Abstract
The archaeal L7Ae and eukaryotic 15.5kD protein homologs are members of the L7Ae/15.5kD protein family that characteristically recognize K-turn motifs found in both archaeal and eukaryotic RNAs. In Archaea, the L7Ae protein uniquely binds the K-loop motif found in box C/D and H/ACA sRNAs, whereas the eukaryotic 15.5kD homolog is unable to recognize this variant K-turn RNA. Comparative sequence and structural analyses, coupled with amino acid replacement experiments, have demonstrated that five amino acids enable the archaeal L7Ae core protein to recognize and bind the K-loop motif. These signature residues are highly conserved in the archaeal L7Ae and eukaryotic 15.5kD homologs, but differ between the two domains of life. Interestingly, loss of K-loop binding by archaeal L7Ae does not disrupt C′/D′ RNP formation or RNA-guided nucleotide modification. L7Ae is still incorporated into the C′/D′ RNP despite its inability to bind the K-loop, thus indicating the importance of protein–protein interactions for RNP assembly and function. Finally, these five signature amino acids are distinct for each of the L7Ae/L30 family members, suggesting an evolutionary continuum of these RNA-binding proteins for recognition of the various K-turn motifs contained in their cognate RNAs.
Keywords: box C/D snoRNA, snoRNP, kink-turn, L7Ae/L30 proteins, RNA–protein interaction
INTRODUCTION
Members of the L7Ae/L30 protein family are found in both Archaea and Eukarya (Koonin et al. 1994; Klein et al. 2001; Boulon et al. 2008). Two archaeal L7Ae/L30 proteins, designated rpL7Ae and rpL30e, are components of the large ribosomal subunit (Ban et al. 2000). Archaeal L7Ae also functions as a core protein of the box C/D and H/ACA sRNP nucleotide modification complexes (Ban et al. 2000; Kuhn et al. 2002; Omer et al. 2002; Rozhdestvensky et al. 2003). Members of the L7Ae/L30 protein family are more numerous in eukaryotes. Nhp2p and 15.5kD are core proteins of the box H/ACA and box C/D snoRNPs, respectively. In addition, 15.5kD also functions as a core component of the spliceosomal U4 snRNP (Vidovic et al. 2000; Watkins et al. 2000; Wang and Meier 2004). Rpp38p, an RNP core protein that contains an L7Ae/L30-like domain, is a constituent of the endonucleolytic MRP complex (Welting et al. 2004). The rpL30 and rpL7a proteins are components of the large ribosomal subunit; rpL30 is also capable of binding its own messenger RNA to regulate translation (Mao et al. 1999; Russo et al. 2005). SBP2 is an mRNA-binding protein and contains a domain homologous to the L7Ae/L30 proteins. SBP2 recognizes mRNAs possessing the SECIS RNA element important for selenocysteine incorporation into selenoproteins (Allmang et al. 2002; Clery et al. 2007).
A defining feature of the L7Ae/L30 protein family is the ability to bind folded RNA motifs known as kink-turns or K-turns (Mao et al. 1999; Ban et al. 2000; Kuhn et al. 2002; Clery et al. 2007). The archetypical K-turn consists of an asymmetric bulge flanked by two RNA duplexes, designated stems I and II. As its name implies, a kink-turn establishes a sharp bend between the two RNA duplexes. Tandem, sheared G·A base pairs that hydrogen-bond across the asymmetric bulge are critical for folding this RNA element. K-turns were first described in the archaeal large 50S ribosomal subunit where they are recognized by a number of ribosomal proteins, including L7Ae (Ban et al. 2000; Klein et al. 2001). In Archaea, K-turns are also found in the box C/D and H/ACA guide sRNAs (Dennis et al. 2001; Kuhn et al. 2002; Rozhdestvensky et al. 2003). In Eukarya, the K-turn motif appears to be more prevalent and is found not only in ribosomal RNAs and box C/D snoRNAs, but also in U4 snRNA and certain mRNAs (Mao et al. 1999; Vidovic et al. 2000; Clery et al. 2007). A K-turn variant known as the K-loop is also found in certain RNAs, most notably the box C/D and box H/ACA RNAs (Nolivos et al. 2005). This motif is structurally similar to the K-turn, but stem I is replaced with a short loop (Li and Ye 2006).
Two homologous L7Ae/L30 family members, eukaryotic 15.5kD and archaeal L7Ae, initiate box C/D RNP assembly by binding K-turns in the box C/D RNAs (Kuhn et al. 2002; Omer et al. 2002; Tran et al. 2003). A total of four core proteins (15.5kD, Nop58p, Nop56p, and fibrillarin) bind the box C/D snoRNAs to assemble the eukaryotic snoRNP. Three archaeal proteins (L7Ae, Nop56/58p, and fibrillarin) are homologous to the four eukaryotic proteins and correspondingly assemble a complex with the archaeal sRNAs (Gagnon et al. 2008). These ribonucleoproteins function as nucleotide modification enzymes, guiding the 2′-O-methylation of designate nucleotides in numerous target RNAs. Guide sequences within the box C/D RNAs base pair with complementary sequences in the target RNA to determine the specific nucleotide for modification while the RNA-associated proteins catalyze the methyltransferase reaction (Kiss-Laszlo et al. 1998; Omer et al. 2002; Tran et al. 2003).
Interestingly, archaeal L7Ae and eukaryotic 15.5kD have distinct RNA-binding specificities (Tran et al. 2003; Charron et al. 2004; Hamma and Ferré-D'Amaré 2004; Oruganti et al. 2005; Clery et al. 2007). Eukaryotic 15.5kD protein recognizes only the K-turn motif, whereas archaeal L7Ae binds both the K-turn and the variant K-loop motifs. This differential binding is puzzling as the crystal structures of both proteins bound to K-turn or K-loop RNAs have revealed essentially superimposable three-dimensional structures (Vidovic et al. 2000; Hamma and Ferré-D'Amaré 2004; Moore et al. 2004; Li and Ye 2006; Liu et al. 2007; Soss and Flynn 2007). Accordingly, the distinct RNA-binding specificities among these family members are not evident by simple inspection of their protein structures.
We have analyzed the differential RNA-binding capabilities of archaeal L7Ae and eukaryotic 15.5kD and have determined that five amino acid differences account for their subtle, yet distinct RNA-binding specificities. These five amino acid residues are well conserved in each kingdom, but are distinctly different between the archaeal and eukaryotic homologs. Substitution of these “signature” amino acid residues in L7Ae with the corresponding 15.5kD residues preserved K-turn binding but abrogated specific K-loop recognition, thus mimicking eukaryotic 15.5kD protein binding. Nevertheless, this mutated L7Ae protein was still incorporated into a functional C′/D′ RNP assembled on the K-loop motif, thus demonstrating the importance of core protein–protein interactions for box C/D RNP assembly. Furthermore, the signature amino acids for each L7Ae/L30 protein family member are unique and conserved. We propose that each protein family member may have divergently evolved to recognize its specific K-turn motif in the respective cognate RNAs.
