Abstract
Survivin has been identified as an apoptosis inhibitor and a key regulator of mitosis. A common polymorphism (-31G>C) at the survivin promoter has been extensively studied in various cancers and reported to influence survivin expression. We hypothesize that polymorphisms in the survivin promoter are associated with clinical outcomes of patients with ovarian cancer. In this study, we genotyped all of five common and independent (r2 < 0.25 for all LD) survivin promoter polymorphisms (-1547A/G [rs3764383], -644C/T [rs8073903], -625C/G [rs8073069], -241C/T [rs17878467], and -31G/C [rs9904341]) in 168 patients with primary epithelial ovarian cancer, using the polymerase chain reaction-restriction fragment length polymorphism method. We found that -1547A/G and -31G/C were significantly associated with age of disease onset. Compared with patients with the -1547GG genotype, the -1547AA genotype showed a significantly younger age of disease onset (58.8 years vs. 70.1 years; P = 0.001); the -31CC genotype had a decrease, though not significant, in the age of disease onset, compared with patients with the -31GG genotype (57.1 years vs. 62.8 years; P = 0.058). The numbers of -1547A and -31C alleles were associated with a decrease in age of onset in an allele-dose response manner (Ptrend = 0.001 and 0.026, respectively). However, no association was found between survivin polymorphisms and patients' prognosis, except for -625C/G SNP in 37 patients with a persistent disease. The findings suggest that the promoter variants of survivin may have an effect on age of onset of ovarian cancer. Validation studies with larger sample sizes are warranted.
Keywords: BIRC5, genetic polymorphism, apoptosis, ovarian cancer, survival
Introduction
Survivin (also known as BIRC5) is one of the first reported inhibitors of apoptosis proteins (IAPs), which is an important family of proteins that regulate apoptosis [1, 2]. It is developmentally regulated and expressed during cell differentiation in both humans and mice [3, 4]. Suvivin is usually expressed in embryonic tissues and homozygous survivin deletion results in early embryonic death, showing its essential role in cell development, differentiation, and homeostasis [3, 5]. Suvivin has also been reported to be selectively expressed in cancer cells but not in normal tissues [4]. The overexpression of survivin was found to be associated with disease development, recurrence, and prognosis in various malignancies, including cancers of the bladder, cervix, head and neck, prostate, skin, and ovaries [6–19]. The underlying mechanisms of survivin expression have been explored in various ways.
The survivin gene is located on human chromosome 17q25. It has a TATA-less promoter with a canonical CpG island, three cell-cycle dependent elements (CDE), one cell cycle homology region (CHR) [20], and numerous Sp1 sites in the 5′ flanking region in both humans and mice [21, 22]. The deletion of CDE and CHR in the survivin promoter region leads to loss of cell-cycle dependent expression needed for the basal transcriptional requirements of survivin expression [21, 22]. The results of several studies on the regulation between the survivin expression and methylation status of the CpG islands in the promoter have been conflicting [21, 23–25]. Genetic variation has been implicated in the alteration of gene expression, especially the regulatory polymorphisms located in promoter regions [26, 27]. The genetic variant -31G/C in the survivin promoter region has been identified to be associated with overexpression of survivin at both protein and mRNA levels in cancer cells [26, 27]. This polymorphism has also been studied for its effect on risk of gastric, urothelial, colorectal, cervical, and lung cancers [26, 28–32]. However, few studies have investigated the effects of genetic variants in the survivin promoter region on clinical outcomes of cancers [26].
Survivin has been implicated in the regulation of microtubule dynamics [22, 33–36]. The functional study of survivin in the mitotic phase has revealed changes in the duration of prometaphase and metaphase stages and formation of mitotic spindles in different lengths according to the survivin status. In addition, survivin showed to significantly reduce pole-to-pole distance in metaphase cells and to stabilize microtubules [34]. Survivin may influence microtubule dynamics and microtubule stability via interactions with microtubule-associated protein or motor proteins participating in spindle dynamics [37, 38].
Taxanes, one of the standard chemotherapeutic agents for the treatment of ovarian cancer, block microtubule disassembly, to which survivin is related [39]. Zaffaroni et al. reported a direct link between survivin expression and sensitivity to taxol [40]. Athanassiadou et al. reported that the survivin levels were associated with adverse clinical features in ovarian cancer patients, implying a role of this gene in ovarian cancer progression [41].
Therefore, in this study, we investigated associations between potentially functional single nucleotide polymorphisms (SNPs) of survivin and clinical outcomes in 168 ovarian cancer patients, who had chemotherapies including taxanes. Because there are no reported common (minor allele frequency [MAF] ≥ 5%) non-synonymous SNPs in this gene, we genotyped all of five common SNPs in the promoter region of survivin, including -1547A/G (rs3764383), -644C/T (rs8073903), -625C/G (rs8073069), -241C/T (rs17878467), and -31G/C (rs9904341) and evaluated their associations with clinical outcomes of patients with primary epithelial ovarian cancer.
