections and immune evasion by the polyomavirus, SV40 (Grassmann and Jeang 2008; Cullen 2009). Murid herpesvirus 4 (MuHV-4), commonly known as Murine gammaherpesvirus 68 (γHV68), is a member of the gammaherpesvirus family of viruses. Gammaherpesviruses consist of lymphotropic herpesviruses that contain large double-stranded DNA genomes and are associated with a variety of malignancies and lymphoproliferative diseases. Gammaherpesvirus infections are both host-specific and lifelong in duration. γHV68 is a natural pathogen of wild rodents and is similar to its genomic organization and disease associations to the human pathogens Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV), making it a relevant small animal model for gammaherpesvirus (γHV) pathogenesis (Blaskovic et al. 1980; Simas and Efstathiou 1998; Speck and Virgin 1999). Confirmed roles for gammaherpesvirus miRNAs during infection include autoregulation of the viral DNA polymerase and the LMP1 signaling molecule during EBV infection, in addition to regulation of the cellular transcripts BACH1 and THBS1 by KSHV and PUMA and CXCL11 by EBV (Cullen 2009). Currently there are no confirmed targets associated with the γHV68 encoded miRNAs, nor investigations of associated activity.
EBV and KSHV both produce maturely processed miRNAs that are abundantly expressed during latency and processed from RNA pol II transcribed pri-miRNA transcripts (Pfeffer et al. 2004; Cai and Cullen 2006; Cai et al. 2006). γHV68 is the only herpesvirus examined to date that has been predicted to generate its pri-miRNA transcript through RNA polymerase III (pol III) transcription. In collaboration with the Tuschl laboratory, we previously identified γHV68 miRNAs from total RNA isolated from the latently infected mouse tumor cell line known as S11 (Pfeffer et al. 2005). After their identification, it was noted that each γHV68 miRNA gene was located immediately downstream from a previously proposed viral tRNA-like (vtRNA) element in the left end of the γHV68 genome (Pfeffer et al. 2005). These proposed vtRNA sequences have previously been shown to (1) contain RNA pol III, type 2 promoter elements; (2) lack any upstream TATA boxes; (3) contain up to 75% sequence homology with known tRNA genes; and (4) be transcribed during all phases of the viral life-cycle, both latent and lytic (Bowden et al. 1997; Simas et al. 1999; Ebrahimi et al. 2003). Collectively, this information led to the hypothesis that the vtRNAs and the viral miRNAs were transcribed by RNA pol III as one dicistronic transcript (Pfeffer et al. 2005). Evidence for transcription of a pri-miRNA by RNA pol III has been limited to only two recently identified examples: miRNAs produced by RNA pol III transcription of Alu elements in mammalian genomes, and the miRNAs produced from the adenovirus VAI gene (Borchert et al. 2006; Sano et al. 2006). A synthetic pol III-derived tRNA–shRNA dicistronic transcript has been described (Scherer et al. 2007), but the existence of a naturally occurring tRNA–miRNA dicistronic transcript with potential to generate biologically active miRNAs has never been demonstrated. Furthermore, the predicted pol III-derived dicistronic transcript provides an atypical substrate for the miRNA processing machinery.
Interestingly, a previous investigation centered on building a mammalian miRNA expression atlas failed to detect maturely processed γHV68 miRNAs from lytically infected mouse fibroblast cells (Supplemental Table S6; Landgraf et al. 2007). This is surprising because the predicted pri-miRNAs of γHV68 are transcribed during all stages of infection (Bowden et al. 1997; Ebrahimi et al. 2003). Here we demonstrate for the first time that γHV68 pri-miRNAs, processed intermediates, and mature miRNAs exist within lytically infected cells in vitro, as well as acutely and latently infected tissue samples ex vivo. We also determine that the γHV68 miRNAs are biologically active in their ability to down-regulate the expression of genes containing target sequences for the viral miRNAs. Furthermore, we experimentally prove that the γHV68 miRNAs are transcribed by RNA pol III by detecting maturely processed viral miRNAs within RNA isolated from infected cells treated with α-amanitin. Additionally, detailed sequence analysis of the left end of the γHV68 genome revealed that each of the γHV68 pri-miRNA transcripts is initiated from previously uncharacterized RNA pol III type 2-like promoter elements containing a unique configuration of three overlapping A-box promoter elements coupled with one B-box promoter element. Deletion of this unique RNA pol III promoter resulted in loss of γHV68 miRNA detection and biological activity. Finally, we show that despite common pol III promoter elements, the maturely processed miRNAs of γHV68 accumulate with distinct abundance patterns and biological activities during lytic infection. Together, these data demonstrate the existence of novel RNA pol III promoter elements giving rise to a series of unique viral transcripts that are processed into biologically functional miRNAs.
RESULTS
Northern blot analysis of γHV68 miRNA-associated transcription during lytic infection
Nearly all well-characterized miRNAs are derived from pol II transcripts. However, the γHV68 miRNAs have been suggested to be transcribed by RNA pol III, based on published observations of vtRNA transcripts in the immediate proximity of the miRNA encoding region of the γHV68 genome (Bowden et al. 1997; Pfeffer et al. 2005). These viral transcripts, which we refer to simply as the γHV68 pol III transcripts, numbered 1 through 8 beginning at the left end of the viral genome, are unique RNA molecules containing both tRNA-like and miRNA encoding domains. RNA transcripts with this conformation have not been identified in eukaryotic cells or in other viral genomes. In collaboration with the Tuschl laboratory, we previously analyzed RNA from a latently infected tumor cell line and found mature viral miRNAs and small RNA transcripts by Northern analysis consistent with pol III-derived transcripts (Pfeffer et al. 2005). However, we found no evidence of mature miRNAs from lytically infected cells despite the fact that abundant transcription of the viral pol III transcripts has been previously demonstrated during lytic infection (Bowden et al. 1997; Ebrahimi et al. 2003; Landgraf et al. 2007). Due to this apparent discrepancy, we sought to further examine γHV68 miRNA production within lytically infected cells.
The transcriptional start and stop sites of the γHV68 pol III transcripts have yet to be experimentally mapped. However, using the canonical rules of RNA pol III transcription, we were able to predict plausible transcriptional start and stop sites that would yield the γHV68 pri-miRNA transcripts. RNA pol III type 2 promoters, typically used in the transcription of cellular tRNA genes, are found in all eight of the γHV68 pol III genes and have been predicted to be used in the transcription of these genes (Bowden et al. 1997; Paule and White 2000; Pfeffer et al. 2005). Transcription of genes using the RNA pol III type 2 promoter begins with the recognition of two internal promoter elements known as the A box (TRGYNNARNNG) and the B box (RGTTCRANTCC) by multisubunit transcription factor complexes, which together recruit the RNA pol III enzyme to initiate transcription 7 nucleotides (nt) upstream of the A box promoter element (Paule and White 2000; Hamada et al. 2001). RNA pol III transcription continues until the first run of four or more thymidine residues. Applying these rules, we predicted the start and stop sites of transcription of all eight of the γHV68 pol III transcripts and the possible secondary structures associated with the γHV68 pol III transcripts.
Similar to our previous report from the latently infected tumor cell line, we determined that the γHV68 pol III transcripts contain three distinct structural domains consisting of a tRNA-like domain at the 5′ end followed by two stem–loop domains (stem–loops 1 and 2) that contain the mature miRNA sequences of γHV68 (Pfeffer et al. 2005). Using the γHV68 pol III-1 transcript as a representative example, a predicted pol III transcript 196 nt in length can be broken down into the three aforementioned domains with the tRNA-like domain occupying the first 73 nt of the RNA molecule, followed by stem–loop 1, 57 nt in length, and stem–loop 2, 66 nt in length (Fig. 1A,B). Viral pol III transcripts of similar lengths and secondary structures were also predicted for the seven other γHV68 pol III transcripts (Supplemental Fig. 1).
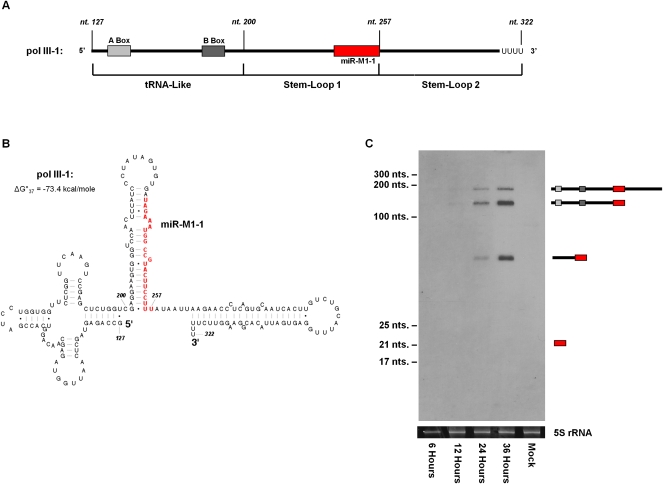
FIGURE 1.
