Abstract
The thi-box riboswitch regulates gene expression in response to the intracellular concentration of thiamine pyrophosphate (TPP) in archaea, bacteria, and eukarya. To complement previous biochemical, genetic, and structural studies of this phylogenetically widespread RNA domain, we have characterized its interaction with TPP by isothermal titration calorimetry. This shows that TPP binding is highly dependent on Mg2+ concentration. The dissociation constant decreases from ∼200 nM at 0.5 mM Mg2+ concentration to ∼9 nM at 2.5 mM Mg2+ concentration. Binding is enthalpically driven, but the unfavorable entropy of binding decreases as Mg2+ concentration rises, suggesting that divalent cations serve to pre-organize the RNA. Mutagenesis, biochemical analysis, and a new crystal structure of the riboswitch suggest that a critical element that participates in organizing the riboswitch structure is the tertiary interaction formed between the P3 and L5 regions. This tertiary contact is distant from the TPP binding site, but calorimetric analysis reveals that even subtle mutations in L5 can have readily detectable effects on TPP binding. The thermodynamic signatures of these mutations, namely decreased favorable enthalpy of binding and small effects on entropy of binding, are consistent with the P3–L5 association contributing allosterically to TPP-induced compaction of the RNA.
Keywords: allostery, calorimetry, crystallography, riboswitch, thiamine pyrophosphate
INTRODUCTION
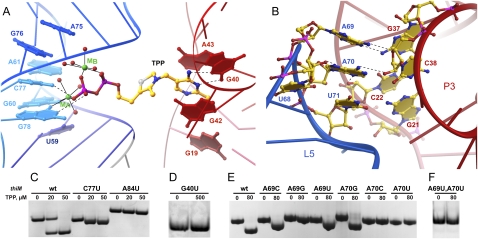
The thi-box riboswitch is an mRNA element that regulates gene expression in response to the intracellular concentration of thiamine pyrophosphate (TPP) (Miranda-Ríos et al. 2001; Winkler et al. 2002; Miranda-Ríos 2007). At present, it is the riboswitch with the broadest known phylogenetic distribution (Kubodera et al. 2003; Sudarsan et al. 2003; Bocobza et al. 2007; Cheah et al. 2007) and has been shown to be a target of an antibacterial compound (Sudarsan et al. 2005). Crystal structures have been reported of the aptamer domains of the TPP-responsive riboswitches thiM from Escherichia coli (Edwards and Ferré-D'Amaré 2006; Serganov et al. 2006) and thiC from Arabidopsis thaliana (Thore et al. 2006), both in complex with TPP. Superposition of these structures reveals nearly identical folds with minor variations due to insertions and deletions in nonconserved regions. The thi-box RNA is h-shaped (Fig. 1), with the TPP bridging the “pyrimidine sensor helix” (the coaxial stack of the P1-P2-P3 paired regions) and the “pyrophosphate sensor helix” (the P4-P5 stack) (Thore et al. 2006). The aminopyrimidine ring of TPP is recognized by residues of junction J3/2 of the riboswitch, where it stacks between G42 and A43 and forms hydrogen bonds with G40 and the 2′-OH of G19 (Fig. 2A). The pyrophosphate is recognized by residues 59–61 of J4/5 and 75–78 of J5/4 as a divalent cation chelate (Fig. 2A). Direct contacts to nonbridging oxygens of the β-phosphate of TPP are made by N4 of C77 and N1 of G78, but all other pyrophosphate–RNA contacts are mediated by the bound cations or their waters of hydration (Edwards and Ferré-D'Amaré 2006; Serganov et al. 2006; Thore et al. 2008).
FIGURE 1.
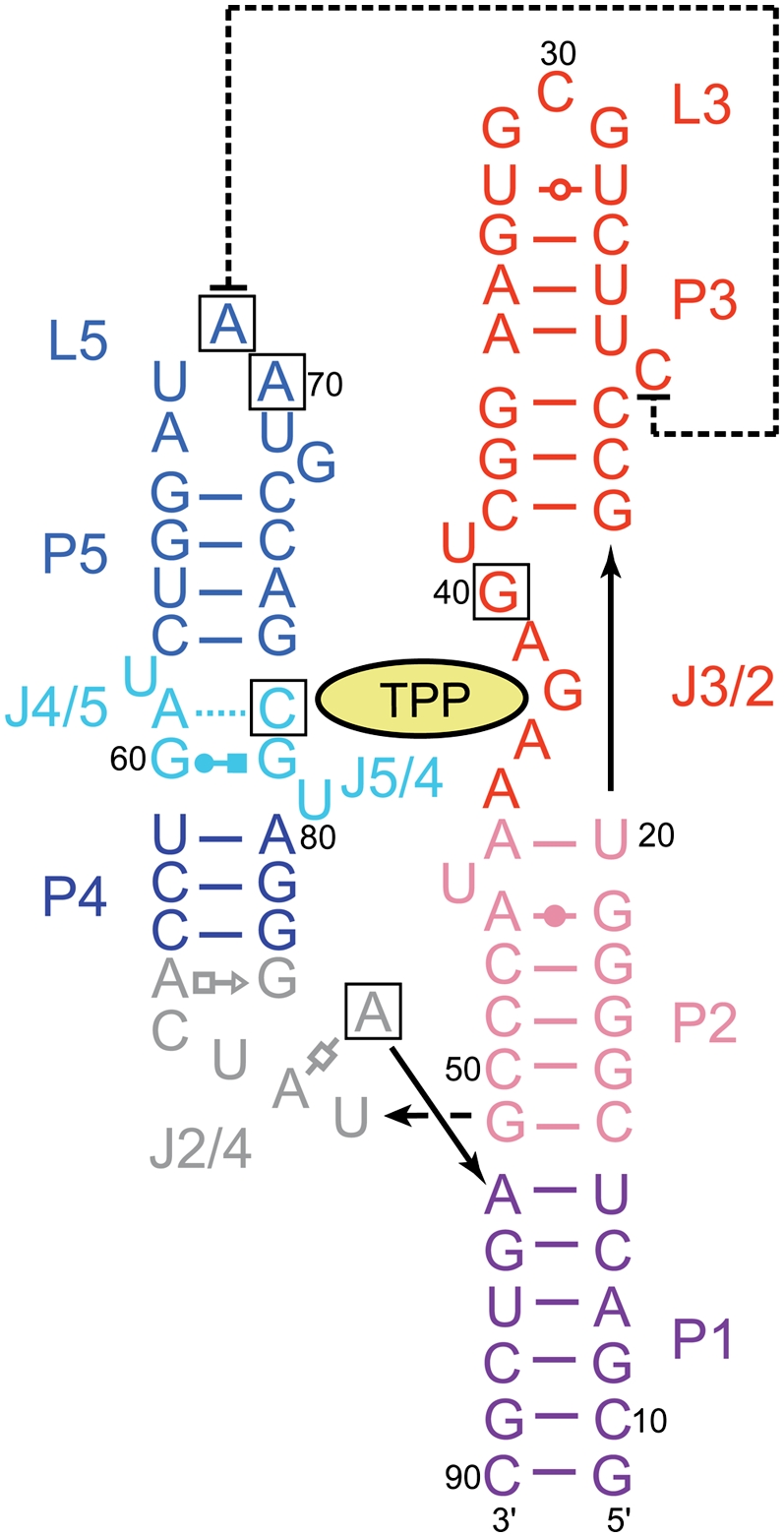
Sequence and secondary structure of the E. coli thiM riboswitch, based on the structure given by Serganov et al. (2006). (Black boxes) Site-directed mutations examined in this study, (lines with arrowheads) connectivity. Noncanonical pairs are denoted with Leontis–Westhof (Leontis and Westhof 2001) symbols. (Dashed line) Tertiary stacking interaction.