RESULTS
Archaeal L7Ae and eukaryotic 15.5kD homologs possess unique “signature” amino acids that flank a conserved RNA-binding region
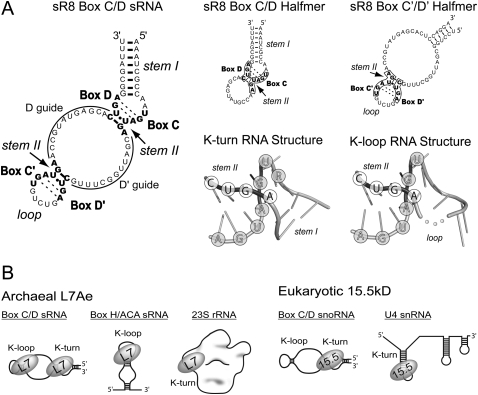
The defining characteristic of the L7Ae/L30 proteins is their ability to bind K-turn RNA motifs (Fig. 1A). Prominent members of this protein family are archaeal L7Ae and eukaryotic 15.5kD. Strikingly, L7Ae recognizes both K-turn and K-loop motifs, whereas 15.5kD binds only the K-turn (Fig. 1B). To understand the molecular basis of the differential K-turn/K-loop recognition, we began by comparing the primary sequences of the archaeal L7Ae and eukaryotic 15.5kD homologs. Alignment of 15 L7Ae and 15 15.5kD proteins revealed a highly conserved RNA-binding region (RBR) (Fig. 2A). Specific amino acids in this region interact with the K-turn RNA and are well conserved across both kingdoms (Vidovic et al. 2000; Hamma and Ferré-D'Amaré 2004; Moore et al. 2004; Li and Ye 2006; Liu et al. 2007; Soss and Flynn 2007). In contrast, the N-terminal and C-terminal regions of the archaeal and eukaryotic homologs are more divergent and also vary in length.
FIGURE 1.
Structure of sR8 box C/D sRNA and RNA-binding specificity of the archaeal L7Ae and eukaryotic 15.5kD homologs. (A) Sequence and structure of the M. jannaschii sR8 box C/D and the box C/D (K-turn) and C′/D′ (K-loop) RNA halfmers (Tran et al. 2003). Stem I and the equivalent loop, as well as stem II, are indicated for each motif. Conserved sR8 box nucleotides are indicated in bold. D and D′ guide sequences are complementary to target RNAs and guide 2′-O-methylation of designate nucleotides in target RNAs. Corresponding box C/D and C′/D′ RNA halfmers are schematically presented at the right with corresponding C, D, C′, and D′ box sequences enclosed. Corresponding tertiary structures of the K-turn and K-loop determined by X-ray crystallography are shown (Vidovic et al. 2000; Li and Ye 2006). Stem I and the equivalent loop, as well as stem II, are indicated, and the nucleotides of the conserved boxes are encircled. An unresolved nucleotide in the K-loop RNA is represented by dots connecting the RNA backbone. (B) Schematic presentation of the archaeal L7Ae and eukaryotic 15.5kD homologs binding their cognate RNA targets.
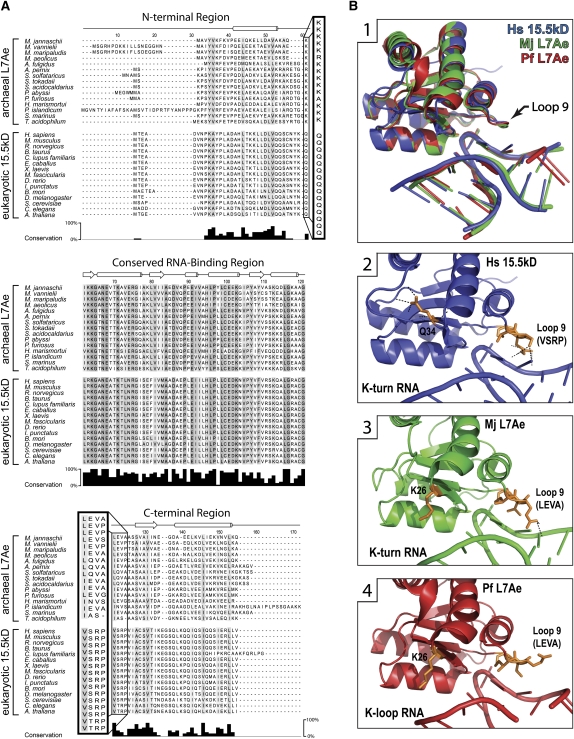
FIGURE 2.
Archaeal L7Ae and eukaryotic 15.5kD proteins possess conserved signature amino acids distinct for archaeal and eukaryotic homologs. (A) Sequence alignment of archaeal L7Ae and eukaryotic 15.5kD proteins. Fifteen archaeal and 15 eukaryotic homologs were aligned with amino acid conservation indicated as shades of gray on the sequences. The level of conservation is depicted in the bar graph below the alignment. Protein secondary structure is indicated above the alignment as tubes (α-helices), arrows (β-sheets), or lines (loops). Protein sequences are divided into three structural regions consisting of a conserved RNA-binding region (RBR) flanked by less conserved N-terminal and C-terminal regions. Signature amino acids that flank the RBR are boxed and highlighted. (B1) Crystal structures of 15.5kD and L7Ae proteins bound to K-turn and K-loop motifs. Human 15.5kD (Hs) (Liu et al. 2007), archaeal L7Ae from M. jannaschii (Mj) (Moore et al. 2004), archaeal L7Ae from P. furiosus (Pf) (Li and Ye 2006), and the K-turn or K-loop motifs are overlaid. (B2–B4) Regions of the individual proteins interacting with K-turn or K-loop motifs. (Orange) Signature amino acids; (black dots) their side chain hydrogen-bonding network.
Comparison of archaeal L7Ae and eukaryotic 15.5kD primary sequences revealed five amino acids that are well conserved within the archaeal L7Ae and eukaryotic 15.5kD homologs, but are distinct and unique for each domain of life (Fig. 2A). These residues have been designated “signature” amino acids. Flanking the RBR on the N-terminal side is a highly conserved lysine residue for the archaeal L7Ae proteins (K26 of Methanocaldococcus jannaschii). This residue in the eukaryotic 15.5kD proteins is a highly conserved glutamine (Q34 of mouse/human). Flanking the C-terminal side of the RBR, four signature amino acids are contained in the Loop 9 regions of the homologs. The highly conserved L7Ae Loop 9 residues leucine–glutamate–valine–alanine (LEVA) are chemically and structurally distinct from the corresponding eukaryotic 15.5kD residues, valine–serine–arginine–proline (VSRP). Their placement near the RBR suggested a potential role in influencing the RNA-binding specificity of each homolog.