Northern blot analysis of γHV68 pol III-1 during lytic infection. (A) Schematic of the predicted γHV68 pol III-1 transcript. RNA pol III type 2 internal promoter elements are shown as solid gray boxes. γHV68 miRNA miR-M1-1 is represented as a solid red box. γHV68 genomic coordinates are labeled in italics above the gene diagram. (B) Predicted secondary structure of the γHV68 pol III-1 transcript containing the lowest ΔG°37 value using the RNAstructure 4.6 software (Mathews et al. 2004). Nucleotides comprising miR-M1-1 are shown in red. (C) Northern blot of γHV68 pol III-1 from total RNA isolated from infected 3T3 cells collected at 6, 12, 24, and 36 h post infection. γHV68 pol III-1 transcript products are diagrammed to the right of the Northern blot. Ethidium bromide-stained 5S rRNA shown below serves as a loading control.
To determine the existence and size of the γHV68 pol III-1 transcript, we performed Northern blot analysis for γHV68 pol III-1 from total RNA collected during lytic infection of 3T3 cells at 6, 12, 24, and 36 h post-infection. Using a 5′-biotinylated probe designed to anneal perfectly to miR-M1-1, γHV68 pol III-1 expression was first clearly detected at 24 h post-infection as three bands ranging in size from ∼200 to ∼50 nt (Fig. 1C). The exact size of the bands on the blot was calculated by gel migration comparison with size standard markers (Supplemental Fig. 2) and was determined to be 192, 130, and 51 nt in length. Based on these band sizes, the top band likely represents the full-length predicted γHV68 pol III-1 transcript, while the middle band represents a molecule containing the tRNA-like and stem–loop 1 portion of the γHV68 pol III-1 transcript. The bottom band represents only the stem–loop 1 portion of the γHV68 pol III-1 transcript, previously predicted to be the pre-miRNA molecule further processed to generate the mature miR-M1-1 miRNA (Pfeffer et al. 2005). Consistent with previous cloning and sequencing analysis, a band at 22 nt representing the mature form of miR-M1-1 was not detected by Northern analysis of lytically infected cells (Landgraf et al. 2007).
There are two potential reasons why mature miRNAs were not detected by Northern blot. The first is that maturely processed miR-M1-1 is simply not made during lytic infection. The second is that miR-M1-1 is made, but at levels below our detection limit by Northern blot. We addressed these issues by determining the limit of detection of our Northern blot and by using dicer substrate (DS) mimics to confirm dicer processing and miRNA activity within 3T3 and 293 cells.
MicroRNAs undetectable by Northern blotting are biologically active
To determine our ability to detect maturely processed miRNAs via Northern blotting, independent of viral infection and transcriptional regulation, we transfected both 3T3 and 293 cells with DS mimics of the γHV68 miRNAs miR-M1-1 and miR-M1-5 (Fig. 2A). Design and implementation of these DS mimics was performed as previously described for enhanced RNAi activity (Kim et al. 2005). Transfecting cells with DS mimics should result in the processing of the RNA mimic molecules from an initial size of 27 nt to biologically functional molecules 21 nt in length (Kim et al. 2005). Surprisingly, Northern blot analysis of total RNA harvested from DS transfected cells did not show the expected accumulation of a processed, 21-nt-long RNA product (Supplemental Fig. 3A,B). Northern blotting of titrated concentrations of untransfected DS mimics demonstrated that our Northern blot sensitivity is near 10 fmol, or ∼6 × 109 total molecules (Supplemental Fig. 3C).
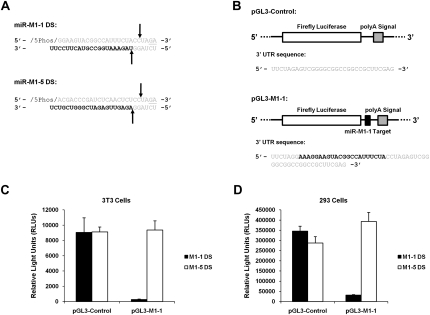
FIGURE 2.
Dicer substrate (DS) processing, detection, and biological activities. (A) Schematic of miR-M1-1 and miR-M1-5 DSs. The entire molecule consists of ribonucleic acid nucleotides except for the two underlined bases that are deoxyribonucleic acid nucleotides. Arrows represent predicted dicer cut sites on the molecule. The bold bases represent the mature miRNA molecule. (B) Schematic of the pGL3-Luciferase plasmid constructs. pGL3-Control contains a short 3′UTR sequence with no miRNA target sequences, shown below the pGL3 diagram. pGL3-M1-1 contains the perfect complement sequence of γHV68 miRNA miR-M1-1 in its 3′UTR, shown in bold below the pGL3-M1-1 diagram, and serves as a target to gauge miR-M1-1 activity in transfected cells. (C,D) Dual luciferase reporter analysis from cotransfection experiments with the DS molecules and pGL3 reporter plasmids in 3T3 and 293 cells. pGL3 reporter and M1-1 DS cotransfections are shown as solid black bars, while pGL3 reporter and M1-5 DS cotransfections are shown as solid white bars. Data are shown as total firefly luciferase activity normalized to the renilla luciferase transfection control readings.
To measure possible biological activity from the DS miRNA mimics, we transfected DS mimics into 3T3 and 293 cells with the pGL3-Control or pGL3-M1-1 target firefly luciferase reporter plasmid (Fig. 2B). The pGL3-M1-1 target plasmid contains a perfect target site for miR-M1-1 miRNA binding and allows for quantification of miR-M1-1 miRNA activity in the luciferase assay. Cells were also transfected with the pRL-TK renilla luciferase plasmid as a transfection control to normalize the firefly transfection results. Upon cotransfection with the DS and the pGL3 reporter plasmids, expression from the pGL3-M1-1 plasmid in 3T3 cells was decreased by 97% (35-fold reduction) relative to the cotransfection with the pGL3-Control plasmid (Fig. 2C). Upon cotransfection with the DS and the reporter plasmids, expression from the pGL-M1-1 plasmid in 293 cells was down 91% (11-fold reduction) relative to the pGL3-Control plasmid (Fig. 2D). Cotransfection with the miR-M1-5 DS had no effect on the amount of luciferase expression from the pGL3-Control or the pGL3-M1-1 firefly luciferase plasmids in either cell type, as expected.
These data demonstrate that even though the maturely processed form of the DS was undetectable by Northern blot following transfection, biological activity specific to the DS molecules is present in the transfected cells. One formal, but unlikely, explanation of these results is that unprocessed DS may down-regulate the expression of the pGL3-M1-1 reporter. However, the more likely explanation is that the DS molecules are processed and biologically active, but at levels too low to be detected by Northern blot. To directly determine whether the DS molecules are maturely processed, we used reverse ligation mediated RT-PCR (RLM-RT-PCR) to detect mature forms of miRNAs.
Identification of mature miRNAs using RLM-RT-PCR
To identify mature miRNAs we used a RLM-RT-PCR assay that allows for the amplification and sequencing of mature miRNA products in as little as 200 ng of total RNA. The RLM-RT-PCR procedure is centered on the fact that the T4 RNA ligase will ligate any two pieces of single-stranded nucleic acid as long as one strand contains a 5′ monophosphate end and the other a 3′ hydroxyl end (Romaniuk and Uhlenbeck 1983). Mature miRNAs, direct products of Dicer cleavage, are single-stranded molecules that contain a 5′ monophosphate and a 3′ hydroxyl group that make them suitable substrates for this reaction (Elbashir et al. 2001; Bartel 2004). A synthetic linker oligo of known sequence was ligated onto small RNAs isolated from cells to create a platform long enough for RT-PCR. We designed two different linker oligos: one RNA linker oligo containing hydroxyl groups at both the 5′ and 3′ ends of the linker molecule for RLM-RT-PCR detection of miRNAs arising from the 3′ stem of a precursor miRNA stem–loop (as is the case for γHV68 miR-M1-1), and one RNA linker oligo containing a 5′ monophosphate used for RLM-RT-PCR of miRNAs arising from the 5′ stem of a precursor miRNA stem–loop (as is the case for γHV68 miR-M1-5). Using the miRNA sequence as one primer binding site and the RNA linker oligo as the other primer binding site, RT-PCR analysis of the reverse ligated RNA products was conducted. RT-PCR products were then cloned and sequenced to verify ligation of mature miRNAs to the linker oligo (Fig. 3A).
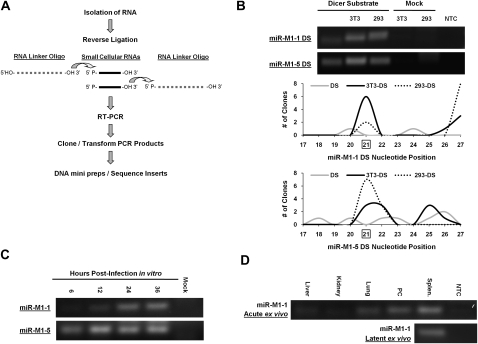
FIGURE 3.