FIGURE 2.
Importance for TPP binding of structural features of the thi-box riboswitch. (A) Detail of the TPP binding pocket, with magnesium ion-dependent recognition of the pyrophosphate binding (left) and recognition of the aminopyrimidine by base pairing with G40 and stacking interactions. (Dashed lines) Hydrogen bonds, (solid lines) inner-sphere coordination. (B) P3–L5 interactions. (C) EMSA analysis shows that the mutation C77U is permissive for binding, whereas mutations A84U and G40U abolish binding (D). (E,F) EMSA analysis of mutants in positions 69 and 70 shows that only A69C, A69U, and A70G are permissive of binding. RNA concentration was 40 μM in C–F.
Three sets of phylogenetically conserved interactions stabilize the side-by-side packing of the pyrimidine and pyrophosphate sensor helices in the TPP-bound structures of both the bacterial and the plant thi-box riboswitches (Edwards and Ferré-D'Amaré 2006; Serganov et al. 2006; Thore et al. 2008). First, stacking and base-pairing interactions around J2/4 form the three-helix junction that connects the two helical stacks. J2/4 is formed by stacking of noncanonical base pairs A56•G83 and A53•A84, which dock into the minor groove of G•C pairs in the adjacent P2. There is continuous stacking between P1 and P2, and between noncanonical pairs in J2/4 and P4. Second, as described above, the bound TPP bridges the two helical stacks. Third, residues 69 and 70 from loop L5, which folds into a UNRN U-turn (Quigley and Rich 1976) motif (residue 68 being the first U), dock into the minor groove of P3 (Fig. 2B). Either monovalent (Serganov et al. 2006) or divalent (Edwards and Ferré-D'Amaré 2006) cations have been observed to bind to the RNA in the vicinity, possibly stabilizing this P3–L5 interaction.
Lang et al. (2007) synthesized a number of E. coli thiM riboswitch variants, in each of which a single nucleobase was replaced with 2-aminopurine. They employed these constructs and fluorescence spectroscopy to monitor the rate constants for conformational rearrangement induced by TPP binding in different parts of the RNA. From these studies, they deduced that TPP-induced folding of the riboswitch is not concerted. Instead, the P3–L5 interaction occurs first, the pyrophosphate- and aminopyrimidine-binding pockets form nearly at the same time or very shortly thereafter, and the three-helix junction region (and P1) folds last (Lang et al. 2007).
To complement the kinetic analyses of Lang et al. (2007), we have characterized the same bacterial riboswitch employing a combination of methods. We performed site-directed mutagenesis based on the crystal structures and qualitatively characterized the TPP binding activity of the mutant RNAs using an electrophoretic mobility shift assay (EMSA). Then, we analyzed the dependence on Mg2+ concentration of TPP binding to the riboswitch by isothermal titration calorimetry (ITC). These studies show that TPP binding to the riboswitch is enthalpically driven, and that there is a steep dependence on Mg2+ concentration. Furthermore, together with ITC analysis of thiamine monophosphate (TMP) binding to the riboswitch, these calorimetric analyses suggest that higher Mg2+ concentrations serve to pre-organize the RNA. Comparison of previous structures of the E. coli thiM riboswitch (Edwards and Ferré-D'Amaré 2006; Serganov et al. 2006) reveals a discrepancy in the structure of the L3 loop that is possibly due to crystal packing interactions. Because of the proximity of L3 to the functionally critical P3–L5 tertiary interactions, we determined a third independent crystal structure of the E. coli riboswitch, allowing us to establish the most likely structure of P3 in the absence of crystal packing interactions. This set the stage for a combined mutational and ITC analysis of the two critical nucleotides 69 and 70 at the center of the P3–L5 interaction. Our work corroborates the functional importance of structurally characterized features of the TPP-bound state of the riboswitch and provides a thermodynamic framework to understand metabolite binding and the metabolite-induced folding of the widely distributed TPP riboswitch.
RESULTS AND DISCUSSION
Mutational analysis of structural features of the thi-box-TPP complex
We developed an EMSA assay that allows rapid qualitative characterization of TPP binding by the thi-box riboswitch (Fig. 2C; Materials and Methods). Under our experimental conditions, the wild-type RNA displays an increased electrophoretic mobility when incubated in the presence of saturating TPP concentrations prior to native gel electrophoresis. This increase in electrophoretic mobility is consistent with TPP binding-induced compaction of the RNA determined in solution by small-angle X-ray scattering (SAXS) experiments (N Baird and A Ferré-D'Amaré, in prep.).