Material and methods
Study subjects
This study included 168 patients registered at The University of Texas M. D. Anderson Cancer Center from 2000 to 2007. Patients had been staged according to the International Federation of Gynecology and Obstetrics (FIGO) surgical staging system, and their primary epithelial ovarian cancer was histopathologically confirmed. After the surgery, patients had been treated with chemotherapy, a combination of platinum (carboplatin, cisplatin) and taxanes (taxol, docetaxel). Patients who had a previous cancer history or multiple primary neoplasms at the time of diagnosis were excluded from this study. The Gynecologic Cancer Tumor Bank at M. D. Anderson Cancer Center provided peripheral blood samples of these patients. The institutional review board approved this study, and guidelines for the protection of human subjects were followed.
Data collection
We collected demographic and clinicopathologic data on patients from medical records. Demographic characteristics included age at diagnosis and race/ethnicity, and clinicopathologic characteristics included tumor stage, cell type and grade, optimality of the primary debulking operation, chemotherapy regimen, number of chemotherapies, disease recurrence, and response of tumors to chemotherapy. Optimal debulking or cytoreductive surgery is defined as the largest residual tumor nodule measuring 1 cm or less, according to Gynecologic Oncology Group [42]. The response evaluation criteria in solid tumors (RECIST) [20] were used to define the response of tumors to treatment.
Overall survival (OS) and progression-free survival (PFS) were calculated as the date of disease diagnosis to the date of death or last contact or the date of recurrence or progression, respectively. Disease recurrence was defined as the reappearance of any lesion that had previously disappeared or the appearance of a new lesion that was histopathologically confirmed by a biopsy. Information about the date of last contact and status of the patient at last contact was obtained from the M. D. Anderson Tumor Registry and Social Security Death Index, when this information was missing from the medical records.
SNP selection and genotyping
All five reported common SNPs in the survivin promoter region were selected from the National Center for Biotechnology Information SNP database (http://www.ncbi.nlm.nih.gov/snp): -1547A/G (rs3764383), -644C/T (rs8073903), -625C/G (rs8073069), -241C/T (rs17878467), and -31G/C (rs9904341). Genomic DNA was extracted from all samples using the classic method with phenol and chloroform. Genotyping analyses were performed using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
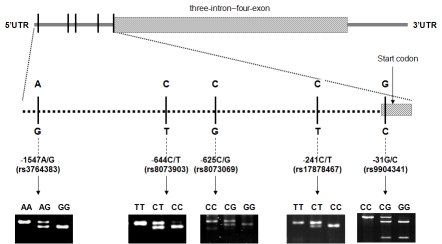
The PCR conditions consisted of an initial melting step of 95°C for 5 min, followed by 35 cycles of denaturation (95 °C for 30 s), annealing (52°C - 66 °C for 45 s, according to SNPs), and extension (72 °C for 1 min), and a final extension step of 72 °C for 10 min. Detailed information on primers, restriction enzymes, and the PCR product sizes are summarized in Table 1. The digested products were checked on a 3% MetaPhor agarose gel containing ethidium bromide. The gene structure, locations of the five SNPs and the genotype patterns were shown in Figure 1. Two people checked the patterns of the digested products, and any uncertain genotyping results were repeated for quality control, and the concordance was 100%.
Table 1.
Conditions for polymerase chain reaction–restriction fragment length polymorphism analysis of the survivin promoter
| Position and base change | Primers | Annealing Temperature (°C) | PCR products (bp) | Enzyme | Digested PCR products (bp) |
|---|---|---|---|---|---|
| -1547A/G (rs3764383) | FP 5'-GCCCGATGCATTTAAATAAAAGA-3' | 52 | 118 | HincII | AA: 118 AG: 118/96/22 |
| RP 5'-GCAGAGAGTGAATGTTAAAGTTAA-3' | GG: 96/22 | ||||
| -644C/T (rs8073903) | FP 5'-AGGTCGTGCAGTCAACGATGT-3' | 52 | 89 | StyI | TT: 89 CT: 89/66/23 |
| RP 5'-CAGACGGGCATGAAGGACCCATG-3' | CC: 66/23 | ||||
| -625C/G (rs8073069) | FP 5'-TGTTCATTTGTCCTTCATGCGC-3' | 61 | 125 | BstUI | CC: 125 CG: 125/104/21 |
| RP 5'-CCAGCCTAGGCAACAAGAGCAA-3' | GG: 104/21 | ||||
| -241C/T (rs17878467) | FP 5'-GATTACAGGCGTGAGCCACT-3' | 61 | 128 | HaeII | TT: 128 CT: 128/104/24 |
| RP 5'-GTGTGCCGGGAGTTGTAGTC-3' | CC: 104/24 | ||||
| -31G/C (rs9904341) | FP 5'-CGTTCTTTGAAAGCAGTCGAG-3' | 66 | 329 | EcoO109I | CC: 329 CG: |
| RP 5'-TGTAGAGATGCGGTGGTCCT-3' | 329/234/92 GG: 234/92 |
bp=base pair; FP=forward primer; RP=reverse primer.
