Abstract
The stromal tissue, made of extracellular matrix and mesenchymal cells, is vital for the functional design of all complex tissues. Fibroblasts are key components of stromal tissue and play a crucial role during organ development, wound repair, angiogenesis and fibrosis. We have previously reported the identification of a novel WD-domain protein, STRAP1 that inhibits transforming growth factor-β (TGF-β) signaling and enhances tumorigenicity via TGF-β-dependent and TGF-β-independent mechanisms. Here, we report, for the first time, that deletion of STRAP from Mouse Embryonic Fibroblasts (MEFs) results in a loss of mesenchymal morphology. These cells lose their spindle shape and exhibit features of an epithelial morphology. Gene expression profiling has confirmed that deletion of STRAP affects expression of sets of genes important for diverse functions including cell-cell adhesion and cell polarization, and up-regulates E-cadherin expression leading to the formation of adherens junctions, subsequent localization of β-catenin to the cell membrane and downregulation of the mesenchymal markers like LEF1 (lymphoid enhancer-binding factor 1). Upregulation of WT1 (Wilms tumor homolog 1) in STRAP null MEFs plays a role in E-cadherin induction. Finally, stable expression of STRAP in these cells results in a loss of WT1 and E-cadherin expression, and a reversal from epithelial to the mesenchymal morphology. Thus, these results provide a novel TGF-β-independent function of STRAP and describe a mechanism for the role of STRAP in the maintenance of mesenchymal morphology.
Keywords: Serine threonine receptor associated protein, E-cadherin, epithelial-to-mesenchymal transition, mesenchymal-to-epithelial transition, mouse embryonic fibroblast, Wilms Tumor homolog 1
1. Introduction
Epithelial and mesenchymal cells represent two distinct cell lineages that have unique gene expression profiles and functions specific to that cell type. Compared to differentiated epithelial cells, mesenchymal cells do not establish intercellular junctions in a stable manner mostly through suppression of E-cadherin expression and this imparts them with a higher capacity to detach in response to low shear forces such as within lymphatic vessels and venules. A similar process also decreases the adhesive force in epithelial cells during normal embryonic development and during carcinogenesis. Epithelial cells located at the periphery of a tumor frequently exhibit a substantial downregulation of epithelial markers along with a loss of intercellular junctions and other features of epithelial cells, accompanied by expression of a mesenchymal set of genes [1]. This switch is referred to as epithelial-to-mesenchymal transition (EMT). Practically, it is often difficult to classify cell phenotypes into either extremes such as mesenchymal or epithelial and a relative shift from one phenotype to the other holds more importance. A recent study reported phenotypic alterations of the metastatic T24/TSU-Pr1 bladder carcinoma line that could express markers of both epithelial and mesenchymal type [2]. A new term ‘metastable phenotype’ has been coined by Savagner et al. for such cells that continue to express attributes of both epithelial and mesenchymal phenotypes [3]. Such mixed phenotypes are being reported more frequently now [4].
EMT provides a mechanism for epithelial cells to overcome the physical barrier of intercellular junctions and thus switch to a more motile phenotype. In many types of carcinomas, presence of EMT correlates with poor histologic differentiation, loss of tissue integrity and metastasis. Diverse developmental signaling pathways such as epidermal growth factor (EGF), transforming growth factor-β (TGF-β), hedgehog, Wnt/β-catenin, Notch and integrin signaling may play a role in the events leading to EMT [5]. Interestingly, a reverse process where mesenchymal cells acquire an epithelial phenotype has also been defined and termed as mesenchymal-to-epithelial transition (MET) [6]. MET occurs during normal embryogenesis as well as during re-establishment of metastatic cells at distant sites. E-cadherin is known to be upregulated or re-expressed in most of these instances [7, 8, 9]. In vitro, stable expression of E-cadherin alone has been shown to force the fibroblasts to adopt an epithelial morphology [10]. Expression of E-cadherin sequesters β-catenin and p120-catenin to the cell membrane and helps deactivate the mesenchymal cell program [11, 12]. Conversely, absence of E-cadherin frees up β-catenin that can translocate to the nucleus and activate transcription of a number of mesenchymal transcription factors like including c-myc, LEF1, CyclinD1, cdc2 [13,14,15] . Nuclear localization of β-catenin is therefore frequently used as a marker of EMT and indicates a poor prognosis in cancer [16].
E-cadherin thus acts as a master regulator of epithelial phenotype and a continuous downregulation of E-cadherin is required in mesenchymal cells [17]. This can be achieved through transcriptional as well as post-transcriptional mechanisms. A number of transcription factors such as Snail, Slug, ZEB1, SIP1, E2A, and WT1 are known to regulate E-cadherin expression [18, 19, 20, 21, 22]. Interestingly, many of these transcription factors that are important for EMT during embryonic development were later found to play a role in EMT during cancer progression [23]. Yet most of the reports regarding these factors have only studied their roles during EMT and little is known about the molecular mechanisms important for the maintenance of mesenchymal morphology of fibroblasts themselves. Apart from E-cadherin itself, overexpression of WT1 in NIH3T3 fibroblasts has been shown to induce epithelialization with E-cadherin upregulation and formation of adherens junctions [24]. Accordingly, absence of MET in kidney is one of the main features in the development of Wilm’s tumor, a rare embryonal malignancy which often has deregulated WT1 expression [25]. More recently, overexpression of Versican, a type of proteoglycan, in NIH3T3 fibroblasts and deletion of Prkar1a from MEFs has been shown to induce MET [26].
We have previously reported the identification of a novel WD domain protein, STRAP that can bind with TGF-β type I and type II receptors (TβRI & TβRII) and inhibit downstream TGF-β signaling through interaction with Smad7 [27, 28]. The common function of WD domain proteins is to provide a suitable scaffold for coordinating multiprotein complex assemblies and thus, regulate a variety of cellular processes like signal transduction, transcriptional regulation, programmed cell death, RNA synthesis/processing, chromatin assembly, cell cycle progression and vesicular trafficking [29]. Likewise, STRAP has been implicated in a wide array of cellular functions. Our previous studies suggest that STRAP is upregulated in several cancers and functions as an oncogene [30]. Apart from its role in TGF-β pathway, STRAP interacts with PDK1 to positively modulate PDK1 activity [30]. We have shown that STRAP induces extracellular signal-regulated kinase (MEK/ERK) activity in a TGF-β-independent manner [31], and binds with EWS and inhibits the interaction between EWS with p300, resulting in a downregulation of EWS target genes [32]. In a different role, STRAP seems to be involved in the assembly of the SMN complex which is necessary for mRNA splicing, through its interaction with Gemin6 and Gemin7 as well as SmB, SmD2 and SmD3 components of the complex [33, 34]. In a relevant interesting finding, STRAP along with EWS was shown to be present in the kinesin driven mRNA transport granules in the dendrites of murine neurons and also shown to bind with NFX1 and MAP1B to help export of mRNA out of the nucleus [35]. Homozygous deletion of STRAP gene in mice resulted in embryonic lethality between embryonic day (E) 10.5 and 12.5 due to the defects in angiogenesis, cardiogenesis, somitogenesis, neural tube closure and embryonic turning [36]. A recent study suggests that STRAP may be a predictive marker of 5-fluorouracil (5-FU)-based adjuvant chemotherapy benefit in colorectal cancer [37]. This wide variety of functions of STRAP suggests a broader role for it in tumorigenesis and development. However, nothing is known about of the function of STRAP in morphological changes of cells and mechanisms involved in it. Here, we demonstrate, for the first time, that loss of STRAP expression induces a mesenchymal-to-epithelial transition through upregulation of E-cadherin. Furthermore, STRAP-mediated downregulation of WT1 may play a role in the regulation of E-cadherin and subsequently in the maintenance of mesenchymal morphology.
