Abstract
MicroRNAs (miRNAs) regulate gene expression post-transcriptionally through binding specific sites within the 3′ untranslated regions (UTRs) of their target mRNAs. Numerous investigations have documented repressive effects of miRNAs and identified factors required for their activity. However, the precise mechanisms by which miRNAs modulate gene expression are still obscure. Here, we have examined the effects of multiple miRNAs on diverse target transcripts containing artificial or naturally occurring 3′ UTRs in human cell culture. In agreement with previous studies, we report that both the 5′ m7G cap and 3′ poly(A) tail are essential for maximum miRNA repression. These cis-acting elements also conferred miRNA susceptibility to target mRNAs translating under the control of viral- and eukaryotic mRNA-derived 5′ UTR structures that enable cap-independent translation. Additionally, we evaluated a role for the poly(A)-binding protein (PABP) in miRNA function utilizing multiple approaches to modulate levels of active PABP in cells. PABP expression and activity inversely correlated with the strength of miRNA silencing, in part due to antagonism of target mRNA deadenylation. Together, these findings further define the cis- and trans-acting factors that modulate miRNA efficacy.
Keywords: miRNA, IRES, PABP, Paip2, poly(A)
INTRODUCTION
MicroRNAs (miRNAs) are small, 22–24-nucleotide (nt), noncoding RNAs that regulate gene expression post-transcriptionally. In vertebrates, the products of an estimated 500–1000 miRNA genes have the potential to target ∼30% of the transcript pool, indicating prominent roles in gene expression regulation (Lewis et al. 2005; Kim and Nam 2006; Bushati and Cohen 2007). miRNAs loaded into the RNA-induced silencing complex (RISC) bind to partially complementary sites in the 3′ UTR of target mRNAs. While the effects of miRNAs on gene expression have been clearly documented, the specific molecular mechanism(s) of their action remain elusive. This is illustrated by variable results obtained by analyses of two “founding” miRNAs, lin-4 and let-7. An early study suggested that lin-4 in Caenorhabditis elegans represses target mRNA translation after the initiation phase without causing significant target degradation (Olsen and Ambros 1999). However, a subsequent report described significant mRNA degradation as a consequence of lin-4 activity (Bagga et al. 2005). In mammalian cell culture systems, let-7 appeared to repress transfected reporters with engineered let-7 sites at the level of translation initiation, coupled with moderate target decay (Pillai et al. 2005). However, another study implicated nascent polypeptide chain proteolysis as the regulatory step in let-7-mediated translation repression (Nottrott et al. 2006).
Despite these conflicting reports, there is increasing evidence that imperfect miRNA–mRNA base pairing can elicit significant target mRNA degradation in some contexts. For example, miRNA-targeted mRNAs may be deadenylated (Wakiyama et al. 2007) and subsequently degraded (Behm-Ansmant et al. 2006; Giraldez et al. 2006; Wu et al. 2006; Eulalio et al. 2009). Thus, mRNA decay may occur simultaneously with, or independently of, translation repression. Alternatively, the latter could trigger secondary deadenylation and target mRNA degradation. Understanding how cis-acting signals within mRNAs modulate miRNA-mediated effects on protein synthesis may help unravel mechanisms responsible for these events. For example, the inability of miRNAs to repress uncapped, IRES-containing mRNAs suggests a requirement for the m7G cap in miRNA function (Humphreys et al. 2005; Pillai et al. 2005). Moreover, experiments in various mammalian and insect cell-free extracts implicate the m7G cap as a crucial cis-regulatory element that confers susceptibility to miRNAs (Mathonnet et al. 2007; Thermann and Hentze 2007; Wakiyama et al. 2007). In contrast to the cap, a potential role for the poly(A) tail in miRNA-mediated repression is less clear as the diverse systems used to study miRNA function have yielded opposing conclusions (Humphreys et al. 2005; Pillai et al. 2005; Wang et al. 2006; Wu et al. 2006; Wakiyama et al. 2007; Eulalio et al. 2008).
The roles of PABP in translation and mRNA metabolism have been extensively studied (Mangus et al. 2003). Through simultaneously binding the poly(A) tail and eIF4G, PABP is believed to approximate mRNA termini and enhance translation initiation (Sachs 2000; Kahvejian et al. 2005; Amrani et al. 2008). PABP may also modulate the termination phase via interaction with the release factor eRF3 (Hoshino et al. 1999). PABP is regulated by at least two interacting partners known as Paip1 and Paip2 (Craig et al. 1998; Khaleghpour et al. 2001a). Paip1 is similar in structure to a central domain of eIF4G and enhances translation through bridging of eIF3 and PABP (Martineau et al. 2008), while Paip2 exerts an inhibitory function through displacing PABP from the poly(A) tail and interfering with PABP–eIF4G interaction (Khaleghpour et al. 2001b; Karim et al. 2006).
We comprehensively examined mechanistic aspects of miRNA-mediated translation repression using multiple empirical approaches. In particular, we analyzed effects of miRNAs on translation and decay of diverse target transcripts, including endogenous mRNAs and artificial reporter constructs with variable modes of initiation. Moreover, we evaluated the effects of modulating PABP activity on miRNA-mediated repression. Our results extend previous studies implicating canonical cis-acting elements in miRNA function and define a novel role for PABP in controlling miRNA efficacy.
