Abstract
An X-ray structure of the lactose permease of Escherichia coli (LacY) in an inward-facing conformation has been solved. LacY contains N- and C-terminal domains, each with six transmembrane helices, positioned pseudosymmetrically. Ligand is bound at the apex of a hydrophilic cavity in the approximate middle of the molecule. Residues involved in substrate binding and H+ translocation are aligned parallel to the membrane at the same level and may be exposed to a water-filled cavity in both the inward- and outward-facing conformations, thereby allowing both sugar and H+ release directly into either cavity. These structural features may explain why LacY catalyzes galactoside/H+ symport in both directions utilizing the same residues. A working model for the mechanism is presented that involves alternating access of both the sugar- and H+-binding sites to either side of the membrane.
Keywords: membrane transport, membrane protein structure, transport mechanism
INTRODUCTION
The mechanism of energy transduction in biological membranes is an important, fascinating problem. It has been recognized for some time that the driving force for a variety of seemingly unrelated phenomena (e.g., secondary active transport, oxidative phosphorylation, and rotation of the bacterial flagellar motor) is a bulk-phase, transmembrane electrochemical ion gradient. However, insight into the molecular mechanisms by which free energy stored in such gradients is transduced into work or into chemical energy has just begun. On the other hand, gene sequencing and analyses of deduced amino acid sequences indicate that many biological machines involved in energy transduction—membrane transport in particular—fall into families encompassing proteins from archaea to the mammalian central nervous system (http://www.tcdb.org/), thereby raising the possibility that the members may have common basic structural features and mechanisms. In addition, many of these proteins play important roles in human disease (e.g., cystic fibrosis, resistance to antibiotics and chemotherapeutic drugs, gastric ulcer, glucose/galactose malabsorption and some forms of drug abuse), as well as the mechanism of action of a number of drugs.
Transport involves substrate-specific membrane proteins that catalyze equilibration and/or uphill translocation of solute across a membrane. These proteins are called a variety of interchangeable names—symporters, cotransporters, transporters, carriers, or permeases. Three main categories of systems are involved in active transport, and each utilizes a distinct energy source: (a) The phosphoenolpyruvate:sugar phospho-transferase system (PTS) is a multicomponent system that catalyzes vectorial phosphorylation of various sugars and sugar alcohols in certain bacteria exclusively (40, 76). Thus, in Escherichia coli, for example, glucose and certain other sugars and sugar alcohols are translocated across the membrane by PTS-catalyzed phosphorylation (i.e., glucose appears on the cytoplasmic side of the membrane as glucose-6-phosphate). The glucose PTS also plays a key role in the phenomenon of catabolite repression. (b) Transporters of the ATP-binding cassette (ABC) system (19) are found in both prokaryotes and eukaryotes and have a common global organization with two integral membrane components, each of which has multiple-transmembrane helices, and two cytoplasmic components, each of which has one ATP-binding cassette (19, 61). ABC transporters may be in the form of a monomer or various combinations of fused components. These systems utilize the energy released from the hydrolysis of ATP to drive accumulation or efflux unidirectionally against a concentration gradient. Many bacterial ABC systems also require a binding protein on the outside surface of the membrane. (c) Transporters that utilize an electrochemical ion gradient (46, 120) are also known as secondary transporters. Primary systems consist of pumps that generate an electrochemical ion gradient. Most ion-coupled transport proteins are composed of 12 to 14 membrane-spanning helices (http://www.tcdb.org/), and for those studied intensively, a single polypeptide utilizes the free energy released from the energetically downhill movement of a cation (mainly H+ or Na+) in response to an electrochemical ion gradient to catalyze transport of substrate against concentration gradient. However, neurotransmitter reuptake transporters often utilize anions, as well as cations (51, 70). Unlike the PTS, with ion-gradient-driven transport, substrate enters the cell in an unchanged form. Mechanistically and energetically, only the ion-gradient-driven permeases catalyze solute/ion symport in both directions across the membrane (influx and efflux), unlike the other systems described.
With regard to ion-gradient-driven permeases, the chemiosmotic hypothesis of Peter Mitchell (66–68) has been supported strongly by the experiments of West (112) and West & Mitchell (113, 114) and demonstrated quantitatively in bacterial membrane vesicles (41–43). Thus, accumulation of a wide variety of solutes against a concentration gradient is driven by an electrochemical H+ gradient (Δμ̄H+) composed of an electrical component (ΔΨ; interior negative) and/or a pH gradient (ΔpH; interior alkaline). This review focuses on the lactose permease of E. coli (LacY), a galactoside/H+ symporter, as this membrane protein is arguably the most intensively studied secondary transporter.
BACKGROUND
The existence of a transport system for galactosidic sugars was first inferred in 1955 by Cohen & Rickenberg (10) and subsequently found to be part of by the famous lac operon (69). In addition to regulatory loci, the lac operon contains three structural genes: (a) the Z gene encoding β-galactosidase, a cytosolic enzyme that catalyzes cleavage of lactose into galactose and glucose once it enters the cell; (b) the Y gene encoding LacY, the galactoside/H+ symporter or lactose permease; and (c) the A gene encoding galactoside transacetylase, a cytosolic enzyme that catalyzes acetylation of mainly thio-β-d-galactopyranosides with acetyl-CoA as the acetyl donor and has an unknown physiological function.
The lacY gene was the first gene encoding a membrane transport protein to be cloned into a recombinant plasmid (100) and sequenced (6). This success in the early days of molecular biology opened the study of secondary active transport at the molecular level. LacY has been used as a paradigm for secondary transport proteins to explore the mechanism of energy transduction. Overexpression of lacY was combined with use of a highly specific photoaffinity probe (49) and functional reconstitution into proteoliposomes (21, 71, 72, 107). It was then shown (62, 107) that LacY catalyzes all the translocation reactions typical of the transport system in vivo with comparable turnover numbers. Thus, the product of the lacY gene is responsible solely for all the translocation reactions catalyzed by LacY.
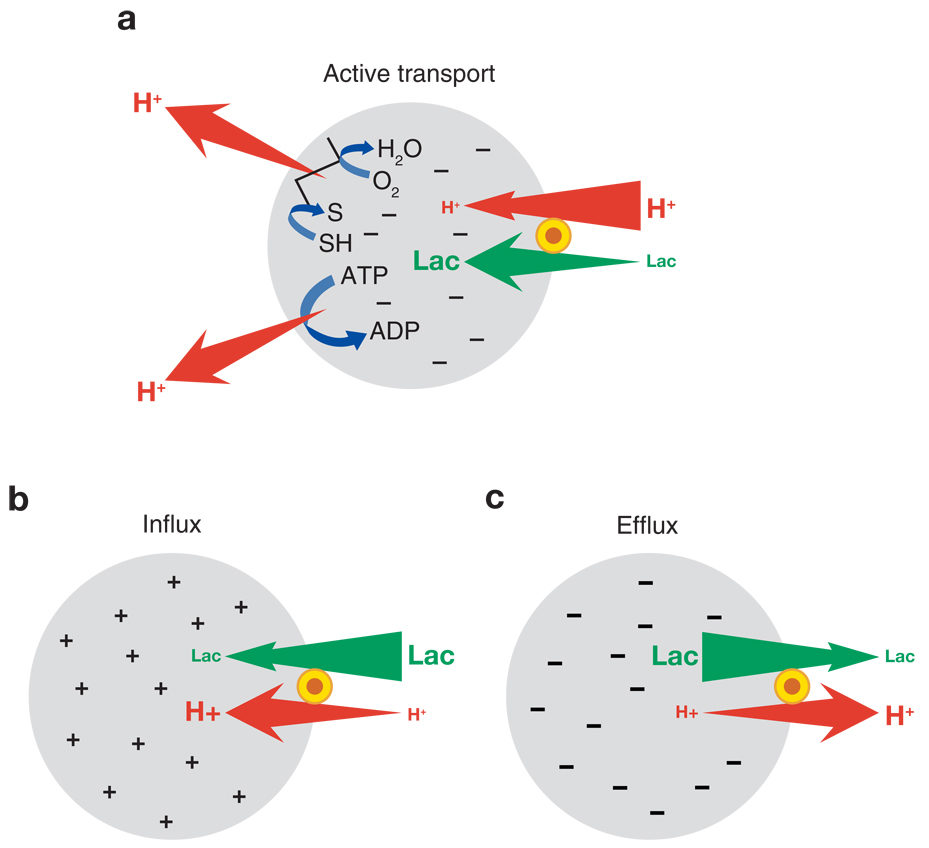
LacY belongs to the major facilitator superfamily (MFS) (http://www.tcdb.org/), a large group of transport proteins thought to be evolutionarily related. LacY is selective for disaccharides containing a d-galactopyranosyl ring, as well as d-galactose, but has no affinity for d-glucopyranosides or d-glucose (92). LacY carries out the coupled stoichiometric translocation of a galactoside with an H+ (i.e., galactoside/H+ symport), utilizing the free energy released from downhill translocation of H+ to drive accumulation of galactosides against a concentration gradient (Figure 1a). Notably, in the absence of Δμ̄H+, LacY also catalyzes the converse reaction, utilizing free energy released from downhill translocation of sugar to drive up-hill translocation of H+ with generation of Δμ̄H+, the polarity of which depends upon the direction of the substrate concentration gradient (Figure 1b,c). In the absence of substrate, LacY does not translocate H+; however, LacY catalyzes exchange or counterflow of sugar without translocation of H+. Because substrate gradients by themselves generate a Δμ̄H+ of either polarity, it seems intuitively likely that the primary driving force for turnover is binding and dissociation of sugar on either side of the membrane.
Figure 1.
Lactose/H+ symport. In the absence of substrate, LacY does not translocate H+ in the presence of Δμ̄H+. (a) Free energy released from the downhill movement of H+ is coupled to the uphill accumulation of lactose. (b, c) Substrate gradients generate electrochemical H+ gradients, the polarity of which depends upon the direction of the substrate concentration gradient.
To obtain insight into the mechanism of transport, it is essential to identify side chains that are crucial, to delineate their function and relationship to one another, and to obtain structural and dynamic information. The use of molecular biological techniques to engineer LacY for site-directed biochemical and biophysical approaches has provided important information about mechanism, as well as structure, and these methods have been applied to the study of many other membrane proteins. Moreover, an X-ray structure of LacY has been solved recently to a resolution of 3.5 Å (2). The structure provides critical information regarding the overall fold, the sugar-binding site, and the position of residues involved in H+ translocation. Importantly, the structure confirms many conclusions derived from biochemical and biophysical studies and also reveals a number of unexpected, novel findings. Remarkably, at the same time, the structure of another member of the MFS, the phosphate/glycerol-3-P antiporter (GlpT), was solved at 3.3 Å (59). The folds of LacY and GlpT are similar (1, 59), supporting the notion that all members of the MFS may have similar structures.
LacY IS FUNCTIONAL AS A MONOMER
One particularly difficult problem to resolve with hydrophobic membrane proteins is their functional oligomeric state. Although indirect early evidence (43) suggested that oligomerization might be important for activity, LacY was shown to be functional as a monomer by rotational diffusion measurements with eosinylmaleimide-labeled LacY or by freeze-fracture electron microscopy. Using the former approach, Dornmair et al. (15) utilized fluorescence anisotropy to demonstrate that purified, reconstituted LacY exhibits the rotational diffusion constant of a 46.5-kDa particle and that the diffusion constant is not altered in the presence of Δμ̄H+. Regarding the microscopic approach, purified LacY was reconstituted into proteoliposomes under conditions where the protein is fully functional and shown to be a monomer in the absence or presence of Δμ̄H+ (13). Moreover, the initial rate of Δμ̄H+-driven lactose transport in proteoliposomes varies linearly with the ratio of LacY to phospholipid. If more than a single molecule of LacY is required for active lactose transport, a sigmoidal relationship should be observed, particularly at low LacY/phospholipid ratios.
Notwithstanding evidence that LacY is functional as a monomer, certain paired in-frame deletion mutants complement functionally (4). Although cells expressing the deletions individually do not catalyze active transport, cells simultaneously expressing specific pairs of deletions catalyze transport up to 60% as well as cells expressing wild-type LacY, and it is clear that the phenomenon occurs at the protein and not at the DNA level. Remarkably, complementation is observed only with pairs of LacY molecules containing large deletions separated by at least two hypothetical transmembrane helices and not with missense mutations or point deletions. Although the mechanism of complementation is unclear, it is likely related to the phenomenon whereby independently expressed N- and C-terminal fragments of LacY interact to form a functional complex (3, 121, 122). In any case, the observation that certain pairs of deletion mutants complement functionally rekindled concern regarding the functional oligomerization state of wild-type LacY.