Detection of mature miRNAs using RLM-RT-PCR. (A) Schematic of the RLM-RT-PCR process for the detection of mature miRNAs. (B) RLM-RT-PCR of miRNAs miR-M1-1 and miR-M1-5 produced from the dicer substrates in transfected cells. Total RNA from the indicated cell types was used as template for the RLM-RT-PCR procedure. RT-PCR products were resolved by gel electrophoresis and visualized by ethidium bromide. Histogram representation of sequence analysis of the RT-PCR products from the dicer substrates is shown below the agarose gels. Each dicer substrate RLM-RT-PCR product in B was cloned, and 10 clones of each product were sequenced. The histograms show the position where the dicer substrate molecule ligated to the RNA linker oligo. Nucleotide position 21 is the predicted dicer cleavage site for these molecules and is boxed. Clones that yielded primer dimer products are not represented in the histogram. The gray lines represent clones sequenced from the untransfected dicer substrates. The solid black line represents clones sequenced from dicer substrates transfected into 3T3 cells. The dotted black line represents clones sequenced from dicer substrates transfected into 293 cells. (C) RLM-RT-PCR analysis of mature γHV68 miR-M1-1 and miR-M1-5 from γHV68 infected samples. Lytic in vitro RLM-RT-PCR products were amplified from RNA samples of 3T3 cells infected at an MOI = 5 with total RNA isolated at 6, 12, 24, and 36 h post infection. (D) Acute and latent ex vivo RLM-RT-PCR products were amplified from RNA of C57BL/6J mouse tissues collected at day 5 post-infection (acute) or day 16 post-infection (latent). All RLM-RT-PCR products were resolved and visualized using ethidium bromide staining. Bands were confirmed to be mature miR-M1-1 miRNAs by cloning and sequencing. Splen. indicates splenocytes cells; PC, peritoneal cells.
Applying the RLM-RT-PCR strategy to stock DS, we found some RLM-RT-PCR products using both the miR-M1-1 DS and the miR-M1-5 DS primer sets for amplification (Fig. 3B). Cloning and sequencing of the products from the untransfected miR-M1-1 DS (Fig. 3B, left lane) revealed a predominant product of 35 base pairs (bp) (six of 10 clones sequenced) (products detailed in Supplemental Table 1), smaller than the 38 bp corresponding to a properly processed DS molecule. Sequence analysis of this product revealed that it was a primer dimer generated during the RT-PCR reaction. However, with the untransfected miR-M1-5 (Fig. 3B, left lane), a wide variety of inserts ranging from RNA molecules shorter or longer than a properly processed DS were also cloned. Of the 10 clones sequenced from the miR-M1-5 DS RLM-RT-PCR, none of the inserts revealed an RNA molecule of the proper size to be a mature miRNA as expected from this untransfected sample. No product was detected in the mock transfected samples or the no template control (NTC) samples (Fig. 3B). Importantly, mature miRNA products can be recovered from cells transfected with either DS.
RLM-RT-PCR sequence analysis of both the miR-M1-1 DS and the miR-M1-5 DS transfected samples in either the 3T3 or the 293 cell type show processing of the DS to generate perfectly processed miRNA molecules as predicted. The clones were not homogeneous in their representation of the mature miRNA product, and some sequences revealed the presence of RNAs longer than the mature form of the DS. This is not entirely surprising however, due to the fact that these long RNAs are detected via Northern blotting (Supplemental Fig. 3A,B). Graphical representation of all of the RLM-RT-PCR clones sequenced is shown in Figure 3B and summarized in Supplemental Table 1. In addition to the longer RNAs in the population of RNAs amplified, RLM-RT-PCR confirmed mature miRNAs of the correct size and sequence in the DS transfected samples. Therefore the biological activity associated with the DS transfection corresponds to the presence of maturely processed miRNA products.
Additionally, with input of 200 ng for RLM-RT-PCR versus 20 μg of total RNA for the Northern blot, it is clear that the RLM-RT-PCR strategy is very efficient at detecting mature miRNAs. Because the γHV68 mature miRNAs were never detected by Northern blot in spite of our demonstration that the γHV68 miRNAs are biologically active in the cell, we determined that the limit of detection of our Northern blot using biotinylated probes was 5000–10,000 mature miRNAs per cell. RLM-RT-PCR allowed us to detect mature miRNAs with at least 100-fold higher sensitivity than Northern blotting.
Using this sensitive RLM-RT-PCR assay, we then analyzed mature miRNAs produced in lytically infected 3T3 cells (Fig. 3C) as well as in infected tissues (Fig. 3D) from both acute and latent stages from C57BL/6J mice infected with γHV68. Maturely processed miR-M1-1 detected by RLM-RT-PCR was confirmed by cloning and sequencing the RT-PCR product. During lytic infection of 3T3 cells, miR-M1-1 was first definitively detected at 24 h post infection and appears to remain constant to the point of cell lysis at 36 h post-infection. In contrast to miR-M1-1, γHV68 miRNA miR-M1-5 is readily detectable as early as 6 h post-infection with an apparent maximum expression level reached at 12 h post-infection.
Mature γHV68 miR-M1-1 was also detected within ex vivo tissue samples from mice infected with γHV68. At day 5 post-infection, during acute γHV68 infection, miR-M1-1 was differentially expressed within different organs with the highest level of expression of miR-M1-1 in the spleen followed by peritoneal cells and lung. Infected spleen samples were also found to contain mature miR-M1-1 at day 16 post infection, during latent γHV68 infection and at a time when only one cell in 100 is estimated to be infected (Flaño et al. 2002; Willer and Speck 2003).
RLM-RT-PCR analysis revealed that γHV68 mature miRNA expression can be readily detected in a variety of infection conditions where Northern blot analysis does not reveal mature miRNAs. Additionally, the RLM-RT-PCR detection of mature miRNAs from cells transfected with DS mimics is consistent with the demonstration of functionality of these DS mimics by luciferase reporters (Fig. 2C,D). With a robust and convenient system in place to detect mature miRNA expression from virally infected cells, we next sought to determine whether we could detect all 10 of the previously annotated γHV68 miRNAs as well as demonstrate biological activity of the viral miRNAs using the pGL3 luciferase reporter system during in vitro lytic infection.
Lytically infected 3T3 cells contain maturely processed γHV68 miRNAs that are biologically active
To determine whether the γHV68 miRNAs are processed into mature miRNA products during primary infection in vitro, we infected 3T3 cells at a multiplicity of infection (MOI) of 5 and isolated the total cellular RNA 42 h post-infection for RLM-RT-PCR analysis. We used primer sets specific to each individual γHV68 miRNA and to one cellular control miRNA (mmu-miR-21) shown previously to be expressed in 3T3 cells (Hu et al. 2008). We detected each γHV68 mature miRNA from lytically infected 3T3 cells, with the exception of miR-M1-2 (Fig. 4A). Of the γHV68 miRNAs detected, it was apparent that not every viral miRNA was expressed or detected to equal levels. This is somewhat surprising for pol III transcripts, which are presumably made consistently and equivalently. However, this differential detection is consistent with the fact that the viral miRNAs were initially cloned with varying frequency (Pfeffer et al. 2005). In the initial identification of the γHV68 miRNAs, miR-M1-1, miR-M1-5, miR-M1-7-5p, and miR-M1-7-3p were the four most abundantly cloned miRNAs from the γHV68 latently infected cell line S11 (Usherwood et al. 1996; Pfeffer et al. 2005). Our RLM-RT-PCR results indicated that the most abundant γHV68 miRNAs during primary infection of 3T3 cells were miR-M1-1, miR-M1-3, miR-M1-6, miR-M1-7-5p, and miR-M1-9. The relative differences in miRNA abundance in this study in comparison to previous work could be due to the different sources of RNA, that is, latently infected tumor cells versus lytically infected cells. It has been shown from studies of other gammaherpesviruses, that the state of viral infection, whether lytic or latent, can impact the overall levels of viral miRNAs in infected cells (Cai et al. 2005, 2006; Gottwein et al. 2006). As expected, there was no detection of mature miRNAs from the cells infected with γHV68Δ9473, a mutant virus lacking the left end 9.5 kb of the γHV68 genome, including the viral miRNA coding region (Clambey et al. 2002). Both sets of virally infected cells clearly show signs of lytic infection based on the presence of the lytic viral transcript ORF 50 in both infected cell populations (Fig. 4B).
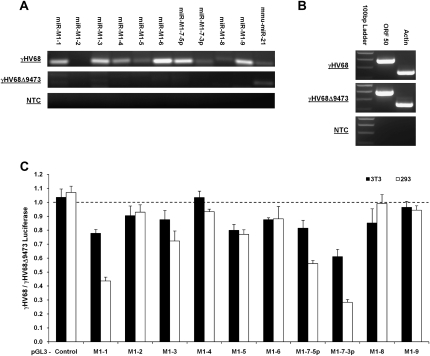
FIGURE 4.
Detection and confirmation of biological activity of the γHV68 miRNAs during lytic infection in vitro. (A) RLM-RT-PCR for miRNAs in 3T3 cells infected with either γHV68 or γHV68Δ9473. mmu-miR-21 serves as an endogenous control for miRNA expression in 3T3 cells. RT-PCR products were resolved by gel electrophoresis and visualized by ethidium bromide. (B) RT-PCR for ORF 50, a marker of lytic infection for 3T3 cells infected with γHV68. Actin serves as a RT-PCR control for endogenous transcription in 3T3 cells. (C) Dual luciferase reporter analysis following infection with either the γHV68 or γHV68Δ9473 viruses. pGL3 luciferase reporter target is labeled below the graph. All results are displayed as a ratio of firefly luciferase expression in γHV68 infected cells divided by firefly luciferase expression in γHV68Δ9473 infected cells. Firefly luciferase readings are all normalized to the renilla luciferase transfection control. Readings obtained from 3T3 infected cells are shown in solid black bars; 293 cells, in solid white bars. Readings below the dashed line represent repression by the corresponding viral miRNA in γHV68 infected cells.