2. Materials and Methods
2.1 Cell culture and plasmids
Wild type and STRAP null mouse embryonic fibroblasts (MEFs) and NIH3T3 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), antibiotics, and glutamine (GIBCO BRL). STRAP null MEFs were used to generate clones stably re-expressing STRAP using the STRAP pBabe Puro retroviral vector and clones were selected in 0.75 μg/ml puromycin. The E-cadherin luciferase construct (−178/+92) was a gift from Dr. Amparo Cano (Universidad Autónoma de Madrid, Instituto de Investigaciones Biomédicas Alberto Sols, Madrid, Spain). Plasmids expressing A and B isoforms of murine WT1 were obtained from Dr. Jerry Pelletier (McGill University, Montreal, Quebec, Canada).
2.2 Recombinant adenovirus
A STRAP expressing adenovirus was generated through homologous recombination between a linearized transfer vector pAD-Track and the adenoviral backbone vector pAD-Easy [38]. pAD-STRAP contained the full length murine STRAP cDNA with a carboxy-terminal HA or Flag-tag. In addition to the STRAP transgene the virus encoded the green fluorescent protein (GFP) transcribed from a second independent CMV promoter. GFP expression was used to monitor viral infection efficiency. An adenovirus coding for GFP only (pAD-GFP) was used as a control in all experiments. For both adenoviruses, a titer of 200 MOI was used in all the experiments to infect the cells for 8 hours in a serum free medium. The cells were then kept in a serum containing medium for 60 hours where not specifically mentioned.
2.3 Immunoblot analysis
For immunoblotting, whole-cell lysates were prepared from early passage MEFs, separated by 10% SDS/PAGE, transferred to nitrocellulose membrane (Biorad), and probed with primary antibodies from the following sources: Santa Cruz Biotechnologies (vimentin, WT1 and fibronectin), and BD Biosciences (E-cadherin, beta-catenin and N-cadherin) as described in Halder et al., 2006 [30]. Binding of primary antibodies was detected after incubation with species-specific secondary antibodies using chemiluminescence reagents (Perkin-Elmer).
2.4 Reverse Transcription-PCR Analyses
Total RNA was isolated from each cell line using Trizol method and RT–PCR amplification was carried out using MMLV reverse transcriptase. The RNA samples (2 μg) were retrotranscribed into cDNA using oligo-dT primers in a total volume of 20 μl containing 5 mM MgCl2, 1 mM dNTPs (Boheringer), 1 U RNase inhibitor (Perkin Elmer) and 2.5 U MuLV reverse transcriptase (Perkin Elmer) at 37° for 1 hour. Amplification by PCR was carried out using 2μl of the cDNA with the Red Taq polymerase according to the manufacturer’s protocol. The thermal cycles were: denaturing at 94°C for 1 min, annealing at 54°C for 1 min, and extension at 72°C for 1 min. The cDNA was amplified for 28 cycles. The primer pairs of E-cad, WT1, LEF1, Snail, SIP1, Slug, E2A, Twist, S100A and GAPDH are shown in table 1. GAPDH was amplified in each sample as an internal control. All experiments were repeated at least three times.
Table 1.
Primers used for RT-PCR
| Gene | Upstream Primer | Downstream Primer | Size (bp) |
|---|---|---|---|
| E-cadherin | ggaatccttggagggatcctc | gtcgtcctcgccaccgccgtacat | 560 |
| WT1 | cagagagcaaggcaccag | taagagcccagtgctagtg | 221 |
| LEF1 | ccaactttccggaggaggc | gtaggagggtcccttgttgtac | 313 |
| Snail | cagctggccaggctctcggt | gcgagggcctccggagca | 370 |
| Slug | ccaaggatcacagtggttca | cagtgcagctgcttgtgttt | 843 |
| ZEB1 | cgccaacaagcagactattc | tgaggcctcttacctgtgtg | 295 |
| SIP1 | agtccaatgcagcacttaggt | ttcatgctgatgcaggggaat | 490 |
| S100A | aggagctactgaccagggagct | tcattgtccctgttgctgtcc | 103 |
| E2A | tacccctccgccaagacc | ttgggggataaggcactg | 535 |
| Twist | cgggtcatggctaacgtg | cagcttgccatcttggagtc | 190 |
| GAPDH | accacagtccatgccatcac | tccaccaccctgttgctga | 452 |
2.5 FITC-Phalloidin staining
After fixation in 3.7% fresh paraformaldehyde in PBS for 15 min, cells were washed twice with PBS, and permeabilized with 0.1% Triton X-100 in PBS for 8 min. After treatment with blocking solution (1% BSA and 0.1% Triton X-100 in PBS) for 10 min, the cells were stained with FITC-phalloidin (1 μg/ml) in blocking solution for 20 min in a dark room at room temperature to localize F-actin. The slides were washed twice with PBS, each for 10 min. Incubation and washing were performed in parallel for all wells on a slide. A coverslip was mounted on the slide with Vectashield H-1000 (Vector Laboratories, Burlingame, CA). Actin was visualized with a fluorescence microscope (Olympus BHT, Tokyo, Japan).
2.6 Electron microscopy
Cells were washed with 6.8% saccharose in 0.1 M cacodylate buffer, pH 7.4, at room temperature and fixed in 2% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, at room temperature for 30 min. The cells were rinsed three times in the same buffer with 6.8% sucrose solution and subsequently postfixed in 2% OsO4/3% K4Fe(CN)6 in 0.2 M cacodylate buffer (pH 7.4) at 4°C for 1 h. After rinsing in 0.1 M cacodylate buffer, pH 7.4, and dehydration in a graded alcohol series, the cells were embedded in Epon 812 and polymerized for at 58°C for 64 h. Finally, ultrathin sections (60 nm) were cut and stained with uranyl acetate and lead citrate. The sections were examined using a Philips CM 12 electron microscope operating at 80 kV, and micrographs were taken.
2.7 Fluorescence Microscopy
Cells cultured on glass coverslips were fixed with ice-cold methanol in PBS for 10 min at 4°C, followed by permeabilization with 0.1% Triton X-100 in PBS at room temperature for 5 min. Blocking incubations were performed in PBS containing 3% BSA at room temperature for 1 h. After extensive washes with PBS, cells were incubated with the first antibody at room temperature for 2 h. After washing with PBS, cells were then incubated with the corresponding secondary antibody at room temperature for 1 h. After another round of extensive washes in PBS, the coverslips were mounted in a drop of mounting medium (Vectashield). The antibodies used were as follows: mouse monoclonal anti-E-cadherin and mouse monoclonal anti-β-catenin antibody from BD Biosciences, and Alexa Fluor-596 goat anti-mouse from Molecular Probes, Eugene, OR.
2.8 Reporter Assays
NIH3T3 cells, wild type and STRAP null embryonic fibroblasts were plated in 12-well plates. After 30 h, luciferase constructs (0.5 μg/well where not mentioned) along with expression plasmids for WT1 (100 ng) and/or STRAP (2 doses of 150 and 450 ng) were transfected into the cells using Lipofectamine and Plus reagent following the manufacturers protocol. After approximately 48 hours, cells were lyzed and luciferase assays were performed using a luminometer (BD bioscience) according to the manufacturer’s protocol. Transfection of each construct was performed in triplicate in each assay and a total of three assays were performed on three separate days. All wells were transfected with 25 ng of beta-galactosidase to serve as a control for the transfection efficiency. Ratios of luciferase readings to beta-gal readings were taken for each experiment and triplicates were averaged. Bars represent the averages of the normalized values with error bars indicating the standard deviation.