RESULTS
A m7G cap and poly(A) tail confer susceptibility to miRNA repression
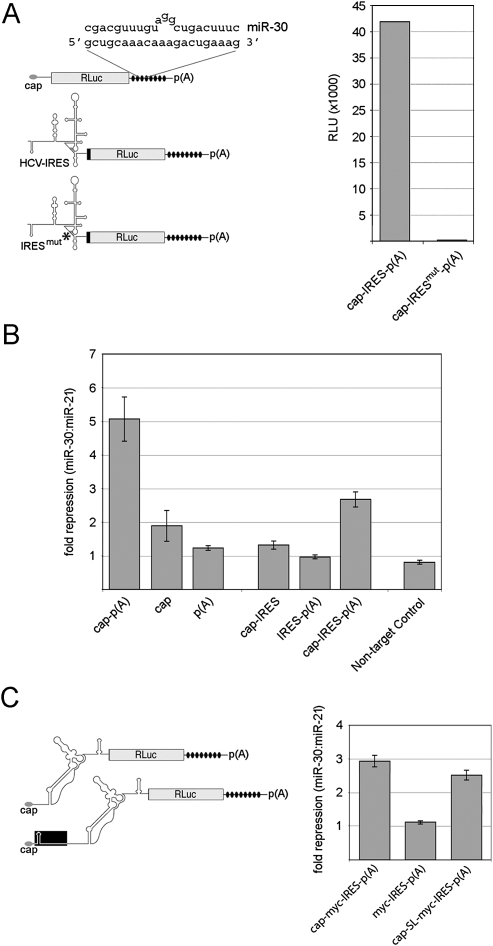
We derived a series of Renilla luciferase (RLuc) reporter constructs containing eight synthetic miR-30 recognition sites tandemly arrayed within the 3′ UTR (Fig. 1A) analogous to those previously described (Zeng et al. 2003). Base pairing between miR-30 and target mRNA results in a 3-nt bulge due to imperfect complementarity. In order to examine the relative importance of cis-acting elements involved in miR-30-mediated repression, we co-transfected in vitro transcribed reporter mRNAs into 293T cells with synthetic miR-30 or miR-21 duplex. miR-21 has been previously utilized as a negative control in miRNA studies (Zeng and Cullen 2003; Zeng et al. 2003). The assay was carried out essentially as described using mRNAs with or without a 5′ cap and 3′ poly(A) tail (Humphreys et al. 2005). RLuc levels were measured 10 h post-transfection, and repression levels were calculated (Fig. 1B). Reporter mRNA bearing both cap and poly(A) tail was repressed approximately fivefold by miR-30. In contrast, translation of uncapped, polyadenylated mRNA was insensitive to miR-30, while capped, unpolyadenylated mRNA exhibited twofold repression. These findings are consistent with previous reports indicating that a 5′ cap is required for sensitivity to miRNA activity while the presence a poly(A) tail, though not absolutely required, confers elevated repression by miRNAs.
FIGURE 1.
An m7G-cap and poly(A) tail are required for maximum miRNA-mediated repression. (A) (Left panel) Schematic representation of reporter mRNAs utilized. Each reporter contains eight tandem miR-30 target sites (black ovals) in the 3′ UTR and either ∼30 nt of vector-derived sequence, the HCV IRES, or a mutated HCV IRES lacking subdomain IIIf (asterisk; IRESmut) in the 5′ UTR. The predicted base-pairing between miR-30 and a single target site is shown. (Right panel) Raw RLU values for capped and polyadenylated HCV IRES reporter mRNAs after transfection into 293T cells. (B) miR-30 repression levels of transfected mRNA constructs. The indicated mRNAs were co-transfected with either miR-21 (nontargeting) or miR-30 (targeting) duplex RNA, and the repression levels were calculated. The nontarget control construct lacks miR-30 target sites. Error bars represent calculated values for standard deviation. (C) Analysis of c-myc IRES-containing constructs. (Left panel) Schematic depiction of c-myc IRES RLuc mRNAs. A stem–loop (SL) was inserted adjacent to the 5′ end of the cap-SL-myc-IRES-p(A) construct, and ∼200 nt of vector sequence separates the SL and IRES. (Right panel) Repression levels for individual reporters. Transfections were performed in triplicate in three separate experiments.
We extended our analyses to miR-30 target constructs containing the HCV IRES, which mediates initiation through direct recruitment of the 40S ribosomal subunit (Pestova et al. 1998). As a negative control, an HCV IRES variant construct was established that contains a 12-nt deletion of subdomain IIIf (Fig. 1A, IRESmut), a region critically involved in binding the 40S subunit (Kieft et al. 2001). Transfection of IRESmut reporter mRNA generated background levels of RLuc activity (Bradrick et al. 2006; data not shown), even when appended with a 5′ m7G cap and poly(A) tail (Fig. 1A), indicating that RLuc expression from this construct results from IRES-dependent translation. In contrast to cap- and poly(A)-dependent protein synthesis, HCV IRES-mediated translation was not susceptible to miR-30, in agreement with previous observations (Fig. 1B; Pillai et al. 2005).
Although HCV IRES-mediated translation is refractory to miRNA function, it is not clear whether the presence of an active IRES precludes repression in the context of a capped and polyadenylated mRNA. An HCV reporter mRNA appended with canonical terminal features may be considered a “hybrid” transcript that possesses an active IRES, as well as the capability of assembling eIF4E, PABP, and associated factors. We sought to determine whether addition of an m7G cap and/or poly(A) tail to HCV reporter mRNA modifies miRNA susceptibility. These modifications had no effect on overall RLuc production by the HCV construct (data not shown). Neither cis-element individually affected repression, but a combination of both m7G and poly(A) rendered the HCV reporter mRNA susceptible to miR-30 (Fig. 1B). Importantly, the HCV IRES-containing mRNA consistently displayed reduced sensitivity to miR-30 compared with mRNA initiating in a strictly cap-dependent manner (about threefold versus fivefold).
We widened our analysis to include constructs containing the 5′ UTR of the c-myc mRNA, which harbors RNA elements known to facilitate cap-independent translation under conditions where cap-dependent translation is abrogated (Johannes and Sarnow 1998). Importantly, in contrast to its HCV counterpart, the c-myc 5′ UTR is devoid of out-of-frame upstream start codons that might be recognized by scanning ribosomes, an event that would not be detected by RLuc measurements. Two distinct constructs were derived that contain artificial miR-30 recognition sites and the 5′ UTR of the P2 c-myc transcript. One construct was engineered to contain a stem–loop (SL) structure previously shown to block scanning when tested in a cell-free translation system (Hundsdoerfer et al. 2005) and transfected HeLa cells (Kaiser et al. 2008) (Fig. 1C). As with HCV IRES constructs, the uncapped c-myc IRES reporter was not affected by co-transfected miR-30 duplex (Fig. 1C). However, appendage of a 5′ cap rendered the c-myc reporter mRNAs susceptible to miR-30, even when scanning was hindered by the SL. These observations provide additional evidence supporting the notion that the presence of a cap and poly(A) tail is sufficient to confer susceptibility to miRNA-mediated repression.
Significant target mRNA degradation is associated with susceptibility to miR-30 repression
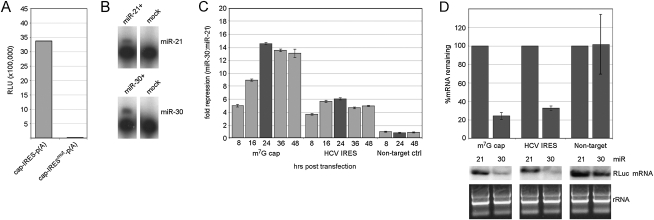
It is difficult to reliably assess integrity of transfected RNA (Barreau et al. 2006). Therefore, we cannot attribute the observed differential effects of miRNA activity to either translation or mRNA degradation mechanisms. In order to investigate a role for mRNA turnover in miRNA function, we conducted experiments with RNA Pol II reporter constructs containing either vector-derived 5′ UTR sequence or the HCV IRES. The HCV IRESmut construct failed to produce RLuc after transfection, similar to results obtained from RNA transfection experiments (Fig. 2A). This confirms that RLuc translation via the HCV IRES is strictly cap-independent in the context of an RNA pol II-synthesized transcript.