To resolve the problem, Sahin-Tóth et al. (91) engineered a fusion protein that contains two LacY molecules covalently linked in tandem (LacY dimer). The covalently linked LacY dimer is inserted into the membrane in a functional state, and negative dominance is not observed by either mutation or chemical modification of either half of the dimer. In order to test the caveat that oligomerization between dimers might account for the findings, a LacY dimer containing different deletion mutants, which complement each other when expressed as untethered molecules, did not catalyze lactose accumulation. Therefore, it is unlikely that LacY dimers oligomerize.
Finally, single-Cys replacement mutants in the transmembrane helices or loops of LacY dimerize in a stochastic manner (18, 31, 99). Moreover, the asymmetric unit of the LacY crystal is composed of an artificial dimer with two molecules oriented in opposite directions (2). Taken as a whole, the results argue strongly that LacY functions as a monomer.
PRIMARY AND SECONDARY STRUCTURE
Primary
LacY is composed of 417 amino acid residues and has a molecular mass of 46,517 Da. However, like most hydrophobic membrane proteins, LacY electrophoreses with a relative molecular mass of ∼33,000 Da. Electrospray ionization–mass spectrometry (ESI-MS) has been applied successfully to LacY (54, 57, 111, 115, 116), as well as other hydrophobic membrane proteins. The molecular weight reconstruction from ESI-MS of LacY with a 6-His affinity tag at the C terminus reveals that the purified protein is homogeneous and that the computed mass is within 0.01% of that calculated from the DNA sequence with a formyl group on the initiating methionine. Although the formyl group is normally removed from native LacY (17), overexpression may saturate the deformylase.
Secondary
The α-helical content of LacY was examined by circular dichroism (85%–90%) (20), laser Raman (∼70%) (108), and attenuated total reflectance–Fourier transform spectroscopy (ATR-FTIR) (∼70%) (56, 75). Interestingly, relative to the X-ray structure, which has a helical content of 86% (80% within the membrane), circular dichroism is the most accurate of the spectroscopic methods used.
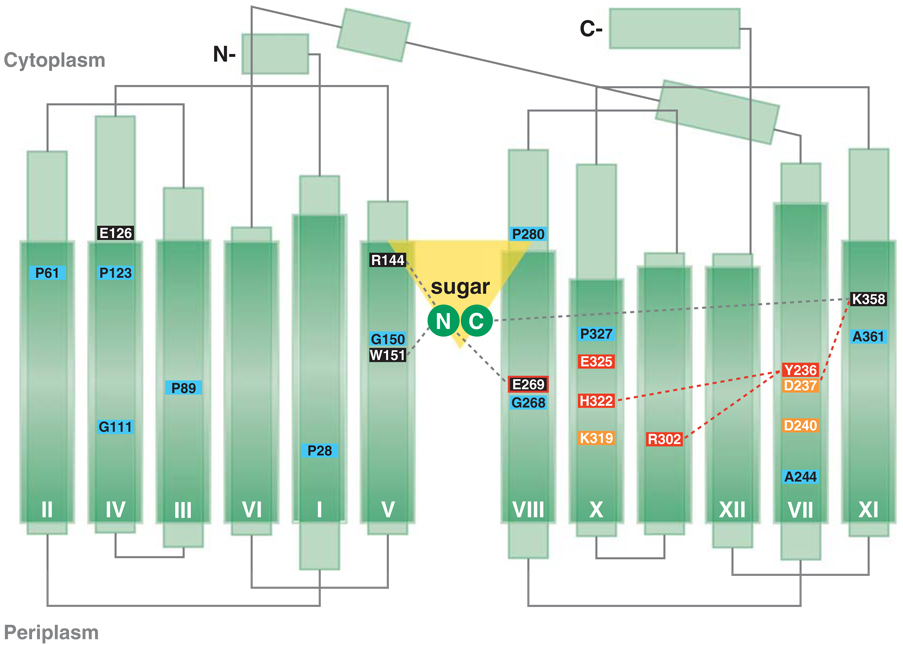
Hydropathy analysis was first applied, predicting 12 hydrophobic transmembrane domains (20). The number and orientation of each individual transmembrane domain was studied experimentally by phoA fusion analysis (7), which is consistent with 12 transmembrane domains traversing the membrane in zigzag fashion connected by relatively hydrophilic loops with both N and C termini on the cytoplasmic face. The overall topology model has been confirmed by the three-dimensional structure (Figure 2).
Figure 2.
Secondary structure model of LacY derived from the X-ray structure. The helices (rectangles) traverse the membrane in a zigzag fashion connected by hydrophilic domains external to the membrane. The helices are drawn to scale according to the X-ray structure, and dark green areas represent the portions of the helices that are within the membrane. The loops depict connectivity only. Residues at the kinks in the transmembrane helical domains are shown as light blue rectangles; residues in black rectangles are involved in substrate binding; residues in red are involved in H+ translocation. Glu-269 (black rectangle bordered in red) is involved in both substrate binding and H+ translocation. Salt-bridged residues are shown as orange rectangles. The large hydrophilic cavity is designated by an inverted yellow triangle, and sugar is depicted by two green circles, with N and C representing those moieties that interact with the N- and C-terminal halves of LacY, respectively.
Generally, the boundaries of the transmembrane domains are difficult to predict. With LacY specifically, the secondary structure model was modified because of the identification of charge pairs between certain helices. Initially, hydropathy analysis placed Asp-237 in loop VII/VIII. However, second-site suppression analyses (52) indicated that Asp-237 interacts with Lys-358 (helix XI), a conclusion supported strongly by site-directed mutagenesis, chemical modification (16, 86), and site-directed spin labeling (101). In addition, Asp-240, which was also originally placed in loop VII/VIII, is charge-paired with Lys-319 in helix X (16, 58, 86). Therefore, the whole region from Cys-234 to Phe-247 was moved into transmembrane VII, which approximates the X-ray structure very well (2). Glu-126 and Arg-144 were also initially placed in loop IV/V. However, studies utilizing single amino acid deletions (117, 119), as well as site-directed spin labeling (124), indicate that both residues are in the helices IV and V.
Helix Packing and Functionally Irreplaceable Side Chains
Use of molecular biology approaches to engineer LacY for site-directed biochemical and biophysical studies has provided important information about its structure and mechanism (46). A possible helix packing model was proposed from about 100 distance constraints obtained from thiol cross-linking experiments and engineered Mn(II)-binding sites (97). Additional methods that approximate distance were also utilized qualitatively, among which are site-directed mutagenesis with the explicit purpose of engineering LacY for site-directed biochemical and biophysical techniques. In most instances, construction of mutants begins with a cassette lacY gene (EMBL X-56,095) containing a unique restriction site about every 100 bp that encodes LacY with a single Cys residue at virtually every position (cysteine-scanning mutagenesis) (27). With a library of mutants containing a single Cys residue at almost every position, it is a simple operation to construct paired Cys replacement mutants by restriction fragment replacement. Furthermore, by inserting the biotin acceptor domain from a Klebsiella pneumoniae oxalacetate decarboxy-lase or 6 to 10 His residues in either the middle cytoplasmic loop or at the C terminus, the mutant proteins are readily purified in a single step by monovalent avidin affinity or metal affinity chromatography (12, 96), respectively. Most of the methodology involves site-directed measurements—pyrene excimer fluorescence (38), engineered divalent metal-binding sites (paired His residues) (34, 35, 39), electron paramagnetic resonance (11), and thiol cross-linking (47). Although important global aspects of the structure are not revealed (e.g., the large inward-facing hydrophilic cavity or helix packing at the cytoplasmic face; see below), many of the local interactions (29, 32) are detected by using the techniques described. In view of the X-ray structure of LacY, it is not surprising that global aspects of the structure are not revealed by these approaches. However, many of the techniques are useful for dynamic information, which is difficult to obtain from a crystal structure.
Functional analysis of mutants at virtually every position has led to the following three observations (27, 46, 84). (a) Only six side chains are irreplaceable with respect to active transport: Glu-126 (helix IV) and Arg-144 (helix V), which are crucial for substrate binding; Glu-269 (helix VIII), which is likely involved in both substrate binding and H+ translocation; and Arg-302 (helix IX),His-322 (helix X), and Glu-325 (helix X), which play irreplaceable roles in H+ translocation. (b) Residues such as Trp-151 and Tyr-236 are important, but not irreplaceable. (c) Substrate-induced changes in the reactivity of side chains with various chemical modification reagents, site-directed fluorescence, and spin labeling suggest widespread conformational changes during turnover.
TERTIARY STRUCTURE
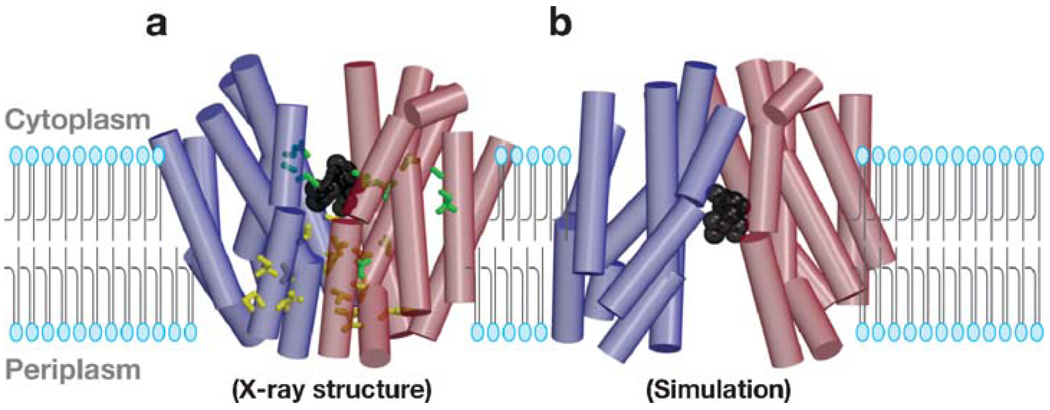
The first X-ray structure of LacY was obtained with a thermostable mutant (C154G) that binds sugar as well as the wild-type, but exhibits little or no transport activity (64, 96). Therefore it was surmised that the C154G mutant might be suitable for crystallization because it strongly favors a single conformation. This notion came to fruition, and a crystal structure was obtained in collaboration with Jeff Abramson and So Iwata at Imperial College London (2).