Our point-mutant RNAs (Fig. 1) displayed varying degrees of impairment in TPP binding. The N4 exocyclic amine of C77 makes a hydrogen bond with a nonbridging phosphate oxygen of the pyrophosphate of the bound TPP (Fig. 2A). Consistent both with the importance of this interaction and with the structural observation that this hydrogen bond is one of several interactions between the RNA and the pyrophosphate moiety of TPP, the C77U mutant maintains the ability to gel-shift in the presence of TPP, but its gel-shift is smaller than that of wild-type RNA (Fig. 2C). The nucleobase of A84 engages in multiple interactions at the three-helix junction. It forms a Hoogsteen-face pair with A53, its N1 hydrogen bonds to the 2′-OH of C50, and its N3 hydrogen bonds to the N2 of G16. In agreement with the importance of these interactions, the A84U mutant loses the ability to gel-shift in the presence of TPP. In addition, the electrophoretic mobility of this mutant is substantially less than that of the wild-type or the C77U mutant RNAs, suggesting that the A84U mutant RNA is not fully folded and that the formation of the junction is critical for correct folding (Fig. 2C). The nucleobase of G40 recognizes the aminopyrimidine ring of TPP by base pairing with it using its sugar edge (Fig. 2A). Thus, it is not surprising that the mutation G40U abolishes TPP binding (Fig. 2D). A G40C mutation has previously been shown to abolish TPP binding in vitro and riboswitch activity in vivo (Ontiveros-Palacios et al. 2008). We prepared all possible single mutations of A69 and A70, the central nucleotides of L5, and find that while position 69 tolerates both pyrimidines but not G, position 70 can only be mutated to G while maintaining TPP binding (Fig. 2E). The importance of the P3–L5 interaction (Lang et al. 2007) is corroborated by the double A69U, A70U mutant, which is inactive in TPP binding (Fig. 2F). Overall, these EMSA studies qualitatively confirm the importance of crystallographically observed TPP–RNA and RNA–RNA interactions, and emphasize the importance of the specific nucleotide sequence of L5 for TPP binding by the riboswitch.
Thermodynamics of ligand binding to the thiM aptamer domain
Circular dichroism (CD) studies on the E. coli thiA TPP-responsive riboswitch by Yamauchi et al. (2005) revealed that the aptamer domain of that RNA is markedly dependent on Mg2+ to bind to its cognate metabolite. Thus, its Kd for TPP was found to be 200 nM at a Mg2+ concentration of 0.2 mM, 36 nM at a Mg2+ concentration of 0.5 mM, and 20 nM at Mg2+ concentrations of 1.0 mM and higher. The riboswitch has a specific requirement for Mg2+, as molar concentrations of Na+, or 1 mM Ca2+ in the presence of 10 mM Na+, did not support TPP binding comparable to that seen even at 0.2 mM Mg2+ concentration. These studies also revealed thermodynamic linkage (Wyman 1948) between TPP and Mg2+ binding to the riboswitch: The apparent Kd for Mg2+ of the RNA in the absence of TPP is 0.17 mM, and Hill analysis of this Mg2+ binding revealed no cooperativity (Yamauchi et al. 2005).
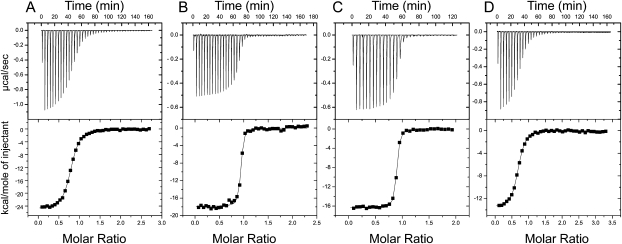
We employed ITC to deconvolute the enthalpy and entropy of TPP binding to the E. coli thiM riboswitch aptamer domain (Fig. 3; Table 1; Materials and Methods). At a Mg2+ concentration of 0.5 mM, the Kd for TPP is 198 nM. Binding is enthalpically favored (ΔH = −24 kcal mol−1) and entropically disfavored (−TΔS = 15 kcal mol−1). The unfavorable entropy of binding likely includes contributions from lost degrees of translational freedom (Murphy et al. 1994), organization of the RNA and TPP, and ion fixation. Binding becomes considerably tighter at 2.5 mM Mg2+ concentration (Kd ∼9 nM), and the entropic penalty is decreased markedly (ΔH = −18 kcal mol−1, −TΔS = 6 kcal mol−1). Increasing the Mg2+ concentration further to 10 mM has little impact on Kd, although the entropic penalty decreases slightly (ΔH = −16 kcal mol−1, −TΔS = 5 kcal mol−1). Direct characterization of dissociation constants of this order (∼9 nM) by ITC is experimentally problematic due to the steepness of the binding transition under conditions in which sufficient heat for measurement is evolved (Fig. 3B,C). However, the Kd values we determined are in good agreement with those measured for the E. coli thiA aptamer domain by CD (Yamauchi et al. 2005) and the E. coli thiM aptamer domain by in-line probing (Winkler et al. 2002). We also corroborated our direct ITC Kd measurements with competitive binding ITC experiments (Feig 2007) with TMP (data not shown).
FIGURE 3.
ITC analysis of TPP and TMP binding by the wild-type thiM thi-box aptamer domain. Representative TPP titrations at 0.5 mM (A), 2.5 mM (B), and 10.0 mM (C) Mg2+ concentration. (D) TMP titration at 10 mM Mg2+ concentration.
TABLE 1.
ITC analysis of ligand binding by wild-type E.coli thiM riboswitch
Calorimetric characterization of ligand binding has previously been reported for two other riboswitches. The aptamer domain of the guanine-responsive riboswitch of the xpt-pbuX operon of Bacillus subtilis was found to bind to hypoxanthine, a physiologic ligand, with a Kd of 3 μM, a strongly favorable enthalpy (ΔH = −35 kcal mol−1), and a strongly unfavorable entropy (−TΔS = 28 kcal mol−1) at 1 mM Mg2+ concentration. The Kd was found to decrease to 1.3 μM at 5 mM Mg2+ concentration, and to 730 nM at 20 mM Mg2+ concentration, with the enthalpy remaining favorable and the entropy unfavorable (Batey et al. 2004). Structural, mutational, and heat capacity change analyses support an induced fit of the RNA upon ligand binding that depends on formation of distal loop–loop interactions in addition to encapsulation of the purine ligand at its binding site (Gilbert et al. 2006). ITC analysis of ligand binding by the tetracycline aptamer at 10 mM Mg2+ concentration demonstrated that this artificial riboswitch binds its cognate ligand with a Kd of 0.8 nM, again, with a favorable enthalpy (ΔH = −23 kcal mol−1) and an unfavorable entropy (−TΔS = 10.5 kcal mol−1; Müller et al. 2006). Mutational and structural characterization suggest that this RNA is partially pre-organized, and that tetracycline binding is accompanied by local folding events in proximity to the ligand (Müller et al. 2006; Xiao et al. 2008).