To determine the biological activity of the γHV68 miRNAs, 3T3 or 293 cells were lytically infected at an MOI of 5 following cotransfection with a pGL3 firefly luciferase reporter plasmid containing a target site specific to a single γHV68 miRNA and the pRL-TK renilla luciferase transfection control plasmid. To specifically discern biological activity of the viral miRNAs, the cotransfected cells were either infected with wild-type γHV68 or γHV68Δ9473. Interestingly, this analysis demonstrated that not all of the γHV68 miRNAs confer robust down-regulation of their respective targets in the luciferase reporter system (Fig. 4C). Luciferase readout is displayed as a ratio of pGL3 luciferase reporter expression from the γHV68 infection divided by pGL3 luciferase reporter expression from the γHV68Δ9473 infection. Therefore the pGL3 luciferase reporter plasmids with the lowest expression ratio are the most strongly down-regulated by γHV68 infection. Only miRNAs miR-M1-1, miR-M1-7-5p, and miR-M1-7-3p have the ability to reduce their luciferase reporter target expression by 50% or more. This could be due to the fact that the cells are succumbing to lytic infection prior to our ability to detect strong viral miRNA activity, in contrast to latently infected cells where miRNA accumulation is possible. These data demonstrated mature and functional viral miRNAs within primary infected cells, a condition in which both pol II and pol III transcription has been reported from the miRNA coding region of the virus genome. With these methods to detect and gauge the biological activity of the γHV68 mature miRNA products, we next sought to definitively determine whether production of the γHV68 miRNAs was dependent on RNA pol III transcription as previously predicted (Pfeffer et al. 2005).
The γHV68 miRNA primary transcripts are transcribed by RNA pol III
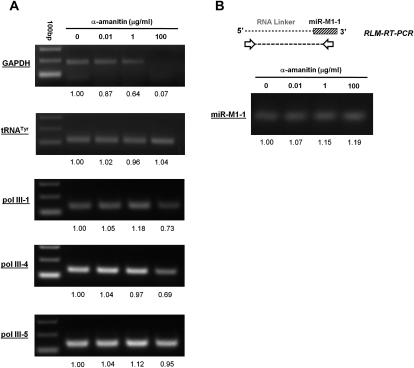
To experimentally determine whether the pri-miRNA transcripts of γHV68 are transcribed by RNA pol III, we infected α-amanitin-treated 293T cells with γHV68 for RT-PCR analysis to measure the abundance of the γHV68 pol III-1, pol III-4, and pol III-5 transcripts. α-Amanitin is widely used to distinguish the transcriptional activities of RNA pol II and III due to the fact that RNA pol II transcription is inhibited at relatively low doses of α-amanitin, whereas RNA pol III transcription is not inhibited. To control for the efficacy of α-amanitin, we tested transcription of a well-characterized RNA pol II gene, GAPDH, and RNA pol III gene, tRNATyr, by RT-PCR using total RNA from α-amanitin-treated 293T cells as template. The primer sets used for RT-PCR amplification of GAPDH and tRNATyr were designed to include intronic regions of these transcripts to identify only newly synthesized RNA made during treatment with α-amanitin. This experimental approach was directly modeled on the previous report that established that miRNA transcription is via pol II transcription (Lee et al. 2004). RNA pol II transcription was confirmed to be inhibited in mock infected 293T cells treated with α-amanitin at a concentration of 100 μg/mL for 24 h as shown by a lack of GAPDH transcription (Fig. 5A). This dose of α-amanitin did not inhibit RNA pol III activity as shown by the tRNATyr transcript (Fig. 5A). Therefore, treating 293T cells with α-amanitin at a concentration of 100 μg/mL for 24 h was sufficient to distinguish between RNA transcription of RNA pol II genes and RNA pol III genes.
FIGURE 5.
Demonstration of RNA pol III transcription of γHV68 miRNAs. 293 cells were infected with γHV68 and treated with α-amanitin at the indicated concentration. (A) RT-PCR analysis, with relative abundance of each RT-PCR product determined by densitometry analysis reported below the gels. (B) RLM-RT-PCR of miR-M1-1 products were cloned and sequenced to confirm the identity of the mature form of miR-M1-1. Primers for the cellular controls of the GAPDH and tRNATyr genes were used as previously described (Lee et al. 2004).
RT-PCR analysis of total RNA isolated from infected 293T cells treated with α-amanitin revealed no inhibition of transcription of the γHV68 pol III-5 transcript as expected for a RNA pol III transcript (Fig. 5A). Using this same total RNA, RT-PCR analysis of infected 293T cells treated with α-amanitin did reveal slight inhibition of both γHV68 pol III-1 and pol III-4. Therefore, to further validate RNA pol III transcription of the pri-miRNA viral transcripts in γHV68 infection, the abundance of maturely processed miR-M1-1 was determined by RLM-RT-PCR analysis from total RNA of the infected α-amanitin-treated cells. Shown in Figure 5B, the overall levels of mature miR-M1-1 were unchanged throughout the course of the α-amanitin treatment. It is important to note that α-amanitin treatment began 1 h after the initiation of infection, meaning that no previously accumulated viral transcripts play a role in this analysis. Together, these data demonstrate that biogenesis of both the pri-miRNA pol III transcripts and the mature processed miRNAs associated with them are resistant to α-amanitin treatment, proving that transcription of the γHV68 pri-miRNAs is indeed by RNA pol III during infection.
The γHV68 miRNAs are predicted to be processed from RNA pol III transcripts containing novel RNA pol III type 2-like promoters
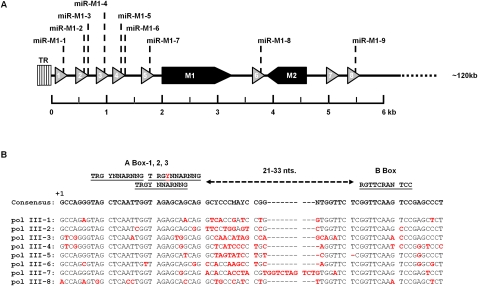
Previously published reports have predicted that the expression of the γHV68 miRNAs would be linked to eight closely spaced RNA pol III genes that contain putative tRNA-like elements (Fig. 6A; Pfeffer et al. 2005). These predictions were based on the prior identification of γHV68 tRNA-like genes immediately upstream of each miRNA (Bowden et al. 1997). Each γHV68 tRNA-like gene was shown to contain promoter elements consisting of internal A and B boxes, characteristic of RNA pol III type 2 promoters known to be associated with the transcription of tRNA genes (Paule and White 2000). Interestingly, more detailed examination of the RNA pol III type 2 promoter elements within the γHV68 miRNA coding locus revealed that each pol III type 2 promoter is more complex than previously appreciated. Rather than having a single A box promoter element present in each promoter, consistent with canonical pol III type 2 promoters, each promoter contained a set of three overlapping A boxes followed by a single B box. Spacing between any of the A boxes and the B box is within acceptable distance to allow for RNA pol III transcription of the γHV68 RNA pol III genes using any of the available A box promoter elements present (Fig. 6B; Paule and White 2000; Schramm and Hernandez 2002). In addition to previous reports of pol II run through transcripts, poorly characterized pol II transcripts, and vtRNAs, the presence of multiple A boxes raised the possibility that transcripts from these genes could vary from previous predictions (Bowden et al. 1997; Dutia et al. 2004). Therefore, to determine whether the γHV68 miRNAs are derived from RNA pol III transcription, we set out to delete the RNA pol III type 2-like promoters to determine their requirement for the production of viral miRNAs.
FIGURE 6.
Features of the γHV68 RNA pol III coding locus. (A) The left end 6 kb of the γHV68 genome containing the γHV68 RNA pol III transcripts 1–8 is shown. γHV68 RNA pol III transcripts are represented as triangles shaded in gray. Two protein coding transcripts, M1 and M2, are shaded in black. TR indicates terminal repeat region. (B) The RNA pol III promoter sequences for each γHV68 pol III gene. All γHV68 pol III transcripts are aligned to the γHV68 consensus sequence of all γHV68 RNA pol III promoter regions, which is indicated above the alignments. Minimal spacing distance between the A box and B box is shown as 21–33 nt. The red letter within the A box-2 consensus sequence represents a nucleotide position that is not conserved in the γHV68 pol III promoter regions. The red letters within the γHV68 pol III transcript sequences are nucleotide positions that deviate from the γHV68 pol III consensus sequence.