Figure 1.
Survivin gene structure, locations of the five SNPs and the genotype patterns.
Statistical analysis
Statistical analysis was performed using the Chi-square test and analysis of variance (ANOVA) analysis for categorical and the for continuous variables. The Proc Allele procedure in the SAS/Genetics program (SAS Institute Inc., Cary, NC, USA) was used to calculate linkage disequilibrium (LD). The Kaplan-Meier method and the log-rank test were used to estimate OS and PFS. The Cox proportional hazards regression model was used to analyze individual prognostic factors. All statistical tests were two-sided, a P value of 0.05 was considered statistically significant, and all analyses were performed using the Statistical Analysis System/Genetics software (SAS version 9.13; SAS Institute Inc.).
Results
Demographic and clinical characteristics
Demographic and clinicopathologic characteristics of the 168 patients included in this study are summarized in Table 2. Nearly 85% of the patients were older than 50 years, with a mean age of 60.8 ± 10.8 years for all patients. One-hundred-thirty-six patients (80.9%) were non-Hispanic white, nine (5.4%) Black, 15 (8.9%) Hispanic, six (3.6%) Asian, and two (1.2%) were others. One-hundred-fifty-one patients (91.6%) had an advanced disease with 127 patients (76.0%) diagnosed at stage III and 26 patients (15.6%) diagnosed at stage IV. Most of the patients had high grade (159, 96.4%) and serous cell type (137, 81.5%), and 106 patients (71.6%) had received an optimal debulking operation as primary surgery.
Table 2.
Patient demographic and clinicopathologic characteristics
| Characteristic | No. of patients |
|---|---|
| Age at Diagnosis (years) | 168 |
| < 50 | 26 |
| 50 – 70 | 107 |
| > 70 | 35 |
| Race/Ethnicity | 168 |
| Non-Hispanic White | 136 |
| Black | 9 |
| Hispanic | 15 |
| Asian | 6 |
| Other | 2 |
| Surgical stagea | 167 |
| I | 7 |
| II | 7 |
| III | 127 |
| IV | 26 |
| Tumor Gradea | 165 |
| 1 | 6 |
| 3 | 159 |
| Histology | 168 |
| Serous | 137 |
| Mucinous | 3 |
| Endometrioid | 2 |
| Clear cell | 1 |
| Brenner | 3 |
| Mixed | 22 |
| Optimality of primary operationa | 148 |
| No | 42 |
| Yes | 106 |
| Peritoneal cytologya | 134 |
| Negative | 16 |
| Positive | 118 |
Missing patient information: 1 for surgical stage; 3 for tumor grade; 20 for optimality of primary operation; 34 for peritoneal cytology.
Correlation of survivin genotype with age of onset of ovarian cancer
The genotype distributions of the five SNPs among these patients are listed in Table 3. Since we have 80.9% patients were non-Hispanic white and few cases in other minority groups, and we didn't find the difference of the effects of genotypes on age onset and survival among the whites and other groups; therefore, we combine all of the samples together for the following presentation. The allele frequencies were similar to those reported in the National Center for Biotechnology Information database (Table 3). The LD tests further confirmed that these five SNPs were not in LD, with a range of r2 between 0 - 0.25, suggesting that these SNPs were indeed independent in this study population. When we stratified the age of disease onset by these genotypes, we found that patients with the -1547AA genotype (rs3764383) showed a significantly younger age of disease onset, compared with patients with the -1547GG genotype (58.8 ± 11.2 years vs. 70.1 ± 5.5 years; P = 0.001) (Table 3), and there was an A allele dose-response between age of onset and -1547A/G genotypes (trend test: P = 0.001) (Figure 2A). We also found that patients with the -31CC genotype (rs9904341) showed a decrease, though not significant, in the age of disease onset, compared with patients with the -31GG genotype (57.1 ± 12.8 years vs. 62.8 ± 10.5 years; P = 0.058) (Table 3); furthormore, the trend for the number of the C allele in the genotypes (i.e., the -31GG, -31GC, and -31CC genotypes) was statistically significant (P = 0.026) (Figure 2B). When we analyzed data for patients with a combination of these two SNPs (-1547A/G + -31G/C), we found that 26 patients with both the -1547AA and -31CC genotypes had the youngest age of onset of ovarian cancer (57.0 ± 13.0 years), and there was a trend of an increase in age of disease onset as the number of these (i.e., A and C) alleles decreased in the combined genotypes (Ptrend = 0.003) (Figure 2C). Whereas, these trends in age of onset of ovarian cancer were not observed for the other three SNPs (-241C/T, -625C/G, and -644C/T) (Table 3).
Table 3.