Like in the cases of the guanine and tetracycline riboswitches (Batey et al. 2004; Müller et al. 2006), ligand binding by the thi-box riboswitch is enthalpically driven and Mg2+ ion-dependent. In our ITC experiments, TPP was dissolved in buffer solutions with the same Mg2+ concentration as the RNA solutions. Therefore, the heats of binding detected are not due to chelation of TPP by Mg2+. In control experiments, we determined that the heat of dilution is very small under our experimental conditions (by titrating TPP dissolved in buffer with Mg2+ into buffer with the same Mg2+ concentration; data not shown). In addition, to control for slight Mg2+ concentration mismatches between the TPP and RNA solutions, we carried out experiments in which TPP in buffer without Mg2+ was titrated into buffer solutions containing as much as 10 mM Mg2+, but without riboswitch RNA. These titrations evolved little heat (data not shown). The small heat of chelation of TPP by Mg2+ is also consistent with the thermodynamic linkage (Wyman 1948) between Mg2+ and TPP binding to the riboswitch reflected in the CD experiments of Yamauchi et al. (2005). Similarly to the behavior of the guanine riboswitch, TPP binding by the thi-box riboswitch becomes pronouncedly tighter at physiologic (∼1 mM) (Batey et al. 2004) Mg2+ concentration. However, unlike the guanine riboswitch, where the unfavorable entropy drops only modestly between low and high Mg2+ concentrations (−TΔS drops from 34 kcal mol−1 at 0.25 mM Mg2+ to 25 kcal mol−1 at 20 mM Mg2+; Batey et al. 2004), the thi-box riboswitch exhibits a sharp reduction in the unfavorable entropy as Mg2+ is raised to physiologic levels (Table 1). We interpret this as an indication that the thiM riboswitch aptamer domain becomes considerably pre-organized (Draper 2004) at physiologic or higher Mg2+ concentrations prior to the addition of TPP. Consistent with this, TPP binding results in a substantially less favorable enthalpy of binding as the Mg2+ concentration is increased (Table 1). This interpretation is also in agreement with SAXS studies showing that the thi-box riboswitch aptamer domain compacts, even in the absence of TPP, as Mg2+ concentration is raised above 2.5 mM (N Baird and A Ferré-D'Amaré, in prep.).
Crystallographic structure determination of the E. coli thiM riboswitch in complex with TMP revealed that the single phosphate of this metabolite is recognized by the RNA in a manner analogous to how it recognizes the pyrophosphate of TPP. Namely, the phosphate of TMP is coordinated by two divalent cations, and these, and their hydration waters, largely mediate recognition of the phosphate by the RNA (Edwards and Ferré-D'Amaré 2006). Because TMP is shorter than TPP, the RNA adopts a conformation in which the pyrophosphate and pyrimidine sensor helices are closer to each other than in the cognate (TPP) complex. In addition to this global accommodation, comparative analysis of the complexes of the riboswitch with TPP, TMP, and two unnatural ligands of lower affinity revealed an increase in static disorder of nucleotides that form the pyrophosphate binding site as the number of phosphates of the ligand decreased from two to one to zero. We characterized TMP binding by the E. coli thiM riboswitch (Fig. 3D; Table 1) and observe binding parameters consistent with these structural findings. The Kd for TMP is ∼260 nM at a Mg2+ concentration of 10 mM, and the enthalpy of binding is less favorable than that for TPP (−15.7 kcal mol−1 versus −13.6 kcal mol−1). In contrast, the unfavorable entropy of binding is similar between TPP and TMP binding (−TΔS = 4.5 kcal mol−1). The lower affinity for TMP likely results from the molecular strain resulting from the additional compaction of the RNA needed to bring the two helical stacks closer together, and the less favorable enthalpy reflects the lower degree of complementarity between the TMP and the RNA, as manifest crystallographically in increased static disorder (Edwards and Ferré-D'Amaré 2006). Since the degree of pre-organization of the RNA induced by physiologic or higher Mg2+ should be independent of the ligand, the entropies of binding to TMP or TPP are similar. The affinity of the riboswitch for TMP at lower Mg2+ concentrations was too weak to determine by ITC. Because physiologic Mg2+ concentration is ∼1 mM (Batey et al. 2004), the 30-fold discrimination between TPP and TMP shown by the riboswitch at a Mg2+ concentration of 10 mM is probably a lower bound for its in vivo selectivity.
Structural plasticity of P3 and L3
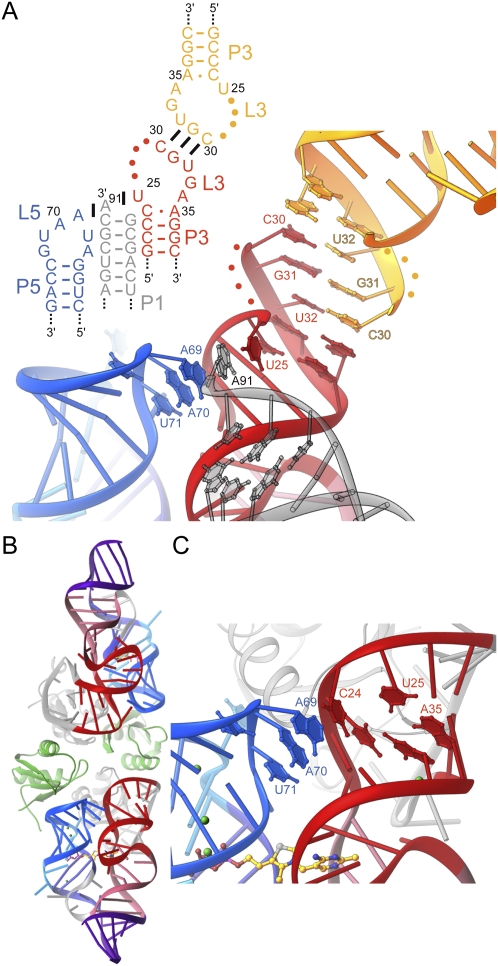
Two crystal structures of the E. coli thiM riboswitch aptamer domain in complex with TPP have been reported (Edwards and Ferré-D'Amaré 2006; Serganov et al. 2006). Overall, the structures superimpose very closely, with the exception of the distal portion of P3 and of L3. In the crystal form of Serganov et al. (2006), C24 is extruded from P3, and its nucleobase stacks on top of A69 from L5 (Fig. 1). In this structure, L3 is formed by residues 29–31, which adopt a U-turn like conformation with their bases exposed. This allows C30 and G31 to form Watson–Crick pairs with the same residues of a second RNA in the asymmetric unit of the crystal. In contrast, in the crystal form of Edwards and Ferré-D'Amaré (2006), C24 base pairs with A35, and therefore does not stack on A69. Instead, the 3′-terminal nucleotide A91 of a symmetry-related molecule stacks on this nucleotide from L5. Furthermore, this symmetry-related A91 in turn stacks below U25 from the reference molecule, which is extruded from P3. L3 is much larger, comprising nucleotides 25–34, with residues 26–29 disordered, and 30–33 forming Watson–Crick pairs with a second symmetry-related molecule (Fig. 4A). Essential to this intermolecular arrangement of three riboswitch RNAs is an unnatural terminal 2′,3′-cyclic phosphate of the crystallization construct, which was generated by ribozyme cleavage (Ferré-D'Amaré and Doudna 1996) during transcription (T Edwards and A Ferré-D'Amaré, unpubl.). Because the thiM aptamer domain behaves as a monomer in solution (N Baird and A Ferré-D'Amaré, in prep.), it is unlikely that the specific nucleotide arrangement observed in the crystal form of Edwards and Ferré-D'Amaré (2006) occurs in vivo. However, given that A69 is critical for riboswitch function (Fig. 2E), it is important to establish whether stacking of C24 on A69, made possible by extrusion of the former from P3 (Fig. 1), is a feature of the riboswitch structure that is not contingent on specific crystal packing interactions.