Available crystal structures of the archaeal L7Ae and eukaryotic 15.5kD proteins bound to K-turn or K-loop motifs have revealed that the Loop 9 signature residues are in close vicinity to the RNA to affect binding (Fig. 2B, panel 1; Vidovic et al. 2000; Hamma and Ferré-D'Amaré 2004; Moore et al. 2004; Li and Ye 2006; Liu et al. 2007; Soss and Flynn 2007). Loop 9 is positioned near stem II and the corresponding loop of K-turn and K-loop RNA motifs, respectively. Analysis of protein–RNA hydrogen-bond distances within the crystal structures indicated that residues of 15.5kD Loop 9 make four polar contacts with the K-turn RNA (Fig. 2B, panel 2). In contrast, L7Ae Loop 9 makes only two polar contacts with the K-turn through its glutamate residue (LEVA) and, surprisingly, no contact with the K-loop (Fig. 2B, panels 3,4). Indeed, the L7Ae Loop 9 glutamate is rotated 180° away from the RNA when L7Ae is bound to a K-loop. This may be due to the upward angle of the loop, which could produce steric hindrance or charge repulsion and consequently push a negatively charged residue away from the RNA. Previous analyses of L7Ae and 15.5kD bound to K-turn RNAs indicated that the first three of these Loop 9 amino acids are potentially involved in the differential K-loop recognition of L7Ae and 15.5kD (Hamma and Ferré-D'Amaré 2004). It was noted that the positively charged arginine of 15.5kD (VSRP) interacts with the phosphate backbone of the K-turn stem II, an interaction not made by the corresponding L7Ae valine (LEVA). The fifth signature amino acid, L7Ae's K26, protrudes from the protein surface and does not appear sufficiently close to interact with RNA (Fig. 2B, panels 3,4). The side chain of 15.5kD's corresponding Q34 amino acid is rotated inward to make three intra-protein polar contacts (Fig. 2B, panel 2), thus altering the structure of Loop 10.
Replacement of the archaeal L7Ae signature amino acids with the corresponding eukaryotic 15.5kD residues results in loss of K-loop binding
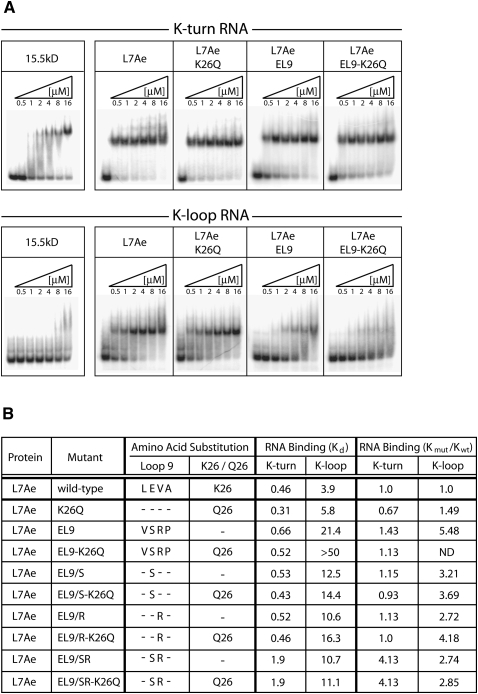
The five signature amino acids of M. jannaschii L7Ae were systematically replaced with the corresponding mouse 15.5kD residues to assess their contribution to the differential RNA-binding specificities of the two proteins. RNA-specificity was assayed by binding the substituted L7Ae protein to 5′-radiolabeled box C/D (K-turn) and C′/D′ (K-loop) RNA halfmers derived from M. jannaschii sR8 sRNA (Fig. 1A). Assembled RNP complexes were resolved on native polyacrylamide gels (Fig. 3A). Electrophoretic mobility-shift assays (EMSA) confirmed the differential RNA-binding specificities of the wild-type archaeal L7Ae and eukaryotic 15.5kD proteins for the K-turn and K-loop motifs. Replacement of L7Ae amino acid K26 with the corresponding eukaryotic Q26 amino acid (L7Ae-K26Q) had little effect on L7Ae's binding to both K-turn and K-loop RNAs. In contrast, replacement of the four signature L7Ae Loop 9 amino acids with the corresponding eukaryotic residues (L7Ae-EL9) greatly reduced L7Ae binding to the K-loop but not the K-turn RNA. Loss of K-loop binding was more pronounced when the K26Q mutation was included, yielding an L7Ae protein substituted with all five eukaryotic signature amino acids (L7Ae EL9–K26Q). Thus, replacement of all five archaeal L7Ae signature amino acids with the corresponding eukaryotic residues produced an L7Ae protein that mimicked the binding characteristics of the eukaryotic 15.5kD homolog.
FIGURE 3.
Replacement of L7Ae signature amino acids with corresponding 15.5kD residues confers eukaryotic RNA-binding specificity to the archaeal homolog. (A) Electrophoretic mobility-shift assays (EMSA) of wild-type and substituted L7Ae proteins possessing eukaryotic signature amino acids bound to K-turn and K-loop motifs. Radiolabeled RNA halfmers possessing either the K-turn or K-loop motif (Fig. 1A) were incubated with increasing concentrations of wild-type or substituted L7Ae. RNA–protein complexes were resolved on native polyacrylamide gels and visualized by autoradiography. The protein species are indicated above the panels. (B) Comparative binding affinities of the wild-type and mutant L7Ae proteins for the K-turn and K-loop motifs estimated from EMSA analysis. Ratios of mutant L7Ae to wild-type L7Ae binding affinities (Kmut/Kwt) are calculated for both the K-turn and K-loop. A binding affinity of >50 μM and Kmut/Kwt ratio of >10 indicates no specific binding as determined by EMSA analysis.
EMSA analysis was then used to determine the relative binding affinities of the various L7Ae protein mutants (Fig. 3B). This approach does not measure equilibrium binding between RNA and protein as does a filter-binding assay (Kuhn et al. 2002), but can nevertheless be useful in determining the comparative affinities of the various L7Ae mutants with wild-type protein. For these analyses, a binding affinity of >50 μM indicates no specific binding as determined by EMSA analysis. Initial assessment of the Loop 9 substitutions in the presence or absence of the K26Q mutation was consistent with the observed gel-shift profiles. The K26Q mutation alone had little effect on K-turn and K-loop binding affinities, respectively, when compared with wild-type L7Ae. When L7Ae was substituted with eukaryotic Loop 9 residues (L7Ae EL9), its K-loop affinity was reduced. L7Ae affinity for the K-loop was essentially lost when all five signature amino acid substitutions were incorporated (L7A EL9–K26Q). However, each of these mutant proteins still recognized the K-turn motif with comparable binding affinities.