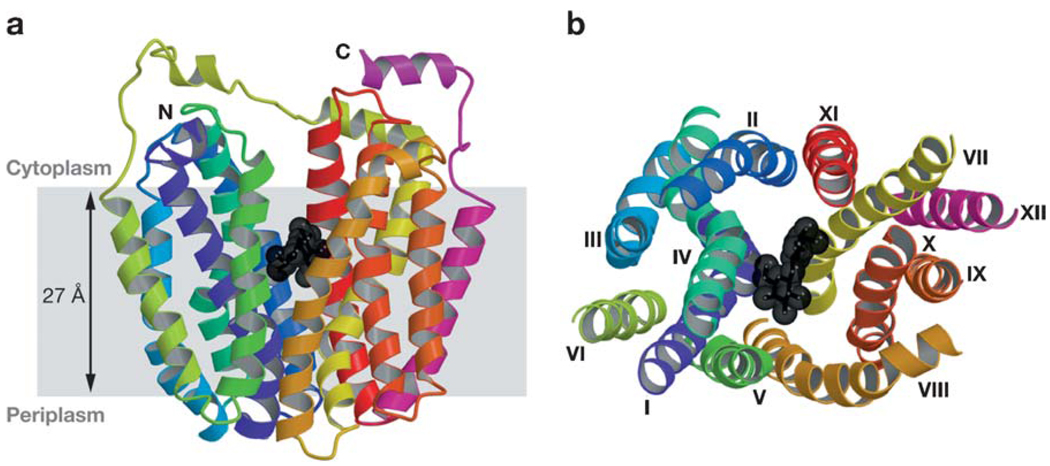
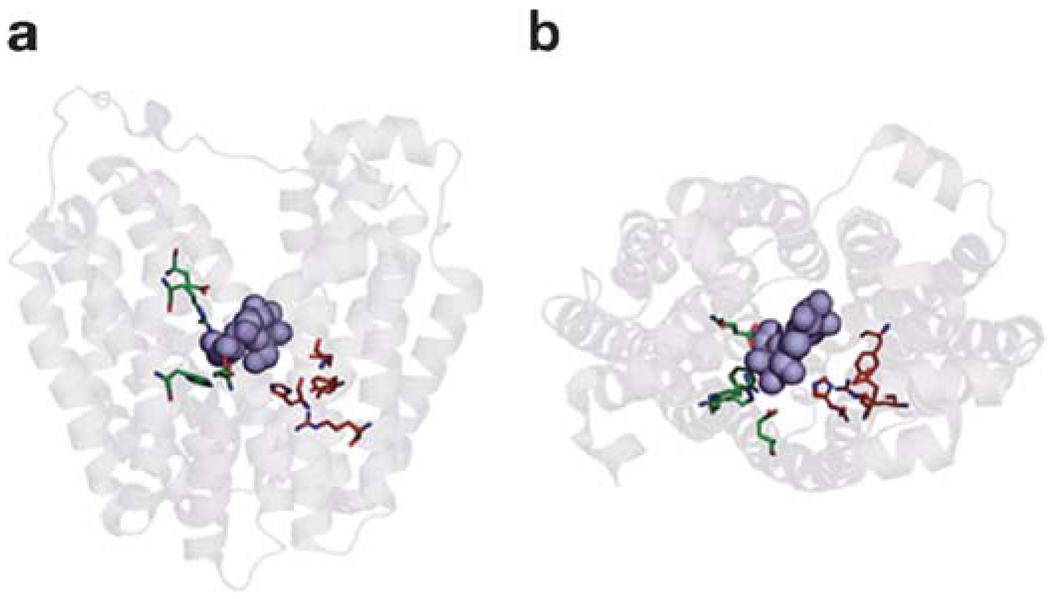
In a side view (Figure 3a), the monomer is heart-shaped with an internal cavity open on the cytoplasmic side, and the largest dimensions of the molecule are 60 × 60 Å. In a cytoplasmic view (Figure 3b), the molecule has a distorted oval shape, with dimensions of 30 × 60 Å. Remarkably, the structure features a large interior hydrophilic cavity, which is open only to the cytoplasmic side, with dimensions of 25 × 15 Å, indicating that the structure represents the inward-facing conformational state of LacY (Figure 3a,b). Within the cavity, a single sugar-binding site is observed at the apex of the cavity in the approximate middle of the molecule.
Figure 3.
The figure is based on the C154G mutant structure with bound TDG. (a) Ribbon representation of LacY viewed parallel to the membrane. The 12 transmembrane helices from the N and C termini are colored from dark blue (N-terminal bundles) to red (C-terminal bundles), and TDG is represented by black spheres. (b) Ribbon representation of LacY viewed along the membrane normal from the cytoplasmic side. For clarity, the loop regions have been omitted. The color scheme is the same as in panel (a), and the 12 transmembrane helices are labeled with Roman numerals. Reproduced, with permission, from Reference 2.
The monomer has 12 transmembrane helices as described above. The molecule is organized as two six-helix bundles connected by a long loop between helices VI and VII (loop VI–VII, Figure 2). The N- and C-terminal six-helix domains have the same topology and exhibit twofold pseudosymmetry. Similar to GlpT (59) and OxlT (17), within each domain, twofold pseudosymmetry is observed, indicating that the N- and C-terminal domains have the same genetic origin, as postulated by Pao et al. (74). The hydrophilic cavity is lined with helices I, II, IV, and V of the N-terminal domain and helices VII, VIII, X, and XI of the C-terminal domain. Helices III, VI, IX, and XII are embedded largely in the bilayer and not exposed to solvent, as suggested by Guan et al. (31).
Many of the helices are distorted (Figure 3). The distorted helices and the large hydrophilic cavity provide a ready explanation for the high rate of backbone hydrogen/deuterium exchange observed with LacY, as determined by ATR-FTIR (90% within 10 to 20 min relative to ∼50% in 2 to 3 h with the potassium channel SliK) (55). In addition to large-scale conformation changes, the H-bonds should be unstable in distorted helices and the hydrophilic cavity should allow access of deuterium to much of the backbone amide groups. Therefore, it is unlikely that the two six-helix domains move as rigid bodies.
Most recently, wild-type LacY has yielded crystals after more than a decade of effort (L. Guan & H.R. Kaback, unpublished data). Although the crystals of wild-type LacY are difficult to obtain because of rapid aggregation, a structure of unmodified, native LacY was obtained at a resolution of 4 Å. Importantly, the overall fold of wild-type LacY exhibits an inward-facing conformation and is similar to that of the C154G mutant. Because wild-type GlpT and LacY, as well as C154G LacY, have similar overall folds and are in the inward-facing conformation, this conformation likely reflects the lowest free-energy form. However, it is not clear whether this is the preferred form in the membrane or in a crystal lattice. In any event, the hydrophilic cavity is sufficiently large that it is visualized by freeze-fracture electron microscopy of proteoliposomes reconstituted with purified LacY (13, 14) or in filamentous arrays of LacY (60).
THE SUGAR-BINDING SITE
Indirect Studies
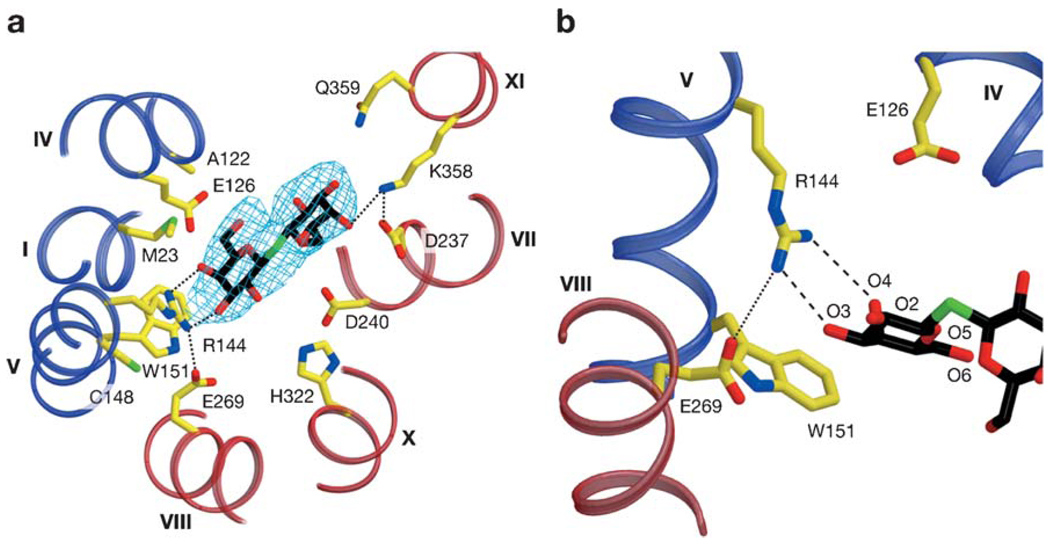
Galactoside transport is inactivated by N-ethylmaleimide (NEM), and LacY can be selectively labeled with radioactive NEM by substrate protection against alkylation (22). The substrate-protected residue was shown to be Cys-148 (5), thus providing initial evidence that the substrate-binding site may involve helix V. However, when Cys-148 is replaced with various side chains, neither binding nor active transport is abolished (37). From the X-ray structure (see below), it is apparent that Cys-148 is near the galactosyl end of the ligand but does not make direct contact with sugar (Figure 4). Thus the inactivating effect of alkylation is due to a steric effect, and conversely, binding of a galactoside protects against alkylation. A similar steric effect is observed when Ala-122 (helix IV) is replaced with Cys. However, it is remarkable that alkylation of mutant A122C causes LacY to become specific for binding and transport of the monosaccharide galactose (32, 53).
Figure 4.
Substrate-binding site of LacY. Possible H-bonds and salt bridges are represented by broken blue lines. (a) Residues involved in TDG binding viewed along the membrane normal from the cytoplasmic side. TDG is depicted as a stick model. (b) Close up of the N-terminal domain of the TDG-binding site. Reproduced, with permission, from Reference 2.
Cysteine-scanning mutagenesis reveals that replacement of Glu-126 (helix IV) or Arg-144 (helix V) with neutral amino acyl side chains abolishes transport, and activity is not observed with double-neutral substitutions or when the residues are interchanged (24). Only mutants E126D and R144K exhibit any activity whatsoever. Mutant E126D accumulates lactose at a lower rate to a normal steady state, whereas mutant R144K transports lactose at a negligible rate to a miniscule steady state (24). In addition, lactose-induced H+ translocation is observed at a slow rate with E126D permease, but not with any of the other Glu-126 or Arg-144 mutants.
Glu-126 and Arg-144 are postulated to form a charge pair. Replacement of either residue with Ala in a LacY mutant containing a single-Cys residue at position 148 markedly decreases NEM labeling of Cys-148, but the double-Ala mutant labels normally. Thus, an unpaired charge causes a conformational perturbation that decreases the reactivity of Cys-148, whereas double-neutral replacement with Ala, replacement of Arg-144 with Lys, or interchanging Glu-126 and Arg-144 has no such effect (105). Further evidence for charge pairing was obtained by spontaneous disulfide cross-linking (118) and site-directed spin labeling (125). Direct binding assays show that none of the Arg-144 mutants, including R144K, binds lactose or the high-affinity substrate analogue p-nitrophenyl α-d-galactopyranoside (NPG), and that neutral replacements for Glu-126 do not bind NPG or β-d-galactopyranosyl 1-thio-β-d-galactopyranoside (TDG), but mutant E126D exhibits significant binding of TDG (93). In addition, there is no substrate protection against NEM labeling when Glu-126 and Arg-144 are interchanged. As a whole, the results demonstrate that a carboxyl group at position 126 and a guanidino group at position 144 are absolute requirements for substrate binding and suggest that the two residues may be charge-paired. Recent studies using ESI-MS also support the interaction of Arg-144 and Glu-126 by covalent modification of the guanidino group with the Arg-specific reagent butane 2,3-dione (BD) (109). The reactivity of Arg-144 with BD is low and reduced further in the presence of ligand. Interestingly, replacement of Glu-126 with Ala results in an increase in the reactivity of Arg-144, consistent with a charge pair between Arg-144 and Glu-126 in the absence of sugar that is disrupted upon ligand binding.
Trp-151, two turns of helix V from Arg-144, also plays an important role in substrate binding (29), although mutants W151Y and W151F catalyze active lactose transport with time courses similar to those of the wild-type. Mutant W151F or W151Y binds NPG and TDG relatively poorly, but surprisingly, there is relatively little change in the kinetics of lactose transport. In addition, amino acid replacements with an alkyl side chain exhibit little or no transport and no significant binding affinity.
The fluorescent properties of Trp-151 from a fully functional mutant devoid of all other Trp residues were investigated (103). The steady-state fluorescence spectrum of Trp-151 and fluorescence quenching experiments with water-soluble quenchers demonstrate that Trp-151 is in a hydrophilic environment. Furthermore, substrate binding leads to a blue shift in the fluorescence spectrum and reduction in accessibility to polar quenchers, indicating that Trp-151 becomes less exposed to aqueous solvent. In addition, the phosphorescence spectrum of Trp-151 is red-shifted in the presence of substrate, indicating a direct stacking interaction between the galactopyranosyl and indole rings. Finally, studies with N-bromosuccinimide (NBS) show that in the presence of ligand reaction with Trp-151 is protected (104).
Glu-269 (helix VIII), another irreplaceable residue, is also critical for substrate recognition and may be involved in H+ translocation as well (see below) (23, 33, 90, 102, 105, 110). Neutral replacements abolish lactose transport, and replacement with Asp leads to decreased affinity for TDG or NPG. Significant transport of TDG is observed, and there is about a threefold increase in H+/TDG stoichiometry (23, 102). Furthermore, Glycine-scanning mutagenesis of mutant E269D shows that it can be rescued with respect to binding and all translocation reactions, suggesting that positioning of the car-boxyl group at position 269 in the binding site is critical (110). Analysis of the CNBr fragment containing Glu-269 by ES-MSI demonstrates that this carboxyl group is protected by substrate against reaction with hydrophobic carbodiimides. Together with other evidence, the findings suggest that Glu-269 may H-bond with the O3 of the d-galactopyranosyl ring (111).