FIGURE 4.
Structural plasticity of P3. (A) Schematic secondary structure and cartoon representation of the P3–L5 region of the riboswitch in its crystalline context, in the structure given by Edwards and Ferré-D'Amaré (2006). Interactions formed with symmetry-related molecules in the crystal (gray and yellow) are shown. Note intercalation of A91 from a symmetry-related molecule between U25 and A69. (B) Cartoon representation of the new E. coli thiM aptamer domain crystal structure. The two RNA molecules in the asymmetric unit are colored as in Figure 1. (Gray) Cognate U1A-RBDs, (green) two additional U1A-RBDs that do not make specific RNA-binding interactions. (C) Detail of P3–L5 interaction in the new crystal structure. Note extrusion of C24 from P3, and its stacking on A69.
To resolve this question, we determined the structure at 3.1 Å resolution of a new crystallization construct of the thiM riboswitch aptamer domain (Table 2; Materials and Methods). In order to eliminate crystal packing interactions of L3, we replaced this loop with the binding site for the spliceosomal protein U1A (Ferré-D'Amaré and Doudna 2000a). Comparison of the structures of unrelated RNAs determined with and without the U1A crystallization module, such as the hairpin ribozyme (Rupert and Ferré-D'Amaré 2001; Ferré-D'Amaré 2004; Alam et al. 2005) and the glmS ribozyme-riboswitch (Klein and Ferré-D'Amaré 2006; Cochrane et al. 2007; Klein et al. 2007b) have demonstrated that placement of the U1A binding site distal to the active site of RNAs has minimal structural impact on their core structures. Our new structure comprises two independent thiM riboswitch aptamer domains in the asymmetric unit, and four copies of the RNA-binding domain (RBD) of the U1A protein. Two of these are binding their respective cognate sites, and two are not making specific RNA contacts (Fig. 4B). As expected, the structure of the core of the riboswitch is indistinguishable from those of previous structures. Importantly, in both RNA molecules in the asymmetric unit, C24 is extruded from P3, and stacks on A69, in a manner indistinguishable from that seen in the Serganov et al. (2006) crystal structure (Fig. 4C). Therefore, the C24–A69 stacking interaction is likely to be an authentic structural feature of the E. coli thiM riboswitch–TPP complex. Collectively, the crystallographic studies also highlight an interesting asymmetry in the P3–L5 interaction: While L5 appears to be insensitive to crystal packing interactions, P3 is quite labile. Thus, it is possible that P3 and L3 are poorly structured in the free state of the riboswitch, and undergo folding as part of TPP binding. This may contribute to the unfavorable entropy of TPP binding.
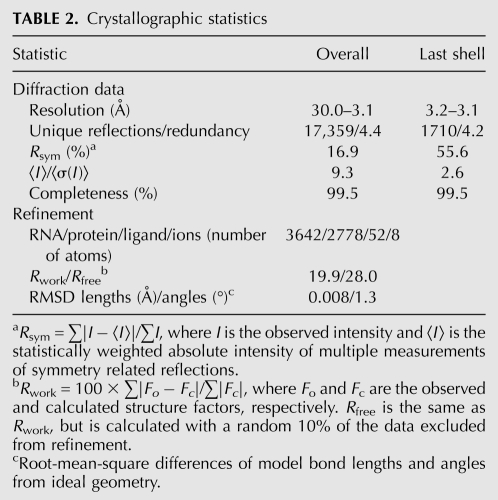
TABLE 2.
Crystallographic statistics
Thermodynamics of mutant L5–P3 interactions
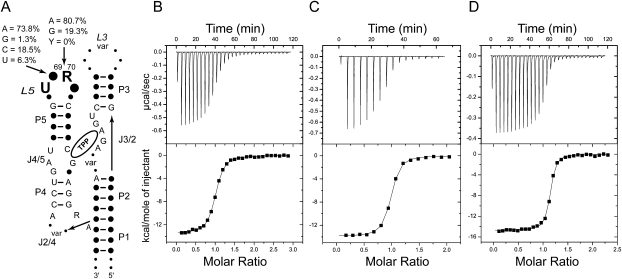
Analysis of 500 representative thi-box riboswitch structures in the Rfam database (Gardner et al. 2009) shows that while L3 is highly variable, L5 has a pattern of strong sequence conservation (Fig. 5A). More than 90% of sequences have U68 at the first position of the eponymous (Quigley and Rich 1976) U-turn. A thi-box riboswitch with a U68A mutation has been shown to be inactive in vivo (Ontiveros-Palacios et al. 2008). Position 69 is usually (∼74%) an adenosine, although cytosine is quite frequent (∼19%), with uridine being observed occasionally (∼6%). Position 70 is invariably a purine, with adenosine being predominant (∼81%). Position 71 has no clear sequence preference. Our EMSA results (Fig. 2E), discussed above, are consistent with this conservation pattern, as A69G, A70C, and A70U all abolish the ability of the thiM aptamer domain to gel-shift in the presence of TPP. To provide additional insight into the role of positions 69 and 70 in TPP binding by the riboswitch, we analyzed the mutants that retain in vitro activity by ITC.
FIGURE 5.
ITC analysis of the L3–P5 interaction. (A) Schematic representation of phylogenetic sequence conservation of the thi-box riboswitch aptamer domain. (Solid circles, “var”) Variable positions. A summary of the analysis of 500 sequences from the Rfam database (Gardner et al. 2009) is given for positions 69 and 70. (B) Representative ITC experiment analyzing binding of TPP to E. coli thiM aptamer domain mutant A69C. (C) TPP titration of mutant A69U. (D) TPP titration of mutant A70G. All experiments were performed in the presence of 10 mM Mg2+.