Additional L7Ae mutagenesis determined the contribution of individual signature amino acids for K-loop binding where only one or two signature amino acids were replaced (Fig. 3B). Previous L7Ae crystal structures have suggested that the Loop 9 amino acids glutamate and valine may have critical roles in K-loop binding (Fig. 2B). Therefore, these residues were replaced as either single or double substitutions with the corresponding 15.5kD amino acids (serine and arginine). Neither substitution, alone or in combination, had a substantial effect on K-turn binding (Fig. 3B). However, binding affinity for the K-loop RNA was reduced. Substitution of L7Ae lysine 26 to the eukaryotic glutamine residue did not affect K-loop binding except when all four Loop 9 residues were also substituted. Relative mutant to wild-type L7Ae binding affinities were calculated as Kmut/Kwt ratios (Fig. 3B). Collectively, these results demonstrated that complete loss of L7Ae binding to the K-loop motif requires replacement of all five signature amino acids with the corresponding eukaryotic amino acids.
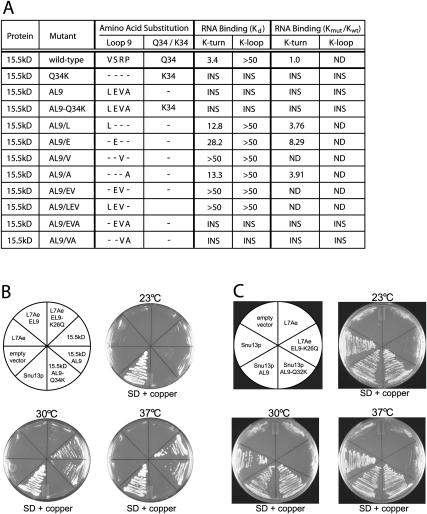
Replacement of eukaryotic 15.5kD signature amino acids with corresponding archaeal L7Ae residues reduces both K-turn and K-loop binding in vitro
The five signature amino acids of mouse 15.5kD were systematically replaced with the corresponding M. jannaschii L7Ae residues to determine if these substitutions could confer K-loop binding (Fig. 4A). Unfortunately, many of the substituted 15.5kD recombinant proteins were insoluble. This was most evident when more than two amino acids were substituted. Single amino acid substitutions did produce soluble 15.5kD, except for the Q34K mutation. Notably, replacement of 15.5kD valine or proline with L7Ae leucine or alanine, respectively, yielded proteins with diminished K-turn binding affinities. Substitution of the eukaryotic serine and arginine residues with the archaeal glutamate and valine residues also reduced K-turn binding. This is consistent with the L7Ae mutational analyses, indicating that these two internal amino acids are critical for K-turn/K-loop recognition. For sufficiently soluble 15.5kD mutants, the relative binding affinities for K-turn and K-loop motifs were calculated with respect to wild-type 15.5kD protein (Fig. 4A). Again, a binding affinity of >50 μM and Kmut/Kwt ratios of >10 indicated no specific binding as determined by EMSA analysis.
FIGURE 4.
Replacement of 15.5kD signature amino acids with the corresponding archaeal L7Ae residues reduces K-turn/K-loop recognition. (A) Binding affinities of wild-type and signature amino acid-substituted 15.5kD proteins for the K-turn and K-loop motifs were approximated from EMSA analysis. (INS) Insoluble recombinant protein. Ratios of mutant 15.5kD to wild-type 15.5kD binding affinities (Kmut/Kwt) are calculated for both the K-turn and K-loop. A binding affinity of >50 μM and Kmut/Kwt ratio of >10 indicates no specific binding as determined by EMSA analysis. (B) The mouse 15.5kD gene but not the archaeal L7Ae gene rescues the depleted yeast SNU13 gene. The endogenous SNU13 gene in yeast strain YGALSNU13 was replaced by an SNU13 gene under control of a GAL1 promoter. This strain was transformed with plasmid vectors encoding wild-type or substituted 15.5kD or L7Ae proteins whose expression is under control of a copper-inducible promoter. The ability of the mouse or archaeal homolog to rescue lost Snu13p was assessed by growth in glucose medium (SD) containing copper. Expressed wild-type 15.5kD and L7Ae proteins and substituted 15.5kD and L7Ae possessing replacement signature amino acids are indicated in the sectored plate with growth of the various strains shown at three different temperatures. (C) Growth of yeast expressing endogenous Snu13p core protein substituted with archaeal L7Ae signature amino acids. The sectored plate indicates the substituted Snu13p protein with L7Ae controls.
Additional in vivo experiments in yeast assessed the ability of wild-type and substituted M. jannaschii L7Ae and mouse 15.5kD proteins to replace wild-type Snu13p (yeast 15.5kD protein homolog). Yeast strain YGALSNU13 (Dobbyn and O'Keefe 2004) with the SNU13 gene under control of the GAL1 promoter was transformed with plasmids encoding L7Ae and 15.5kD constructs whose transcription was under control of a copper-inducible CUP1 promoter (Labbe and Thiele 1999). Consequently, growth of the transformed yeast on glucose media containing copper was dependent on function of the expressed 15.5kD or L7Ae proteins. The mutant L7Ae and 15.5kD proteins assayed contained all four Loop 9 amino acid substitutions and either wild-type or K26/Q34 substitution (Fig. 4B). Expression of wild-type mouse 15.5kD moderately compensated for Snu13p loss at normal (30°C) and elevated (37°C) growth temperatures, but compensated poorly at reduced (23°C) temperature. Substitution of the four Loop 9 signature amino acids with the archaeal residues resulted in similar growth phenotypes. Interestingly, the additional substitution of the 15.5kD Q34 residue with lysine resulted in no growth at any temperature, suggesting strong effects on either mouse protein stability or function. Neither wild-type L7Ae nor L7Ae possessing the four Loop 9 amino acid substitutions was able to compensate for Snu13p depletion. When L7Ae was substituted with all five eukaryotic signature amino acids, very limited growth was observed at 30°C. Results of yeast growth in liquid media were consistent with growth phenotypes on agar plates (data not shown). Interestingly, replacement of the endogenous Snu13p signature amino acids with the archaeal residues, either Loop 9 substitutions alone or in concert with the K26Q residue, had no observable effect on yeast growth at all three temperatures tested. Collectively, these results indicated that endogenous Snu13p could be substituted with the archaeal signature amino acids and not affect protein stability or function in vivo. In contrast, the eukaryotic mouse protein was only partially able to compensate for Snu13p, suggesting sufficient differences in protein structure to affect function. Clearly, the archaeal L7Ae protein was unable to compensate for loss of Snu13p, indicating that despite superimposable tertiary structures, distinct differences in archaeal and eukaryotic protein structure affect in vivo function.