Genotype frequencies and age of disease onset stratified by survivin genotypes
| Genotype | No. of patients (%) | Age at diagnosis (years, mean ±SD) |
P valuea |
|---|---|---|---|
| -1547A/G (rs3764383) | |||
| AA | 86 (51.2) | 58.8 ± 11.2 | Ref. |
| AG | 69 (41.1) | 61.4 ± 9.8 | |
| GG | 13 (7.7) | 70.1 ± 5.5 | 0.001 |
| G allele frequency | 0.283 | Ptrend = 0.001b | |
| -644C/T (rs8073903) | |||
| TT | 120 (71.4) | 61.0 ± 10.8 | Ref. |
| CT | 37 (22.0) | 60.0 ± 10.6 | |
| CC | 11 (6.6) | 60.5 ± 11.3 | 0.90 |
| C allele frequency | 0.176 | Ptrend = 0.68b | |
| -625C/G (rs8073069) | |||
| GG | 91 (54.2) | 60.9 ± 11.2 | Ref. |
| CG | 30 (17.8) | 59.1 ± 10.8 | |
| CC | 47 (28.0) | 61.4 ± 9.7 | 0.63 |
| C allele frequency | 0.369 | PTrend = 0.61b | |
| -241C/T (rs17878467) | |||
| CC | 106 (63.1) | 61.5 ± 11.1 | Ref. |
| CT | 48 (28.6) | 59.5 ± 10.2 | |
| TT | 14 (8.3) | 58.8 ± 9.7 | 0.43 |
| T allele frequency | 0.226 | Ptrend = 0.13b | |
| -31G/C (rs9904341)c | |||
| GG | 61 (36.5) | 62.8 ± 10.5 | Ref. |
| CG | 78 (46.7) | 60.3 ± 9.8 | |
| CC | 28 (16.8) | 57.1 ± 12.8 | 0.058 |
| C allele frequency | 0.399 | Ptrend = 0.026b |
One-way ANOVA (Analysis of variance) for age differences among 3 genotypes for each SNP.
p values for the trend tests of age at diagnosis among the 3 genotypes for each SNP from a general linear model adjusted for race.
One sample was missing genotype information for -31G/C.
Figure 2.
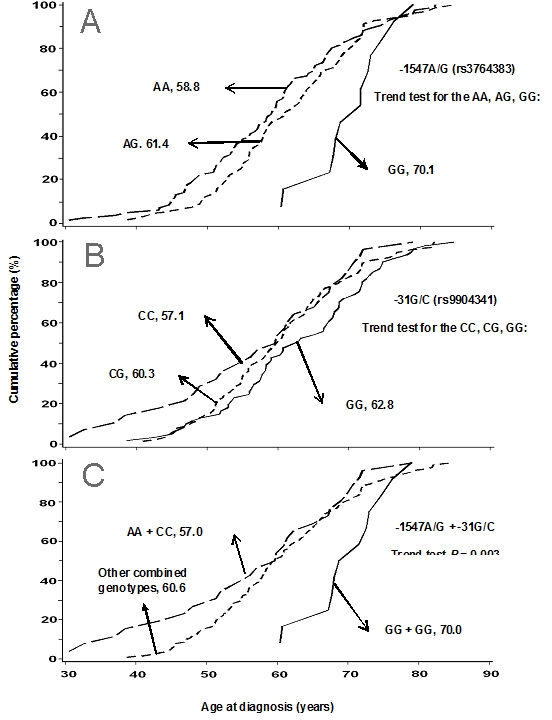
Comparison of age of onset of ovarian cancer by survivin promoter genotypes: (A) 1547A/G, (B) -31G/C, and (C) their combination.
Effects of survivin polymorphisms on OS and PFS
We further investigated the effects of Survivin polymorphisms on the treatment response of patients. We analyzed OS and PFS for patients who were treated with the anti-mitotic chemotherapeutic agents, taxanes, in combination with platinum. In this study population, 133 patients received the taxanes/platinum combination regime as adjuvant therapy after their surgery, and 132 patients were available for evaluation of the drug responsiveness. Among them, 37 patients (37/132, 28.0%) had persistent disease (resistant to taxanes treatment) and 95 patients (95/132, 72.0%) responded to the treatment. The patients with a persistent disease had shorter OS and PFS compared with the treatment-responsive group. It was consistent with what we expected on the basis of common notion, suggesting that the study population, although relatively small in size, may be representative of the overall ovarian cancer patient population. We did not find the correlation between OS or PFS and number of cycle, regimen, and dosage (data not shown). However, when we stratified OS and PFS by the genotypes of the five SNPs, we found that the -625C/G SNP showed a difference in the median PFS, as measured in 37 patients with a persistent disease (GG: 4.7 months, CG: 1.4 months, and CC: 4.1 months; P = 0.035; Figure 3).
Figure 3.