Direct Evidence from the Crystal Structure
Visualization of the substrate-binding site in the X-ray structure demonstrates a primary interaction between the irreplaceable residue Arg-144 (helix V) and the O3 and O4 atoms of the galactopyranosyl ring via a bidentate H-bond (2), as suggested by the biochemical findings discussed above (Figure 4). The irreplaceable residue Glu-126 (helix IV) is in proximity to Arg-144 and may interact with the O4, O5, or O6 atoms of the galactopyranosyl ring via water molecules. A direct interaction between Arg-144 and Glu-126 is not observed in the ligand-bound structure, consistent with the idea that the salt bridge is absent in the presence of ligand (105, 109). Satisfyingly, a hydrophobic interaction between the bottom of the galactopyranosyl ring and the indole ring of Trp-151 (helix V) is confirmed. The C-6 atom of the galactopyranosyl ring also appears to interact hydrophobically with Met-23 (helix I), but mutagenesis of Met-23 has no effect on ligand binding (I. Smirnova & H. R. Kaback, unpublished data).
The binding site in the N-terminal domain is similar to those of many other galactoside- and sugar-binding proteins (69, 75, 95). Glu-269 in helix VIII in the C-terminal domain appears to form a salt bridge with Arg-144 and is in close proximity to Trp-151. Fluorescence studies with NBS suggest an H-bond between the carboxyl group at position 269 and the indole N of Trp-151 in the absence of ligand (104). Thus, contacts between Glu-269 in the C-terminal domain and Arg-144 and Trp-151 in the N-terminal domain may be key to providing the important energetic link between the two helical bundles.
Fewer interactions are observed with the sugar and the C-terminal domain. Helices VII (Asp-237) and XI (Lys-358) (Figure 4), which are symmetrically related to helices I and V, respectively, are also involved in TDG binding. However, these residues likely play only a supporting role relative to the N-terminal primary binding site by providing additional affinity for disaccharide substrates. This explains why the monosaccharide galactose has poor affinity for LacY but behaves like any other substrate with respect to transport and protection of Cys-148 against alkylation (85). It is critical to understand that galactose is the most specific substrate for LacY but has low affinity, which is increased markedly by various adducts, particularly if they are hydrophobic, at the anomeric carbon (85). In contrast to Arg-144 and Glu-126, which are absolutely required, the charge pair between Asp-237 and Lys-358 is interchangeable, and both can be replaced simultaneously by neutral side chains with little effect on activity (16, 86, 88). Therefore, the interaction of Lys-358 with ligand cannot be absolutely required for galactoside binding, although the charge pair between Lys-358 and Asp-237 is important for efficient insertion of LacY into the membrane (16, 26). The essential portion of the substrate-binding site with respect to specificity is in the N-terminal domain, and the residues in the C-terminal domain that interact with the adduct on the anomeric carbon of galactopyranoside increase affinity but have little to do with specificity. Furthermore, although the C2, C3, and C6 OH groups on the galactopyranosyl ring play roles in H-bonding, the C4 OH is unequivocally the most important determinant for specificity (92). The hydrophobic interaction between the galactopyranosyl ring and Trp-151 is likely to orient the ring so that important H-bonds can be realized (29).
As discussed above, highly specific photo-labeling of LacY with NPG (49) was useful for following the protein during purification (71). Subsequent proteolysis experiments (28) demonstrate that the photolabel is in the C-terminal half of LacY. However, because it is clear that all the determinants for sugar specificity are in the N-terminal half of the molecule, there is an apparent conundrum. The nongalactosyl end of NPG (which is photo-activated) is proximal to helices VII and XI (i.e., the C-terminal half of the molecule). Thus, although NPG labels LacY in a highly specific manner, misleading information is obtained regarding the important part of the binding site and may represent a general caveat regarding the use of photoaffinity probes.
RESIDUES INVOLVED IN H+ TRANSLOCATION AND COUPLING
Biochemistry
One fundamentally important problem is the identification of the residues involved in H+ binding and the mechanism of coupling between sugar and H+ translocation. As opposed to an initial proposal that sugar binds to unprotonated LacY (44), current evidence indicates that LacY is protonated prior to ligand binding (90). Most recently, it has been shown directly ( J. Vazquez-Ibar, S. Schuldiner & H.R. Kaback, unpublished data) that addition of TDG to a concentrated solution of purified, detergent-solubilized LacY induces no change in pH, whereas a positive control with the antiporter EmrE under identical conditions releases 1 H+/mol EmrE upon addition of tetraphenylphosphonium (98).
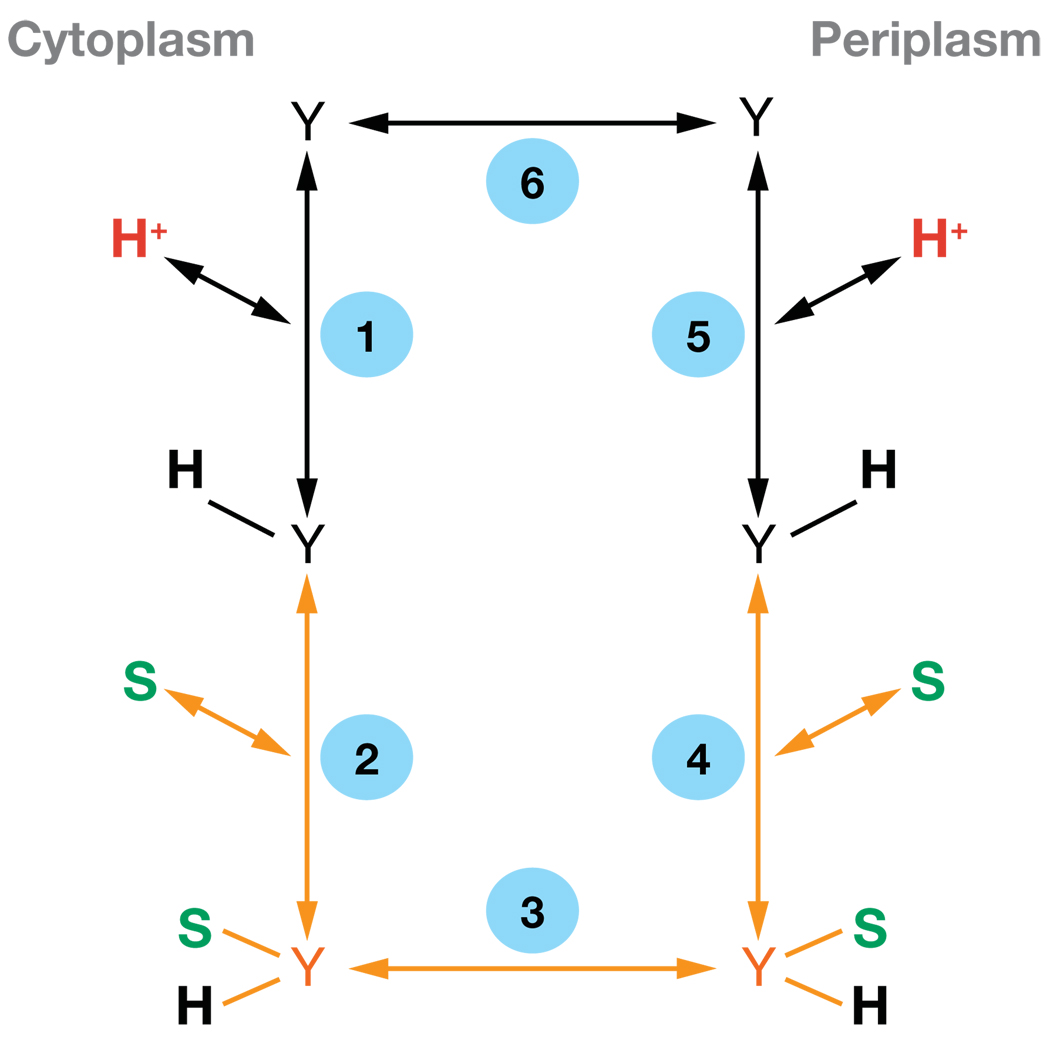
It is difficult to study the mechanism of H+ translocation. However, in addition to Δμ̄H+-driven active transport, LacY catalyzes other modes of translocation that are important for coupled H+ translocation. Because individual steps in the overall translocation cycle cannot be delineated by studying Δμ̄H+-driven active transport, LacY-mediated efflux down a chemical gradient, equilibrium exchange, and entrance counterflow are used to probe the mechanism (48, 50). Efflux, exchange, and counterflow with wild-type LacY are explained by a simple kinetic scheme (Figure 5). H+-coupled efflux consists of five steps: [1] binding of H+ and [2] binding of lactose to LacY at the inner surface of the membrane; [3] a conformational change in LacY that results in translocation of lactose and H+ to the outer surface of the membrane; [4] release of substrate; [5] release of H+; [6] a conformational change corresponding to return of unloaded LacY to the inner surface of the membrane. Alternatively, during exchange and counterflow LacY does not deprotonate and only steps 1,2, and 3 are involved.
Figure 5.
Kinetic scheme for galactoside/H+ symport, exchange, and counterflow. Y represents LacY, and S is substrate (lactose). Steps involved in exchange and counterflow (steps 2,3, and 4) are in orange. During downhill symport in the absence of Δμ̄H+, the rate-limiting step is deprotonation (step 1 or 5); in the presence of Δμ̄H+, dissociation of sugar is rate limiting (step 2 or 4).
Many enzyme reactions involve H+ transfer in the rate-limiting step, and as a result, these reactions may exhibit a solvent isotope effect when studied in deuterium oxide (D2O). In brief, such reactions proceed slower in D2O because of differences in the zero-point stretch vibrations of bonds to protium relative to deuterium (36, 94). With right-side-out (RSO) vesicles or proteoliposomes reconstituted with purified LacY, over threefold slowing of the rate of H+-coupled downhill lactose influx or efflux is observed in D2O from pH 5.5 to 7.5, with no effect on Δμ̄H+-driven active transport, exchange, counterflow, or affinity for sugar (9, 50, 106). These and other observations indicate that reactions involved in protonation or deprotonation are not rate determining for Δμ̄H+-driven active transport, exchange, or counterflow, whereas protonation or deprotonation are rate limiting when a lactose gradient drives H+ translocation (Figure 1b,c).
Efflux, exchange, and counterflow are blocked in His-322 mutants, and replacement with Asn or Gln results in a 50-fold decrease in affinity for TDG (90). However, the mutants catalyze lactose influx down a concentration gradient at a slow rate (73, 77, 78) without H+ translocation. In contrast, Glu-325 mutants are specifically defective in all steps involving net H+ translocation but catalyze lactose exchange and counterflow as well as or better than wild-type LacY (43). Thus, Glu-325 is required for deprotonation (step 4). Glu-325 mutations mimic D2O or mAb 4B1 (8, 9), but affinity is unaffected by neutral replacements, D2O, or mAb 4B1. Similarly, replacement of Arg-302 with Ala or Ser maintains exchange and counterflow, although active transport is completely inhibited (63, 89). Extensive mutagenesis and functional characterization reveal that neutral replacements for Glu-269 cause LacY to become defective in all translocation reactions. Even replacement of Glu-269 with Asp yields LacY that hardly catalyzes active lactose transport, efflux down a concentration gradient, or equilibrium exchange. However, as discussed above, mutant E269D accumulates TDG with an increase in H+/TDG stoichiometry (23, 102) and markedly decreased affinity for TDG (33, 90, 105, 110). Therefore, Glu-269 is probably involved in H+ translocation and sugar binding, as well as coupling between sugar and H+ translocation.
Interestingly, when Tyr-236 (helix VII) is replaced with Phe, active transport and efflux are abolished, and the mutant catalyzes significant equilibrium exchange (84). In contrast, mutants with Cys (27) or Ala (S. Frillingos & H.R. Kaback, unpublished data) in place of Tyr-236 catalyze significant active transport. A possible interpretation of this behavior is that in the Y236C or Y236A mutants, water replaces the OH group of Tyr. It is intriguing that the X-ray structure (see below) shows Tyr-236 to be within H-bond distance of Arg-302 and His-322.