Loss of L7Ae K-loop binding does not affect L7Ae incorporation into the C′/D′ RNP or C′/D′ RNP-guided nucleotide modification
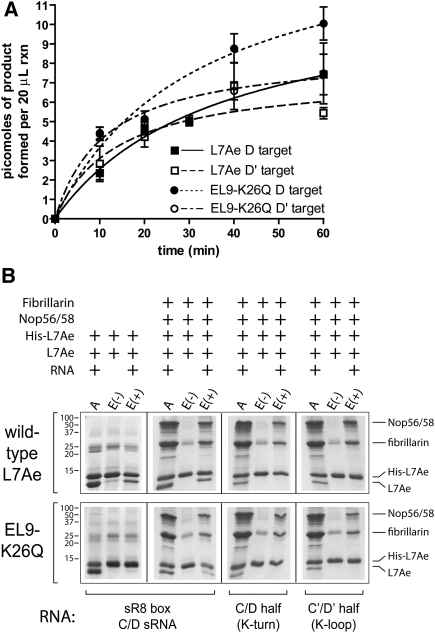
The 2′-O-methylation activity of in vitro assembled archaeal box C/D sRNPs requires symmetric assembly of both box C/D and C′/D′ RNPs possessing all three core proteins (L7Ae, Nop56/58, and fibrillarin) (Tran et al. 2003). Efficient methyltransferase activity of each RNP is also dependent on the juxtapositioning of the partner RNPs within the full-length complex (Tran et al. 2005). We examined the effects of the substituted archaeal L7Ae protein on both sR8 box C/D sRNP assembly and methylation function. The expectation was that assembly of the C′/D′ RNP (K-loop motif) should be disrupted, its methyltransferase activity lost, and the methyltransferase activity of the partner box C/D RNP significantly reduced. Surprisingly, replacement of wild-type L7Ae with the substituted L7Ae EL9–K26Q protein that is unable to bind the K-loop motif did not significantly affect methylation of the D and D′ target RNAs when compared with the wild-type sRNP complex (Fig. 5A). This suggested that despite the loss of L7Ae EL9–K26Q recognition of the K-loop motif, the mutant L7Ae protein was nonetheless incorporated into the C′/D′ RNP, and both RNPs were competent to guide nucleotide methylation.
FIGURE 5.
Archaeal L7Ae protein substituted with eukaryotic 15.5kD signature amino acids is assembled into a catalytically active box C/D sRNP via protein–protein interactions. (A) Nucleotide methylation activity of box C/D sRNPs assembled in vitro with wild-type and signature amino acid–substituted archaeal L7Ae. Box C/D sRNPs were assembled with M. jannaschii sR8 sRNA (Fig. 1A), wild-type Nop56/58 and fibrillarin core proteins, and wild-type (L7Ae) or substituted L7Ae (EL9–K26Q). Methylation activity was assessed by the incorporation of [3H]-CH3 into D and D′ target RNAs. (B) Affinity isolation of sR8 box C/D sRNP assembled with His(6X)-tagged wild-type and substituted L7Ae core proteins. sR8 box C/D sRNP assembly reactions contained a mixture of His-tagged and untagged wild-type or substituted L7Ae core proteins. RNP assembly reactions were applied to an Ni-NTA affinity column (lanes A are loading controls). Eluted core proteins of affinity column–bound assembly reactions assembled with added RNA (lanes E+) or without RNA (lanes E−) are indicated. Bound and eluted proteins were resolved on SDS-polyacrylamide gels and revealed by staining. Components of the individual assembly reactions are indicated above the respective gel lanes, and the specific sR8 sRNAs or RNA halfmers are indicated below. Gel molecular weight markers and specific core proteins are indicated at the sides.
The core protein composition of the sR8 sRNP assembled with L7Ae EL9–K26Q was then examined. A series of RNP assembly experiments was carried out using a mixture of His-tagged and untagged L7Ae core proteins along with untagged Nop56/58 and fibrillarin (Fig. 5B). RNPs were assembled with full-length sR8 sRNA or RNA halfmers possessing either the box C/D motif (K-turn) or C′/D′ motif (K-loop). Subsequent isolation of various core protein combinations incubated with (E+) and without (E−) RNA via Ni-NTA affinity chromatography yielded the tagged L7Ae and other core proteins associated with this tagged protein through their incorporation into assembled RNPs. Therefore, isolation of both tagged and untagged L7Ae via affinity chromatography indicated incorporation of two L7Ae proteins into the sRNP with one L7Ae in both the box C/D and C′/D′ RNPs. Coisolation of Nop56/58 and fibrillarin further indicated assembly of the respective RNP complex with all three core proteins.
Initial control sRNP assemblies with tagged and untagged wild-type L7Ae proteins yielded expected results (Fig. 5B). Both tagged and untagged wild-type L7Ae were purified by Ni-NTA affinity chromatography when incubated with box C/D sR8 sRNA, thus demonstrating the binding of one L7Ae protein at each motif. (Appearance of a nonspecific band near fibrillarin during L7Ae affinity purification should not be confused with fibrillarin.) Isolation of the second, untagged L7Ae was dependent on the presence of sR8 sRNA (E+) in the assembly reaction. When wild-type tagged and untagged L7Ae were bound to sR8 sRNA in the presence of Nop56/58 and fibrillarin, both Nop56/58 and fibrillarin were coisolated, indicating complete box C/D sRNP assembly. Again, their coisolation with tagged L7Ae was dependent on added sR8 sRNA. When tagged and untagged L7Ae were incubated with the respective RNA halfmers, only tagged L7Ae was isolated by affinity chromatography, demonstrating a single L7Ae protein binding to each motif. The coisolation of Nop56/58 and fibrillarin in these incubations containing the respective RNA halfmers indicated complete assembly of the individual box C/D and C′/D′ RNPs with all three core proteins.
An identical series of sRNP assembly and affinity purifications was then carried out utilizing tagged and untagged versions of the substituted L7Ae EL9–K26Q core protein. When tagged and untagged L7Ae EL9–K26Q were incubated with sR8 sRNA and selected with Ni-NTA affinity chromatography, only the tagged version of the protein was isolated. This demonstrated binding of a single L7Ae protein to sR8, presumably at the box C/D motif as expected. However, when the Nop56/58 and fibrillarin core proteins were included in the assembly reaction, both tagged and untagged versions of L7Ae EL9–K26Q were isolated by affinity chromatography. Co-isolation of the second L7Ae protein as well as the Nop56/58 and fibrillarin proteins was dependent on added sR8 sRNA. This indicated that L7Ae EL9–K26Q, although not able to bind the K-loop motif, was nonetheless assembled into the C′/D′ RNP. Control experiments utilizing this same mixture of core proteins incubated with the RNA halfmers possessing either box C/D or C′/D′ motifs confirmed these results. L7Ae EL9–K26Q was coisolated with Nop56/58 and fibrillarin when incubated with the RNA halfmer possessing either the box C/D (K-turn) or C′/D′ (K-loop) motif. Therefore, substituted L7Ae was incorporated into the C′/D′ RNP, despite its inability to bind the K-loop motif. We concluded that incorporation of the L7Ae EL9–K26Q mutant into the C′/D′ RNP involved protein–protein interactions and that these interactions are likely an important factor in box C/D RNP assembly.