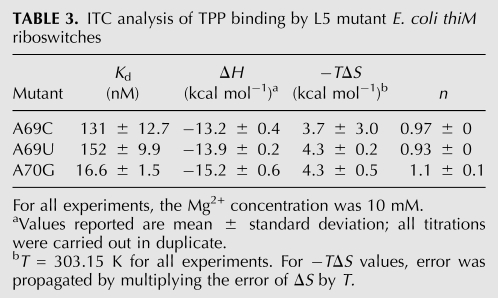
In the crystal structures, the nucleobase of A69 is sandwiched between C24 and A70 and makes two hydrogen bonds in the minor groove of P3: its N1 receives a hydrogen bond from the N2 exocyclic amine of G37, and its N6 exocyclic amine donates a hydrogen bond to the O4′ furanose ring oxygen of C38 (Fig. 2B). The former interaction is sequence specific for both positions 69 and 37. Only a guanosine in position 37 of P3 would place a hydrogen bond donor at this position in the minor groove. The interaction would be lost with an adenosine (which has no exocyclic functional group at purine position 2), and cytosine and uridine would place a carbonyl oxygen at approximately the same position. In addition to hydrogen bonding, the position of the base of A69 brings its C2 into van der Waals contact (3.2 Å) of the furanose ring oxygen of C23. This network of interactions explains why the A69G mutant is inactive (Fig. 2E): a guanosine would have an exocyclic amine at position 2 (clashing with the furanose of C23), a hydrogen bond donor at position N1 (clashing with the N2 amine of G37), and a carbonyl at position 6 (clashing with the furanose of C38). The mutations A69C and A69U are probably tolerated (Fig. 2E) because the smaller pyrimidine bases can occupy the position of the adenosine, but do not reach deep enough into the minor groove of P3 to make favorable or unfavorable interactions. This would also be consistent with the phylogenetic conservation pattern (Fig. 5A). ITC analysis shows that, at 10 mM Mg2+ concentration, the A69C mutant binds TPP with a Kd of 131 nM, and the A69U mutant binds TPP with a Kd of 152 nM (Fig. 5B,C; Table 3). Consistent with pyrimidines being tolerated at position 69 because of lack of steric clashes, the affinity for TPP of the mutants is reduced by 16- to 18-fold and the favorable enthalpy of binding by these mutants is somewhat smaller than that of the wild-type (−13.2 and −13.9 kcal mol−1 for the mutants versus −15.7 kcal mol−1 for the wild type). Conversely, their entropy of binding (−TΔS ∼4 kcal mol−1) is slightly less unfavorable than that of wild type (−TΔS = 4.5 kcal mol−1).
TABLE 3.
ITC analysis of TPP binding by L5 mutant E. coli thiM riboswitches
The nucleobase of A70 makes two sequence-independent hydrogen bonds (Fig. 2B). First, its N6 exocyclic amine donates a hydrogen bond to the 2′-OH of C38. Second, its N3 imine receives a hydrogen bond from the 2′-OH of C22. Because both hydrogen bond-partners in P3 are ribose hydroxyls, any sequence would be tolerated in this step of P3. The A70G mutant would be able to receive a hydrogen bond at its N3. Although it would have a carbonyl rather than an amine at position 6, its partner, the hydroxyl of C38, would be able to function as a donor rather than as an acceptor. By similar arguments, it might be rationalized that both U and C would be tolerated at this position (both pyrimidines have a hydrogen bond acceptor, their O2 carbonyls, at approximately the same location as the N3 of the purines, and they would be too short to reach the 2′-OH of C38). The inactivity of the A70C and A70U mutants (Fig. 2E) and the lack of pyrimidines at this position in phylogeny (Fig. 5A) are explained by a third hydrogen bonding interaction of the nucleobase of A70. Position N7 of A70 receives a hydrogen bond (2.6 Å) from the 2′-OH of U68 of the U-turn motif. The exocyclic functional groups of either pyrimidine (N4 for cytosine and O4 for uridine) are likely to clash with the 2′-OH, and therefore preclude formation of the U-turn. This would have the consequence of disrupting the P3–L5 interaction. ITC analysis of TPP binding (at 10 mM Mg2+ concentration) by the A70G mutant (Fig. 5D; Table 3) is consistent with both the presence of guanine at this position in many thi-box riboswitches (Fig. 5A) and in biochemical activity of the RNA (Fig. 2E). The mutant RNA binds TPP with an affinity that is only twofold lower than that of wild type, and both its enthalpy and entropy of binding are comparable to those of the wild-type RNA. Thus, our thermodynamic analysis confirms that guanine is well tolerated at this position.
Our mutational analysis (Fig. 2E,F) demonstrates that alteration of the P3–L5 interaction, which does not participate directly in forming the TPP binding site, can abrogate riboswitch function. Our ITC characterization of three L5 mutants that retain the ability to gel-shift in the presence of TPP show that even relatively subtle point mutations in nucleotides that form this tertiary interaction can have readily detectable effects on the thermodynamics of riboswitch–TPP interaction (Table 3). This complements the kinetic analysis of thiM riboswitch function by Lang et al. (2007), who found that it is precisely the P3–L5 interaction that forms first when the riboswitch encounters its cognate metabolite. The functional importance of this distal interaction has parallels in other well-characterized h-shaped biological RNAs. For instance, mutational, thermodynamic, and kinetic characterization of the guanine riboswitch has demonstrated that formation of tertiary interactions that are distant from the metabolite binding pocket but help form the parallel arrangement of helices present in the guanine-bound form of the RNA are essential for function (Gilbert et al. 2006). Perhaps most dramatic is the discovery that loop–loop interactions distal from the active site are essential for the hammerhead ribozyme to function efficiently (De la Peña et al. 2003; Khvorova et al. 2003; Canny et al. 2004) and to fold into an h-shaped arrangement in which the substrate is presented in a chemically reactive conformation to a fully assembled active site (Martick and Scott 2006).