FIGURE 2.
miR-30 mediated repression of Pol II-driven reporter constructs. (A) Raw RLU values for the wild-type and mutant HCV IRES Pol II constructs. (B) Expression of miR-21 (upper panel) and miR-30 (lower panel) was evaluated by primer extension analysis. miR-21 or miR-30 expression plasmids were transfected into 293T cells, and primer extension was performed on RNAs harvested 24 h later. No miRNA was expressed in mock-transfected samples. (C) 293T cells were co-transfected with miR-21 or miR-30 plasmids and the indicated reporter construct. Cells were harvested for analysis at the indicated time points. (Darker shaded bars) Time points used for characterization of target mRNA integrity. A plasmid encoding FLuc was used as a transfection control. (D) Quantitative RT-PCR (qRT-PCR; upper panel) for RLuc mRNA was normalized to endogenous GAPDH mRNA levels. Target mRNAs co-transfected with miR-21 are set to 100%. Northern blots (lower panel) for RLuc mRNAs were performed as described in the Materials and Methods. rRNAs from corresponding samples are shown for loading control. qRT-PCR experiments were repeated on at least three separate occasions.
Prior to analyses of RNA pol II reporter constructs, levels of endogenous miR-21 and miR-30 were examined in 293T cells using primer extension (Fig. 2B). Both miRNAs were readily detectable only after transfection of plasmids encoding the respective pre-miRNAs, indicating low levels of endogenous miR-30/21 in 293T cells. We subsequently evaluated repression of target mRNAs by co-transfection of miRNA-encoding and miR-30 target plasmids. As an internal control for transfection variability, a plasmid encoding firefly luciferase (FLuc) was included in co-transfections. RLuc and FLuc levels were measured at intervals spanning 8 to 48 h post-transfection, and target mRNA abundance was determined by Northern blot and quantitative (q)RT-PCR for samples exhibiting the highest levels of repression (24 h post-transfection) (Fig. 2C). As with transfected mRNAs, Pol II-generated reporter transcripts were susceptible to miR-30-mediated repression independent of their mode of initiation (Fig. 2C). Repression levels were uniformly stronger in these assays and divergent between HCV and conventional target mRNAs. Specifically, repression of the HCV construct reached a maximum of about sixfold at 16 h post-transfection, while the cap-dependent reporter was repressed ∼14-fold by 24 h (Fig. 2C), similar to the ratio observed in RNA transfection experiments (Fig. 1B). Significant decay of target mRNAs was clearly evident from both qRT-PCR and Northern blot analyses (Fig. 2D). Quantitative measurements indicated an ∼70% decline in target mRNA abundance upon repression by miR-30 (Fig. 2D). Interestingly, the levels of induced decay for the cap-dependent and HCV IRES-containing transcripts were indistinguishable. Thus, apparently identical rates of target mRNA decay accompanied clearly divergent levels of repression.
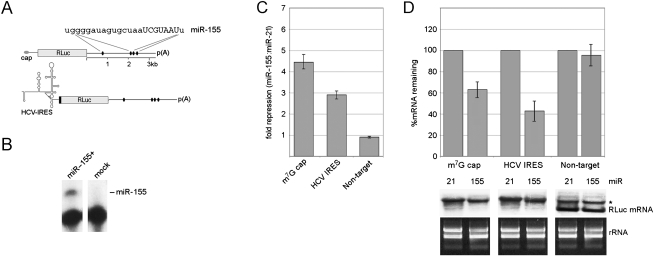
Artificial 3′ UTRs with eight identical miR-30 sites in tandem are not representative of endogenous target mRNAs that typically contain fewer, more heterogeneous, and spatially dispersed recognition sites. Therefore, we expanded our analyses to include an authentic 3′ UTR targeted by a distinct miRNA (miR-155) that is absent in 293T cells (Fig. 3A). BACH1 is a transcription factor whose mRNA is known to be regulated by miR-155 (Gottwein et al. 2007). The BACH1 mRNA 3′ UTR is 3.3 kb in length and contains four miR-155 target sites. We exchanged the artificial 3′ UTRs in our conventional and HCV IRES-driven reporter constructs with the authentic BACH1 3′ UTR (Fig. 3A) and then measured the effects of miR-155 on RLuc expression and mRNA degradation (Fig. 3B,C). The magnitudes of RLuc repression and RNA decay were weaker than those obtained with miR-30. Nevertheless, miR-155 targeting of the BACH1 3′ UTR differentially affected RLuc production by the cap-dependent and HCV IRES mRNAs despite similar levels of decay. Thus, our observations do not depend on a specific miRNA:target combination.
FIGURE 3.
miR-155 represses cap- and IRES-driven targets containing the authentic BACH1 3′ UTR. (A) Schematic of reporter constructs containing the BACH1 3′ UTR with four predicted miR-155 sites (black ovals). The sequence of mature miR-155 is shown with the seed sequence in capital letters. (B) Expression of miR-155 was evaluated in 293T cells by primer extension. (C) miR-155 repression levels of the indicated constructs. (D) qRT-PCR (upper panel) and Northern blot (lower panel) analyses were performed. (*) Background band consistently observed with the nontarget control.
Modulation of PABP expression and activity alters the extent of miRNA repression
Since the presence of a poly(A) tail on targeted mRNA elevated miRNA-mediated repression in our empirical system, we chose to investigate a potential role for PABP in this process. We focused on PABP because of its critical roles in regulating mRNA translation and decay (Mangus et al. 2003). Moreover, the endogenous PABP inhibitor, Paip2, provides a useful tool for in vitro and in vivo studies of PABP.