L7Ae/L30 protein family members have unique signature amino acids
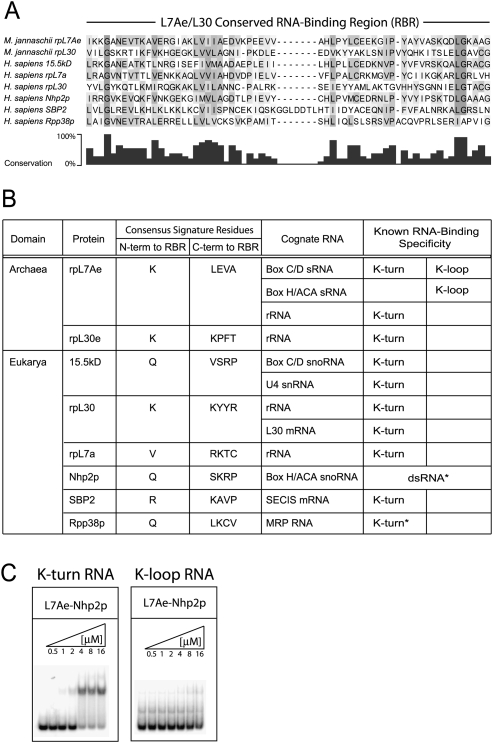
Archaeal L7Ae and eukaryotic 15.5kD homologs are members of the L7Ae/L30 protein family. Other family members include archaeal ribosomal protein rpL30e and eukaryotic proteins rpL7a, rpL30, Nhp2p, SBP2, and Rpp38p. All family members possess a well-conserved RNA-binding region (RBR) (Fig. 6A). Analyses of the signature amino acids for each of these proteins revealed the conservation of these residues in each member of this protein family (Fig. 6B; Supplemental Table S1). Notably, the signature amino acids appear to be unique for each L7Ae/L30 family member (Fig. 6B). This distinctiveness suggests a fine-tuning of RNA-binding specificity for recognition of the respective cognate RNA's K-turn motif. Attempts to assess the evolutionary relationship of these family members using the conserved RBR yielded phylogenetic trees with low accuracy due to the short sequence of the RBR and the small number of known L7Ae/L30 family members (data not shown).
FIGURE 6.
Members of the L7Ae/L30 protein family share a conserved RNA-binding region but possess distinct signature amino acid residues. (A) Alignment of the conserved RNA-binding region (RBR) of the eight L7Ae/L30 protein family members from M. jannaschii and human. Conserved and similar amino acids are indicated in shades of gray and in the bar graph below the alignment. (B) Signature amino acids and cognate RNAs of the archaeal and eukaryotic L7Ae/L30 protein family members. The consensus sequence of signature amino acids for each protein was determined from available family member homologs in the NCBI database (Supplemental Table S1). The known RNA-binding specificity of each protein for K-turn and K-loop motifs is indicated. Asterisks for Nhp2p and Rpp38p denote putative RNA-binding specificities. (C) Archaeal L7Ae substituted with the signature amino acids of Nhp2p results in loss of K-loop binding and weakened K-turn binding. Binding of substituted archaeal L7Ae with Nhp2p signature amino acids (L7Ae–Nhp2p) to the sR8 box C/D and C′/D′ halfmers was assessed by EMSA analysis.
All of the L7Ae/L30 proteins bind RNA, but unique aspects are noted for two of the proteins. First, archaeal L7Ae is the only protein that recognizes both K-turn and K-loop motifs. Second, eukaryotic Nhp2p is the only member that does not specifically recognize either the K-turn or K-loop motif, although it does exhibit double-stranded RNA binding (Henras et al. 2001; Wang and Meier 2004). We replaced archaeal M. jannaschii L7Ae signature amino acids with those of Nhp2p. Notably, this substituted protein (L7Ae–Nhp2p) was no longer able to bind a K-loop RNA, and L7Ae's affinity for the K-turn was reduced (Fig. 6C). This is consistent with a role of signature amino acids for K-turn/K-loop binding and the observation that Nhp2p assembly into the eukaryotic box H/ACA RNPs, whose RNA lacks both K-turn or K-loop motifs, occurs primarily through protein–protein interactions (Henras et al. 2001; Wang and Meier 2004).
DISCUSSION
The differential binding of archaeal L7Ae and eukaryotic 15.5kD to K-turn and K-loop motifs is striking as both proteins exhibit practically superimposable folded structures when bound to their cognate RNAs. Comparative L7Ae and 15.5kD sequence and structural analyses coupled with amino acid replacement experiments have identified five “signature” amino acids that enable the archaeal L7Ae homolog to recognize and bind the K-loop motif. These signature amino acids are highly conserved in both the archaeal L7Ae and eukaryotic 15.5kD proteins, but are distinct for each domain of life. Moreover, these signature amino acids are highly conserved and distinct for each member of the L7Ae/L30 protein family, suggesting a fine-tuning or adaptation of RNA-binding specificities of each family member for the specific K-turn motifs found in the respective cognate RNA(s).
Members of the L7Ae/L30 family possess an RNA-binding region (RBR) that bestows K-turn recognition. Each protein binds a specific K-turn in its respective cognate RNA, thus establishing a unique ribonucleoprotein (RNP) complex. These distinct RNPs form within a given cell even when multiple L7Ae/L30 proteins are present recognizing different K-turn motifs in different cognate RNAs. Some family members, such as Rpp38p and SBP2, possess other functional domains important for protein–protein interactions in their respective RNP complex (Allmang et al. 2002; Welting et al. 2004). These RNP protein–protein interactions help to assemble these specific RNP complexes with distinct L7Ae/L30 proteins recognizing specific K-turns of the cognate RNA. However, we suspect that a critical factor in specific cognate RNA recognition may well be the unique signature amino acids characteristic of each L7Ae/L30 protein family member. This would therefore suggest that each L7Ae/L30 protein has evolved its signature amino acids to facilitate the recognition of distinct K-turn motifs in different cognate RNAs.
The differential binding of the archaeal L7Ae versus eukaryotic 15.5kD homologs for the K-loop motif has until now been an enigma. Previous analyses of L7Ae bound to a K-loop RNA implicated the Loop 9 residues as potentially involved in K-loop recognition (Hamma and Ferré-D'Amaré 2004). These four of the five signature amino acids are adjacent to the RBR and in close proximity to the bound RNA motifs. The conserved L7Ae Loop 9 amino acids L, E, and V and their eukaryotic replacements V, S, and R were thought to account for L7Ae's recognition of the K-loop and 15.5kD's inability to bind this same RNA element. We have now demonstrated that five amino acids, including the L, E, V residues of Loop 9, are important for L7Ae recognition of the K-loop motif. This does not mean that the signature amino acids alone are sufficient for K-loop binding, but clearly they are critical for its recognition by L7Ae.