Deletion of the RNA pol III promoter elements eliminates γHV68 miRNA production
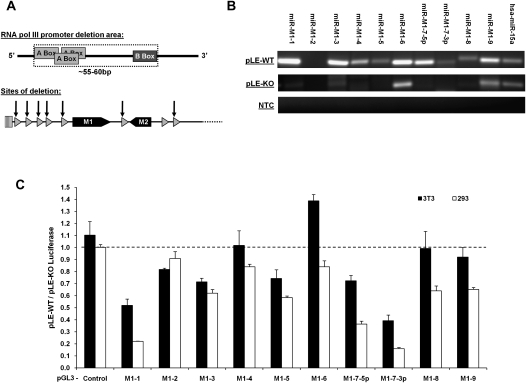
To corroborate our α-amanitin results, we sought to independently confirm that the transcription of the γHV68 miRNAs is by RNA pol III transcription. Using the pHV68-Left End wild-type (pLE-WT) plasmid containing the left ∼6 kb of the γHV68 genome as a template for site-directed mutagenesis, we deleted the RNA pol III promoter regions containing the three A boxes and the B box from every γHV68 pol III gene (except pol III-7, which is not documented to be associated with any immediate downstream miRNAs) to generate the pHV68-Left End miRNA KO (pLE-KO) plasmid (Fig. 7A). This plasmid was designed to be incapable of producing γHV68 miRNAs if RNA pol III transcription is required for their production. The miRNA sequences themselves were not deleted in this plasmid to allow for the possible production of the miRNAs that might be processed from any overlapping transcript.
FIGURE 7.
Confirmation of the production of the γHV68 miRNAs by RNA pol III using the Left End plasmids. (A) Schematic of the γHV68 RNA pol III promoter deletion strategy. (B) RLM-RT-PCR for miRNAs in 293 cells transfected with either pLE-WT or the pLE-KO plasmids. hsa-miR-15a serves as an endogenous control for miRNA expression in 293 cells. RT-PCR products were resolved by gel electrophoresis and visualized by ethidium bromide. (C) Results from the dual luciferase reporter analysis following transfection with either the pLE-WT or pLE-KO plasmid. pGL3 luciferase reporter target is labeled below the graph. All results are displayed as a ratio of firefly luciferase expression in the pLE-WT transfected cells divided by firefly luciferase expression in the pLE-KO transfected cells. Firefly luciferase readings are all normalized to the renilla luciferase transfection control. Readings obtained from 3T3 transfected cells are shown in solid black bars; 293 cells, in solid white bars. Readings below the dashed line represent repression by the corresponding viral miRNA in pLE-WT transfected cells.
To determine whether the pLE-KO plasmid lacked the ability to make the γHV68 miRNAs, 293 cells were transfected with either the pLE-WT or pLE-KO, and total RNA was harvested 42 h post-transfection for RLM-RT-PCR analysis of mature miRNA production. As with virus infection, transfection of the pLE-WT plasmid resulted in varying abundance of mature miRNA products and failed to produce mature miR-M1-2 miRNA product (Fig. 7B). In the 293 total RNA sample, the human miRNA miR-15a was used as an endogenous control based on previous work showing that this miRNA is expressed and processed within 293 cells (Landgraf et al. 2007). The RNA isolated from cells transfected with the pLE-KO plasmid did not contain mature viral miRNAs, providing independent confirmation that RNA pol III transcribes these viral miRNAs (Fig. 7B). However, RLM-RT-PCR amplification products were present within the miR-M1-6 and miR-M1-9 lanes from the pLE-KO samples. We determined that these amplification products were nonspecific and unrelated to viral miRNAs by cloning and sequencing. Sequencing of the corresponding RLM-RT-PCR products from the pLE-WT transfection yielded mature miR-M1-6 or miR-M1-9 in 100% of the clones sequenced.
We noted that miR-M1-8 yielded a RLM-RT-PCR product slightly larger than the other γHV68 miRNA products. This was seen with γHV68 infection as well (Fig. 4A). Upon cloning and sequencing of the miR-M1-8 RLM-RT-PCR product, it was revealed that these products were on average 3 nt longer than their expected size based on the published sequence of miR-M1-8. This was due to two factors. First, in all 11 clones analyzed, the 3′ end processing of the miRNA molecule was 1–3 nt beyond the previously published report. Second, in addition to the lengthened 3′ end of the miRNA molecule, five of the 11 clones analyzed also contained one to five nontemplated uracil residues at the 3′ end of the miRNA molecule, suggesting that the miR-M1-8 miRNA is subject to post-transcriptional and/or post-processing modification in virally infected and transfected cells.
To determine the biological activity associated with the γHV68 miRNAs produced from the pLE-WT plasmid, 3T3 or 293 cells were cotransfected with the pLE-WT or pLE-KO plasmids and the pGL3 luciferase reporter plasmids and the pRL-TK transfection control plasmid. Again, these experiments showed that not all of the γHV68 miRNAs confer a robust down-regulation of their respective targets in the luciferase reporter system (Fig. 7C). Luciferase readout is displayed as a ratio of pGL3 luciferase reporter expression from the pLE-WT transfections divided by the pGL3 luciferase reporter expression from the pLE-KO transfections. The pGL3 luciferase reporter plasmids with the lowest expression ratio are the most strongly down-regulated. As in the infected cells, miRNAs miR-M1-1, miR-M1-7-5p, and miR-M1-7-3p have the greatest ability to reduce their target reporter expression with a maximum repression approaching 85%.
DISCUSSION
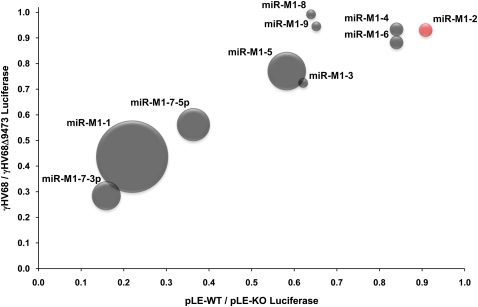
Early research related to miRNA biogenesis indicated that RNA pol II was uniquely responsible for transcribing pri-miRNA transcripts (Lee et al. 2004). However, recent studies have shown that RNA pol III can also transcribe naturally occurring pri-miRNAs (Borchert et al. 2006). Currently it is estimated that over 20% of all human miRNAs could be transcribed by RNA pol III, primarily from Alu repeat regions of the human genome (Borchert et al. 2006). Alu genes, like tRNAs, contain the same general RNA pol III type 2 promoter as the γHV68 pol III transcripts (Häsler et al. 2007) but lack the overlapping A box promoter elements seen in the γHV68 genome. Here we have shown that γHV68 RNA pol III transcription is directly associated with the production of the γHV68 miRNAs. Infection of cells treated with the RNA pol II inhibitor, α-amanitin, still allowed for the production of the viral pri-miRNA and mature viral miRNA molecules. We also demonstrated the requirement for the unique γHV68 RNA pol III promoters in the generation of the γHV68 miRNAs through promoter deletions. The miRNA biogenesis levels and biological activity of the pLE-WT plasmid closely mimic what is seen during γHV68 infection, indicating that all regulatory elements for miRNA generation are contained within this 6-kb genomic region and that the biological effects of the viral miRNAs can be uncoupled from virus infection and associated cytopathic effects. In this study, the pLE-WT plasmid resulted in the same pattern of miRNA biological activity as γHV68 infection, but the effect was stronger, presumably due to sustained miRNA expression in the absence of cytopathic effect. The direct correlation between the activity of the γHV68 miRNAs produced during infection and the γHV68 miRNAs produced from the pLE-WT plasmid during transfection is represented within Figure 8.
FIGURE 8.
Correlation of viral miRNA expression with viral miRNA biological activity. Shown is a graphical representation of γHV68 miRNA activity during γHV68 infection and pLE-WT transfection of 293 cells in relation to the cloning frequency demonstrated for each viral miRNA within a latently infected tumor cell line (Pfeffer et al. 2005). The y-axis represents the γHV68 to γHV68Δ9473 firefly luciferase ratio within infected 293 cells established in Figure 4C. The x-axis represents the pLE-WT to pLE-KO luciferase ratio within transfected 293 cells established in Figure 7C. The size of each data point represents the cloning frequency of each miRNA within the latently infected tumor cell line (Pfeffer et al. 2005). The color of each data point represents our ability to identify the maturely processed γHV68 miRNA product from γHV68 infected or pLE-WT transfected 293 cells. Gray data points are positive for our ability to detect these miRNAs; the red data point for miR-M1-2 indicates that we were unable to detect this miRNA during γHV68 infection or pLE-WT transfection. The correlation coefficient for the biological activity of all 10 γHV68 miRNAs between viral infection of 293 cells compared with transfection of the pLE-WT plasmid within 293 cells is R2 = 0.833. The correlation coefficient for the biological activity of the γHV68 miRNAs (except miR-M1-8 and miR-M1-9, which did not give appreciable biological activity within lytic infection of 293 cells) is R2 = 0.9627.
The role of the newly described RNA pol III type 2-like promoters, containing overlapping and triplicated A boxes, in relation to miRNA biogenesis is not known and is currently being investigated. Interestingly, the γHV68 RNA pol III transcripts were originally identified by their homology with tRNA pol III promoters and predicted to give rise to vtRNA transcripts (Bowden et al. 1997). This raises the possibility that other annotated tRNA genes from cellular genomes may contain this novel RNA pol III promoter element and that these features could signify the presence of a pri-miRNA transcript embedded within the predicted tRNA transcript. This hypothesis is currently under investigation.