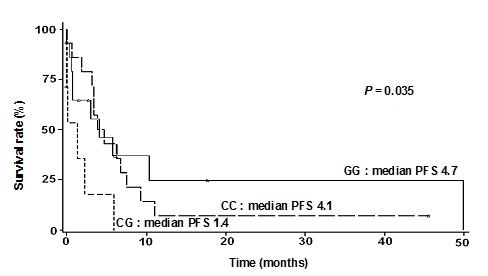
Progression-free survival was affected by the survivin promoter SNP -625C/G (rs8073069).
Discussion
Among the five common polymorphisms in the survivin promoter region, -31G/C (rs9904341) has been the most extensively studied, because it is located at the CDE/CHR repressor elements and thus may be functional. It has been shown that this SNP was associated with increased survivin expression at both the mRNA and protein levels, as well as an aberrant cell cycle-dependent transcription [27]. In our study, we found that the variant -31C allele was associated with the early age of onset of ovarian cancer in an allele-dose response manner, which further suggested a role of genetic variation in the survivin promoter in the etiology of ovarian cancer. We also found that another promoter SNP -1547A/G was associated with age of onset of ovarian cancer. However, the functional significance of this SNP remains unclear and needs to be investigated in future studies.
Several studies have reported that genetic variants may be correlated with the age of disease onset in patients with brain, prostate, gastric, breast, and ovarian cancers [43–51]. To our knowledge, our study is the first to find that survivin promoter polymorphisms may be associated with the age of onset of ovarian cancers. These findings suggest that genetic variations in the survivin promoter may play an important role in the pathophysiological changes of the ovaries.
When we analyzed OS and PFS for patients who were treated with platinum and taxanes combination chemotherapies and investigated the effects of survivin polymorphisms on the treatment response, only the -625C/G SNP showed a difference in the median PFS. The patients with -625C allele showed a relatively poor survival as measured in 37 patients with a persistent disease. Even though the number of patients available for the analysis is low, it may imply a relation of this SNP with the treatment response and prognosis. In an Korean study on survivin polymorphisms and risk of lung cancer, it was found that -625G allele contributed to the decreased risk of lung cancer in the haplotype analysis [29]. A Chinese research group reported that the - 625CC genotype was associated with significantly increased risk of esophageal squamous cell carcinoma and that the -625C allele was found to contributue to the high risk haplotype and enhance the survivin expression in esophageal cancer patients [52, 53]. There are no reports on similar studies on ovarian cancer; however, our finding on this SNP was considered preliminary, and further larger studies are needed to validate the significance of this SNP and its interaction with treatment.
Several studies reported that wild-type p53 represses anti-apoptotic survivin expression at both protein and mRNA levels [54–56]. Recently, it was reported that esophageal tumors with p53 gene mutations showed higher levels of mRNA expression of survivin than tumors with the wild-type p53 [52]. Therefore, interactions of genetic variants in survivin with other genetic variants, such as those in the p53 gene, should be investigated as well.
In conclusion, this pilot study provides evidence of an association between SNPs in the survivin promoter and age of onset of ovarian cancer. However, rigorous studies with larger sample sizes and studies of biological functions of these SNPs are needed to validate the role of survivin promoter SNPs in the development of ovarian cancer and their associations with clinicopathologic factors, especially age of disease onset of cancers.
Acknowledgments
This research was supported in part by a National Institutes of Health Ovarian Specialized Programs of Research Excellence grant (P50 CA08363) to G. B. Mills, a BLANTON-DAVIS Ovarian Cancer Research Development Award to L-E. Wang, a grant from the National Cancer Institute (R01 CA131274) to Q. Wei, and a Cancer Center Core grant from the National Cancer Institute to M. D. Anderson (CA16672).