Of particular interest is identification of the fifth signature amino acid involved in K-loop recognition, initially revealed here by comparative sequence analysis. Surprisingly, this fifth amino acid, L7Ae lysine 26 or 15.5kD glutamine 34, is spatially removed from the RNA ligand and therefore would not be expected to play a role in K-loop recognition by simple inspection of its position in the overall structure of the L7Ae/15.5kD homologs. However, amino acid replacement has clearly shown a role in K-loop recognition. Indeed, effective disruption of L7Ae's ability to recognize the K-loop requires substitution of all five of the signature amino acids, although a gradual loss in binding strength is observed with the systematic replacement of these residues. This fifth signature amino acid does flank the N-terminal side of the RBR but is removed from the bound RNA. Thus, this residue likely functions in K-loop binding by playing a role in L7Ae/15.5kD protein structure and flexibility (Hamma and Ferré-D'Amaré 2004; Soss and Flynn 2007). L7Ae K26 and 15.5kD Q34 both reside in Loop 2 but appear to have different interactions within the protein (Fig. 2). For 15.5kD, Q34's side chain makes three intra-protein contacts with Loop 10, significantly altering local structure. No obvious intra-protein interactions are established by L7Ae's K26. The hydrogen-bonding network and unique local structure established by Q34 is likely important for 15.5kD structure and stability. Consistent with this, mutating Q34 to lysine rendered the mouse 15.5kD protein insoluble when expressed as a recombinant protein and nonfunctional when expressed in yeast and combined with the moderately tolerated Loop 9 mutations. Our observations suggest that flexibility in protein structure can play an important role in RNA-binding specificity.
Attempts to substitute the signature amino acids of eukaryotic 15.5kD with the archaeal residues were met with limited success. Initial expression of recombinant proteins revealed few with sufficient solubility for K-turn and K-loop binding assessment. Most substituted proteins were insoluble, indicating sensitivity of the mouse 15.5kD protein to structural changes induced by signature amino acid changes. The soluble proteins with single or double amino acid substitutions revealed no recognition of the K-loop motif but also disruption of K-turn binding. This suggested that the signature amino acids can also impact K-turn recognition of the 15.5kD protein. In vivo experiments were more successful but could not provide a direct demonstration of K-loop binding for either the substituted mouse 15.5kD or yeast Snu13p proteins. The ability of substituted Snu13p to replace wild-type Snu13p with no obvious growth effects suggested that the amino acid substitutions did not have a significant impact on either protein stability or function. The more limited ability of the mouse 15.5kD protein to accept the archaeal residues and sustain growth may well reflect changes in overall protein structure that impede interactions with other substrates, other proteins, and/or RNAs in the yeast cell.
Finally, the complete assembly of catalytically active box C/D and C′/D′ RNPs using archaeal L7Ae substituted with eukaryotic signature amino acids was unexpected given the assumption that L7Ae should not recognize the K-loop motif and thus not be incorporated into the C′/D′ RNP. However, pull-down experiments clearly showed that L7Ae recognition of the K-loop was not requisite for assembly of a C′/D′ RNP with full core protein composition. This implicates core protein–protein interactions as playing an important role in RNP assembly.
This is certainly the case for assembly of the box H/ACA RNPs (Wang and Meier 2004). Our observations raise interesting questions concerning the structure of the eukaryotic box C/D snoRNP complex, given that the 15.5kD protein does not bind the K-loop RNA motif in vitro (Cahill et al. 2002; Szewczak et al. 2002; Tran et al. 2003). Does the eukaryotic C′/D′ RNP contain a 15.5kD protein in vivo, maintained through protein–protein interactions, thereby establishing box C/D and C′/D′ RNPs each possessing three core proteins? Whether the basic structure of the archaeal and eukaryotic box C/D RNPs has been evolutionarily conserved in terms of RNP organization and core protein composition awaits further investigation of the eukaryotic box C/D snoRNP's fine structure.
MATERIALS AND METHODS
Protein purification and RNA synthesis
Genes for M. jannaschii L7Ae, fibrillarin, and Nop56/58 and mouse 15.5kD were cloned as previously described (Tran et al. 2003). L7Ae, fibrillarin, and 15.5kD were expressed with N-terminal His(6X) tags, and the Nop56/58 protein was untagged. Mutations in the L7Ae and 15.5kD proteins were performed using Stratagene QuickChange site-directed mutagenesis kits. Recombinant proteins were expressed in Rosetta (DE3) cells (Novagen) and purified by Ni-NTA affinity chromatography or cation-exchange chromatography (Tran et al. 2003) and dialyzed into Buffer D (20 mM HEPES at pH 7.2, 3 mM MgCl2, 100 mM NaCl, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol). N-Terminal His(6x) tags were removed by digestion with bovine thrombin (Tran et al. 2003). The sR8 guide sRNA and box C′/D′ halfmer (K-loop) RNA were transcribed in vitro from a PCR-generated DNA template and purified by denaturing polyacrylamide gel electrophoresis as previously described (Tran et al. 2003; Gagnon et al. 2006). Box C/D halfmer (K-turn) RNA was purchased from Dharmacon, Inc.
Electrophoretic mobility-shift assays
RNA–protein complexes were assembled and analyzed by electrophoretic mobility-shift assays as previously described (Tran et al. 2003). Briefly, 5′-radiolabeled RNA was incubated with wild-type/mutant L7Ae protein (8 min at 70°C) or wild-type/mutant mouse 15.5kD protein (30 min at 30°C). Assembled RNPs were resolved on native, phosphate-buffered 6% polyacrylamide gels and visualized using a Molecular Dynamics PhosphorImager. The quantities of bound RNA (i.e., RNP complexes) and free RNA were determined, and dissociation constants were calculated with Prism 4.0 software using a rectangular hyperbolic binding model.
Protein and RNP affinity copurification
Analysis of protein–protein and protein–RNA interactions by affinity copurification was performed as previously described (Tran et al. 2003; Gagnon et al. 2006; Zhang et al. 2006). Briefly, His(6X)-tagged wild-type or mutant L7Ae was incubated at 70°C with untagged L7Ae wild-type or mutant protein with or without the indicated RNAs. Additionally, untagged Nop56/58 or fibrillarin core proteins were included in reactions where indicated. Reactions were incubated with Ni-NTA Resin, stringently washed, and gently eluted with imidazole-containing buffer. Elution fractions were acetone-precipitated and resolved on 16% SDS-polyacrylamide gels.
In vitro 2′-O-methylation assay
In vitro 2′-O-methylation assays for assembled box C/D sRNPs were carried out as previously described (Tran et al. 2003; Gagnon et al. 2006). Aliquots were removed at selected time points, TCA-precipitated, and washed, and incorporation of [3H]–CH3 into D or D′ target RNAs was determined by scintillation counting. Incorporation of [3H]–CH3 into target RNAs previously methylated at the target nucleotide served as baseline controls.