2.9 Microarray Analysis
To characterize the gene expression profile of wild type and STRAP null MEFs, RNA was isolated from these cells using the Trizol method and further purified using Qiagen RNeasy kit according to the protocol from the manufacturer (Qiagen, Valencia, CA, USA). Microarray was done using GeneChip 430 Mouse 2.0 Array from Affymetrix that contains 45,000 probes for analyzing 39,000 variants of 34,000 mouse genes and signal intensity was detected according to supplier’s instructions.
3. Results
3.1 STRAP Deletion Induces an Epithelial-like Morphological Conversion through upregulation of E-cadherin
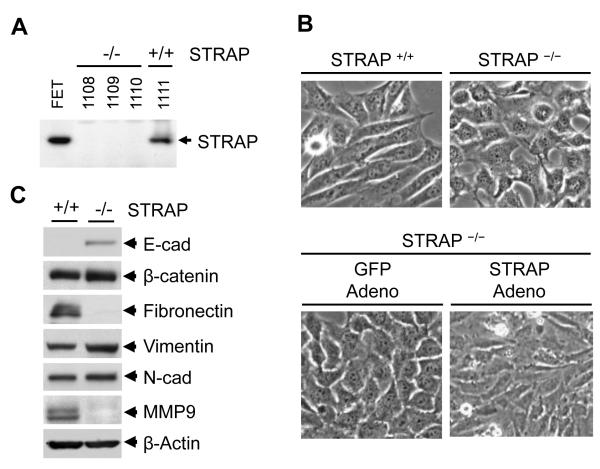
We have previously reported a role for STRAP in inhibition of TGF-β signaling [28]. We and others have also shown that STRAP is upregulated in lung, colon and breast cancer [30, 40]. To determine the role of STRAP deletion in mouse embryonic fibroblasts (MEFs), we used STRAP null and wild type MEFs that were generated during microarray coupled gene trap mutagenesis to find out PDGF target genes. The gene trap insertion results in embryonic lethality between embryonic day E10.5 and E12.5 due to loss of STRAP [36]. We first confirmed the loss of STRAP expression in STRAP null MEFs by western blot analysis (Fig 1A). Interestingly, we observed an obvious morphological change in all three STRAP null MEFs, whereas the wild type MEFs maintained mesenchymal morphology, as expected. STRAP null MEFs aggregated and formed epithelial islands with an increased capacity for adhesion to the tissue culture plates (Fig 1B, top panel). Two other MEF cell lines isolated from different STRAP null embryos showed similar morphological changes (data not shown). To confirm that the cell adhesion and morphological change were due to STRAP loss, we generated and tested HA and Flag-tagged STRAP adenovirus as well as the GFP adenovirus (Fig 4A). Transient expression of adenoviral HA or Flag-tagged STRAP reverted the morphology of these null MEFs back to the original fibroblastoid type (Fig 1B, bottom panel). The morphological changes in STRAP null MEFs suggested an alteration in intercellular adhesion. Then we analyzed the expression of different epithelial and mesenchymal markers including E-cadherin, beta-catenin, fibronectin, cytokeratin and vimentin by western blot analyses. Our data showed that E-cadherin expression was significantly upregulated in STRAP null MEFs. Conversely, fibronectin and MMP9 were downregulated in STRAP null MEFs and total beta-catenin and vimentin showed no change. These data suggest that absence of STRAP lead to a partial epithelial transition or a metastable phenotype in MEFs.
Figure 1. Effect of STRAP on cell morphology.
(A) MEFs, isolated from wild type and STRAP null embryos (36), were tested for STRAP expression. (B) Phase-contrast photomicrographs showing that loss of STRAP results in a loss of fibroblastoid morphology of the MEFs as cells acquire a epithelial morphology (upper panel). Adenoviral transient expression of STRAP can induce epithelial-to-mesenchymal transition (EMT) in STRAP null MEFs while a control GFP adenovirus fails to induce EMT (lower panel). Adenoviral titer: 200 MOI. (C) Lysates prepared from wild type and STRAP null MEFs were used to study markers of epithelial and mesenchymal differentiation. Western analyses shows upregulation of E-cadherin and downregulation of fibronectin and MMP-9 while expression of β-catenin, N-cadherin, vimentin does not change significantly.
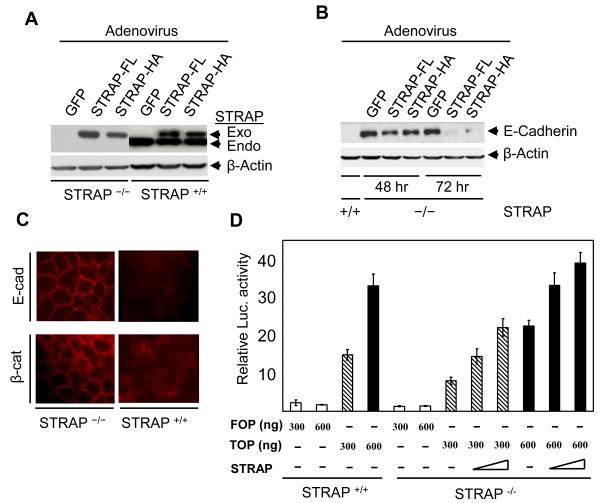
Figure 4. STRAP inversely regulates E-cadherin expression and induces transcriptional activity of β-catenin.
(A) Replication deficient adenoviruses (RDA) that are able to transiently express either FLAG-tagged or HA-tagged STRAP were generated using AdEasy vector system. Exogenous expression of STRAP was comparable to endogenous expression in wild type MEFs. (B) Both HA-tagged and FLAG-tagged STRAP re-expression in STRAP null MEFs repressed E-cadherin expression in a time-dependent manner whereas GFP adenoviral infection did not affect E-cadherin expression. (C) STRAP deletion results in expression and localization of E-cadherin and β-catenin to the cell membrane. Wild type and STRAP null MEFs were grown in chamber slides, washed with PBS and after fixing with 4% paraformaldehyde, they were incubated first with either anti-E-cadherin or anti-β-catenin primary antibody and then with cy3-conjugated mouse secondary antibody. (D) Decrease in β-catenin transcriptional activity by STRAP. Wild type and STRAP null MEFs were cotransfected with two doses (300 and 600 ng) of either wild-type (TOPFlash) or mutant (FOPFlash) reporter plasmid and β-galactosidase plasmid with or without increasing doses of STRAP expression plasmid. The total amount of DNA was kept equal. After 48 h of transfection, luciferase activity was determined and normalized to β-galactosidase activity. The mean of triplicate luciferase values was used for fold expression comparison.