References
- 1.Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 2.Uren AG, Coulson EJ, Vaux DL. Conservation of baculovirus inhibitor of apoptosis repeat proteins (BIRPs) in viruses, nematodes, vertebrates and yeasts. Trends Biochem Sci. 1998;23:159–162. doi: 10.1016/s0968-0004(98)01198-0. [DOI] [PubMed] [Google Scholar]
- 3.Adida C, Crotty PL, McGrath J, Berrebi D, Diebold J, Altieri DC. Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am J Pathol. 1998;152:43–49. [PMC free article] [PubMed] [Google Scholar]
- 4.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 5.Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, Choo KH. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol. 2000;10:1319–1328. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- 6.Karam JA, Lotan Y, Ashfaq R, Sagalowsky AI, Shariat SF. Survivin expression in patients with non-muscle-invasive urothelial cell carcinoma of the bladder. Urology. 2007;70:482–486. doi: 10.1016/j.urology.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Lee JP, Chang KH, Han JH, Ryu HS. Survivin, a novel anti-apoptosis inhibitor, expression in uterine cervical cancer and relationship with prognostic factors. Int J Gynecol Cancer. 2005;15:113–119. doi: 10.1111/j.1048-891X.2005.15011.x. [DOI] [PubMed] [Google Scholar]
- 8.Lo Muzio L, Pannone G, Leonardi R, Staibano S, Mignogna MD, De Rosa G, Kudo Y, Takata T, Altieri DC. Survivin, a potential early predictor of tumor progression in the oral mucosa. J Dent Res. 2003;82:923–928. doi: 10.1177/154405910308201115. [DOI] [PubMed] [Google Scholar]
- 9.Shariat SF, Lotan Y, Saboorian H, Khoddami SM, Roehrborn CG, Slawin KM, Ashfaq R. Survivin expression is associated with features of biologically aggressive prostate carcinoma. Cancer. 2004;100:751–757. doi: 10.1002/cncr.20039. [DOI] [PubMed] [Google Scholar]
- 10.Sui L, Dong Y, Ohno M, Watanabe Y, Sugimoto K, Tokuda M. Survivin expression and its correlation with cell proliferation and prognosis in epithelial ovarian tumors. Int J Oncol. 2002;21:315–320. [PubMed] [Google Scholar]
- 11.Grossman D, McNiff JM, Li F, Altieri DC. Expression and targeting of the apoptosis inhibitor, survivin, in human melanoma. J Invest Dermatol. 1999;113:1076–1081. doi: 10.1046/j.1523-1747.1999.00776.x. [DOI] [PubMed] [Google Scholar]
- 12.Als AB, Dyrskjot L, von der Maase H, Koed K, Mansilla F, Toldbod HE, Jensen JL, Ulhoi BP, Sengelov L, Jensen KM, Orntoft TF. Emmprin and survivin predict response and survival following cisplatin-containing chemotherapy in patients with advanced bladder cancer. Clin Cancer Res. 2007;13:4407–4414. doi: 10.1158/1078-0432.CCR-07-0109. [DOI] [PubMed] [Google Scholar]
- 13.Karam JA, Lotan Y, Karakiewicz PI, Ashfaq R, Sagalowsky AI, Roehrborn CG, Shariat SF. Use of combined apoptosis biomarkers for prediction of bladder cancer recurrence and mortality after radical cystectomy. Lancet Oncol. 2007;8:128–136. doi: 10.1016/S1470-2045(07)70002-5. [DOI] [PubMed] [Google Scholar]
- 14.Marioni G, Bertolin A, Giacomelli L, Marchese-Ragona R, Savastano M, Calgaro N, Marino F, De Filippis C, Staffieri A. Expression of the apoptosis inhibitor protein Survivin in primary laryngeal carcinoma and cervical lymph node metastasis. Anticancer Res. 2006;26:3813–3817. [PubMed] [Google Scholar]
- 15.Piras F, Murtas D, Minerba L, Ugalde J, Floris C, Maxia C, Colombari R, Perra MT, Sirigu P. Nuclear survivin is associated with disease recurrence and poor survival in patients with cutaneous malignant melanoma. Histopathology. 2007;50:835–842. doi: 10.1111/j.1365-2559.2007.02695.x. [DOI] [PubMed] [Google Scholar]
- 16.Preuss SF, Weinell A, Molitor M, Semrau R, Stenner M, Drebber U, Wedemeyer I, Hoffmann TK, Guntinas-Lichius O, Klussmann JP. Survivin and epidermal growth factor receptor expression in surgically treated oropharyngeal squamous cell carcinoma. Head Neck. 2008;30:1318–1324. doi: 10.1002/hed.20876. [DOI] [PubMed] [Google Scholar]
- 17.Troeger A, Siepermann M, Escherich G, Meisel R, Willers R, Gudowius S, Moritz T, Laws HJ, Hanenberg H, Goebel U, Janka-Schaub GE, Mahotka C, Dilloo D. Survivin and its prognostic significance in pediatric acute B-cell precursor lymphoblastic leukemia. Haematologica. 2007;92:1043–1050. doi: 10.3324/haematol.10675. [DOI] [PubMed] [Google Scholar]
- 18.Yi L, Jian W, Chang-yin Z, Jing F, Shuang-yun C, Xuanbing T. Analysis of molecular pathological factors of unfavorable prognosis for young cervical cancer patients. Onkologie. 2007;30:502–506. doi: 10.1159/000107796. [DOI] [PubMed] [Google Scholar]