In vivo assessment of mutant M. jannaschii L7Ae, mouse 15.5kD, and yeast Snu13p function
Yeast strain YGALSNU13 with SNU13 under the control of the GAL1 promoter was a gift from Raymond O'Keefe (Dobbyn and O'Keefe 2004). PCR-amplified genes for wild-type and mutant L7Ae, mouse 15.5kD, and Snu13p proteins were cloned into the BamHI/HindIII restriction sites of plasmid vector pCu415-CUP1, which was kindly provided by Dennis J. Thiele (Labbe and Thiele 1999). Transcription of inserted genes was under the control of the copper-inducible CUP1 promoter. Vector pCu415-CUP1 also carried the LEU2 gene for growth in leucine-deficient media. Yeast transformants were grown on standard glucose (SD) media lacking leucine or standard galactose (SG) media containing leucine. To induce protein expression, copper chloride was added to a concentration of 1 μM. Bathocuproine disulfonic acid (BCS), a copper-specific chelator, was added to a concentration of 100 μM to inhibit protein expression when applicable. Cultures were grown at the indicated temperatures. Cell growth in liquid media was accomplished as described (Fabrizio et al. 1997).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org. Supplemental Table S1 reports signature amino acids of the various L7Ae/L30 proteins from different organisms and the derived consensus signature amino acids for each protein.
ACKNOWLEDGMENTS
We thank Raymond O'Keefe for yeast strain YGALSNU13, Dennis J. Thiele for plasmid pCu415-CUP1, Jim Brown for assistance with phylogenetic analyses, and Beth Tran for her suggestion of the yeast experiments. This work was supported by NSF Grant MCB 0543741 (to E.S.M.) and NIH Grant GM069699 (to B.A.B.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1692310.
REFERENCES
- Allmang C, Carbon P, Krol A. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: Identification of SBP2 amino acids important to SECIS RNA binding. RNA. 2002;8:1308–1318. doi: 10.1017/s1355838202020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz JA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, Rothe B, Pescia C, Robert MC, Kiss T, et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill NM, Friend K, Speckman W, Li ZH, Terns RM, Terns MP, Steitz JA. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 2002;21:3816–3828. doi: 10.1093/emboj/cdf376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron C, Manival X, Clery A, Senty-Segault V, Charpentier B, Marmier-Gourrier N, Branlant C, Aubry A. The archaeal sRNA binding protein L7Ae has a 3D structure very similar to that of its eukaryal counterpart while having a broader RNA-binding specificity. J Mol Biol. 2004;342:757–773. doi: 10.1016/j.jmb.2004.07.046. [DOI] [PubMed] [Google Scholar]
- Clery A, Bourguignon-Igel V, Allmang C, Krol A, Branlant C. An improved definition of the RNA-binding specificity of SECIS-binding protein 2, an essential component of the selenocysteine incorporation machinery. Nucleic Acids Res. 2007;35:1868–1884. doi: 10.1093/nar/gkm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PP, Omer A, Lowe T. A guided tour: Small RNA function in Archaea. Mol Microbiol. 2001;40:509–519. doi: 10.1046/j.1365-2958.2001.02381.x. [DOI] [PubMed] [Google Scholar]
- Dobbyn HC, O'Keefe RT. Analysis of Snu13p mutations reveals differential interactions with the U4 snRNA and U3 snoRNA. RNA. 2004;10:308–320. doi: 10.1261/rna.5970404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Laggerbauer B, Lauber J, Lane WS, Lührmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Zhang X, Agris PF, Maxwell ES. Assembly of the archaeal box C/D sRNP can occur via alternative pathways and requires temperature-facilitated sRNA remodeling. J Mol Biol. 2006;362:1025–1042. doi: 10.1016/j.jmb.2006.07.091. [DOI] [PubMed] [Google Scholar]
- Gagnon KT, Zhang X, Maxwell ES. The Box C/D RNPS: Evolutionarily ancient nucleotide modification complexes. In: Smith HC, editor. The Box C/D RNPS: Evolutionarily ancient nucleotide modification complexes. Wiley; Hoboken, NJ: 2008. pp. 311–339. [Google Scholar]
- Hamma T, Ferré-D'Amaré AR. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure. 2004;12:893–903. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Henras A, Dez C, Noaillac-Depeyre J, Henry Y, Caizergues-Ferrer M. Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res. 2001;29:2733–2746. doi: 10.1093/nar/29.13.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Bork P, Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J, Tran E, Maxwell ES. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002;30:931–941. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe S, Thiele DJ. Copper inducible and repressible promoter systems in yeast. Methods Enzymol. 1999;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- Liu S, Li P, Dybkov O, Nottrott S, Hartmuth S, Lührmann R, Carlomango T, Wahl MC. Binding of the human Prp31 Nop domain to a composite RNA–protein platform in U4 snRNP. Science. 2007;316:115–120. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- Mao H, White SA, Williamson JR. A novel loop–loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat Struct Biol. 1999;6:1139–1147. doi: 10.1038/70081. [DOI] [PubMed] [Google Scholar]
- Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA–protein interactions: Cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- Nolivos S, Carpousis AJ, Clouet-d'Orval B. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 2005;33:6507–6514. doi: 10.1093/nar/gki962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer A, Ziesche S, Ebhardt H, Dennis PP. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc Natl Acad Sci. 2002;99:5289–5294. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oruganti S, Zhang Y, Li H. Structural comparison of yeast snoRNP and spliceosomal protein Snu13p with its homologs. Biochem Biophys Res Commun. 2005;333:550–554. doi: 10.1016/j.bbrc.2005.05.141. [DOI] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo G, Cuccurese M, Monti G, Russo A, Amoresano A, Peitropaolo C. Ribosomal protein L7a binds RNA through two distinct RNA-binding domains. Biochem J. 2005;385:289–299. doi: 10.1042/BJ20040371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soss SE, Flynn PF. Functional implications for a prototypical K-turn binding protein from structural and dynamical studies of 15.5K. Biochemistry. 2007;46:14979–14986. doi: 10.1021/bi701254q. [DOI] [PubMed] [Google Scholar]
- Szewczak LB, DeGregorio SJ, Strobel SA, Steitz JA. Exclusive interaction of the 15.5kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem Biol. 2002;9:1095–1107. doi: 10.1016/s1074-5521(02)00239-9. [DOI] [PubMed] [Google Scholar]
- Tran EJ, Zhang X, Maxwell ES. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 2003;22:3930–3940. doi: 10.1093/emboj/cdg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhang X, Lackey L, Maxwell ES. Conserved spacing between the box C/D and C′/D′ RNPs of the archaeal box C/D sRNP complex is required for efficient 2′-O-methylation of target RNAs. RNA. 2005;11:285–293. doi: 10.1261/rna.7223405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic I, Nottrot S, Hartmuth K, Lührmann R, Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- Wang C, Meier UT. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Lührmann R. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- Welting TJM, van Venrooij WJ, Pruijn GJM. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32:2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Champion EA, Tran E, Brown BA, II, Baserga SJ, Maxwell ES. The coiled-coil domain of the Nop56/58 core protein is dispensable for sRNP assembly but is critical for archaeal box C/D sRNP-guided nucleotide methylation. RNA. 2006;12:1092–1103. doi: 10.1261/rna.2230106. [DOI] [PMC free article] [PubMed] [Google Scholar]