3.2 STRAP null MEFs show polarization at cell organelle level
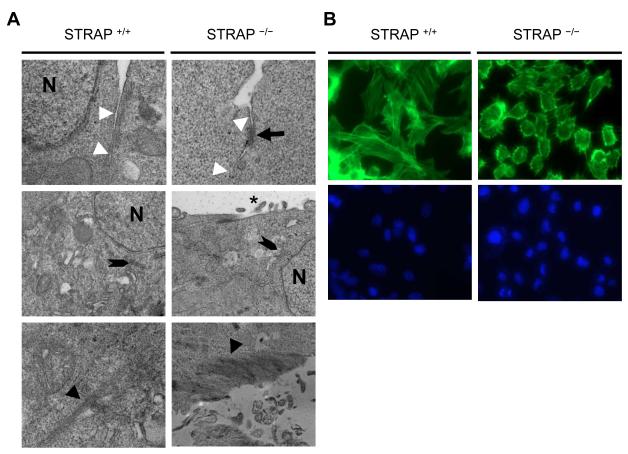
Together with protein expression changes, changes at cell organelle level often accompany phenotypic alterations in cells and tend to have a functional impact on the cell characteristics. Therefore we decided to look at the cell-organelle level to understand more about the changes induced by loss of STRAP using transmission electron microscopy (TEM). TEM confirmed the formation of a tightly packed epithelial monolayer showing a close alignment of the lateral plasma membranes of adjacent STRAP null MEFs. It further revealed that the STRAP null MEFs exhibited an apical-basal polarity, which is a characteristic of epithelial cells. STRAP null MEFs contained numerous electron-dense membranous structures that resembled adherens junctions at basolateral border as indicated by the black arrow. The cell-cell interface is indicated by white arrowheads (Fig 2A upper 2 panels). STRAP null MEFs have an apical distribution of the Golgi complex (shown by notched black arrowheads) relative to the cell nucleus (N) and exhibited microvilli along the apical surface as shown by black an asteric. These features were absent in wild type MEFs (Fig 2A middle 2 panels). TEM also showed the presence of multiple electron-dense cytoplasmic fibers running through the length of wild type MEFs, indicated by black arrowheads. These represent the cytoplasmic stress fibers that are typical for cells of mesenchymal origin. These fibers are contractile actomyosin bundles that are instrumental in the maintenance of the mesenchymal architecture inside cells and facilitate cellular motility. In contrast, these fibers had a more membranous localization in STRAP null MEFs which is typically observed in epithelial cells (Fig 2A lower 2 panels). To confirm this redistribution of actin organization, we stained the MEFs with FITC-phalloidin. As expected, in wild type MEFs, F-actin formed parallel cytoplasmic stress fibers whereas actin was mostly redistributed towards the cell membrane in the STRAP null MEFs (Fig 2 B). These data suggest that STRAP null cells underwent the process of MET not only in regards to changes in gene expression but also showed a consistent change in the cell architecture and distribution of cellular organelles.
Figure 2. Electron microscopic examination indicates STRAP induces EMT through changes at the cell organelle level.
(A) Wild type and STRAP null cells were grown in culture on formvar coated coverslips, then fixed with 2.5 % Glutaraldehyde and stained with lead citrate and geranyl acetate. Thin sections were cut using microtome and the sections were analyzed using electron microscopy. Cell-cell interface, indicated by white arrowheads, were observed for frequency of adherens junction formation indicated by black arrows (upper 2 panels). In the middle 2 panels, localization of the Golgi complex, indicated by notched arrows, was observed for polarization relative to the cell orientation. Cell surfaces were observed for microvilli, indicated by a black asteric. Finally, in the lower panels, cells were analyzed for the pattern and localization of actin (black arrowheads). (N: nucleus). (B) Wild type and STRAP-null MEFs were grown in culture, washed with PBS, fixed with 4% paraformaldehyde and permeabilized with 0.1 % Triton X. The cells were then stained with 2 μg/ml of FITC-conjugated phalloidin to map the actin arrangement and localization (upper panel). Nuclei were stained with DAPI (lower panel).
3.3 STRAP regulates expression of genes critical for multiple cellular signaling pathways and biological processes
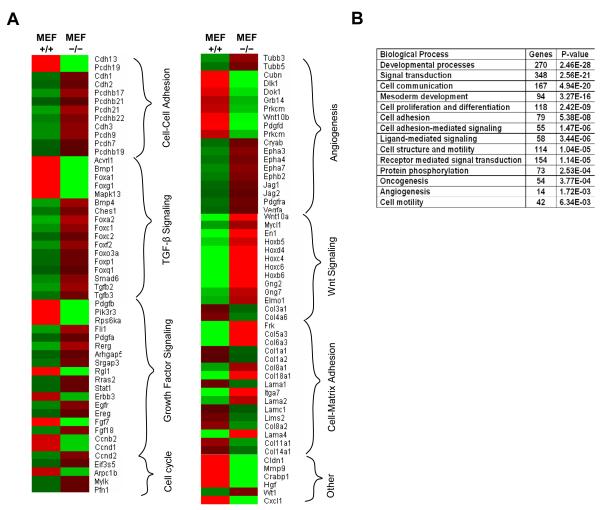
In previous studies, STRAP has been implicated to play role in diverse functions including TGF-β signaling, mRNA splicing, RNA transport, PDK1 activation etc. Here, we noted that STRAP deletion caused partial epithelialization of MEFs. So next we decided to study the overall impact of STRAP in terms of gene expression regulation in MEFs. Analysis of microarray data using PANTHER Prowler analysis tool confirmed that STRAP loss affected sets of genes important for cellular functions like TGF-β signaling, growth factor signaling, cell-cell adhesion, cell-matrix adhesion, cell cycle regulation and Wnt signaling (Fig 3A). Analyses of genes affected by STRAP deletion were also done after regrouping them according to the overall biological processes they are involved with. These results suggest that STRAP deletion affects biological processes such as developmental processes, cell adhesion, signal transduction, mesoderm development, cell motility, angiogenesis, oncogenesis etc (Fig 3B). The cellular functions affected by STRAP, including TGF-β/other growth factor and Wnt signaling as well as cell-cell/cell-matrix adhesion are known to play diverse roles in the regulation of cell morphology.
Figure 3. Differential expression of important groups of genes in wild type and STRAP null MEFs.
(A) Different gene groups important for diverse functions are altered in wild type and STRAP null MEFs. Wild type and STRAP null MEFs were used to isolate mRNA using Trizol method. Microarray was done using GeneChip 430 Mouse 2.0 Array from Affymetrix that contains 45,000 probes for analyzing 39,000 variants of 34,000 mouse genes. Relative expression of selected genes is shown as a heat diagram. Intense red and green colors indicate high and low expression respectively. (B) Biological processes most significantly affected after STRAP deletion were assessed based on the whole array of gene expression changes obtained form the microarray analysis using PANTHER Prowler analysis tool.
3.4 STRAP downregulates E-cadherin from cellular junctions
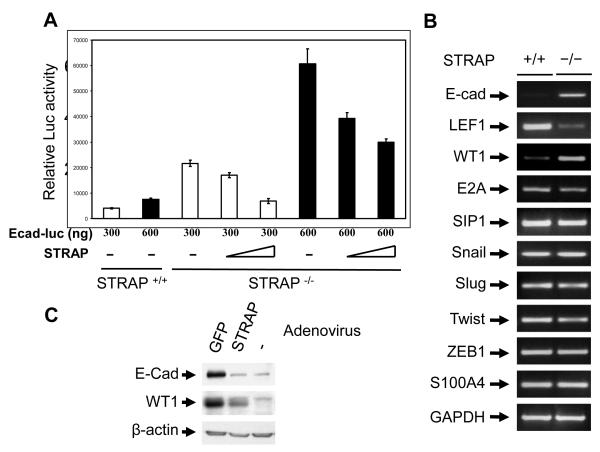
To test the specificity of the effect of STRAP on the regulation of E-cadherin expression, we generated Flag- and HA-tagged STRAP adenovirus and tested its expression in wild type and STRAP null MEFs. We observed that exogenous STRAP expression is similar to or less than the endogenous level (Fig 4A). Next we used these adenoviruses to assess the effect of STRAP re-expression on E-cadherin expression in STRAP null MEFs. Adenoviral re-expression of STRAP led to downregulation of E-cadherin in a time-dependent manner whereas β-gal adenovirus had no effect on E-cadherin (Fig 4B). To further determine whether endogenous STRAP can downregulate E-cadherin from the cellular membrane, we performed immunofluorescence staining and examined subcellular distribution of E-cadherin and β-catenin. The staining pattern showed that E cadherin was absent in wild type MEFs and was seen prominently at the cell-cell junctions in STRAP null MEFs. Hence we speculated that β-catenin would be lost from membranes of wild type MEFs but it would be localized to the membranes in STRAP null MEFs. Indeed, β-catenin was present mainly as a diffused signal in the cytoplasm in wild type MEFs. By contrast, β-catenin was localized predominantly at the cell–cell contacts in STRAP null MEFs (Fig 4C). These results are consistent with the total levels of E-cadherin and β-catenin in these MEFs (Fig 1C). Collectively, these findings suggest that STRAP regulates E-cadherin expression, and in-turn regulates subcellular distribution of β-catenin. This is significant mainly because nuclear β-catenin is considered as an indicator of EMT in cancer cells. The functional impact of this E-cadherin loss on nuclear localization of β-catenin was assessed by luciferase assays using TOPFLASH and FOPFLASH reporters. The FOPFLASH is the control luciferase vector, whereas TOPFLASH has three TCF/LEF/β-catenin complex binding sites [41]. These assays indicated that β-catenin mediated transcription was considerably reduced in STRAP null MEFs when compared to wild type MEFs. Interestingly, transient re-expression of STRAP in null MEFs increased TOPFLASH reporter activity (Fig 4D) to levels comparable to wild type MEFs. Taken together, these data suggest that STRAP induces loss of E-cadherin from the membrane that results in nuclear translocation of β-catenin.