Conclusion
With the exception of the glmS ribozyme-riboswitch (Winkler et al. 2004; Collins et al. 2007; Klein et al. 2007a), riboswitches are thought to modulate gene expression by employing the free energy of binding to their cognate small molecule to adopt a structure that is distinct from that of their ligand-free state. Most commonly, this involves sequestration of an RNA segment into helix P1 of the ligand-bound state of the riboswitch that otherwise is available to interact with the gene expression machinery (for reviews, see Edwards et al. 2007; Henkin 2008; Montange and Batey 2008; Serganov 2009; for a riboswitch that employs a different strategy, see Klein et al. 2009). From this mechanism, it would be expected that mutations in the ligand binding pocket would have pronounced effects on riboswitch function. Indeed, in vivo analysis of such mutations of the thi-box riboswitch has demonstrated that alterations in the TPP binding site can result in inactive or in constitutively active riboswitches (Ontiveros-Palacios et al. 2008).
Our thermodynamic analysis of TPP binding by this riboswitch as a function of Mg2+ concentration is consistent with the view that ligand binding leads to a more compact RNA, and that higher Mg2+ concentrations favor ligand binding by pre-organizing the riboswitch. Our work, together with previous kinetic analyses by Lang et al. (2007), highlights the importance of the P3–L5 interaction that is distal from the TPP binding site for metabolite binding. This allosteric contribution to the function of the riboswitch is reminiscent of the mechanism of action of other h-shaped biological RNAs including riboswitches and ribozymes, and is consistent with riboswitches being generally larger and more structurally complex than in vitro selected RNAs (aptamers) that recognize small molecules with comparable affinity and specificity (Edwards et al. 2007). Riboswitches appear to have evolved to optimize not only their affinity and selectivity, but also their rate of folding (Wickiser et al. 2005a,b). We speculate that allosteric interactions, by modulating the nature of the global rearrangement of the aptamer domain upon metabolite binding, may serve to fine-tune the kinetics of riboswitch function.
MATERIALS AND METHODS
Reagent preparation
The insert of pTB13, a pUC19 derivative that encodes the E. coli thiM thi-box riboswitch preceded by a hammerhead ribozyme and followed by a VS ribozyme substrate stem–loop (Ferré-D'Amaré and Doudna 1996), was prepared by PCR from overlapping synthetic oligonucleotides. Plasmids encoding site-directed mutants were generated using the QuikChange Kit (Stratagene). RNA transcription and purification were as described (Rupert and Ferré-D'Amaré 2004). Expression and purification of U1A-RBD have been described (Ferré-D'Amaré and Doudna 2000a). TPP and TMP were purchased from Fluka and were employed without further purification.
Electrophoretic mobility shift assay
The thi-box RNAs were incubated with TPP or TMP in a buffer containing 5 mM Tris-HCl (pH 8.0), 3 mM MgCl2, 10 mM NaCl, and 100 mM KCl for 30 min at 310 K. Glycerol was added to the RNA samples to a final concentration of 40% (25% for the experiment shown in Fig. 2C) (v/v) prior to loading on native gels (10% polyacrylamide, 0.5× THE, 1 mM MgCl2) that had been pre-run for 30 min. Electrophoresis was carried out at ∼10 W, maintaining the temperature of the gel below 303 K, for ∼2 h. For the experiments shown in Figure 2, C and E, the gel was run for 4 h at ∼20 W. To prevent depletion of Mg2+, the buffers of the anode and cathode chambers were mixed every 30 min. RNA was visualized by toluidine blue staining.
Isothermal titration calorimetry
RNA was dialyzed for 18–24 h against a 100-fold excess of a buffer containing 50 mM HEPES-KOH (pH 7.5), 100 mM NaCl and 0.5-10 mM MgCl2 at 298 K. TPP and TMP were dissolved in the buffer that the RNA was dialyzed against to ensure an exact buffer match to minimize the heat of dilution. The final concentration of the small molecule was 10- to 12-fold higher than that of the RNA sample. RNA concentration was determined by UV spectrometry. Final RNA concentrations of ∼10 μM were used for high-affinity binders, and ∼15 μM for low-affinity binders. Prior to the ITC experiment, all samples were degassed for 10 min at 298 K. ITC experimental parameters were a temperature of 303.15 K, a reference power of 20 μcal sec−1, an initial delay of 60 sec, and titrations consisting of either forty-two 7 μL injections or thirty-one 10 μL injections at an injection rate of 0.5 μL sec−1 with individual injections spaced 240 sec apart. Measurements were carried out on a VP-ITC microcalorimeter (MicroCal) with the exception of measurements on the A69U mutant, which were performed on an ITC200 microcalorimeter (MicroCal). For this mutant, the RNA concentration was ∼23 μM, and instrument parameters were a temperature of 303.15 K, a reference power of 3 μcal sec−1, and an initial delay of 60 sec, and the titration consisted of sixteen 2.48 μL injections at an injection rate of 0.5 μL sec−1 with individual injections spaced 240 sec apart. Data from each experiment were fit to a single-site binding model using Origin ITC software (MicroCal software Inc.).
Crystallization, structure determination, and refinement
A solution comprising 0.1 mM RNA, 0.13 mM U1A-RBD Y31H, Q36R double mutant (Oubridge et al. 1995), 0.5 mM TPP, 100 mM KCl, 5 mM MgCl2, 0.25 mM spermine, 10 mM NaCl, and 5 mM Tris-HCl (pH 8.1) was incubated for 30 min at 310 K. Crystals were grown by vapor diffusion of drops prepared by mixing 1.5 μL of the RNA–protein–TPP complex with 3 μL of a reservoir solution comprising 20% (v/v) PEG 550 MME, 20% MPD, and 80 mM sodium citrate (pH 5.6) at 298 K. Crystals grew as plates with maximum dimensions of 120 × 70 × 20 μm3 within 2 wk, and were flash frozen by mounting in nylon loops and plunging into liquid nitrogen. Diffraction data were collected by the oscillation method at beamline 5.0.2 of the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory, and reduced with the HKL package (Otwinowski and Minor 1997). The crystals have the symmetry of space group P21, with unit cell dimensions a = 52.4 Å, b = 72.0 Å, c = 128.6 Å, and β = 94.6°. From crystal density considerations (Matthews 1968; Ferré-D'Amaré and Doudna 2000b), two RNA–protein complexes were expected to be present in the asymmetric unit.