X-Ray Structure
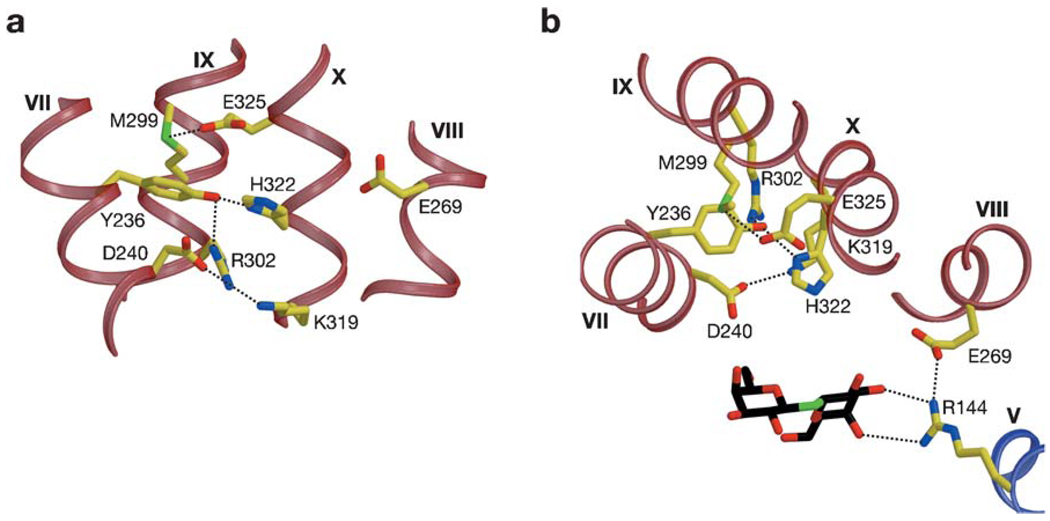
In the structure, a complex salt bridge/H-bond network (Figure 6) composed of residues from helix VII (Tyr-236 and Asp-240), helix X (Lys-319, His-322, and Glu-325), and helix IX (Arg-302) is observed. As discussed above, biochemical analyses indicate that His-322, Glu-325, and Arg-302 are probably directly involved in H+ translocation. Note that Glu-325 is embedded in a hydrophobic milieu formed by Met-299 and Ala-295 (helix IX), Leu-329 (helix X), and Tyr-236 (helix VII), which is consistent with the notion that Glu-325 is protonated in this conformation (46). Therefore, the structure represents the protonated inward-facing conformation with bound substrate. It has been suggested (89) that Arg-302 could interact with Glu-325 to drive deprotonation. On the one hand, in the structure shown, the side chain of Arg-302 is ∼7 Å away from Glu-325, suggesting that a large conformational rearrangement may occur. On the other hand, LacY with two Cys residues at positions 302 and 325 exhibits excimer fluorescence (38), and with two His residues, an Mn(II)-binding site is observed (34). The structural data combined with biochemical/molecular biological studies discussed above provide support for the suggestion that His-322 may be the immediate H+ donor to Glu-325. Because mutants with simultaneous neutral replacements for Asp-240 and Lys-319 maintain significant transport activity (86, 88), it is unlikely that this salt bridge is directly involved in H+ translocation; however, the two residues could be involved in stabilization of the salt bridge/H-bond network. Interestingly, transport is abolished when Asp-240 and Lys-319 are reversed (86) or replaced with Cys residues and then cross-linked (123).
Figure 6.
Residues involved in H+ translocation and coupling. H-bonds are represented by black broken lines. (a) View parallel to the membrane. (b) View along the membrane normal from the cytoplasmic side. Reproduced, with permission, from Reference 2.
The closest distance between this network and the sugar-binding site is more than 6 Å, indicating that the network does not interact directly with the sugar-binding site in the inward-facing conformation. Glu-269 is the only irreplaceable residue in the C-terminal half of LacY that interacts with the N-terminal half and has been postulated to be involved in both substrate and H+ translocation. Glu-269 is in the vicinity of His-322 (closest distance 5.8 Å). On the basis of biophysical studies, it has also been proposed that Glu-269 makes contact with His-322 in another conformation or via a water molecules(s) (33, 38, 39). The results are consistent with the notion that Glu-269 is critical for coupling between sugar and H+ translocation.
Although it is generally thought that H+ translocation across membranes involves movement of H+ through a pathway perpendicular to the plane of the membrane and parallel to the transmembrane helices, surprisingly, this does not appear to be the case for LacY (Figure 7). As shown, the residues involved in H+ translocation in LacY are at about the same level as the residues that form the sugar-binding site (Figure 7a), and they are essentially parallel to the plane of the membrane and perpendicular to the transmembrane helices (Figure 7b). Thus, it seems likely that H+ translocation in LacY may involve a delocalized H+-binding site. By this means, H+ translocation may occur much like sugar translocation, involving alternating access to both binding sites to either side of the membrane. Such a scenario would explain the long-standing problem of how sugar-driven H+ translocation can occur in either direction across the membrane utilizing the same residues.
Figure 7.
Configuration of residues involved in sugar binding (green) and H+ translocation (orange). (a) Viewed parallel to the membrane. (b) Viewed along the membrane normal from the cytoplasmic side.
PROPOSED MECHANISM OF LACTOSE/H+ SYMPORT
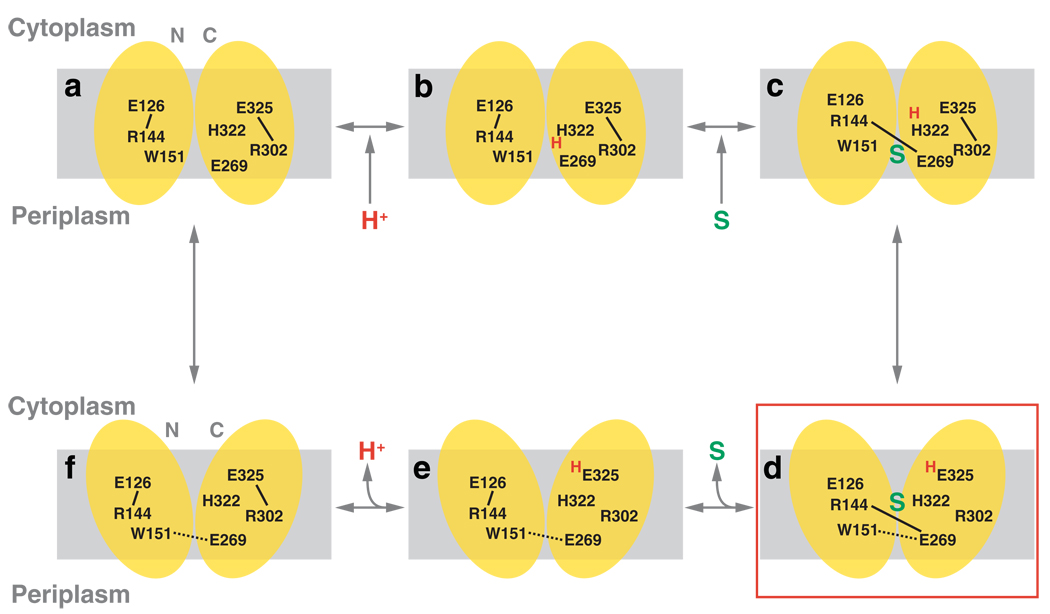
The mechanism of galactoside/H+ symport can be explained by a simple scheme (Figure 8) in which the structure corresponds to the protonated, inward-facing conformation with bound substrate (Figure 8d). LacY in the outward-facing conformation may be unstable (Figure 8a) and protonated immediately as postulated (Figure 8b) (46). In this state, the H+ is shared between Glu-269 and His-322. A galactoside is then recognized initially by Trp-151, Arg-144, and Glu-126, which disrupts the salt bridge between Arg-144 and Glu-126 and brings His-322 in contact with Glu-325. These changes may induce H+ transfer from His-322 to Glu-325 as Glu-269 is recruited to complete the binding site by forming a salt bridge with Arg-144 (Figure 8c). This process may also cause rapid transition to the inward-facing conformation (Figure 8d). Substrate is then released into the cytoplasm (Figure 8e), and the salt bridge between Arg-144 and Glu-126 is reestablished. The H+ is then released from Glu-325 (Figure 8f) probably because of a decrease in pKa caused either by approximation to Arg-302 (46, 89), exposure to solvent in the aqueous cavity (cytoplasmic pH is constant at 7.6), or both. After releasing the H+ inside, transition back to the outward-facing conformation is induced.
Figure 8.
A postulated mechanism for lactose/H+ symport. Key residues are labeled and charge pairs are represented as solid black lines; H-bonds are depicted as broken black lines. The H+ and the substrate are shown in red and green, respectively.
The structure of LacY exhibits a single sugar-binding site at the apex of a hydrophilic cavity open to the cytoplasm, and it has been postulated from the structure (2) that the binding site has alternating access to either side of the membrane during turnover, as shown in Figure 9. However, it is not clear whether Δμ̄H+ changes binding affinity, particularly with LacY, as ΔΨ and ΔpH have quantitatively the same kinetic (83) and thermodynamic (80, 81) effects on transport.
Figure 9.
Possible structural changes between inward- and outward-facing conformations. Transmembrane helices in the N- and C-terminal halves are shown as blue and red cyclinders, respectively. (a) Inward-facing conformation (i.e., the crystal structure) viewed parallel to the membrane. Cys-replacement mutants of residues colored in yellow exhibit increased reactivity with NEM upon substrate binding. (b) Suggested model for outward-facing conformation based on chemical modification and thiol cross-linking. The model was obtained by applying a relatively rigid body rotation of ∼60° (around the axis passing near TDG parallel to the membrane) to the N- and C-terminal domains. Reproduced, with permission, from Reference 2.
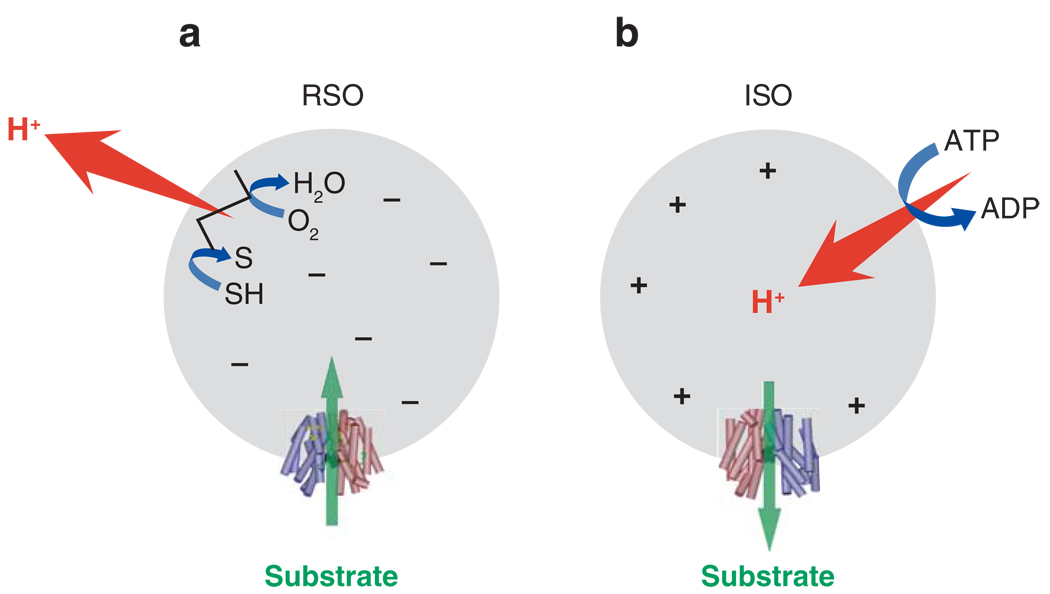
Although substrate protection against alkylation of Cys-148 by NEM is particularly useful for obtaining apparent binding constant values (KD) of LacY for various substrates over a wide range of concentrations (25, 85, 87, 89, 90, 93, 105), it is difficult to obtain true KD values on each side of the membrane for a transport protein in the presence of Δμ̄H+ because the ligands used are translocated across the membrane and may accumulate in RSO vesicles, thereby leading to underestimation of KD. However, in a recent series of experiments (30), lactose or TDG protection of Cys-148 against alkylation by NEM were carried out on ice, which decreases substrate accumulation drastically (45). Under these experimental conditions, in the absence of Δμ̄H+, both ice-cold RSO and ISO (inside-out) vesicles likely equilibrate with the external medium. In the presence of Δμ̄H+ RSO vesicles may still be able to accumulate lactose or TDG two- to threefold, even though the reactions are carried out on ice for only 5 min. Therefore, the measured KD values for RSO vesicles in the presence of Δμ̄H+ may be underestimated by two- to threefold. However, this is unlikely, as ISO vesicles in the presence of ATP generate a Δμ̄H+ of opposite polarity (interior positive and/or acid) (Figure 10) (82) that causes a decrease in the intravesicular concentration of ligand relative to the concentration in the medium (50). Remarkably, results with both lactose and TDG demonstrate that the KD manifested by ISO vesicles exhibits less than a twofold change in the absence or presence of Δμ̄H+. Moreover, the KD values observed with RSO or ISO vesicles in the absence or presence of Δμ̄H+ are similar within experimental error. The results provide a strong indication that Δμ̄H+ has little or no effect on binding affinity, a conclusion that raises a number of interesting considerations regarding the mechanism by which Δμ̄H+ drives accumulation.