3.5 Stable expression of STRAP in null MEFs rescued the mesenchymal phenotype
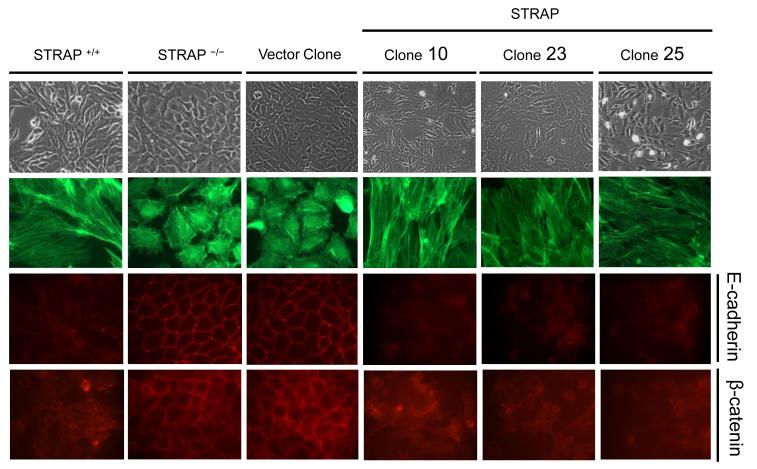
Earlier we showed (Fig 1B) a reversion of STRAP null MEFs from epithelial to a mesenchymal phenotype after transient expression of STRAP. In order to validate this data, we generated stable clones expressing STRAP in null MEFs. pBabe-Puro retroviral vector with mouse STRAP gene was used and the resulting clones were selected in 0.75 μg/ml puromycin The expression of STRAP in these stable clones is shown in Fig 7B. Three independent clones displayed a reversal from the cobblestone-like morphology of STRAP null MEFs to a mesenchymal phenotype (Fig 5). FITC-phalloidin staining revealed that F-actin was organized in parallel stress fibers in these clones (Fig 5) similar to wild type MEFs. Immunofluorescence studies confirmed that E-cadherin expression was almost absent and β-catenin was delocalized from the membrane in STRAP stable clones (Fig 5). No effect on E-cadherin and β-catenin and on morphology was observed in the vector control clone indicating that stable STRAP expression could specifically reverse the MET that occurred in STRAP null MEFs.
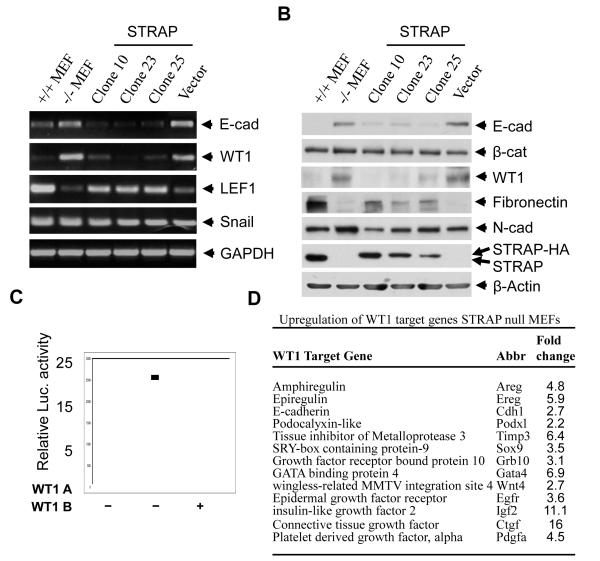
Figure 7. WT1 can transcriptionally induce E-cadherin expression in fibroblasts.
(A) The STRAP stable clones and a vector control clone were used to analyze expression status of E-cadherin and transcriptional regulators of E-cadherin using RT-PCR as described above. STRAP null and wild type MEFs served as negative and positive controls. (B) Western blot analyses were performed to study the expression of E-cadherin, β-catenin, WT1, fibronectin and N-cadherin in STRAP null and wild type MEFs, STRAP stable clones and a vector control clone. STRAP re-expression was verified in the stable clones and β-actin was used as a loading control. (C) Mouse E-cadherin promoter (−178/+92) was transfected in NIH3T3 fibroblasts with either WT1 A or WT1 B expression construct. Luciferase assays were performed and represented as described above. (C) WT1 regulated genes are expressed several fold higher in STRAP null MEFs as indicated by microarray analysis.
Figure 5. Stable STRAP expression in null MEFs restores the mesenchymal phenotype.
STRAP overexpressing clones (#10, 23 and 25) were generated from STRAP null MEFs using STRAP pBabe retrovirus, and selected in 0.75 μg/ml of puromycin. These clones reverted to a mesenchymal morphology whereas the vector control clone did not show any morphological alterations. E-cadherin expression went down from the membrane and β-catenin was delocalized in the STRAP stable clones. Loss of E-cadherin results in actin re-organization from a more cortical form in STRAP null MEFs to parallel stress fibers in STRAP stable clones.