There are other examples of viral RNA pol III transcripts with biological function during infection. Of those described to date, the adenovirus VAI gene has been extensively studied and shares several features with the γHV68 RNA pol III transcripts. The adenovirus VAI RNA is a 160-nt-long transcript of a canonical RNA pol III type 2 promoter. This transcript is produced at very high levels, accumulating up to 108 copies per infected cell (Mathews and Shenk 1991). The VAI RNA has been demonstrated to have several important biological functions. VAI binds to PKR to inhibit activation by viral double-stranded RNAs, bypassing an important innate anti-viral response (Mathews and Shenk 1991). VAI can effect global down-regulation of cellular miRNAs by overwhelming the nuclear-export receptor exportin-5 (Yi et al. 2003) and by suppressing Dicer activity as a competitive substrate (Lu and Cullen 2004; Andersson et al. 2005). This latter effect may be one potential reason for the scarcity of γHV68 mature miRNA relative to γHV68 pre-miRNA by Northern analysis. However, the ability of the VAI RNA to act as a DS does result in cleavage and production of small VA RNAs (svaRNAs). These svaRNAs are associated with RISC complexes during infection (Andersson et al. 2005), and although no target genes for the svaRNA charged RISC complexes have been identified, it is conceivable that this could also be a function of VAI during adenovirus infections.
Another interesting set of genes that share biological characteristics with the γHV68 RNA pol III transcripts are the EBER genes of EBV. Like VAI, the EBER transcripts are 167–172 nt in length and are transcribed at rates up to 5 × 106 copies per infected cell from RNA pol III type 2 promoters (Swaminathan 2008). However, the biological functions of the EBER genes appear to differ from those reported for VAI. Primarily, the EBER RNA molecules remain localized in the nucleus and are not processed into smaller RNAs. However, the exact biological function of the EBER RNAs has been elusive (Swaminathan 2008), and because each of the 10 γHV68 RNA pol III transcripts is not equivalent in their ability to give rise to miRNAs, overlapping biological functions with the EBERs may yet be found. Moreover, based on the physical and functional similarities to the VAI and EBER RNAs, this work has implicated a possible new class of multifunctional RNAs which may function both as regulatory RNAs and as miRNA precursors.
Interestingly, the γHV68 mature miRNAs were previously identified from a latently infected tumor cell line but were not detected in lytically infected cells. This is likely due to the unique opportunity for miRNA accumulation in latently infected tumor cells and the limited sensitivity of Northern blotting. However, we were able to detect mature viral miRNAs in lytically infected cells, indicating that miRNA regulation can play a role throughout all stages of γHV68 infection. The RLM-RT-PCR technique described here is extremely sensitive, with a limit of detection 100-fold below that of Northern blotting, and is capable of robustly detecting cellular miRNAs in the presence of active lytic infection. Sequence analysis of 109 RLM-RT-PCR reactions, cloned directly from unpurified amplification products, from pLE-WT transfected cells and γHV68 infected cells identified that 80% of these represented predicted mature miRNAs. This method has also proven capable of identifying viral miRNAs from total infected tissues ex vivo as far as day 16 post-infection (a time at which only one cell in 100 carries viral genome) (Flaño et al. 2002; Willer and Speck 2003). Overall, RLM-RT-PCR has many potential benefits, including (1) high sensitivity; (2) precise sequence determination of the amplified miRNA product; and (3) specificity, in that a 5′-monophophate, a natural byproduct of mature miRNA formation, is required to drive the T4 RNA ligase reaction.
Our analysis of mature γHV68 miRNAs from virally infected cells or from cells transfected with the pLE-WT plasmid demonstrated an expression pattern that differed from that cloned from the latently infected S11 tumor cell line (Pfeffer et al. 2005). It is not uncommon for the expression pattern associated with viral miRNA production to differ within different cellular contexts for viral infection. In EBV infections, global expression levels of mature EBV miRNAs can vary 25- to 50-fold based on the cell type infected (Pratt et al. 2009). In both EBV and KSHV infections, it has been demonstrated that the infection state of the virus, whether lytic, latent, or in specific stages of latency, can have a profound effect on viral miRNA production (Cai et al. 2006; Gottwein et al. 2006). Additionally, RNA pol III transcription has also been reported to vary based on the cellular context of infection as shown with EBER gene expression (Felton-Edkins et al. 2006). The combined effects of RNA pol III transcription and miRNA processing likely account for the differing levels of miRNA expression we observed in cells lytically infected with γHV68. These effects could also account for the fact that we were never able to detect mature miR-M1-2 in γHV68 infected or pLE-WT transfected cells. Previously, the sequence of the γHV68 miRNAs cloned with low frequency was based on one or two sequences and genomic context, and this RLM-RT-PCR analysis conclusively determined the actual sequence of these 10 viral miRNAs (Pfeffer et al. 2005). Finally, it appears that miR-M1-8 is differentially processed from its primary transcript and post-transcriptionally modified with the addition of nontemplated uracils in a manner that has not been previously described for this miRNA or for any pol III-derived miRNA. In fact, apart from tRNA maturation, post-transcriptional modification of pol III transcripts has not been widely described. Post-transcriptional modification of RNA pol II-derived miRNAs has been previously described, including the addition of nontemplated uracils to the pre-miRNA molecule (Winter et al. 2009). These data suggest that despite distinct transcript origins, pol II- and pol III-derived miRNAs are similar in their potential for post-transcriptional modification.
The biological activity associated with viral miRNA production is a topic of active investigation. Currently it is theorized that viral miRNAs can play key roles during the infection cycle, particularly in the establishment and maintenance of herpesvirus latency (Cullen 2009). Using luciferase reporter analysis, we have shown that several of the γHV68 miRNAs are biologically active beginning in lytic infection. This finding has important implications for the potential of viral miRNAs to shape the course of infection. Specifically the miR-M1-1, miR-M1-7-5p, and miR-M1-7-3p viral miRNAs all show strong biological activity during lytic infection and when generated from the pLE-WT plasmid during transfection. Prior to this work, the biological function of the γHV68 miRNAs was strictly speculative. We have collated γHV68 miRNA expression during latent (Pfeffer et al. 2005) and lytic infection with biological function, as demonstrated in Figure 8 and detailed in Supplemental Table 2. Several γHV68 miRNAs were of low abundance in both states of infection and demonstrated little biological activity. We found strong activity for miR-M1-1, the miRNA cloned with highest frequency from S11 tumor cells and abundant in lytic infection; however, miR-M1-5 was also frequently cloned from S11 tumor cells and abundant in lytic infection, yet showed far more modest biological activity. Therefore, apparent expression levels are not accurate predictors of biological activity and suggest that final activity of these pol III-derived viral miRNAs is subject to complex regulation at multiple levels. Future studies defining the biological targets of the γHV68 miRNAs and their function during various states of infection will yield important insights into the regulation of host and viral gene expression during virus infection and associated disease.
MATERIALS AND METHODS
RNA folding prediction and RNA modeling software
All RNA secondary structures depicted were predicted using the software RNAstructure 4.6 (Mathews et al. 2004). Predicted RNA structures were modeled into jpeg format using the XRNA software found at http://rna.ucsc.edu/rnacenter/xrna/xrna.html.
Viruses and tissue culture
The mouse fibroblast cell line 3T3 (ATCC CRL-1658), was cultured in Dulbecco's modified eagle media (DMEM) supplemented with 10% FBS (Hyclone), 2 mM L-glutamine, 10 U/mL penicillin, and 10 μg/mL streptomycin sulfate. The mouse fibroblast cell line 3T12 (ATCC CRL-164) and human 293T and 293 epithelial cell lines were cultured in DMEM supplemented with 5% FBS, 2 mM L-glutamine, 10 U/mL penicillin, and 10 μg/mL streptomycin sulfate. γHV68 infected or mock infected 293T cells were treated with 0.01 μg/mL, 1 μg/mL, or 100 μg/mL of α-amanitin (Sigma-Aldrich) for 24 h to distinguish between the activities of RNA pol II and III. 293T cells were infected with γHV68 for 1 h prior to the start of the α-amanitin treatment.
γHV68 strain WUMS (ATCC VR-1465) or γHV68Δ9473 were used for all viral infections (Clambey et al. 2002). Virus stocks used for infection were passaged, grown, and titer determined as previously described (Virgin et al. 1997). All in vitro infections were performed at a MOI of 5 plaque forming units (PFU) per cell. C57BL/6J mice (Jackson Laboratory) were used for all in vivo infections. Mice were infected intraperitoneally with 1 × 106 PFU of γHV68 diluted in 500 μL of DMEM supplemented with 5% FBS, 2 mM L-glutamine, 10 U/mL penicillin, and 10 μg/mL streptomycin sulfate. Infected tissues were collected either at day 5 or day 16 post infection. All mouse procedures were performed in accordance with the Institutional Animal Care and Use Committee at the University of Colorado Denver. At day 5, 3 × 108 splenocytes, 3 × 107 peritoneal cells, or ∼50 mg of liver, kidney, and lung tissue were harvested. At day 16, 3 × 108 splenocytes were harvested. Total RNA was isolated from each sample for RLM-RT-PCR analysis of miR-M1-1.