- 19.Lo Muzio L, Pannone G, Staibano S, Mignogna MD, Rubini C, Mariggio MA, Procaccini M, Ferrari F, De Rosa G, Altieri DC. Survivin expression in oral squamous cell carcinoma. Br J Cancer. 2003;89:2244–2248. doi: 10.1038/sj.bjc.6601402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 21.Li F, Altieri DC. Transcriptional analysis of human survivin gene expression. Biochem J. 1999;344 Pt 2:305–311. doi: 10.1042/0264-6021:3440305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Altieri DC. The cancer antiapoptosis mouse survivin gene: characterization of locus and transcriptional requirements of basal and cell cycle-dependent expression. Cancer Res. 1999;59:3143–3151. [PubMed] [Google Scholar]
- 23.Yu J, Zhang H, Gu J, Lin S, Li J, Lu W, Wang Y, Zhu J. Methylation profiles of thirty four promoter-CpG islands and concordant methylation behaviours of sixteen genes that may contribute to carcinogenesis of astrocytoma. BMC Cancer. 2004;4:65. doi: 10.1186/1471-2407-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hattori M, Sakamoto H, Satoh K, Yamamoto T. DNA demethylase is expressed in ovarian cancers and the expression correlates with demethylation of CpG sites in the promoter region of c-erbB-2 and survivin genes. Cancer Lett. 2001;169:155–164. doi: 10.1016/s0304-3835(01)00499-2. [DOI] [PubMed] [Google Scholar]
- 25.Nabilsi NH, Broaddus RR, Loose DS. DNA methylation inhibits p53-mediated survivin repression. Oncogene. 2009;28:2046–2050. doi: 10.1038/onc.2009.62. [DOI] [PubMed] [Google Scholar]
- 26.Gazouli M, Tzanakis N, Rallis G, Theodoropoulos G, Papaconstantinou I, Kostakis A, Anagnou NP, Nikiteas N. Survivin -31G/C promoter polymorphism and sporadic colorectal cancer. Int J Colorectal Dis. 2008. [DOI] [PubMed]
- 27.Xu Y, Fang F, Ludewig G, Jones G, Jones D. A mutation found in the promoter region of the human survivin gene is correlated to overexpression of survivin in cancer cells. DNA Cell Biol. 2004;23:527–537. doi: 10.1089/dna.2004.23.527. [DOI] [PubMed] [Google Scholar]
- 28.Borbely AA, Murvai M, Szarka K, Konya J, Gergely L, Hernadi Z, Veress G. Survivin promoter polymorphism and cervical carcinogenesis. J Clin Pathol. 2007;60:303–306. doi: 10.1136/jcp.2006.037804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang JS, Kim KM, Kang KH, Choi JE, Lee WK, Kim CH, Kang YM, Kam S, Kim IS, Jun JE, Jung TH, Park JY. Polymorphisms in the survivin gene and the risk of lung cancer. Lung Cancer. 2008;60:31–39. doi: 10.1016/j.lungcan.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, Zhu H, Zhou B, Gu H, Yan H, Tang N, Dong H, Sun Q, Cong R, Chen G, Wang B. The Association Between the Survivin C-31G Polymorphism and Gastric Cancer Risk in a Chinese Population. Dig Dis Sci. 2008. [DOI] [PubMed]
- 31.Wang YH, Chiou HY, Lin CT, Hsieh HY, Wu CC, Hsu CD, Shen CH. Association Between Survivin Gene Promoter -31 C/G Polymorphism and Urothelial Carcinoma Risk in Taiwanese Population. Urology. 2008. [DOI] [PubMed]
- 32.Cheng ZJ, Hu LH, Huang SJ. [Correlation of -31G/C polymorphisms of survivin promoter to tumorigenesis of gastric carcinoma] Ai Zheng. 2008;27:258–263. [PubMed] [Google Scholar]
- 33.Altieri DC. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr Opin Cell Biol. 2006;18:609–615. doi: 10.1016/j.ceb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 34.Giodini A, Kallio MJ, Wall NR, Gorbsky GJ, Tognin S, Marchisio PC, Symons M, Altieri DC. Regulation of microtubule stability and mitotic progression by survivin. Cancer Res. 2002;62:2462–2467. [PubMed] [Google Scholar]
- 35.Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 36.O'Connor DS, Wall NR, Porter AC, Altieri DC. A p34(cdc2) survival checkpoint in cancer. Cancer Cell. 2002;2:43–54. doi: 10.1016/s1535-6108(02)00084-3. [DOI] [PubMed] [Google Scholar]
- 37.Andersen SS. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol. 2000;10:261–267. doi: 10.1016/s0962-8924(00)01786-4. [DOI] [PubMed] [Google Scholar]