3.6 Transcriptional upregulation of E-cadherin in STRAP null MEFs through upregulation of WT1
Regulation of the total E-cadherin pool in a cell is a complex process. It has been shown that E-cadherin can be regulated at multiple levels including synthesis, processing and stability of mRNA; synthesis and stability of protein; localization and posttranslational modification and also binding to the catenins. So we next decided to analyze the mechanism responsible for STRAP mediated regulation of E-cadherin. Reporter assays with a mouse E-cadherin promoter luciferase construct showed significant upregulation of E-cadherin promoter activity in the STRAP null MEFs compared to wild type MEFs. This upregulation was suppressed considerably when STRAP was expressed in STRAP null MEFs indicating that STRAP indeed regulates E-cadherin at transcriptional level (Fig 6A). During our analysis of the microarray data, we noticed that one of the known inducers of E-cadherin expression, Wilms tumor 1 (WT1) was significantly upregulated (3.33 fold) in STRAP null MEFs (Fig 3A). On the other hand, zinc finger transcription factors like Snail, Slug, E2A, Twist, SIP1, and ZEB1 are known repressors of E-cadherin expression. We used RT-PCR to analyze the status of the transcriptional regulators of E-cadherin in MEFs. RT-PCR analyses confirmed that E-cadherin and WT1 mRNA were upregulated in STRAP null MEFs, whereas the expression of Snail, Slug, E2A, SIP1, and ZEB1 was not altered in STRAP null MEFs (Fig 6B). This suggests that WT1 might be involved in the upregulation of E-cadherin in STRAP null MEFs. Expression of other mesenchymal markers revealed that LEF1 was downregulated in STRAP null MEFs whereas FSP1 (S100A) remained unchanged (Fig 6B). Free β-catenin is known to go to the nucleus and activate transcription of target genes such as LEF1 together with co-factors like the TCF family transcription factors [41]. This is consistent with our data that β-catenin was localized to the membrane of STRAP null MEFs due to upregulation of E-cadherin. This implies a decrease in the nuclear level of β-catenin that can lead to downregulation of LEF1. Western analyses confirmed upregulation of WT1 in STRAP null MEFs. Transient adenoviral STRAP expression in the STRAP null MEFs reduced expression of both WT1 and E-cadherin (Fig 6C). These data suggest that STRAP-mediated downregulation of WT1 may be involved in the regulation of E-cadherin.
Figure 6. STRAP mediates transcriptional downregulation of E-cadherin through WT1.
(A) STRAP represses E-cadherin promoter activity. Wild type and STRAP null MEFs were cotransfected with two doses (300 and 600 ng) of murine E-cadherin (−178/+92) promoter reporter construct, increasing doses of STRAP expression plasmid and β-galactosidase construct. Luciferase assays were performed and represented as described above. (B) Transcriptional expression of E-cadherin and known regulators of E-cadherin was detected by RT-PCR. Total RNA was extracted from both STRAP null and wild type MEFs using Trizol. After treatment with DNAse 1, cDNA was synthesized from RNA and amplified by PCR for 30 cycles using the primers described in Materials and Methods. PCR products were analyzed by agarose gel electrophoresis. Up-regulation of E-cadherin and WT1 and downregulation of LEF1 was detected in STRAP null MEFs. (C) STRAP null MEFS were grown in culture and infected with STRAP or GFP adenovirus in a serum free media for 6 hours and lysed after another 72 hours. Immunoblotting was used to analyze for E-cadherin and WT1 expression along with appropriate controls.
3.7 Overexpression of STRAP in null MEFs reduces WT1 expression and WT1 activates E-cadherin promoter activity
WT1 has been suggested to induce mesenchymal to epithelial transition in the metanephric mesoderm during the formation of renal parenchyma. It has already been established that WT1 can induce E-cadherin expression in fibroblasts and that stable overexpression of WT1 in fibroblasts induced partial epithelialization [24]. Also, mice with Sertoli cells deficient in WT1 show a loss of adherens junctions [42]. We used STRAP stable clones to test the specificity of the effect of STRAP on the regulation of WT1. WT1 was downregulated in these clones both in mRNA (Fig 7A) and protein (Fig 7B) levels as seen in RT-PCR and western analyses. RT-PCR also showed transcriptional downregulation of E-cadherin and upregulation of LEF1 in the STRAP stable clones (Fig 7A). This is in accordance with the decreased level of membranous β-catenin that can transcriptionally activate LEF1 expression. In western analyses, total level of β-catenin and N-cadherin did not show any appreciable change but fibronectin, an extracellular matrix protein produced by fibroblasts, is re-expressed in STRAP stable clones as compared to parental STRAP null MEFs or the vector control clone (Fig 7B).
The WT1 responsive site has been mapped to the GC rich region at about 40 bp upstream of the transcription start site in the E-cadherin promoter. In luciferase reporter assays, we observed that both WT1 isoforms A and B, successfully induced the E-cadherin promoter activity (Fig 7C). This data suggests that STRAP may play a role in the downregulation of WT1 in wild type fibroblasts and this in turn affects E-cadherin expression. Interestingly, when microarray data from the wild type and STRAP null MEFs was analyzed, STRAP null MEFs showed upregulation of multiple WT1 inducible genes like amphiregulin, epiregulin, IGF2, podocalyxibn-like, SOX9 and TIMP3 suggesting that WT1 is transcriptionally active in these cells (Fig 7D). Taken together, our data indicates that STRAP downregulates WT1 expression in the wild type MEFs to suppress E-cadherin expression, and thus maintains the mesenchymal morphology of these cells.
4. Discussion
Mesenchyme is a loose connective tissue of mesodermal origin. Fibroblasts, a major component of mesenchymal tissue, mainly function to provide structural support to all of the body tissues and organs. During embryogenesis, growth factors secreted by migrating fibroblasts as well as the epithelialmesenchymal interactions in the developing tissues are proposed to be necessary for the optimal organ development. Other functions of fibroblasts include but are not limited to wound healing, inflammation, vasculogenesis, angiogenesis, fibrosis and regulation of self-tolerance [43]. Most of these functions are in part due to the ability of these cells to effectively migrate to the sites of their function. In addition to their role in normal biological processes, fibroblasts can also promote pathological processes. For example, cancer associated fibroblasts are known to accelerate cancer growth by secreting growth factors, promoting neo-angiogenesis, and ECM remodeling. These fibroblastic functions are associated with increased rates of tumor invasion and metastasis.
The so-called mesenchymal morphology of fibroblasts is thought to be of central importance to the much higher migratory ability of these cells. During normal development, the process of EMT provides a means by which epithelial cells can move to their appropriate destination tissue by adopting a motile mesenchymal phenotype. Downregulation of E-cadherin appears to be a common mechanism for the acquisition of mesenchymal morphology, even though the exact mechanism used to downregulate E-cadherin can be context dependent. Some of the mechanisms regulating E-cadherin expression include transcription, stability of mRNA and protein, subcellular localization, and posttranslational modification. Importantly, there is a strong clinicopathological correlation between decreased E-cadherin expression and tumor dissemination [44]. This finding suggests that EMT is a critical step in progression towards invasive and metastatic cancer. A reverse process known as MET, in which cells with a mesenchymal phenotype gain a more adherent epithelial phenotype, is now being proposed as a mechanism for the reestablishment of metastatic cells in the distant organs [7]. Although several studies have identified factors important for the EMT, not much is known about the molecular mechanisms vital for maintenance of the fibroblastoid morphology of the mesenchymal cells like fibroblasts. To the best of our knowledge, there have been no reports describing the role for STRAP in EMT. Apart from E-cadherin itself, only over-expression of a proteoglycan versican or WT1 is known to be able to induce features of epithelial morphology in fibroblasts. Very recently it has been shown that deletion of Prkar1a in MEFs induces MET through upregulation of E-cadherin expression in fibroblasts [45].
Our present study suggests that expression of STRAP is vital for maintenance of fibroblastoid morphology as deletion of STRAP leads to a partial MET in fibroblasts though upregulation of E-cadherin (Fig 1B & 1C). Our electron microscopic studies showed that when compared to wild type MEFs, STRAP null MEFs showed increased adherens junctions, apically located Golgi apparatus, microvilli on their apical surface and a more cortical localization of actin fibers (Fig 2A). These changes are consistent with features of epithelial cells. Furthermore, re-expression of STRAP in null MEFs leads to downregulation of E-cadherin and to a reversal of the MET. Compared to wild type MEFs, expression of E-cadherin in STRAP null MEFs is elevated at both mRNA and protein level (Fig 1C, 3A and 6C). The loss of a mesenchymal marker such as fibronectin was observed but other markers such as N-cadherin, FSP1 and vimentin did not show any change (Fig 1C). This suggests that the process of epithelialization of STRAP null MEFs may be a partial one and fits more with the idea of a ‘metastable phenotype’ described by Savagner et al. [3]. According to this study, cells can express both epithelial and mesenchymal markers when they are in transition.