RNA isolation
Total and/or small (<200 nt) RNA was extracted using the mirVana miRNA Isolation kit (Ambion), using the manufacturer's instructions.
Small RNA Northern blot analysis
To begin, a 12% denaturing acrylamide gel containing a final concentration of 7 M urea was prerun at 60 mA for 45 min. While prerunning the gel, samples consisting of 20 μg of total cellular RNA, the miRNA Marker ladder (New England BioLabs), and the BrightStar Biotinylated RNA Century Plus Markers ladder (Ambion) were each brought to a final total volume of 40 μL using gel loading buffer II (Ambion) and boiled for 5 min at 95°C. After prerunning the gel, the samples were loaded and the gel was run at 30 mA for 60–70 min. The gel was stained with 1 μg/mL of ethidium bromide in 1× TBE for 5 min, and destained in 1× TBE for 2 min. The gel was transferred at 500 mA for 1 h onto Zeta-Probe GT Genomic Tested Blotting Membrane (Bio-Rad) by semi-dry transfer in 1× TBE transfer buffer. The membrane was ultraviolet cross-linked at 1200 × 100 μJ in an ultraviolet Stratalinker 1800 (Stratagene). The membrane was prehybridized in 10mLs of formamide hybridization buffer (KPL) mixed with 50 μL of 20 μg/μL sheared and denatured herring sperm DNA for 1 h at 58°C. During prehybridization, the 5′-biotinylated miR-M1-1 RNA probe (5′-/biotin/AAAGGAAGUACGGCCAUUUCUA-3′) or the 5′-biotinylated miR-M1-5 probe (5′-/biotin/GAGACGACCCGAUCUCAACUCU-3′), was denatured for 10 min at 68°C. Following prehybridization, 100 μL of 50 ng/μL denatured RNA probe was added to the formamide hybridization buffer mixture and the membrane was hybridized with the probe overnight at 58°C.
Following hybridization, the membrane was washed two times with 2× SSPE/0.5% SDS for 15 min at room temperature followed by two washes with 2× SSPE/0.5% SDS for 30 min each at 48°C. This was followed with one wash with 1× SSPE at room temperature for 5 min. The biotin-labeled probe was detected using the KPL blotting kit (no. 54-30-02) as described in the manufacturer's protocol. The AP-SA conjugate was used at a 1:7000 dilution. The membrane was exposed to Blue Lite Autorad Film (ISC BioExpress) from 5 to 30 min.
Plasmids and dicer substrates
The pLE-WT plasmid was constructed by PCR amplification of the left end of the γHV68 genome staring at nucleotide position 10 and ending at nucleotide position 5698 using the HV68 Left End Forward and HV68 Left End Reverse primer set (Supplemental Table 3). Viral genomic DNA was used as the template for the PCR reaction. The PCR consisted of 40 μL of distilled-deionized water, 5 μL of 10× Pfu Buffer, 1 μL of a 10 mM dNTP mix, 1 μL of 100 ng/μL viral genomic DNA template, 1 μL each of 100 ng/μL forward and reverse primer, and 1 μL of 2.5 U/μL Pfu Turbo enzyme (Stratagene). PCR cycling conditions: initial denaturing step of 2 min at 95°C, followed by 30 cycles 30 sec at 95°C, 30 sec at 59°C, and 6 min a 72°C, and a final elongation step of 10 min at 72°C. The ∼5.7-kb PCR product was gel purified and then inserted into the pCR-Blunt II-TOPO plasmid (Invitrogen) using the manufacturer's protocol. The insert was sequence verified to be an exact match of the γHV68 WUMS published sequence.
The pLE-KO plasmid was constructed using site-directed mutagenesis to sequentially delete the RNA pol III A box and B box promoter elements associated with every γHV68 RNA pol III transcript except for γHV68 pol III-7 (not associated with miRNA production) (Pfeffer et al. 2005). Site-directed mutagenesis was performed according to the manufacturer's protocol (Stratagene). After every deletion, the entire HV68 Left End insert was sequenced to confirm proper deletion of the RNA pol III promoter elements without off-target point mutations. Primers used for site-directed mutagenesis are shown in Supplemental Table 3.
The pRL-TK plasmid encodes renilla luciferase under the control of the HSV thymidine kinase promoter and was used as a transfection control in the dual luciferase assays. The pGL3-Control plasmid (Promega) encodes the firefly luciferase gene with transcriptional expression under control of the SV40 promoter. All pGL3 targeting plasmids were constructed by inserting the appropriate miRNA target sequence into the XbaI site in the pGL3-Control plasmid (in the 3′UTR between the stop codon of the firefly luciferase gene and the SV40 polyA tail sequence). Oligos containing the miRNA target site sequences were purchased from Integrated DNA Technologies (IDT) and are listed as XbaI M1-(X) Target Forward or Reverse in Supplemental Table 1. Two micrograms of forward and reverse oligos for each target site were annealed by heating to 95°C for 2 min and then cooled slowly to room temperature. Annealed oligos were run on a 2% TAE gel and gel purified using the QIAquick Gel Extraction Kit (Qiagen) before 23 μL were added to 10 μL of 10× T4 DNA ligase buffer with ATP (New England BioLabs), 1 μL of 10 U/μL T4 polynucleotide kinase (New England BioLabs), and 66 μL water and incubated for 30 min at 37°C . The oligos were then cloned into the pGL3-Control plasmid, and correct clones were confirmed by sequencing.
DS molecules were purchased from IDT. Each DS molecule consists of two oligos. The guide strand oligo, which contains the sequence of the mature miRNA product, is 27 bases long, consisting of only ribonucleic acid nucleotides. The passenger strand oligo is 25 bases in length and contains a 5′ monophosphate followed by 23 ribonucleic acid nucleotides and two deoxyribonucleic acid nucleotides. The sequence of the miR-M1-1 DS is as follows:
Passenger strand, 5′-/5Phos/GGAAGUACGGCCAUUUCUACCUAGA-3′; and
Guide strand, 5′-UCUAGGUAGAAAUGGCCGUACUUCCUU-3′.
The sequence of the miR-M1-5 DS is as follows:
Passenger strand, 5′-/5Phos/ACGACCCGAUCUCAACUCUCCUAGA-3′; and
Guide strand, 5′-UCUAGGAGAGUUGAGAUCGGGUCGUCU-3′.
The underlined bases are DNA; the bold bases represent the predicted, dicer processed mature miRNAs. The DS oligos were resuspended in RNase-Free Duplex Buffer, supplied by IDT, at a stock concentration of 100 μM. After resuspending the oligos, the oligos were annealed together by heating them to 94°C for 2 min and cooling them slowly to room temperature.
Transfections
All transfections for luciferase assays were performed in triplicate in a 24-well plate. All transfections for total RNA isolation were performed in duplicate in a six-well plate. One day prior to transfection, 3T3 or 293 cells were trypsinized and resuspended in their normal growth media, and 500 μL of 1.6 × 105 cells/mL were plated in the 24-well plates and 2 mL of 2 × 105cells/mL were plated in the six-well plates. All transfection experiments were performed using Lipofectamine 2000 (Invitrogen), using the manufacturer's instructions. Transfections in the 24-well plates included 100 ng pRL-TK and 50 ng of pGL3 (control or target) alone or combined with 263 ng of pLE-WT or 252 ng of pLE-KO or 6 pmol of DS per well. Transfections in the six-well plates included 1315 ng of pLE-WT or 1260 ng of pLE-KO or 30 pmol of DS per well. Transfected cells were then incubated until harvested for luciferase assays or total RNA isolation.
Dual luciferase assays
All dual luciferase assays were conducted in triplicate in 24-well plates using the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer's protocol. Forty-two to 48 h post transfection, cells were washed with 250 μL of 1× PBS prior to the addition of 100 μL of 1× passive lysis buffer (PLB), rocked at room temperature for 15 min; lysed cells were pipetted 20×, and 20 μL of lysate transferred into a well of a 96-well plate. Luciferase readings were collected using the Bio-Tek Synergy HT microplate reader (BioTek). The microplate reader dispensed 100 μL of Luciferase Assay Reagent II followed by a 10-sec reading of the sample (Firefly luciferase value) and then dispensed 100 μL of Stop & Glo reagent followed by a 10-sec reading of the sample (Renilla luciferase value). All data were exported as an integral value of the reading taken. The photo multiplier tube sensitivity was set at 110 during most of the dual-luciferase readings.