- 38.Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 39.McGrogan BT, Gilmartin B, Carney DN, McCann A. Taxanes, microtubules and chemoresistant breast cancer. Biochim Biophys Acta. 2008;1785:96–132. doi: 10.1016/j.bbcan.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Zaffaroni N, Pennati M, Colella G, Perego P, Supino R, Gatti L, Pilotti S, Zunino F, Daidone MG. Expression of the anti-apoptotic gene survivin correlates with taxol resistance in human ovarian cancer. Cell Mol Life Sci. 2002;59:1406–1412. doi: 10.1007/s00018-002-8518-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Athanassiadou P, Grapsa D, Athanassiades P, Gonidi M, Athanassiadou AM, Tsipis A, Patsouris E. The prognostic significance of COX-2 and survivin expression in ovarian cancer. Pathol Res Pract. 2008;204:241–249. doi: 10.1016/j.prp.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 42.Ozols RF, Bundy BN, Greer BE, Fowler JM, Clarke-Pearson D, Burger RA, Mannel RS, De-Geest K, Hartenbach EM, Baergen R. Phase III trial of carboplatin and paclitaxel compared with cisplatin and paclitaxel in patients with optimally resected stage III ovarian cancer: a Gynecologic Oncology Group study. J Clin Oncol. 2003;21:3194–3200. doi: 10.1200/JCO.2003.02.153. [DOI] [PubMed] [Google Scholar]
- 43.Arsova-Sarafinovska Z, Matevska N, Petrovski D, Banev S, Dzikova S, Georgiev V, Sikole A, Sayal A, Aydin A, Suturkova L, Dimovski AJ. Manganese superoxide dismutase (MnSOD) genetic polymorphism is associated with risk of early-onset prostate cancer. Cell Biochem Funct. 2008;26:771–777. doi: 10.1002/cbf.1504. [DOI] [PubMed] [Google Scholar]
- 44.Canedo P, Corso G, Pereira F, Lunet N, Suriano G, Figueiredo C, Pedrazzani C, Moreira H, Barros H, Carneiro F, Seruca R, Roviello F, Machado JC. The interferon gamma receptor 1 (IFNGR1) -56C/T gene polymorphism is associated with increased risk of early gastric carcinoma. Gut. 2008;57:1504–1508. doi: 10.1136/gut.2007.143578. [DOI] [PubMed] [Google Scholar]
- 45.El Hallani S, Ducray F, Idbaih A, Marie Y, Boisselier B, Colin C, Laigle-Donadey F, Rodero M, Chinot O, Thillet J, Hoang-Xuan K, Delattre JY, Sanson M. TP53 codon 72 polymorphism is associated with age at onset of glioblastoma. Neurology. 2009;72:332–336. doi: 10.1212/01.wnl.0000341277.74885.ec. [DOI] [PubMed] [Google Scholar]
- 46.Engelmark MT, Ivansson EL, Magnusson JJ, Gustavsson IM, Wyoni PI, Ingman M, Magnusson PK, Gyllensten UB. Polymorphisms in 9q32 and TSCOT are linked to cervical cancer in affected sib-pairs with high mean age at diagnosis. Hum Genet. 2008;123:437–443. doi: 10.1007/s00439-008-0494-8. [DOI] [PubMed] [Google Scholar]
- 47.Khatri RG, Navaratne K, Weil RJ. The role of a single nucleotide polymorphism of MDM2 in glioblastoma multiforme. J Neurosurg. 2008;109:842–848. doi: 10.3171/JNS/2008/109/11/0842. [DOI] [PubMed] [Google Scholar]
- 48.Nicolaiew N, Cancel-Tassin G, Azzouzi AR, Grand BL, Mangin P, Cormier L, Fournier G, Giordanella JP, Pouchard M, Escary JL, Valeri A, Cussenot O. Association between estrogen and androgen receptor genes and prostate cancer risk. Eur J Endocrinol. 2009;160:101–106. doi: 10.1530/EJE-08-0321. [DOI] [PubMed] [Google Scholar]
- 49.Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 50.Taubert H, Bartel F, Greither T, Bache M, Kappler M, Kohler T, Bohnke A, Lautenschlager C, Schmidt H, Holzhausen HJ, Hauptmann S, Wurl P. Association of HDM2 transcript levels with age of onset and prognosis in soft tissue sarcomas. Mol Cancer Res. 2008;6:1575–1581. doi: 10.1158/1541-7786.MCR-07-2150. [DOI] [PubMed] [Google Scholar]
- 51.Araujo AP, Ribeiro R, Pereira D, Pinto D, Sousa B, Catarino R, Medeiros R. Ovarian cancer and genetic susceptibility: association of A61G polymorphism in the EGF gene. Exp Biol Med (Maywood) 2009;234:241–245. doi: 10.3181/0805-RM-146. [DOI] [PubMed] [Google Scholar]
- 52.Yang X, Xiong G, Chen X, Xu X, Wang K, Fu Y, Yang K, Bai Y. Survivin expression in esophageal cancer: correlation with p53 mutations and promoter polymorphism. Dis Esophagus. 2009;22:223–230. doi: 10.1111/j.1442-2050.2008.00885.x. [DOI] [PubMed] [Google Scholar]
- 53.Yang X, Xiong G, Chen X, Xu X, Wang K, Fu Y, Yang K, Bai Y. Polymorphisms of survivin promoter are associated with risk of esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2009. [DOI] [PubMed]
- 54.Grossman D, Kim PJ, Blanc-Brude OP, Brash DE, Tognin S, Marchisio PC, Altieri DC. Transgenic expression of survivin in keratinocytes counteracts UVB-induced apoptosis and cooperates with loss of p53. J Clin Invest. 2001;108:991–999. doi: 10.1172/JCI13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 56.Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirschmeier P, Bishop WR, Nielsen LL, Pickett CB, Liu S. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–2622. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]