Figure 10.
Effect of Δμ̄H+ of opposite polarities on substrate translocation in RSO or ISO vesicles. (a) Δμ̄H+ with RSO vesicles (ΔΨ, interior negative); substrate is accumulated. (b) Δμ̄H+ in ISO vesicles (interior positive and acid); substrate effluxes from the vesicles. Reproduced, with permission, from Reference 30.
In the presence of Δμ̄H+ (interior negative and/or alkaline), wild-type LacY can accumulate lactose against a ∼100-fold concentration gradient. Without a significant decrease in binding affinity on the inside of the membrane, how does Δμ̄H+ drive lactose accumulation against a concentration gradient? Because of the effect of D2O on various translocation reactions, Viitanen et al. (106) have postulated that the rate-limiting step for downhill transport in the absence of Δμ̄H+ is deprotonation, which precedes return of unloaded LacY to the outer surface of the membrane; in contrast, in the presence of Δμ̄H+ dissociation of sugar is rate limiting. Note that the primary kinetic effect of ΔΨ or ΔpH on transport is a dramatic decrease in Km (83). Therefore, it seems reasonable to suggest that Δμ̄H+ enhances the rate of deprotonation on the inner surface of the membrane and thereby allows unloaded LacY to return more rapidly to the outward-facing conformation. Thus, the major effect of Δμ̄H+ on active transport by LacY appears to be kinetic with little or no change in affinity for sugar.
Although biochemical and biophysical studies, as well as a single structure at a resolution of 3.5 Å, can give clues to how the overall conformational change in LacY may be coupled to sugar binding and H+ translocation, many fundamental questions remain. Why is protonation of LacY important for sugar binding? What is the detailed mechanism of coupling between binding and H+ translocation? What is the time of occupancy of LacY in the outward-facing and inward-facing conformations? Therefore, it is essential to obtain higher-resolution structures in different conformations, as well as dynamics, in order to understand fully the mechanism of substrate/ H+ symport by LacY.
LESSONS
It is clear from the studies summarized in this review that local interactions in LacY can be obtained by indirect means. However, it is also apparent that important global aspects of structure are difficult to delineate in this manner. For example, the residues in LacY involved in specificity and sugar binding were identified initially by site-directed mutagenesis and functional studies, and the conclusions are supported largely by the X-ray structure. In contrast, it is highly unlikely that the large, inward-facing hydrophilic cavity, as well as the highly distorted helices, could have been deduced from the indirect techniques utilized.
Although unexpected, the residues involved in H+ translocation are aligned parallel to the plane of the membrane at the same level as the sugar-binding site and may be exposed to a water-filled cavity in both the inward- and outward-facing conformations. Therefore, like the galactosidic sugar, the H+ may be released directly into either cavity during turnover. In any event, it is apparent that H+ translocation through LacY cannot involve a water-filled channel through the molecule. These structural features may also explain why LacY catalyzes lactose/H+ symport in both directions across the membrane utilizing the same residues.
Structural biology of membrane proteins is finally becoming a reality, as evidenced by the increasing number of structures that are now appearing (http://www.mpibp-frankfurt.mpg.de/michel/public/memprotstruct.html). However, it is important to realize that an X-ray structure, even at high resolution, does not provide dynamic information, and with transport proteins like LacY, the time of occupancy in different conformations may be an essential part of the transport mechanism. Therefore, use of molecular biology to engineer membrane proteins for dynamic studies with biochemical and biophysical techniques is critical.
SUMMARY
The lactose permease of E. coli (LacY) couples the free energy released from downhill translocation of H+ in response to an H+ electrochemical gradient to drive the stoichiometric accumulation of d-galactopyranosides against a concentration gradient. An X-ray structure in an inward-facing conformation has been solved, which confirms many conclusions from biochemical and biophysical studies. LacY contains N- and C-terminal domains, each with six transmembrane helices, positioned pseudosymmetrically. A large hydrophilic cavity is exposed to the cytoplasm, and ligand is bound at the twofold axis of symmetry at the apex of the hydrophilic cavity in the approximate middle of the molecule. By combining a large body of experimental data derived from systematic studies of site-directed mutants, residues involved in substrate binding and H+ translocation have been identified, and on the basis of the functional properties of the mutants and the X-ray structure, a working model for the mechanism that involves alternating access of the binding site has been postulated.
Acknowledgements
This work was supported by NIH Grants DK51131, DK069463, GM073210 and GM074929, as well as NSF grant 0450970, to H.R.K.
Glossary
- Electrochemical H+ gradient (Δμ̄H+)
when two aqueous phases are separated by a membrane, the electrochemical potential difference of H+ between the two phases is expressed as Δμ̄H+/F=ΔΨ−2.3RT/FΔpH
- ΔΨ
the electrical potential across the membrane
- ΔpH
the pH; gradient across the membrane
- LacY
lactose permease
- Second-site suppressor analysis
involves selection of a second mutation that corrects (suppresses) the phenotype of an original inactivation mutation, which can be done by random or site-directed mutagenesis
- Cysteine-scanning mutagenesis
the use of site-directed mutagenesis to replace individual residues in a given protein, or a region of that protein, with cysteine
- Excimer fluorescence
an excited-state dimer observed in the fluorescence-emission spectrum, which emits at a higher wavelength than the monomer fluorescence
- Divalent metal-binding site
the simplest consists of two imidazole side chains (His residues) within close proximity; Mn(II) binding is then measured directly by electron paramagnetic resonance
- Hydrogen/deuterium exchange
the rate at which backbone amide protons exchange with deuterium
- NEM
N-ethylmaleimide
- NPG
p-nitrophenyl α-d-galactopyranoside
- TDG
β-d-galactopyranosyl 1-thio-β-d-galactopyranoside
- RSO
right-side-out
- ISO
inside-out
Contributor Information
Lan Guan, Email: LanGuan@mednet.ucla.edu.
H. Ronald Kaback, Email: RKaback@mednet.ucla.edu.
LITERATURE CITED
- 1.Abramson J, Kaback HR, Iwata S. Structural comparison of lactose permease and the glycerol-3-phosphate antiporter: members of the major facilitator superfamily. Curr. Opin. Struct. Biol. 2004;14:413–419. doi: 10.1016/j.sbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 3.Bibi E, Kaback HR. In vivo expression of the lacY gene in two segments leads to functional lac permease. Proc. Natl. Acad. Sci. USA. 1990;87:4325–4329. doi: 10.1073/pnas.87.11.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibi E, Kaback HR. Functional complementation of internal deletion mutants in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1992;89:1524–1528. doi: 10.1073/pnas.89.5.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieseler B, Prinz H, Beyreuther K. Topological studies of lactose permease of Escherichia coli by protein sequence analysis. Ann. N. Y. Acad. Sci. 1985;456:309–325. doi: 10.1111/j.1749-6632.1985.tb14882.x. [DOI] [PubMed] [Google Scholar]
- 6.Büchel DE, Gronenborn B, Müller-Hill B. Sequence of the lactose permease gene. Nature. 1980;283:541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- 7.Calamia J, Manoil C. lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc. Natl. Acad. Sci. USA. 1990;87:4937–4941. doi: 10.1073/pnas.87.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrasco N, Tahara SM, Patel L, Goldkorn T, Kaback HR. Preparation, characterization, and properties of monoclonal antibodies against the lac carrier protein from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1982;79:6894–6898. doi: 10.1073/pnas.79.22.6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrasco N, Viitanen P, Herzlinger D, Kaback HR. Monoclonal antibodies against the lac carrier protein from Escherichia coli. 1. Functional studies. Biochemistry. 1984;23:3681–3687. doi: 10.1021/bi00311a017. [DOI] [PubMed] [Google Scholar]
- 10.Cohen GN, Rickenberg HV. Etude directe de la fixation d’un inducteur de la β-galactosidase par les cellules d’Escherichia coli. Comptes Rendu. 1955;240:466–468. [PubMed] [Google Scholar]
- 11.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem. Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 12.Consler TG, Persson BL, Jung H, Zen KH, Jung K, et al. Properties and purification of an active biotinylated lactose permease from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1993;90:6934–6938. doi: 10.1073/pnas.90.15.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello MJ, Escaig J, Matsushita K, Viitanen PV, Menick DR, Kaback HR. Purified lac permease and cytochrome o oxidase are functional as monomers. J. Biol. Chem. 1987;262:17072–17082. [PubMed] [Google Scholar]
- 14.Costello MJ, Viitanen P, Carrasco N, Foster DL, Kaback HR. Morphology of proteoliposomes reconstituted with purified lac carrier protein from Escherichia coli. J. Biol. Chem. 1984;259:15579–15586. [PubMed] [Google Scholar]
- 15.Dornmair K, Corni AF, Wright JK, Jähnig F. The size of the lactose permease derived from rotational diffusion measurements. EMBO J. 1985;4:3633–3638. doi: 10.1002/j.1460-2075.1985.tb04127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunten RL, Sahin-Tóth M, Kaback HR. Role of the charge pair formed by aspartic acid 237 and lysine 358 in the lactose permease of Escherichia coli. Biochemistry. 1993;32:3139–3145. doi: 10.1021/bi00063a028. [DOI] [PubMed] [Google Scholar]
- 17.Ehring R, Beyreuther K, Wright JK, Overath P. In vitro and in vivo products of E. coli. lactose permease gene are identical. Nature. 1980;283:537–540. doi: 10.1038/283537a0. [DOI] [PubMed] [Google Scholar]
- 18.Ermolova N, Guan L, Kaback HR. Intermolecular thiol cross-linking via loops in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2003;100:10187–10192. doi: 10.1073/pnas.1434239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fetsch EE, Davidson AL. Maltose transport through the inner membrane of Escherichia coli. Front. Biosci. 2003;8:d652–d660. doi: 10.2741/1048. [DOI] [PubMed] [Google Scholar]
- 20.Foster DL, Boublik M, Kaback HR. Structure of the lac carrier protein of Escherichia coli. J. Biol. Chem. 1983;258:31–34. [PubMed] [Google Scholar]
- 21.Foster DL, Garcia ML, Newman MJ, Patel L, Kaback HR. Lactose-proton symport by purified lac carrier protein. Biochemistry. 1982;21:5634–5638. doi: 10.1021/bi00265a038. [DOI] [PubMed] [Google Scholar]
- 22.Fox CF, Kennedy EP. Specific labeling and partial purification of the M protein, a component of the β-galactoside transport system of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1965;54:891–899. doi: 10.1073/pnas.54.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franco PJ, Brooker RJ. Functional roles of Glu-269 and Glu-325 within the lactose permease of Escherichia coli. J. Biol. Chem. 1994;269:7379–7386. [PubMed] [Google Scholar]
- 24.Frillingos S, Gonzalez A, Kaback HR. Cysteine-scanning mutagenesis of helix IV and the adjoining loops in the lactose permease of Escherichia coli: Glu126 and Arg144 are essential. Biochemistry. 1997;36:14284–14290. doi: 10.1021/bi972314d. [DOI] [PubMed] [Google Scholar]