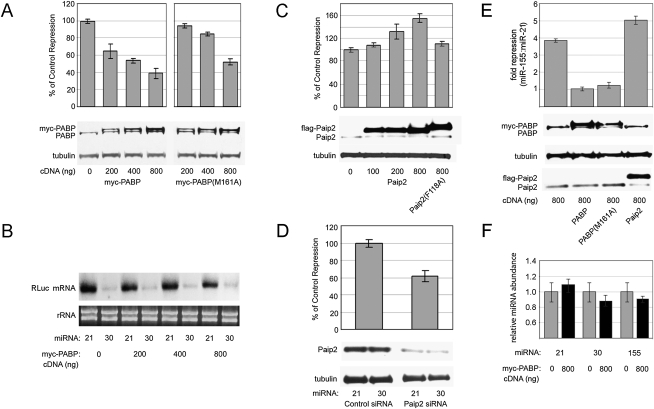
We initially overexpressed different amounts of myc-tagged PABP in 293T cells and analyzed repression of co-transfected miR-30 reporter construct by exogenous miR-30 or control miR-21. Western blot analysis revealed a dose-dependent increase in total PABP by 24 h after transfection with increasing amounts of expression plasmid (Fig. 4A). Coincident with PABP overexpression, miR-30-mediated repression was significantly reduced in a dose-responsive manner (Fig. 4A), reaching 40% of control in cells transfected with 800 ng of PABP cDNA. Thus, elevated levels of PABP appear to counteract miRNA function. PABP modulates miRNA efficacy, but does not prevent repression entirely, similar to other trans-acting factors involved in miRNA repression (Weinmann et al. 2009; Zipprich et al. 2009). Unexpectedly, PABP overexpression had hardly any effect on target mRNA abundance (Fig. 4B), possibly indicating diminished translation repression as its predominant effect.
FIGURE 4.
Changes in PABP expression and activity modulate miRNA repression. (A) miR-30-mediated repression was evaluated in 293T cells transfected with increasing amounts of myc-PABP expression plasmid. The repression level in cells transfected with control vector and miR-30 plasmids was set to 100% and then compared with repression data from cells similarly transfected with miR-30 and PABP (wild-type and M161A) expression plasmids. The total amount of transfected cDNA was kept constant between samples. Western blot analyses were carried out to determine PABP and tubulin levels. Myc-tagged PABP is visible as a distinct band that migrates slightly above endogenous PABP. (B) Northern blot analysis of target mRNA after myc-PABP overexpression. (C) miR-30 activity was evaluated in the context of Paip2 overexpression. Cells were transfected with increasing amounts of Flag-Paip2 plasmid, and repression of miR-30 target mRNA was examined as in A. Flag-Paip2 migrates slightly above endogenous Paip2. (D) miR-30 repression levels during knockdown of Paip2 along with Western blots for Paip2 and tubulin are shown. (E) miR-155 repression of the BACH1 3′ UTR reporter upon PABP, PABP (M161A), and Paip2 overexpression. (F) qPCR of the indicated miRNAs after control or myc-PABP expression plasmid transfection. This experiment was repeated three times, and a representative experiment is shown.
To more clearly define the mechanism(s) whereby PABP affects miRNA repression, we examined a mutant form of PABP (M161A) that cannot bind eIF4G (Kahvejian et al. 2005). This interaction is a crucial function of PABP as it approximates the 5′ and 3′ termini of mRNAs and enhances translation initiation (Kahvejian et al. 2005). Similar to wild-type PABP, the M161A mutant diminished miR-30-mediated repression (to ∼55% of control with 800 ng of PABP [M161A]) (Fig. 4A), but was significantly less effective at all quantities transfected despite similar expression levels (Fig. 4A). This result suggests that a PABP–eIF4G interaction is partly responsible for the antagonistic effect of PABP on miRNA repression.
Ectopic overexpression of translation factors could potentially induce pleiotropic effects on cellular translation rate and physiology that may confound interpretation of our experiments. Global cellular translation measured by 35S-methionine incorporation and translation of the co-transfected FLuc control reporter were not significantly changed in cells with elevated levels of PABP (data not shown). Moreover, there were no changes in cell proliferation rate or morphology during the course of our experiments.
A role for PABP in modulating miRNA efficacy was further probed by manipulating Paip2, which negatively regulates PABP's interactions with poly(A) and eIF4G (Khaleghpour et al. 2001b; Karim et al. 2006). Transfection of plasmids encoding wild-type Paip2 or a variant containing a single amino acid substitution (F118A) that ablates PABP interaction resulted in substantial Paip2 overexpression compared with endogenous levels (Fig. 4C). Increased expression of wild-type Paip2 produced the opposite effect of PABP, elevating repression by miR-30 ∼50%. In contrast, the F118A mutant had negligible effects on miRNA function. We also depleted Paip2 in 293T cells by RNA interference and analyzed effects on target mRNA repression (Fig. 4D). In accordance with results obtained in PABP/Paip2 overexpression experiments, we observed decreased miRNA efficacy as a consequence of Paip2 knockdown. Similar assays in which PABP was targeted for knock-down resulted in co-depletion of Paip2, as previously observed (Yoshida et al. 2006), and showed no effect on miRNA silencing (data not shown). Taken together, these results indicate that PABP expression and activity modulates the extent of miRNA-mediated repression.
To determine if PABP/Paip2 overexpression also influences repression of endogenous miRNA targets, we again used reporters containing the miR-155 targeted BACH1 3′ UTR. The effects of PABP overexpression on miR-155 repression of BACH1 were comparable to miR-30-mediated repression of artificial target 3′ UTRs (Fig. 4E). These results indicate that the effects observed are not due to a specific miRNA:target combination.
A recent report concluded that the cytoplasmic poly(A)polymerase GLD-2 stabilizes the mature form of miR-122 (but not several other miRNAs assayed, including miR-21 used in our study) by the addition of a single adenosine to the 3′UTR (Katoh et al. 2009). Therefore, it is conceivable that PABP may influence miRNA abundance. We tested levels of mature miRNAs -21, -30, and -155 using quantitative PCR with transfection of control or myc-PABP expression plasmids at the maximum amount employed (Fig. 4F). We did not observe a significant change in expression levels of the miRNAs used in this study upon ectopic PABP expression (Fig. 4F).
Increasing PABP expression or activity diminishes repression of an endogenously targeted mRNA
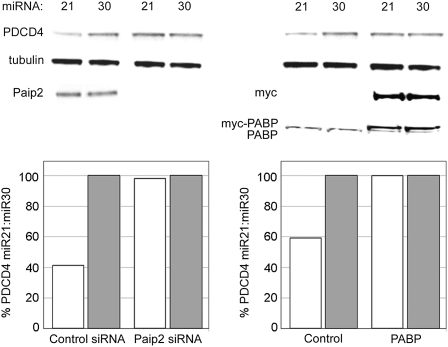
Next, we determined whether manipulation of PABP abundance or activity might affect an endogenous mRNA subject to miRNA regulation. For this purpose, we examined the effect of miR-21 on expression of programmed cell death 4 (Fig. 5, PDCD4), an empirically verified miR-21 target mRNA that contains a single 3′ UTR recognition site (Asangani et al. 2008; Chen et al. 2008; Frankel et al. 2008; Lu et al. 2008; Zhu et al. 2008). As reported previously in HEK-293 cells (Frankel et al. 2008), ectopic expression of miR-21 in 293T cells resulted in decreased steady-state abundance of PDCD4 protein, ranging between 40% and 60% of levels observed in miR-30-transfected cells (Fig. 5). Overexpression of PABP or depletion of Paip2 abrogated the repressive effect of miR-21 on PDCD4 protein levels. This confirms modulation of miRNA activity by PABP observed with artificial 8× tandem target sites and the authentic BACH1 3′ UTR for an endogenous miR-21 target. Assays of PDCD4 mRNA levels using qRT-PCR (data not shown) confirmed earlier reports that miR-21 targeting of PDCD4 mRNA does not significantly alter transcript abundance (Lu et al. 2008). Moreover, we did not observe substantial effects on PDCD4 mRNA levels with ectopic PABP expression or Paip2 depletion (data not shown).