The structure was determined by molecular replacement using the core of the thiM riboswitch structure of Edwards and Ferré-D'Amaré (2006) as the search model and the program PHASER (McCoy et al. 2007). The top solution was manually rebuilt (Jones et al. 1991), subjected to rigid body, simulated annealing, energy minimization, and tightly restrained individual isotropic B-factor refinement (Brünger et al. 1998). The resulting electron density maps revealed the location of two U1A binding loops and their bound U1A-RBDs. Further refinement of the model, comprised of two complete RNA molecules and two U1A RBDs, stalled with an Rfree of 39%. Inspection of residual electron density maps revealed the presence of two additional U1A-RBDs. Upon inclusion of these and refinement, the Rfree dropped smoothly to 32%. Addition of two TPP molecules and their chelated Mg2+ ions followed by rounds of refinement and manual rebuilding produced the current crystallographic model, which has a cross-validated σA coordinate precision of 0.6 Å (Table 2). Structure figures were prepared with RIBBONS (Carson 1997). Ramachandran analysis (Laskowski et al. 1993) shows that 99.7% of amino acid residues are in the most favored and additionally allowed regions; there are no residues with disallowed backbone conformations. Atomic coordinates and structure factor amplitudes have been deposited with the PDB with accession code 3KOJ.
ACKNOWLEDGMENTS
We thank the staff of ALS beamline 5.0.2 and J. Bolduc for assistance with synchrotron and home laboratory diffraction data collection, respectively, and N. Baird, T. Hamma, D. Klein, J. Miranda-Ríos, J. Pitt, J. Posakony, A. Roll-Mecak, and B. Stoddard for discussions. T.E.E. was a Damon Runyon Fellow, and A.R.F.-D. is an Investigator of the Howard Hughes Medical Institute. This work was supported by grants from the Damon Runyon Cancer Research Foundation (DRG-1844-04 to T.E.E.) and the NIH (GM063576 to A.R.F.-D.)
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1847310.
REFERENCES
- Alam S, Grum-Tokars V, Krucinska J, Kundracik ML, Wedekind JE. Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn. Biochemistry. 2005;44:14396–14408. doi: 10.1021/bi051550i. [DOI] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes & Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, et al. Crystallography and NMR system: A new software system for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- Carson M. Ribbons. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker R. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the glmS ribozyme bound to its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes & Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TE, Ferré-D'Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Edwards TE, Klein DJ, Ferré-D'Amaré AR. Riboswitches: Small-molecule recognition by gene regulatory RNAs. Curr Opin Struct Biol. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Feig AL. Applications of isothermal titration calorimetry in RNA biochemistry and biophysics. Biopolymers. 2007;87:293–301. doi: 10.1002/bip.20816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR. The hairpin ribozyme. Biopolymers. 2004;73:71–78. doi: 10.1002/bip.10516. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Doudna JA. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Doudna JA. Crystallization and structure determination of a hepatitis delta virus ribozyme: Use of the RNA-binding protein U1A as a crystallization module. J Mol Biol. 2000a;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Doudna JA. Methods to crystallize RNA. In: Beaucage SL, et al., editors. Current protocols in nucleic acid chemistry. Wiley; New York: 2000b. pp. 7.6.1–7.6.10. [DOI] [PubMed] [Google Scholar]
- Gardner P, Daub J, Tate J, Nawrocki E, Kolbe D, Lindgreen S, Wilkinson A, Finn R, Griffiths-Jones S, Eddy S, et al. Rfam: Updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. (Database): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Henkin T. Riboswitch RNAs: Using RNA to sense cellular metabolism. Genes & Dev. 2008;22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena S. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Ferré-D'Amaré AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Been MD, Ferré-D'Amaré AR. Essential role of an active-site guanine in glmS ribozyme catalysis. J Am Chem Soc. 2007a;129:14858–14859. doi: 10.1021/ja0768441. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Wilkinson SR, Been MD, Ferré-D'Amaré AR. Requirement of helix P2.2 and nucleotide G1 for positioning of the cleavage site and cofactor of the glmS ribozyme. J Mol Biol. 2007b;373:178–189. doi: 10.1016/j.jmb.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D, Edwards T, Ferré-D'Amaré A. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RJ, Macarthur MW, Moss DS, Thornton JM. PROCHECK: A program to check stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Ríos J. The THI-box riboswitch, or how RNA binds thiamin pyrophosphate. Structure. 2007;15:259–265. doi: 10.1016/j.str.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Miranda-Ríos J, Navarro M, Soberón M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montange RK, Batey RT. Riboswitches: Emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- Müller M, Weigand JE, Weichenrieder O, Suess B. Thermodynamic characterization of an engineered tetracycline-binding riboswitch. Nucleic Acids Res. 2006;34:2607–2617. doi: 10.1093/nar/gkl347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KP, Xie D, Thompson KS, Amzel M, Freire E. Entropy in biological binding processes: Estimation of translational entropy loss. Protein Struct Funct Genet. 1994;18:63–67. doi: 10.1002/prot.340180108. [DOI] [PubMed] [Google Scholar]
- Ontiveros-Palacios N, Smith AM, Grundy FJ, Soberon M, Henkin TM, Miranda-Ríos J. Molecular basis of gene regulation by the THI-box riboswitch. Mol Microbiol. 2008;67:793–803. doi: 10.1111/j.1365-2958.2007.06088.x. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Teo C-H, Fearnley I, Nagai K. Crystallization of RNA-protein complexes II. The application of protein engineering for crystallization of the U1A protein-RNA complex. J Mol Biol. 1995;249:409–423. doi: 10.1006/jmbi.1995.0306. [DOI] [PubMed] [Google Scholar]
- Quigley GJ, Rich A. Structural domains of transfer RNA molecules. Science. 1976;194:796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Rupert PB, Ferré-D'Amaré AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Rupert PB, Ferré-D'Amaré AR. Crystallization of the hairpin ribozyme: Illustrative protocols. Methods Mol Biol. 2004;252:303–311. doi: 10.1385/1-59259-746-7:303. [DOI] [PubMed] [Google Scholar]
- Serganov A. The long and the short of riboswitches. Curr Opin Struct Biol. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Barrick D, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Thore S, Frick C, Ban N. Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch. J Am Chem Soc. 2008;130:8116–8117. doi: 10.1021/ja801708e. [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005a;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005b;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Wyman J. Heme proteins. Adv Protein Chem. 1948;4:407–531. doi: 10.1016/s0065-3233(08)60011-x. [DOI] [PubMed] [Google Scholar]
- Xiao H, Edwards TE, Ferré-D'Amaré AR. Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch. Chem Biol. 2008;15:1125–1137. doi: 10.1016/j.chembiol.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Miyoshi D, Kubodera T, Nishimura A, Nakai S, Sugimoto N. Roles of Mg2+ in TPP-dependent riboswitch. FEBS Lett. 2005;579:2583–2588. doi: 10.1016/j.febslet.2005.03.074. [DOI] [PubMed] [Google Scholar]