- 25.Frillingos S, Kaback HR. Probing the conformation of the lactose permease of Escherichia coli by in situ site-directed sulfhydryl modification. Biochemistry. 1996;35:3950–3956. doi: 10.1021/bi952601m. [DOI] [PubMed] [Google Scholar]
- 26.Frillingos S, Sahin-Tóth M, Lengeler JW, Kaback HR. Helix packing in the sucrose permease of Escherichia coli: properties of engineered charge pairs between helices VII and XI. Biochemistry. 1995;34:9368–9373. doi: 10.1021/bi00029a012. [DOI] [PubMed] [Google Scholar]
- 27.Frillingos S, Sahin-Tóth M, Wu J, Kaback HR. Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 28.Goldkorn T, Rimon G, Kaback HR. Topology of the lac carrier protein in the membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1983;80:3322–3326. doi: 10.1073/pnas.80.11.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guan L, Hu Y, Kaback HR. Aromatic stacking in the sugar binding site of the lactose permease. Biochemistry. 2003;42:1377–1382. doi: 10.1021/bi027152m. [DOI] [PubMed] [Google Scholar]
- 30.Guan L, Kaback HR. Binding affinity of lactose permease is not altered by the H+ electrochemical gradient. Proc. Natl. Acad. Sci. USA. 2004;101:12148–12152. doi: 10.1073/pnas.0404936101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan L, Murphy FD, Kaback HR. Surface-exposed positions in the transmembrane helices of the lactose permease of Escherichia coli determined by intermolecular thiol cross-linking. Proc. Natl. Acad. Sci. USA. 2002;99:3475–3480. doi: 10.1073/pnas.052703699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan L, Sahin-Tóth M, Kaback HR. Changing the lactose permease of Escherichia coli into a galactose-specific symporter. Proc. Natl. Acad. Sci. USA. 2002;99:6613–6618. doi: 10.1073/pnas.102178299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He MM, Kaback HR. Interaction between residues Glu269 (helix VIII) and His322 (helix X) of the lactose permease of Escherichia coli is essential for substrate binding. Biochemistry. 1997;36:13688–13692. doi: 10.1021/bi9715324. [DOI] [PubMed] [Google Scholar]
- 34.He MM, Voss J, Hubbell WL, Kaback HR. Use of designed metal binding sites to study helix proximity in the lactose permease of Escherichia coli. 2. Proximity of helix IX (Arg302) with helix X (His322 and Glu325) Biochemistry. 1995;34:15667–15670. doi: 10.1021/bi00048a010. [DOI] [PubMed] [Google Scholar]
- 35.He MM, Voss J, Hubbell WL, Kaback HR. Use of designed metal-binding sites to study helix proximity in the lactose permease of Escherichia coli. 1. Proximity of helix VII (Asp237 and Asp240) with helices X (Lys319) and XI (Lys358) Biochemistry. 1995;34:15661–15666. doi: 10.1021/bi00048a009. [DOI] [PubMed] [Google Scholar]
- 36.Jenks WP. Catalysis in Chemistry and Enzymology. New York: McGraw-Hill; 1969. [Google Scholar]
- 37.Jung H, Jung K, Kaback HR. Cysteine 148 in the lactose permease of Escherichia coli is a component of a substrate binding site. I. Site-directed mutagenesis studies. Biochemistry. 1994;33:12160–12165. doi: 10.1021/bi00206a019. [DOI] [PubMed] [Google Scholar]
- 38.Jung K, Jung H, Wu J, Privé GG, Kaback HR. Use of site-directed fluorescence labeling to study proximity relationships in the lactose permease of Escherichia coli. Biochemistry. 1993;32:12273–12278. doi: 10.1021/bi00097a001. [DOI] [PubMed] [Google Scholar]
- 39.Jung K, Voss J, He M, Hubbell WL, Kaback HR. Engineering a metal binding site within a polytopic membrane protein, the lactose permease of Escherichia coli. Biochemistry. 1995;34:6272–6277. doi: 10.1021/bi00019a003. [DOI] [PubMed] [Google Scholar]
- 40.Kaback HR. The role of the phosphoenolpyruvate-phosphotransferase system in the transport of sugars by isolated membrane preparations of Escherichia coli. J. Biol. Chem. 1968;243:3711–3724. [PubMed] [Google Scholar]
- 41.Kaback HR. Molecular biology and energetics of membrane transport. J. Cell Physiol. 1976;89:575–593. doi: 10.1002/jcp.1040890414. [DOI] [PubMed] [Google Scholar]
- 42.Kaback HR. The lac carrier protein in Escherichia coli: from membrane to molecule. J. Membr. Biol. 1983;76:95–112. doi: 10.1007/BF02000610. [DOI] [PubMed] [Google Scholar]
- 43.Kaback HR. Molecular biology of active transport: from membranes to molecules to mechanism. Harvey Lect. 1989;83:77–103. [PubMed] [Google Scholar]
- 44.Kaback HR. A molecular mechanism for energy coupling in a membrane transport protein, the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1997;94:5539–5543. doi: 10.1073/pnas.94.11.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaback HR, Barnes EM., Jr Mechanisms of active transport in isolated membrane vesicles. II. The mechanism of energy coupling between d-lactic dehydrogenase and β-galactoside transport in membrane preparations from Escherichia coli. J. Biol. Chem. 1971;246:5523–5531. [PubMed] [Google Scholar]
- 46.Kaback HR, Sahin-Tóth M, Weinglass AB. The kamikaze approach to membrane transport. Nat. Rev. Mol. Cell Biol. 2001;2:610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- 47.Kaback HR, Wu J. From membrane to molecule to the third amino acid from the left with the lactose permease of Escherichia coli. Q. Rev. Biophys. 1997;30:333–364. doi: 10.1017/s0033583597003387. [DOI] [PubMed] [Google Scholar]
- 48.Kaczorowski GJ, Kaback HR. Mechanism of lactose translocation in membrane vesicles from Escherichia coli. 1. Effect of pH on efflux, exchange, and counterflow. Biochemistry. 1979;18:3691–3697. doi: 10.1021/bi00584a009. [DOI] [PubMed] [Google Scholar]
- 49.Kaczorowski GJ, Leblanc G, Kaback HR. Specific labeling of the lac carrier protein in membrane vesicles of Escherichia coli by a photoaffinity reagent. Proc. Natl. Acad. Sci. USA. 1980;77:6319–6323. doi: 10.1073/pnas.77.11.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaczorowski GJ, Robertson DE, Kaback HR. Mechanism of lactose translocation in membrane vesicles from Escherichia coli. 2. Effect of imposed delta psi, delta pH, and delta mu H+ Biochemistry. 1979;18:3697–3704. doi: 10.1021/bi00584a010. [DOI] [PubMed] [Google Scholar]
- 51.Kanner BI, Kavanaugh MP, Bendahan A. Molecular characterization of substrate-binding sites in the glutamate transporter family. Biochem. Soc. Trans. 2001;29:707–710. doi: 10.1042/0300-5127:0290707. [DOI] [PubMed] [Google Scholar]
- 52.King SC, Hansen CL, Wilson TH. The interaction between aspartic acid 237 and lysine 358 in the lactose carrier of Escherichia coli. Biochem. Biophys. Acta. 1991;1062:177–186. doi: 10.1016/0005-2736(91)90390-t. [DOI] [PubMed] [Google Scholar]
- 53.Kwaw I, Zen KC, Hu Y, Kaback HR. Site-directed sulfhydryl labeling of the lactose permease of Escherichia coli: helices IV and V that contain the major determinants for substrate binding. Biochemistry. 2001;40:10491–10499. doi: 10.1021/bi010866x. [DOI] [PubMed] [Google Scholar]
- 54.le Coutre J, Kaback HR. Structure-function relationships of integral membrane proteins: membrane transporters vs channels. Biopolymers. 2000;55:297–307. doi: 10.1002/1097-0282(2000)55:4<297::AID-BIP1003>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 55.le Coutre J, Kaback HR, Patel CK, Heginbotham L, Miller C. Fourier transform infrared spectroscopy reveals a rigid alpha-helical assembly for the tetrameric Streptomyces lividans K+ channel. Proc. Natl. Acad. Sci. USA. 1998;95:6114–6117. doi: 10.1073/pnas.95.11.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.le Coutre J, Narasimhan LR, Patel CK, Kaback HR. The lipid bilayer determines helical tilt angle and function in lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1997;94:10167–10171. doi: 10.1073/pnas.94.19.10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.le Coutre J, Whitelegge JP, Gross A, Turk E, Wright EM, et al. Proteomics on full-length membrane proteins using mass spectrometry. Biochemistry. 2000;39:4237–4242. doi: 10.1021/bi000150m. [DOI] [PubMed] [Google Scholar]
- 58.Lee JL, Hwang PP, Hansen C, Wilson TH. Possible salt bridges between transmembrane α-helices of the lactose carrier of Escherichia coli. J. Biol. Chem. 1992;267:20758–20764. [PubMed] [Google Scholar]
- 59.Lemieux MJ, Song J, Kim MJ, Huang Y, Villa A, et al. Three-dimensional crystallization of the Escherichia coli glycerol-3-phosphate transporter: a member of the major facilitator superfamily. Protein Sci. 2003;12:2748–2756. doi: 10.1110/ps.03276603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li J, Tooth P. Size and shape of the Escherichia coli lactose permease measured in filamentous arrays. Biochemistry. 1987;26:4816–4823. doi: 10.1021/bi00389a032. [DOI] [PubMed] [Google Scholar]
- 61.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 62.Matsushita K, Patel L, Gennis RB, Kaback HR. Reconstitution of active transport in proteoliposomes containing cytochrome o oxidase and lac carrier protein purified from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1983;80:4889–4893. doi: 10.1073/pnas.80.16.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Menick DR, Carrasco N, Antes L, Patel L, Kaback HR. lac permease of Escherichia coli: arginine-302 as a component of the postulated proton relay. Biochemistry. 1987;26:6638–6644. doi: 10.1021/bi00395a012. [DOI] [PubMed] [Google Scholar]
- 64.Menick DR, Sarkar HK, Poonian MS, Kaback HR. Cys154 is important for lac permease activity in Escherichia coli. Biochem. Biophys. Res. Commun. 1985;132:162–170. doi: 10.1016/0006-291x(85)91002-2. [DOI] [PubMed] [Google Scholar]
- 65.Merritt EA, Sarfaty S, Feil IK, Hol WG. Structural foundation for the design of receptor antagonists targeting Escherichia coli heat-labile enterotoxin. Structure. 1997;5:1485–1499. doi: 10.1016/s0969-2126(97)00298-0. [DOI] [PubMed] [Google Scholar]
- 66.Mitchell P. Molecule, group and electron transport through natural membranes. Biochem. Soc. Symp. 1963;22:142–168. [Google Scholar]
- 67.Mitchell P. Translocations through natural membranes. Adv. Enzymol. 1967;29:33–87. doi: 10.1002/9780470122747.ch2. [DOI] [PubMed] [Google Scholar]
- 68.Mitchell P. Chemiosmotic Coupling and Energy Transduction. Bodmin, UK: Glynn Res. Ltd; 1968. [Google Scholar]
- 69.Müller-Hill B. The lac Operon: A Short History of A Genetic Paradigm. Berlin/New York: de Gruyter; 1996. [Google Scholar]
- 70.Nelson N. The family of Na+/Cl− neurotransmitter transporters. J. Neurochem. 1998;71:1785–1803. doi: 10.1046/j.1471-4159.1998.71051785.x. [DOI] [PubMed] [Google Scholar]
- 71.Newman MJ, Foster DL, Wilson TH, Kaback HR. Purification and reconstitution of functional lactose carrier from Escherichia coli. J. Biol. Chem. 1981;256:11804–11808. [PubMed] [Google Scholar]
- 72.Newman MJ, Wilson TH. Solubilization and reconstitution of the lactose transport system from Escherichia coli. J. Biol. Chem. 1980;255:10583–10586. [PubMed] [Google Scholar]
- 73.Padan E, Sarkar HK, Viitanen PV, Poonian MS, Kaback HR. Site-specific mutagenesis of histidine residues in the lac permease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1985;82:6765–6768. doi: 10.1073/pnas.82.20.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–32. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patzlaff JS, Moeller JA, Barry BA, Brooker RJ. Fourier transform infrared analysis of purified lactose permease: A monodisperse lactose permease preparation is stably folded, alpha-helical, and highly accessible to deuterium exchange. Biochemistry. 1998;37:15363–15375. doi: 10.1021/bi981142x. [DOI] [PubMed] [Google Scholar]
- 76.Postma PW, Lengeler JW, Jacobson GR, editors. Echerichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington, DC: ASM; 1996. p. 1149. [Google Scholar]