FIGURE 5.
miR-21 targeting of endogenous PDCD4 is ablated when active cellular PABP is increased. 293T cells were transfected with control or Paip2 siRNAs and then transfected with miR-21 or miR-30 ∼20 h later (left). After another 20 h incubation, cells were lysed and protein levels evaluated by Western blot. Effects of PABP overexpression were similarly evaluated (right). Quantification of PDCD4 normalized to tubulin levels in Western blots is shown (lower panels).
Exogenous PABP expression interferes with miRNA-mediated target deadenylation
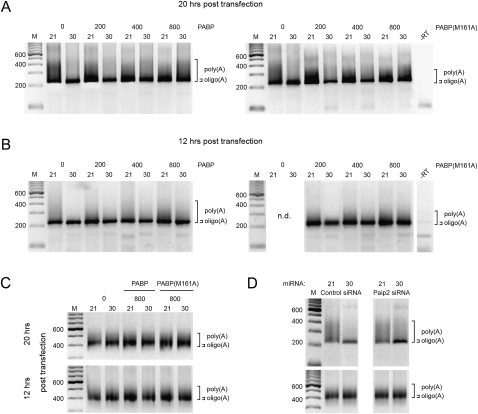
It is widely accepted that deadenylation is a mechanism by which miRNAs repress gene expression (Giraldez et al. 2006; Wakiyama et al. 2007; Eulalio et al. 2009), and PABP has well characterized regulatory roles in deadenylation (Mangus et al. 2003). Accordingly, we hypothesized that PABP may reduce miRNA repression by interfering with deadenylation of targeted constructs. We tested the effects of ectopic PABP and PABP (M161A) expression on target mRNA deadenylation using a PCR-based method for poly(A) tail analyses (Fig. 6). The ligase-mediated poly(A) tail (LM-PAT) assay is an established method for measuring poly(A) tail length (Salles et al. 1999) and has been used to assess miRNA target deadenylation (Clancy et al. 2007). We analyzed the miR-30 tandem 8× target mRNA under repressive and control conditions using LM-PAT. The discrete amplified DNA fragment of ∼220 base pairs is representative of mRNAs with a short oligo(A) tail, while smearing above this fragment indicates increasing poly(A) tail length (Fig. 6). Deadenylation of the target mRNA was clearly evident with miR-30, when compared with the nontargeting miR-21 (Fig. 6A,B,D). Reflecting its effects on miRNA repression levels (Fig. 4), overexpression of both PABP and PABP (M161A) diminished target mRNA deadenylation in a dose-dependent manner (Fig. 6A,B). but did not affect an endogenous control (GAPDH) mRNA (Fig. 6C). We observed similar effects at both 20 h (Fig. 6A) and 12 h (Fig. 6B) post-transfection. Importantly, depletion of Paip2 (Fig. 6D) reduced target mRNA deadenylation in a similar manner (Fig. 6D). Thus, the decrease in miRNA efficacy observed with PABP and PABP (M161A) overexpression and Paip2 depletion is accompanied by significant reduction in target mRNA deadenylation.
FIGURE 6.
The effect of miR-30 on target mRNA adenylation status after manipulation of active PABP. The LM-PAT assay was used to test poly(A) tail length (see Materials and Methods for details). 293T cells were transfected under conditions described in Figure 4, A or D. (A) The effects of PABP and PABP (M161A) ectopic overexpression on the poly(A) tail length of the miR-30 8× target mRNA 20 h post-transfection. (B) miR-30 8× target mRNA assayed as in A at 12 h post-transfection. (C) The adenylation status of a control mRNA (GAPDH) was assayed with the highest amount (800 ng) of PABP and PABP (M161A) cDNA transfected at 20 h and 12 h post-transfection. (D) LM-PAT assays of miR-30 8× target mRNA (upper panel) and control mRNA (GAPDH; lower panel) after knockdown of Paip2.
DISCUSSION
Mechanisms by which miRNAs influence gene expression are complex, involving a host of trans-acting factors and cis-elements that mediate translational repression and/or mRNA decay. The results presented here confirm and extend previous work implicating the m7G cap as a critical determinant in miRNA-mediated repression (Humphreys et al. 2005; Mathonnet et al. 2007; Thermann and Hentze 2007). Experiments with transfected reporter mRNAs also suggest that the 3′ poly(A) tail enhances susceptibility to repression. Indeed, while translation of mRNA containing the HCV IRES was immune to miRNA silencing in RNA transfection experiments, the presence of both a 5′ m7G-cap and poly(A) tail rendered the corresponding RNA pol II-derived transcript similarly sensitive to miRNA-induced decay as a non-IRES construct. Nevertheless, capped and polyadenylated HCV reporter mRNA was consistently less sensitive to repression in both RNA and DNA transfection experiments. Our data show that an active IRES within an mRNA bearing conventional termini does not protect against miRNA repression, at least at the level of RNA decay. Indeed, as equal levels of decay are accompanied by different levels of translation repression, HCV IRES function per se may not be affected by miRNA activity. Thus, repression of HCV IRES reporter mRNA bearing a cap and poly(A) tail may result exclusively from mRNA decay. Since both the m7G-cap and poly(A) are required for full repression of IRES or cap-dependent mRNAs, indirect contact between 5′ and 3′ ends via m7G-cap/poly(A)-dependent interaction between PABP and eIF4G may be negatively regulated by RISC.
The apparent central role for the 5′ m7G-cap in conferring miRNA susceptibility suggests that eIF4E is targeted for inhibition through disruption of either its cap- or eIF4G-binding capability. Indeed, Kiriakidou et al. (2007) proposed that the core RISC component Argonaute 2 (Ago2) associates with the m7G-cap through a motif homologous to eIF4E. Subsequent analyses have challenged Ago2 homology with eIF4E and its putative cap-binding activity, as Ago2 present in cell lysates is not readily enriched by cap-affinity chromatography (Eulalio et al. 2008; Kinch and Grishin 2009; SS Bradrick and M Gromeier, unpubl.). However, it remains possible that Ago2 associates with the m7G-cap indirectly through a presently unidentified RISC-associated factor. Further in-depth proteomic characterization of Ago2 and cap-binding protein complexes may yield insight into a role for the m7G-cap in miRNA function.