In an effort to understand the mechanism of E-cadherin upregulation, we studied expression of known regulators of E-cadherin expression in wild type and STRAP null MEFs. RT-PCR analyses showed that both cell types expressed similar levels of Snail, Slug, ZEB1, SIP1, Twist and E2A mRNA. However, STRAP null MEFs consistently showed elevated expression of WT1 and reduced expression of the mesenchymal marker, LEF1 (Fig 6B and C). It is interesting to note that WT1 is crucial for induction of MET in metanephric mesoderm during embryonic development [46]. Additionally, stable WT1 expression in NIH3T3 fibroblasts has been shown to induce partial epithelialization with formation of adherens junctions [24]. STRAP null MEFs demonstrated upregulation of WT1 expression at both mRNA (Fig 6C) and protein (Fig 7B) levels. Reporter assays using a murine E-cadherin promoter indicated that both WT1 isoforms A and B could induce E-cadherin promoter activity (Fig 7C), suggesting a role for WT1 in regulating E-cadherin expression in STRAP null MEFs. STRAP was able to repress E-cadherin promoter activity in STRAP null cells in a dose dependent manner (Fig 6A) and this is in agreement with the lower E-cadherin message and protein levels in STRAP null cells. Interestingly, microarray analysis of the STRAP null MEFs showed a robust upregulation of multiple WT1 regulated genes including amphiregulin, epiregulin, podocalyxin-like, TIMP3, SOX9 and IGF2 in addition to E-cadherin (Fig 7D). Furthermore, transient adenovirus mediated re-expression of STRAP was able to downregulate WT1 and E-cadherin in STRAP null MEFs (Fig 6C) as well as rescue the mesenchymal phenotype (Fig 1B).
The role of STRAP in the maintenance of mesenchymal morphology was confirmed by generating stable clone re-expressing HA-tagged STRAP in STRAP null MEFs. In accordance with transient STRAP expression, stable STRAP expression results in a reversion to a mesenchymal morphology (Fig 5). This morphological change was accompanied by suppression of WT1 and E-cadherin expression (Fig 7A and B). These cells show delocalization of β-catenin from the cell membrane and reorganization of cortical actin into parallel stress fibers. These results are in agreement with nuclear translocation of β-catenin and with the increase in TOP-FLASH activity (Fig 4D). This may lead to downregulation of LEF1 in STRAP null cells (Fig 6B). This was further supported by the obesrvation that stable expression of STRAP in null cells increased the level of LEF1 (Fig 7A). RT-PCR analyses also confirmed downregulation of both WT1 and E-cadherin mRNA in the STRAP stable clones.
Soriano et al have found that deletion of STRAP in mice leads to an embryonic lethal phenotype between days (E) 10.5 to 12.5. The STRAP null embryos have defects in processes such angiogenesis, cardiogenesis, gut rotation, somitogenesis and neural tube closure. It is unclear whether these defects are due to some intrinsic defects in the cells of these tissues as a result of STRAP deletion or due to the absence of proper stroma and fibroblast function. Further work will be needed to arrive at a conclusion. However, analysis of the microarray data from wild type and STRAP null MEFs revealed an alteration of a number of genes important for functions like cell-cell adhesion, cell motility and mesoderm development. Additionally, STRAP deletion also significantly alters the expression of genes (Fig 3A and B) important for embryonic development, signal transduction, cell communication and angiogenesis, which support the previously published biological functions of STRAP [28, 31, 36]. We also speculate that the balance of EMT versus MET in different tissues may be controlled by cell and tissue type–specific factors including STRAP and therefore, outcomes of such studies will depend on the exact tissue/cell type chosen for that study as previously suggested by the group showing MET in Prkar1a-/- MEFs [45].
EMT allows cancer cells to become more motile and invasive. We reported that STRAP expression is increased in several cancers including 60% of colorectal, 78% of lung and 46 % of breast carcinomas [30, 40]. Ectopic expression of STRAP in different cell lines promotes cellular proliferation, induces anchorage-independent growth and increases tumorigenicity during in vitro and in vivo experiments [30]. It is possible that STRAP overexpression may help tumor cells downregulate E-cadherin in co-operation with other factors known to induce EMT, thereby contributing to the increased migratory and invasive ability of these cells. Further work would be needed in this area to determine whether STRAP can play such a role in tumor cells.
In summary, we have shown, for the first time, that deletion of STRAP in murine fibroblasts is sufficient to cause MET through upregulation of WT1 and subsequently E-cadherin. Re-expression of STRAP in these null cells leads to a loss of WT1 and E-cadherin expression, and a reversal from epithelial to the mesenchymal morphology. Whether STRAP plays a role in EMT in epithelial cancer cells and whether the defects in STRAP null mice are from perturbation of cell phenotypes in local tissues or due to defect in stromal fibroblasts remains to be seen.
ACKNOWLEDGEMENT
We thank the Vanderbilt Microarray Shared Resource facility and the Vanderbilt Electron Microscopy Core Lab. We thank Amparo Cano and Jerry Pelletier for providing the plasmids. This study was supported by R01 CA95195 and CA113519, NCI SPORE grant in lung cancer (5P50CA90949, project# 4) and Veterans Affairs Merit Review Award (to P.K.D).