Reverse ligation mediated RT-PCR
Twenty micrograms of total isolated RNA were combined with 100 ng of the RNA Linker Oligo or the 5′P-RNA Linker Oligo (Supplemental Table 3). The RNA Linker Oligos were designed for minimal secondary structure to facilitate ligation by the T4 RNA ligase enzyme and subsequent RT-PCR. The total isolated RNA and the RNA Linker Oligo mixture was combined with 40 U of RNasin (Promega), 10 U of T4 RNA Ligase (New England BioLabs), and 5 μL of 10× T4 RNA Ligase reaction buffer; brought to 50 μL total volume with DEPC-treated RNase-free water; and incubated for 30 min at 37°C. After ligation, the reaction was heat inactivated for 15 min at 65°C. RNA was precipitated with 5 μL of NaOAc (pH 5.2) and 125 μL of 100% ethanol, incubated at −80°C for a minimum of 20 min, and pelleted at 16,170g for 15 min at 4°C. The pellet was washed with 200 μL of 70% ethanol, air dried 2–5 min, and resuspended in 100 μL of DEPC-treated RNase-free water. Two hundred nanograms of the ligated RNAs were used as template for RT-PCR amplification with the following cycling conditions: 30 min at 50°C, 15 min at 95°C, 40 amplification cycles of 30 sec at 94°C followed by 30 sec of 48°C, followed by 15 sec at 72°C, and a single final incubation for 2 min 30 sec at 72°C. Eighteen microliters of the RT-PCR reaction were then run on a 3% TAE agarose gel, and products were visualized by ethidium bromide. For sequence confirmation of mature miRNA products, 4 μL from the remaining 7 μL RT-PCR reaction were used for cloning using the TOPO TA Cloning Kit for Sequencing (Invitrogen).
RT-PCR amplification
Amplifications were conducted using the primers listed in Supplemental Table 3. RT-PCR amplification was performed in a 25 μL reaction mix using the OneStep RT-PCR Kit (Qiagen) containing a final concentration of each primer at 0.5 μM. Ten microliters (200 ng) of RNA template were treated with RQ1 RNase-Free DNase (Promega) prior to addition to the RT-PCR reaction mix. RT-PCR reactions were performed as follows: 30 min at 50°C (reverse transcription); 15 min at 95°C; 40 amplification cycles of 30 sec at 94°C followed by 30 sec of 48°C–56°C (dependent on the Tm of the primer set) and 1 min at 72°C, and a single final incubation for 10 min at 72°C.
Densitometry analysis of α-amanitin RT-PCR products
All densitometry analysis was conducted with the Kodak Molecular Imaging Software, version 4.0. Briefly, band intensities were measured, and relative band intensities were determined via the following equation: relative PCR product amount = (treated sample intensity − background intensity)/(untreated sample intensity − background intensity). All untreated samples were equal to 1, and all treated samples are expressed as a relative ratio of PCR product versus the untreated sample.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank David Bentley and Eric Clambey for their critical reading of and advice on this manuscript. We acknowledge all members of the van Dyk laboratory for discussions and comments on the manuscript. We thank Erin Buckingham and Lisa Williams for γHV68 infected ex vivo tissue samples. We declare no conflict of interest. This work was supported by NIH grant CA103632 to L.F.v.D. and by a pilot grant award to L.F.v.D. from the Colorado CFAR P30 AI054907.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1873910.
REFERENCES
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Andersson MG, Haasnoot PCJ, Xu N, Berenjian S, Berkhout B, Akusjärvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Blaskovic D, Stanceková M, Svobodová J, Mistríková J. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 1980;24:468. [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Bowden RJ, Simas JP, Davis AJ, Efstathiou S. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J Gen Virol. 1997;78:1675–1687. doi: 10.1099/0022-1317-78-7-1675. [DOI] [PubMed] [Google Scholar]
- Cai X, Cullen BR. Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. J Virol. 2006;80:2234–2242. doi: 10.1128/JVI.80.5.2234-2242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schäfer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clambey ET, Virgin HW, Speck SH. Characterization of a spontaneous 9.5-kilobase-deletion mutant of murine gammaherpesvirus 68 reveals tissue-specific genetic requirements for latency. J Virol. 2002;76:6532–6544. doi: 10.1128/JVI.76.13.6532-6544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Viral and cellular messenger RNA targets of viral microRNAs. Nature. 2009;457:421–425. doi: 10.1038/nature07757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutia BM, Roy DJ, Ebrahimi B, Gangadharan B, Efstathiou S, Stewart JP, Nash AA. Identification of a region of the virus genome involved in murine gammaherpesvirus 68-induced splenic pathology. J Gen Virol. 2004;85:1393–1400. doi: 10.1099/vir.0.79908-0. [DOI] [PubMed] [Google Scholar]
- Ebrahimi B, Dutia BM, Roberts KL, Garcia-Ramirez JJ, Dickinson P, Stewart JP, Ghazal P, Roy DJ, Nash AA. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J Gen Virol. 2003;84:99–109. doi: 10.1099/vir.0.18639-0. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton-Edkins ZA, Kondrashov A, Karali D, Fairley JA, Dawson CW, Arrand JR, Young LS, White RJ. Epstein-Barr virus induces cellular transcription factors to allow active expression of EBER genes by RNA polymerase III. J Biol Chem. 2006;281:33871–33880. doi: 10.1074/jbc.M600468200. [DOI] [PubMed] [Google Scholar]
- Flaño E, Kim I, Woodland DL, Blackman MA. Gamma-herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J Exp Med. 2002;196:1363–1372. doi: 10.1084/jem.20020890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cai X, Cullen BR. Expression and function of microRNAs encoded by Kaposi's sarcoma-associated herpesvirus. Cold Spring Harb Symp Quant Biol. 2006;71:357–364. doi: 10.1101/sqb.2006.71.004. [DOI] [PubMed] [Google Scholar]
- Grassmann R, Jeang K. The roles of microRNAs in mammalian virus infection. Biochim Biophys Acta. 2008;1779:706–711. doi: 10.1016/j.bbagrm.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- Hamada M, Huang Y, Lowe TM, Maraia RJ. Widespread use of TATA elements in the core promoters for RNA polymerases III, II, and I in fission yeast. Mol Cell Biol. 2001;21:6870–6881. doi: 10.1128/MCB.21.20.6870-6881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häsler J, Samuelsson T, Strub K. Useful ‘junk’: Alu RNAs in the human transcriptome. Cell Mol Life Sci. 2007;64:1793–1800. doi: 10.1007/s00018-007-7084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Ren G, Liu JL, Zhao ZA, Yu YS, Su RW, Ma XH, Ni H, Lei W, Yang ZM. MicroRNA expression and regulation in mouse uterus during embryo implantation. J Biol Chem. 2008;283:23473–23484. doi: 10.1074/jbc.M800406200. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kim D, Behlke MA, Rose SD, Chang M, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Poy MN, Stoffel M. Strategies to determine the biological function of microRNAs. Nat Genet. 2006;38(Suppl):S14–S19. doi: 10.1038/ng1799. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom K, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes & Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Shenk T. Adenovirus virus-associated RNA and translation control. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- Paule M, White R. Survey and summary: Transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Pratt ZL, Kuzembayeva M, Sengupta S, Sugden B. The microRNAs of Epstein-Barr virus are expressed at dramatically differing levels among cell lines. Virology. 2009;386:387–397. doi: 10.1016/j.virol.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P, Uhlenbeck O. Joining of RNA molecules with RNA ligase. Methods Enzymol. 1983;100:52–59. doi: 10.1016/0076-6879(83)00045-2. [DOI] [PubMed] [Google Scholar]
- Sano M, Kato Y, Taira K. Sequence-specific interference by small RNAs derived from adenovirus VAI RNA. FEBS Lett. 2006;580:1553–1564. doi: 10.1016/j.febslet.2006.01.085. [DOI] [PubMed] [Google Scholar]
- Scherer LJ, Frank R, Rossi JJ. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007;35:2620–2628. doi: 10.1093/nar/gkm103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes & Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Simas J, Efstathiou S. Murine gammaherpesvirus 68: A model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 1998;6:276–282. doi: 10.1016/s0966-842x(98)01306-7. [DOI] [PubMed] [Google Scholar]
- Simas J, Swann D, Bowden R, Efstathiou S. Analysis of murine gammaherpesvirus-68 transcription during lytic and latent infection. J Gen Virol. 1999;80:75–82. doi: 10.1099/0022-1317-80-1-75. [DOI] [PubMed] [Google Scholar]
- Speck S, Virgin H. Host and viral genetics of chronic infection: A mouse model of gamma-herpesvirus pathogenesis. Curr Opin Microbiol. 1999;2:403–409. doi: 10.1016/s1369-5274(99)80071-x. [DOI] [PubMed] [Google Scholar]
- Swaminathan S. Noncoding RNAs produced by oncogenic human herpesviruses. J Cell Physiol. 2008;216:321–326. doi: 10.1002/jcp.21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usherwood E, Stewart J, Nash A. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J Virol. 1996;70:6516–6518. doi: 10.1128/jvi.70.9.6516-6518.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin H, Latreille P, Wamsley P, Hallsworth K, Weck K, Dal Canto A, Speck S. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer DO, Speck SH. Long-term latent murine gammaherpesvirus 68 infection is preferentially found within the surface immunoglobulin D-negative subset of splenic B cells in vivo. J Virol. 2003;77:8310–8321. doi: 10.1128/JVI.77.15.8310-8321.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara I, Cullen B. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen B. Recognition and cleavage of primary microRNA transcripts. Methods Mol Biol. 2006;342:49–56. doi: 10.1385/1-59745-123-1:49. [DOI] [PubMed] [Google Scholar]