In addition to examination of cis-acting mRNA elements, we investigated a role for trans-acting PABP in modulating miRNA function. Paip2 knockdown and PABP overexpression, distinct approaches with the similar outcome of increased active PABP, both diminished miRNA repression. Concordantly, elevating levels of endogenous Paip2 led to enhanced miRNA efficacy. Since analysis of diverse artificial and natural target mRNAs yielded similar outcomes, our results suggest that PABP may be considered a general modifier of miRNA function.
How might PABP protect from miRNA-mediated repression? One possibility is that PABP activity and abundance correlate broadly with the general rate of cellular protein synthesis, and efficacy of miRNA repression is heightened or depressed proportionally. However, we failed to observe significant changes in protein synthesis rate (tested with reporter assays and 35S methionine incorporation) due to manipulation of PABP or Paip2 expression. This is in agreement with in vivo PABP knockdown studies that did not reveal significant reduction in bulk protein synthesis (Yoshida et al. 2006). miRNAs can regulate target mRNAs by affecting their translation rate, mRNA steady state level, or some combination thereof. Our data suggest that PABP modulates miRNA activity at both the level of mRNA deadenylation and translation inhibition. The M161A form of PABP is capable of binding poly(A), but does not interact with eIF4G or stimulate translation initiation in vitro (Kahvejian et al. 2005). Nevertheless, PABP (M161A) retains the ability to antagonize deadenylation of miRNA-targeted mRNA and to reduce the overall strength of miRNA repression. Thus, the intermediate effect of PABP (M161A) on repression suggests that the overall protective effect of PABP is mediated in part by its interaction with eIF4G.
During preparation of this manuscript, two reports were published detailing a requirement for PABP in miRNA-directed deadenylation mediated by an RNA-independent interaction between PABP and GW182 (Fabian et al. 2009; Zekri et al. 2009). Zekri et al. (2009), similar to our observations, found that ectopic overexpression of PABP decreased the extent of miRNA-mediated repression, although deadenylation of target mRNA was not examined. Our interpretation that eIF4G–PABP interaction may be targeted by miRNAs is in agreement with recent findings obtained with in vitro PABP depletion and PABP (M161A) reconstitution (Fabian et al. 2009) or PABP–eIF4G binding competition by the RISC component GW182 (Zekri et al. 2009). Based on in vitro depletion and add back assays, Fabian et al. (2009) reported that PABP is required for miRNA-mediated deadenylation, a finding that contradicts diminished repression and inhibition of deadenylation with ectopic PABP and Paip2 knock-down in vivo.
Multiple nonmutually exclusive explanations exist that may account for the diverse observations on PABP's role in deadenylation of miRNA target mRNAs. In our assays and in studies by Zekri et al. (2009), PABP overexpression may preserve poly(A) tails either by stimulating closed-loop mRNA conformation through its interaction with eIF4G or by enhanced shielding of poly(A) tails. Accordingly, PABP has been described as a general antagonist of deadenylation in multiple experimental systems (Wormington et al. 1996; Wang et al. 1999; Voeltz et al. 2001; Wilusz et al. 2001). Alternatively, since PABP binds components of the CCR4–NOT1 deadenylase complex (Zekri et al. 2009), elevated levels of free PABP may saturate the complex and effectively inhibit its function.
Distinct empirical systems may reveal different properties of the intricate network regulating miRNA activity. No single approach can be considered unambiguous, and reconciling these differences will require comprehensive analyses in multiple experimental systems. Despite differences regarding its precise effects, our study and prior investigations (Fabian et al. 2009; Zekri et al. 2009) clearly indicate that PABP and factors regulating its function play prominent roles in mediating post-transcriptional gene regulation via miRNAs.
MATERIALS AND METHODS
Cell culture and plasmid constructs
293T cells were maintained in DMEM supplemented with 10% FBS and nonessential amino acids. pCMV-miR-21 and pCMV-miR-30 were obtained from B. Cullen (Duke University) and have been described previously (Zeng et al. 2003). The 8× miR-30 site's 3′ UTR was PCR-amplified from pCMV-target-30B (Zeng et al. 2003) with primers (1) 5′-gagcggccgcccagtgccaaggtct-3′ and (2) 5′-gctctagagttaccatgggttaacgg-3′ and inserted into pTNT (Invitrogen) containing an RLuc cassette (Bradrick et al. 2006) using NotI and XbaI. An XhoI fragment of the resulting plasmid was then cloned into pCI (Promega), containing an enhancer/promoter and optimized chimeric intron for efficient expression after DNA transfection and a T7 promoter for in vitro transcription. The BACH1 3′ UTR was PCR-amplified with primers (3) 5′-agtggtaccacttgcattcacttccttcaaac-3′ or (4) 5′-agtacgcgtacttgcattcacttccttcaaac-3′ and (5) 5′-agtgcggccgccattgttagaaaaattatattat-3′ and cloned into the existing pCI cassette using KpnI (primer [3]) or MluI (primer [4]) and a NotI restriction site. miR-155 was cloned into pBC12 (Zeng et al. 2003) using PCR amplification with primers (6) 5′-agcctcgatcccagtgaccagattatg-3′ and (7) 5′-tatctcgagggtggcacaaaccaggaagg-3′ from template plasmids provided by B. Cullen (Duke University; Gottwein et al. 2007). Reporter constructs containing the full-length HCV IRES or a subdomain IIIf deleted IRES were described previously (Bradrick et al. 2006). The 5′ UTRs of both constructs were PCR amplified using primers (8) 5′-attctcgaggccagccccctgatg-3′ and (9) 5′-tcggttggggagttgttcatttttga-3′. The PCR products were ligated into pCI containing the RLuc cassette and appropriate 3′ UTR. The 8× miR-30 sites were PCR amplified using primers (10) 5′-cggtcgactctagagttaccatggg-3′ and (11) 5′ataagcttaggtaccacgcgtgaatt-3′ and cloned into c-myc and SL-c-myc (Kaiser et al. 2008) using SalI and HindIII sites.