Glossary
- 1 STRAP
Serine Threonine Receptor-Associated Protein
- TGF-β
Transforming Growth Factor-β
- MEF
mouse embryonic fibroblast
- LEF1
lymphoid enhancer-binding factor 1
- WT1
Wilms tumor homolog 1
- EMT
epithelial-to-mesenchymal transition
- EGF
epidermal growth factor
- MET
mesenchymal-to-epithelial transition
- cdc2
cell division cycle 2
- ZEB1
zinc finger E-box binding homeobox 1
- SIP1
survival of motor neuron protein interacting protein 1
- TβRI & TβRII
TGF-β receptors type I and type II
- PDK1
pyruvate dehydrogenase kinase, isozyme 1
- ERK
extracellular signal-regulated kinase
- EWS
Ewing sarcoma breakpoint region 1 protein
- SMN
survival of motor neurons
- NFX1
nuclear transcription factor, X-box binding 1
- MAP1B
microtubule-associated protein 1B
- 5-FU
5-fluorouracil
- GFP
Green fluorescent protein
- CMV
cytomegalovirus
- RT-PCR
reverse transcription-polymerase chain reaction
- MMLV
Moloney Murine Leukemia Virus
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- PBS
phosphate-buffered saline
- FITC
fluorescein isothiocyanate
- PDGF
platelet-derived growth factor
- MMP9
matrix metallopeptidase 9
- TEM
transmission electron microscopy
- SOX9
SRY-box containing gene 9
- TIMP3
tissue inhibitor of metalloproteinase 3
- IGF2
insulin-like growth factor 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lee JM, Dedhar S, Kalluri R, Thompson EW. J. Cell. Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chaffer CL, Brennan JP, Slavin JL, Blick T, Thompson EW, Williams ED. Cancer Res. 2006;66:11271–11278. doi: 10.1158/0008-5472.CAN-06-2044. [DOI] [PubMed] [Google Scholar]
- [3].Klymkowsky MW, Savagner P. Am. J. Pathol. 2009;174:1588–1593. doi: 10.2353/ajpath.2009.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Laffin B, Wellberg E, Kwak HI, Burghardt RC, Metz RP, Gustafson T, Schedin P, Porter WW. Mol. Cell. Biol. 2008;28:1936–1946. doi: 10.1128/MCB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mimeault M, Batra SK. Annals of Oncology. 2007;18:1605–1619. doi: 10.1093/annonc/mdm070. [DOI] [PubMed] [Google Scholar]
- [6].Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, Thompson EW. J. Cell. Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- [7].Wells A, Yates C, Shepard CR. Clin. Exp. Metastasis. 2008;25:621–628. doi: 10.1007/s10585-008-9167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Baum B, Settleman J, Quinlan MP. Semin. Cell Dev. Biol. 2008;19:294–308. doi: 10.1016/j.semcdb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- [9].Chaffer CL, Thompson EW, Williams ED. Cells Tissues Organs. 2007;185:7–19. doi: 10.1159/000101298. [DOI] [PubMed] [Google Scholar]
- [10].Vanderburg CR, Hay ED. Acta Anatomica. 1996;157:87–104. doi: 10.1159/000147870. [DOI] [PubMed] [Google Scholar]
- [11].Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. J. Cell Sci. 1999;112:1237–1245. doi: 10.1242/jcs.112.8.1237. [DOI] [PubMed] [Google Scholar]
- [12].Nelson WJ, Nusse R. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze’ev A. Proc. Natl. Acad. Sci. U. S. A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vadlamudi U, Espinoza HM, Ganga M, Martin DM, Liu X, Engelhardt JF, Amendt BA. J. Cell Sci. 2005;118:1129–1137. doi: 10.1242/jcs.01706. [DOI] [PubMed] [Google Scholar]
- [15].Bryan RT, Nicholls JH, Harrison RF, Jankowski JA, Wallace DM. J. Urol. 2003;170:1892–1896. doi: 10.1097/01.ju.0000092740.51330.39. [DOI] [PubMed] [Google Scholar]
- [16].Lee JM, Dedhar S, Kalluri R, Thompson EW. J. Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rubinfeld B, Rubins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- [18].Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, De Herreros A. García. Nat. Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- [19].Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F. Mol. Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- [20].Hosono S, Gross I, English MA, Hajra KM, Fearon ER, Licht JD. J. Biol. Chem. 2000;275:10943–10953. doi: 10.1074/jbc.275.15.10943. [DOI] [PubMed] [Google Scholar]
- [21].Savagner P, Yamada LM, Thiery JP. J. Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, Cano A. J. Biol. Chem. 2001;6:27424–27431. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- [23].Moustakas A, Heldin CH. Cancer Sci. 2007;98:1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hosono S, Luo X, Hyink DP, Schnapp LM, Wilson PD, Burrow CR, Reddy JC, Atweh GF, Licht JD. Oncogene. 1999;18:417–427. doi: 10.1038/sj.onc.1202311. [DOI] [PubMed] [Google Scholar]
- [25].Li CM, Guo M, Borczuk A, Powell CA, Wei M, Thaker HM, Friedman R, Klein U, Tycko B. Am. J. Pathol. 2002;160:2181–2190. doi: 10.1016/S0002-9440(10)61166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sheng W, Wang G, La Pierre DP, Wen J, Deng Z, Wong CK, Lee DY, Yang BB. Mol. Biol. Cell. 2006;17:2009–2020. doi: 10.1091/mbc.E05-10-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Datta PK, Chytil A, Gorska AE, Moses HL. J. Biol. Chem. 1998;273:34671–34674. doi: 10.1074/jbc.273.52.34671. [DOI] [PubMed] [Google Scholar]
- [28].Datta PK, Moses HL. Mol. Cell. Biol. 2000;20:3157–3167. doi: 10.1128/mcb.20.9.3157-3167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kashikar ND, Datta PK. In: Encyclopedia of Cancer. second ed Schwab M, editor. Springer-Verlag; pp. 2710–2713. [Google Scholar]
- [30].Halder S, Anumanthan G, Maddula R, Mann J, Chytil A, Gonzalez A, Washington M, Moses H, Beauchamp R, Datta PK. Cancer Res. 2006;66:6156–6166. doi: 10.1158/0008-5472.CAN-05-3261. [DOI] [PubMed] [Google Scholar]
- [31].Seong H, Jung H, Choi H, Kim K, Ha H. J. Biol. Chem. 2005;280:42897–42908. doi: 10.1074/jbc.M507539200. [DOI] [PubMed] [Google Scholar]
- [32].Anumanthan G, Halder SK, Friedman DB, Datta PK. Cancer Res. 2006;66:10824–10832. doi: 10.1158/0008-5472.CAN-06-1599. [DOI] [PubMed] [Google Scholar]
- [33].Grimmler M, Otter S, Peter C, Müller F, Chari A, Fischer U. Hum. Mol. Genet. 2005;14:3099–3111. doi: 10.1093/hmg/ddi343. [DOI] [PubMed] [Google Scholar]
- [34].Carissimi C, Baccon J, Straccia M, Chiarella P, Maiolica A, Sawyer A, Rappsilber J, Pellizzoni L. FEBS Lett. 2005;579:2348–2354. doi: 10.1016/j.febslet.2005.03.034. [DOI] [PubMed] [Google Scholar]
- [35].Tretyakova I, Zolotukhin AS, Tan W, Bear J, Propst F, Ruthel G, Felber BK. J. Biol. Chem. 2005;280:31981–31990. doi: 10.1074/jbc.M502736200. [DOI] [PubMed] [Google Scholar]
- [36].Chen W, Delrow J, Corrin P, Frazier J, Soriano P. Nature Genetics. 2004;36:304–312. doi: 10.1038/ng1306. [DOI] [PubMed] [Google Scholar]
- [37].Buess M, Terracciano L, Reuter J, Ballabeni P, Boulay JL, Laffer U, Metzger U, Herrmann R, Rochlitz C. Neoplasia. 2004;6:813–820. doi: 10.1593/neo.04307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].He TC, Zhou S, da Costa LT, Yu J, Kinzler K, Vogelstein B. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kim CJ, Choi BJ, Song JH, Park YK, Cho YG, Nam SW, Yoo NJ, Lee JY, Park WS. Pathol. Int. 2007;57:178–182. doi: 10.1111/j.1440-1827.2007.02078.x. [DOI] [PubMed] [Google Scholar]
- [40].Matsuda S, Katsumata R, Okuda T, Yamamoto T, Miyazaki K, Senga T, Machida K, Thant AA, Nakatsugawa S, Hamaguchi M. Cancer Res. 2000;60:13–17. [PubMed] [Google Scholar]
- [41].Buess M, Terracciano L, Reuter J, Ballabeni P, Boulay JL, Laffer U, Metzger U, Herrmann R, Rochlitz C. Neoplasia. 2004;6:813–820. doi: 10.1593/neo.04307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rao MK, Pham J, Imam JS, MacLean JA, Murali D, Furuta Y, Sinha-Hikim A, Wilkinson MF. Genes Dev. 2006;20:147–152. doi: 10.1101/gad1367806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Haniffa MA, Collin MP, Buckley CD, Dazzi F. Haematologica. 2009;94:258–263. doi: 10.3324/haematol.13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kopstein L, Christofori G. Cell. Mol. Life Sci. 2006;63:449–468. doi: 10.1007/s00018-005-5296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nadella KS, Jones GN, Trimboli A, Stratakis CA, Leone G, Kirschner LS. Cancer Res. 2008;68:2671–2677. doi: 10.1158/0008-5472.CAN-07-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Davies JA, Ladomery M, Hohenstein P, Michael L, Shafe A, Spraggon L, Hastie N. Hum. Mol. Genet. 2004;13:235–246. doi: 10.1093/hmg/ddh015. [DOI] [PubMed] [Google Scholar]