- 77.Püttner IB, Kaback HR. lac permease of Escherichia coli containing a single histidine residue is fully functional. Proc. Natl. Acad. Sci. USA. 1988;85:1467–1471. doi: 10.1073/pnas.85.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Püttner IB, Sarkar HK, Poonian MS, Kaback HR. lac permease of Escherichia coli: His-205 and His-322 play different roles in lactose/H+ symport. Biochemistry. 1986;25:4483–4485. doi: 10.1021/bi00364a003. [DOI] [PubMed] [Google Scholar]
- 79.Quiocho FA, Vyas NK, editors. Bioorganic Chemistry: Carbohydrates. Oxford, UK: Oxford Univ. Press; 1999. p. 441. [Google Scholar]
- 80.Ramos S, Kaback HR. The relationship between the electrochemical proton gradient and active transport in Escherichia coli membrane vesicles. Biochemistry. 1977;16:854–859. doi: 10.1021/bi00624a007. [DOI] [PubMed] [Google Scholar]
- 81.Ramos S, Schuldiner S, Kaback HR. The electrochemical gradient of protons and its relationship to active transport in Escherichia coli membrane vesicles. Proc. Natl. Acad. Sci. USA. 1976;73:1892–1896. doi: 10.1073/pnas.73.6.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reenstra WW, Patel L, Rottenberg H, Kaback HR. Electrochemical proton gradient in inverted membrane vesicles from Escherichia coli. Biochemistry. 1980;19:1–9. doi: 10.1021/bi00542a001. [DOI] [PubMed] [Google Scholar]
- 83.Robertson DE, Kaczorowski GJ, Garcia ML, Kaback HR. Active transport in membrane vesicles from Escherichia coli: The electrochemical proton gradient alters the distribution of the lac carrier between two different kinetic states. Biochemistry. 1980;19:5692–5702. doi: 10.1021/bi00566a005. [DOI] [PubMed] [Google Scholar]
- 84.Roepe PD, Kaback HR. Site-directed mutagenesis of tyrosine residues in the lac permease of Escherichia coli. Biochemistry. 1989;28:6127–6132. doi: 10.1021/bi00440a060. [DOI] [PubMed] [Google Scholar]
- 85.Sahin-Tóth M, Akhoon KM, Runner J, Kaback HR. Ligand recognition by the lactose permease of Escherichia coli: Specificity and affinity are defined by distinct structural elements of galactopyranosides. Biochemistry. 2000;39:5097–5103. doi: 10.1021/bi0000263. [DOI] [PubMed] [Google Scholar]
- 86.Sahin-Tóth M, Dunten RL, Gonzalez A, Kaback HR. Functional interactions between putative intramembrane charged residues in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1992;89:10547–10551. doi: 10.1073/pnas.89.21.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sahin-Tóth M, Gunawan P, Lawrence MC, Toyokuni T, Kaback HR. Binding of hydrophobic D-galactopyranosides to the lactose permease of Escherichia coli. Biochemistry. 2002;41:13039–13045. doi: 10.1021/bi0203076. [DOI] [PubMed] [Google Scholar]
- 88.Sahin-Tóth M, Kaback HR. Properties of interacting aspartic acid and lysine residues in the lactose permease of Escherichia coli. Biochemistry. 1993;32:10027–10035. doi: 10.1021/bi00089a019. [DOI] [PubMed] [Google Scholar]
- 89.Sahin-Tóth M, Kaback HR. Arg-302 facilitates deprotonation of Glu-325 in the transport mechanism of the lactose permease from Escherichia coli. Proc. Natl. Acad. Sci. USA. 2001;98:6068–6073. doi: 10.1073/pnas.111139698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sahin-Tóth M, Karlin A, Kaback HR. Unraveling the mechanism of lactose per-mease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2000;97:10729–10732. doi: 10.1073/pnas.200351797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sahin-Tóth M, Lawrence MC, Kaback HR. Properties of permease dimer, a fusion protein containing two lactose permease molecules from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1994;91:5421–5425. doi: 10.1073/pnas.91.12.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sahin-Tóth M, Lawrence MC, Nishio T, Kaback HR. The C-4 hydroxyl group of galactopyranosides is the major determinant for ligand recognition by the lactose permease of Escherichia coli. Biochemistry. 2001;43:13015–13019. doi: 10.1021/bi011233l. [DOI] [PubMed] [Google Scholar]
- 93.Sahin-Tóth M, le Coutre J, Kharabi D, le Maire G, Lee JC, Kaback HR. Characterization of Glu126 and Arg144, two residues that are indispensable for substrate binding in the lactose permease of Escherichia coli. Biochemistry. 1999;38:813–819. doi: 10.1021/bi982200h. [DOI] [PubMed] [Google Scholar]
- 94.Schowen RL. Isotope Effects on Enzyme-Catalyzed Reactions. Baltimore, MD: Univ. Park Press; 1977. p. 64. [Google Scholar]
- 95.Sixma TK, Pronk SE, Kalk KH, van Zanten BA, Berghuis AM, Hol WG. Lactose binding to heat-labile enterotoxin revealed by X-ray crystallography. Nature. 1992;355:561–564. doi: 10.1038/355561a0. [DOI] [PubMed] [Google Scholar]
- 96.Smirnova IN, Kaback HR. A mutation in the lactose permease of Escherichia coli that decreases conformational flexibility and increases protein stability. Biochemistry. 2003;42:3025–3031. doi: 10.1021/bi027329c. [DOI] [PubMed] [Google Scholar]
- 97.Sorgen PL, Hu Y, Guan L, Kaback HR, Girvin ME. An approach to membrane protein structure without crystals. Proc. Natl. Acad. Sci. USA. 2002;99:14037–14040. doi: 10.1073/pnas.182552199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Soskine M, Adam Y, Schuldiner S. Direct evidence for substrate-induced proton release in detergent-solubilized EmrE, a multidrug transporter. J. Biol. Chem. 2004;279:9951–9955. doi: 10.1074/jbc.M312853200. [DOI] [PubMed] [Google Scholar]
- 99.Sun J, Kaback HR. Proximity of periplasmic loops in the lactose permease of Escherichia coli determined by site-directed cross-linking. Biochemistry. 1997;36:11959–11965. doi: 10.1021/bi971172k. [DOI] [PubMed] [Google Scholar]
- 100.Teather RM, Müller-Hill B, Abrutsch U, Aichele G, Overath P. Amplification of the lactose carrier protein in Escherichia coli using a plasmid vector. Mol. Gen. Genet. 1978;159:239–248. doi: 10.1007/BF00268260. [DOI] [PubMed] [Google Scholar]
- 101.Ujwal ML, Jung H, Bibi E, Manoil C, Altenbach C, et al. Membrane topology of helices VII and XI in the lactose permease of Escherichia coli studied by lacY-phoA fusion analysis and site-directed spectroscopy. Biochemistry. 1995;34:14909–14917. doi: 10.1021/bi00045a036. [DOI] [PubMed] [Google Scholar]
- 102.Ujwal ML, Sahin-Tóth M, Persson B, Kaback HR. Role of glutamate-269 in the lactose permease of Escherichia coli. Mol. Membr. Biol. 1994;11:9–16. doi: 10.3109/09687689409161024. [DOI] [PubMed] [Google Scholar]
- 103.Vazquez-Ibar JL, Guan L, Svrakic M, Kaback HR. Exploiting luminescence spectroscopy to elucidate the interaction between sugar and a tryptophan residue in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2003;100:12706–12711. doi: 10.1073/pnas.1835645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vazquez-Ibar JL, Guan L, Weinglass AB, Verner G, Gordillo R, Kaback HR. Sugar recognition by the lactose permease of Escherichia coli. J. Biol. Chem. 2004;279:49214–49221. doi: 10.1074/jbc.M407408200. [DOI] [PubMed] [Google Scholar]
- 105.Venkatesan P, Kaback HR. The substrate-binding site in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1998;95:9802–9807. doi: 10.1073/pnas.95.17.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Viitanen P, Garcia ML, Foster DL, Kaczorowski GJ, Kaback HR. Mechanism of lactose translocation in proteoliposomes reconstituted with lac carrier protein purified from Escherichia coli. 2. Deuterium solvent isotope effects. Biochemistry. 1983;22:2531–2536. doi: 10.1021/bi00279a034. [DOI] [PubMed] [Google Scholar]
- 107.Viitanen P, Garcia ML, Kaback HR. Purified reconstituted lac carrier protein from Escherichia coli is fully functional. Proc. Natl. Acad. Sci. USA. 1984;81:1629–1633. doi: 10.1073/pnas.81.6.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vogel H, Wright JK, Jähnig F. The structure of the lactose permease derived from Raman spectroscopy and prediction methods. EMBO J. 1985;4:3625–3631. doi: 10.1002/j.1460-2075.1985.tb04126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Weinglass A, Whitelegge JP, Faull KF, Kaback HR. Monitoring conformational rearrangements in the substrate-binding site of a membrane transport protein by mass spectrometry. J. Biol. Chem. 2004;279:41858–41865. doi: 10.1074/jbc.M407555200. [DOI] [PubMed] [Google Scholar]
- 110.Weinglass AB, Sondej M, Kaback HR. Manipulating conformational equilibria in the lactose permease of Escherichia coli. J. Mol. Biol. 2002;315:561–571. doi: 10.1006/jmbi.2001.5289. [DOI] [PubMed] [Google Scholar]
- 111.Weinglass AB, Whitelegge JP, Hu Y, Verner GE, Faull KF, Kaback HR. Elucidation of substrate binding interactions in a membrane transport protein by mass spectrometry. EMBO J. 2003;22:1467–1477. doi: 10.1093/emboj/cdg145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.West IC. Lactose transport coupled to proton movements in Escherichia coli. Biochem. Biophys. Res. Commun. 1970;41:655–661. doi: 10.1016/0006-291x(70)90063-x. [DOI] [PubMed] [Google Scholar]
- 113.West IC, Mitchell P. Proton-coupled β-galactoside translocation in non-metabolizing Escherichia coli. J. Bioenerg. 1972;3:445. doi: 10.1007/BF01516082. [DOI] [PubMed] [Google Scholar]
- 114.West IC, Mitchell P. Stoichiometry of lactose-H+ symport across the plasma membrane of Escherichia coli. Biochem. J. 1973;132:587–592. doi: 10.1042/bj1320587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Whitelegge JP, Gundersen CB, Faull KF. Electrospray-ionization mass spectrometry of intact intrinsic membrane proteins. Protein Sci. 1998;7:1423–1430. doi: 10.1002/pro.5560070619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Whitelegge JP, le Coutre J, Lee JC, Engel CK, Privé GG, et al. Toward the bilayer proteome, electrospray ionization-mass spectrometry of large, intact transmembrane proteins. Proc. Natl. Acad. Sci. USA. 1999;96:10695–10698. doi: 10.1073/pnas.96.19.10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wolin C, Kaback HR. Estimating loop-helix interfaces in a polytopic membrane protein by deletion analysis. Biochemistry. 1999;38:8590–8597. doi: 10.1021/bi990650j. [DOI] [PubMed] [Google Scholar]
- 118.Wolin CD, Kaback HR. Thiol cross-linking of transmembrane domains IV and V in the lactose permease of Escherichia coli. Biochemistry. 2000;39:6130–6135. doi: 10.1021/bi0001269. [DOI] [PubMed] [Google Scholar]
- 119.Wolin CD, Kaback HR. Functional estimation of loop-helix boundaries in the lactose permease of Escherichia coli by single amino acid deletion analysis. Biochemistry. 2001;40:1996–2003. doi: 10.1021/bi0025767. [DOI] [PubMed] [Google Scholar]
- 120.Wright EM, Turk E. The sodium/glucose cotransport family SLC5. Pflugers Arch. 2004;447:510–518. doi: 10.1007/s00424-003-1063-6. [DOI] [PubMed] [Google Scholar]
- 121.Wrubel W, Stochaj U, Sonnewald U, Theres C, Ehring R. Reconstitution of an active lactose carrier in vivo by simultaneous synthesis of two complementary protein fragments. J. Bacteriol. 1990;172:5374–5381. doi: 10.1128/jb.172.9.5374-5381.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zen KH, McKenna E, Bibi E, Hardy D, Kaback HR. Expression of lactose permease in contiguous fragments as a probe for membrane-spanning domains. Biochemistry. 1994;33:8198–8206. doi: 10.1021/bi00193a005. [DOI] [PubMed] [Google Scholar]
- 123.Zhang W, Guan L, Kaback HR. Helices VII and X in the lactose permease of Escherichia coli: proximity and ligand-induced distance changes. J. Mol. Biol. 2002;315:53–62. doi: 10.1006/jmbi.2001.5206. [DOI] [PubMed] [Google Scholar]
- 124.Zhao M, Zen K-C, Hernandez-Borrell J, Altenbach C, Hubbell WL, Kaback HR. Nitroxide scanning electron paramagnetic resonance of helices IV, V and the intervening loop in the lactose permease of Escherichia coli. Biochemistry. 1999;38:15970–15977. doi: 10.1021/bi991754x. [DOI] [PubMed] [Google Scholar]
- 125.Zhao M, Zen K-C, Hubbell WL, Kaback HR. Proximity between Glu126 and Arg144 in the lactose permease of Escherichia coli. Biochemistry. 1999;38:7407–7412. doi: 10.1021/bi9906524. [DOI] [PubMed] [Google Scholar]