Primer extension
Primer extension was performed as described previously (Zeng et al. 2003). Briefly, confluent 293T cells were transfected with 400 ng of CMV-miR-21 or CMV-miR-30, and after ∼18 h cells were harvested and total RNA was isolated using RNeasy (Qiagen) and treated with DNase. Five micrograms of total RNA were then analyzed using Primer Extension System (Promega). The primers used to specifically detect miR-21 and miR-30 have been described (Zeng et al. 2003). Primer (12) 5′-accccgagcacgattag-3′ was used to detect miR-155.
In vitro transcription, RNA and DNA transfection
DNA constructs were linearized by endonuclease digestion with NotI downstream of the 3′ UTR and transcribed in vitro with T7 RNA polymerase using the MegaScript Kit T7(Ambion). c-myc-8× and SL-c-myc-8× were linearized with MluI. In vitro transcript RNAs were then purified using RNeasy (Qiagen) and treated with DNase. Subsequently, RNAs were treated with guanylyl-transferase (Ambion) to add a m7G-cap and polyadenylated with yeast poly(A) polymerase (USB) where indicated. The integrity and purity of all in vitro transcript RNAs was verified by agarose gel electrophoresis and UV spectrometry of the 260:280 absorbance ratio. 293T cells were co-transfected as described (Humphreys et al. 2005) with 2 nM final concentration of miR-30 duplex (+)5′-cuuucagucggauguuugcagc-3′:(−)5′-gcugcaaacauccgaaugaaag-3′ or miR-21 duplex (+)5′-uagcuuaucagacugauguuga-3′:(−)5′-caacaccagucgaugggcugu-3′ (IDT) and 100 ng of the indicated target construct using Lipofectamine2000 (Invitrogen) according to the manufacturer's instructions. Prior to transfection, single-stranded oligonucleotide RNAs were annealed. RLuc was measured 10 h after RNA transfection using the Renilla Luciferase System (Promega). DNA transfections in Figure 2 were carried out as follows: for reporters with 8× miR-30 target sites, 1 μg of indicated target construct, 0.5 μg of pGL3 (Promega) expressing Firefly Luc (used as a transfection control), and 25 μg of pCMV-miR-21 or -30 were co-transfected into 293T cells grown in 10-cm culture dishes using Lipofectamine2000 (Invitrogen). For BACH1 3′ UTR reporters (Fig. 3), 10 ng of target cDNA were co-transfected with 5 ng of pGL3 and 400 ng of either pCMV-miR-21 or -155 in individual wells of a 24-well plate. Luc was measured 20 h post-transfection. Rluc values were first normalized to FLuc (pGL3), then compared as fold repression (miR-21 transfected/miR-30 transfected).
PABP/Paip2 overexpression and knockdown
A myc tag (5′-gagcagaaactcatctctgaagaggatctg-3′) was cloned into pcDNA5 (Invitrogen) using oligonucleotides. The PABPC1 sequence was then cloned into this myc-tagged construct using primers (13) 5′-gcggatccatgaaccccagtgcccccag-3′ and (14) 5′gcgcggccgcttaaacagttggaacaccggt ggc-3′ using INTEIN-PABP (Bradrick et al. 2007) as a template and BamHI and NotI restriction sites. PABP (M161A) was constructed using overlapping primers containing the mutation. GST-Paip2 has been described (Bradrick et al. 2007). The Paip2A ORF was PCR-amplified and inserted into pcDNA3.1 (Invitrogen). A Flag tag was inserted using primers (15) 5′-ctagcaccatggattacaaggatgacgacgataga-3′ and (16) 5′-agcttcttatcgtcgtcatccttgtaatccatggtg-3′. For the miR-30 8× target co-transfections, 400 ng of miR-21 or miR-30, 10 ng of miR-30 8× target, 5 ng of pGL3, and the indicated amount of myc-PABP, Flag-Paip2, or Flag-Paip2A (F118A) were transfected. Rluc values were first normalized to FLuc (pGL3), and then compared as fold repression (miR-21 transfected/miR-30 transfected). Values were then calculated as percent of control repression. For the PDCD4 analyses, 800 ng of miR-21 or miR-30 along with 400 ng of myc-PABP was transfected and assayed ∼48 h post-transfection. siRNA duplexes for Paip2A (Qiagen) were used as described in the text. Control siRNA is the “All-stars Negative Control Duplex” (Qiagen). Briefly, the siRNAs were transfected using Lipofectamine2000 (Invitrogen) at 100 nM total concentration of all siRNAs (25 nM each siRNA duplex). For all siRNA experiments, 293T cells were seeded onto 24-well plates on day 1. On day 2, cells were transfected with siRNA duplex(es). On day 3, (∼20 h post-transfection), the medium was changed and cells were transfected with 400 ng of miR-21 or miR-30, 10 ng of miR-30 8× target, 5 ng of pGL3 for the miR-30 8× experiments. PDCD4 was analyzed ∼20 h (day 4) after transfection of 1 μg of miR-21 or miR-30 on day 3.
Northern and Western blot analyses
For Northern analyses, total RNA was extracted with RNeasy columns (Qiagen), treated with DNase (Qiagen), and subjected to gel electrophoresis on 1% agarose gels containing glyoxal. The quality of the extracted total RNA samples was verified by agarose gel electrophoresis and UV spectrometry. RNA was transferred from the gel to membrane (Nytran SuperCharge [Whatman]) by passive diffusion. Hybridization with radioactively labeled probe was carried out with ExpressHyb solution (Clontech) according to the manufacturer's protocol. Probes corresponded to portions of the RLuc open reading frame generated by SfuI/XmnI digestion (corresponding to nt 10–538) or PCR amplification with primers (17) 5′-gggcggtaggcgtgtacggtggg-3′ and (18) 5′-gatggcaacatggtttccacg-3′. Probes were labeled with [α32P]-dCTP by random priming using the High Prime system (Roche) according to the manufacturer's suggestions. Each Northern blot assay was repeated at least two times, and a representative experiment is shown. Western blots were carried out essentially as described before (Dobrikova et al. 2006). Western blot quantifications were carried out using the densitometer function of a Fluorchem HD2 (Alpha Innotech). All PDCD4 quantifications were first normalized to the tubulin signal. Primary antibodies used in this study were raised against PABP (Sigma), Paip2A (Sigma), Paip2 (a gift of N. Sonenberg, McGill University), PDCD4 (Rockland Immunochemicals), myc (Sigma), and tubulin (Sigma).
Quantitative RT-PCR
293T cells were transfected as indicated above, lysed with RLT buffer and QIAshredder columns (Qiagen), then purified using RNeasy columns and DNase treated (Qiagen). We performed quantitative RT-PCR using the Plexor one-step qRT-PCR system (Promega). One hundred nanograms of total RNA was used in each reaction. The primer pair to detect target RLuc mRNA was designed to span the pCI intron, and different reverse primers were used to detect m7G-cap-dependent ([19] 5′-caccactgcggaccagttatca-3′) and HCV IRES-driven ([20] 5′-acaggggagtgattcatggtgg-3′) reporters. The forward primer (21) 5′-cgtgaggcactgggcaggt-3′ contains a 5′ dFAM-isoC modification (Promega). GAPDH ReadyMade primers (IDT) were used as an internal control, and all RLuc mRNA measurements were normalized to GAPDH. miRNA qPCR was performed using the Taqman miRNA reverse transcription system (Applied Biosystems) and 200 ng of total RNA per reaction, followed by Taqman PCR on an ABI 7900HT. 293T cells were transfected with 800 ng of control or myc-PABP and 400 ng of the indicated miRNA (see above). miRNA values were first normalized to the endogenous control RNU48. All assays were performed at least three times and measurements were taken in triplicate. A representative experiment is shown. The ΔΔ-Ct method was used to compare samples (Lowe et al. 2003).
Poly(A) tail length assays
We used the LM-PAT assay basically as described to test for poly(A) tail length (Salles et al. 1999; Clancy et al. 2007). Briefly, total RNA was extracted from 293T cells transfected with the indicated constructs (400 ng of miR-21 or miR-30; 10 ng of miR-30 8× target; 5 ng of pGL3 [as a transfection control]; 800 ng of empty vector or PABP or PABP [M161A]) using RNeasy columns (Qiagen) and DNase treated. Paip2 knockdown experiments were performed as previously described. Two micrograms of RNA was then incubated with p(dT)15 in the presence of DNA ligase at 37°C. The temperature was then lowered to 12°C and an oligo(d)T anchor was added to the reaction. Superscript II reverse transcriptase (Invitrogen) was added, and the reaction was incubated at 42°C for at least 2 h. Subsequent PCR reactions were carried out using Luc-specific primer (24) 5′-caaatcgttcgttgagcgagttctcaaaaatg-3′ and the oligo(dT) anchor. The GAPDH-specific primer was (25) 5′-tctcctctgacttcaacagcgaca-3′.
ACKNOWLEDGMENTS
We thank Dr. B. Cullen for reagents and critical discussion of the manuscript. We also thank Dr. W. Xie and Dr. C. Strayer for excellent technical advice. This work was supported by a grant from the Susan G. Komen Foundation and PHS grant CA124756 (to M.G.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1795410.
REFERENCES
- Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Barreau C, Dutertre S, Paillard L, Osborne HB. Liposome-mediated RNA transfection should be used with caution. RNA. 2006;12:1790–1793. doi: 10.1261/rna.191706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradrick SS, Walters RW, Gromeier M. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradrick SS, Dobrikova EY, Kaiser C, Shveygert M, Gromeier M. Poly(A)-binding protein is differentially required for translation mediated by viral internal ribosome entry sites. RNA. 2007;13:1582–1593. doi: 10.1261/rna.556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. MicroRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu W, Chao T, Zhang Y, Yan X, Gong Y, Qiang B, Yuan J, Sun M, Peng X. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272:197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- Clancy JL, Nousch M, Humphreys DT, Westman BJ, Beilharz TH, Preiss T. Methods to analyze microRNA-mediated control of mRNA translation. Methods Enzymol. 2007;431:83–111. doi: 10.1016/S0076-6879(07)31006-9. [DOI] [PubMed] [Google Scholar]
- Craig AW, Haghighat A, Yu AT, Sonenberg N. Interaction of polyadenylated-binding protein with the eIF4G homolog PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- Dobrikova EY, Grisham RN, Kaiser C, Lin J, Gromeier M. Competitive translation efficiency at the picornavirus type 1 internal ribosome entry site facilitated by viral cis and trans factors. J Virol. 2006;80:3310–3321. doi: 10.1128/JVI.80.7.3310-3321.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:1–12. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:1–13. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundsdoerfer P, Thoma C, Hentze MW. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc Natl Acad Sci. 2005;102:13421–13426. doi: 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes & Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Dobrikova EY, Bradrick SS, Shveygert M, Herbert JT, Gromeier M. Activation of cap-independent translation by variant eukaryotic initiation factor 4G in vivo. RNA. 2008;14:2170–2182. doi: 10.1261/rna.1171808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim MM, Svitkin YV, Kahvejian A, De Crescenzo G, Costa-Mattioli M, Sonenberg N. A mechanism or translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc Natl Acad Sci. 2006;103:9494–9499. doi: 10.1073/pnas.0603701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes & Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell. 2001a;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- Khaleghpour K, Kahvejian A, De Crescenzo G, Roy G, Svitkin YV, Imataka H, O'Connor-McCourt M, Sonenberg N. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol. 2001b;21:5200–5213. doi: 10.1128/MCB.21.15.5200-5213.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Kinch LN, Grishin NV. The human Ago2 MC region does not contain an eIF4E-like mRNA cap-binding motif. Biol Direct. 2009;4:2. doi: 10.1186/1745-6150-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lowe B, Avila HA, Bloom FR, Gleeson M, Kusser W. Quantitation of gene expression in neural precursors by reverse-transcription polymerase chain reaction using self-quenched, fluorogenic primers. Anal Biochem. 2003;315:95–105. doi: 10.1016/s0003-2697(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau Y, Derry MC, Wang X, Yanagiya A, Berlanga JJ, Shyu AB, Imataka H, Gehring K, Sonenberg N. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol Cell Biol. 2008;28:6658–6667. doi: 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes & Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Sachs A. Physical and functional interaction between the mRNA cap structure and the poly(A) tail. In: Mathews MB, editor. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 447–466. [Google Scholar]
- Salles FJ, Richoards WG, Strickland S. Assaying the polyadenylation state of mRNAs. Methods. 1999;17:38–45. doi: 10.1006/meth.1998.0705. [DOI] [PubMed] [Google Scholar]
- Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Ongkasuwan J, Standart N, Steitz JA. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes & Dev. 2001;15:774–788. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes & Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Gao M, Jones CL, Wilusz J, Peltz SW. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA. 2001;7:1416–1424. [PMC free article] [PubMed] [Google Scholar]
- Wormington M, Searfoss AM, Hurney CA. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 1996;15:900–909. [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Yoshida K, Kozlov G, Lim NS, De Crescenzo G, Pang Z, Berlanga JJ, Kahvejian A, Gehring K, Wing SS, et al. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 2006;25:1934–1944. doi: 10.1038/sj.emboj.7601079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri L, Huntzinger E, Heimstadt S, Izaurradle E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of miRNA targets and is required for target release. Mol Cell Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo Y-Y. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]