Abstract
The VRK1 protein kinase has been implicated as a pro-proliferative factor. Genetic analyses of mutant alleles of the Drosophila and Caenorhabditis elegans VRK1 homologs have revealed phenotypes ranging from embryonic lethality to mitotic and meiotic defects with resultant sterility. Herein, we describe the first genetic analysis of murine VRK1. Two lines of mice containing distinct gene-trap integrations into the Vrk1 locus were established. Insertion into intron 12 (GT12) spared VRK1 function, enabling the examination of VRK1 expression in situ. Insertion into intron 3 (GT3) disrupted VRK1 function, but incomplete splicing to the gene trap rendered this allele hypomorphic (∼15% of wild-type levels of VRK1 remain). GT3/GT3 mice are viable, but both males and females are infertile. In testes, VRK1 is expressed in Sertoli cells and spermatogonia. The infertility of GT3/GT3 male mice results from a progressive defect in spermatogonial proliferation or differentiation, culminating in the absence of mitotic and meiotic cells in adult testis. These data demonstrate an important role for VRK1 in cell proliferation and confirm that the need for VRK1 during gametogenesis is evolutionarily conserved.
Keywords: gametogenesis, infertility, kinases, proliferation, protein kinase, signal transduction, spermatogenesis, spermatogonia, testis, VRK1
Depletion of the VRK1 protein kinase in mice causes severe infertility characterized by a progressive and profound loss of proliferating spermatogonia.
INTRODUCTION
Spermatogenesis is capable of producing as many as 100 million spermatozoa per day in humans and up to 1.6 billion sperm per day in pigs, making the testes one of the most highly proliferative adult tissues [1]. This high rate of cell division is a vital biological process, and perturbations to its regulation have serious consequences. Errors in spermatogonial mitosis can alter the delicate balance between self-renewal and differentiation needed for long-term gametogenesis, thus resulting in infertility. Mitotic errors may also lead to aneuploidy as well as to chromosomal breakage and instability, all of which are hallmarks of cancer cells. To avoid the propagation of such errors, checkpoints capable of halting cell growth are present throughout the cell cycle. Many of these checkpoints and, indeed, many normal cell-cycle transitions are regulated by dynamic protein phosphorylation. Given the importance of phosphorylation in orchestrating the events of the cell cycle, understanding the vital contributions of protein kinases to the regulated cell proliferation that drives gametogenesis and the disregulated cell proliferation that drives oncogenesis is of fundamental importance.
Recent evidence indicates that the vaccinia-related kinase-1 (VRK1) has roles in regulating gametogenesis and may contribute to tumor formation [2–8]. VRK1 overexpression has been associated with several types of cancer [4], and most recently, the overexpression of VRK1 has been highlighted as part of a gene expression signature that is prognostic of poor clinical outcome in patients with breast cancer [9]. VRK1 is one of three VRK family members that branch from the casein kinase family. The naming of the VRK genes reflects the high degree of homology between the VRK proteins and the B1 protein kinase encoded by vaccinia virus, the prototypic poxvirus and the virus used in the successful vaccination campaign to eradicate smallpox [10]. VRK1 and VRK2 cDNAs were first isolated from a library enriched for fetal-specific genes, and Northern blot analysis indicated that the mRNAs for both transcripts were found at elevated levels in highly proliferative tissues [10]. VRK3 was identified later [11], and its mRNA transcript also appears to be broadly expressed. The three proteins show a significant degree of homology within their catalytic domains, but they contain distinct extracatalytic domains that mediate differing patterns of intracellular localization and may contribute to substrate specificity. VRK1 and VRK2 are active enzymes that localize to the nucleus and intracellular membranes [12], respectively, and VRK3 is a catalytically inactive pseudokinase that likely functions as a scaffold protein [12–14]. These three paralogs are present in the genomes of vertebrates, whereas Drosophila melanogaster and Caenorhabditis elegans each encode a single VRK that has been shown to play essential roles in mitosis and meiosis [15, 16]. VRK orthologs are not present in budding or fission yeast.
VRK1 has been implicated in modulating cell growth via multiple intracellular signal transduction cascades (for review, see [17]). A variety of stress-responsive transcription factors, such as TP53, ATF2, and JUN, can be phosphorylated and stimulated by VRK1 [3, 5, 6]. It has also been reported that VRK1 can phosphorylate histone H3 on Thr3 and Ser10 [18]; these modifications are associated with the condensation of mitotic chromatin. In addition, VRK1 has been shown to phosphorylate CREB, a transcriptional activator that in turn controls the expression of the key cell-cycle regulator cyclin D [19]. Finally, the barrier to autointegration factor (BAF), which plays an important role in nuclear reassembly late in mitosis, was shown to be a highly efficient substrate for VRK1 in vitro and in vivo. Phosphorylation of BAF blocks its ability to bind to DNA and inhibits its interaction with protein partners; in vivo, phosphorylation of BAF is also associated with its exit from the nucleus [12, 20].
Genetic studies examining VRK mutants in D. melanogaster and C. elegans have revealed phenotypes ranging from embryonic lethality in the most severely affected organisms to sterility in animals carrying hypomorphic alleles [15, 16]. Herein, we describe the first genetic analysis of VRK1 in a mammal by characterizing two lines of mice containing gene-trap insertions within the Vrk1 locus. These studies reveal that depletion of VRK1 leads to progressive male infertility as a result of a cessation of spermatogonial proliferation. These findings substantiate results found in lower eukaryotes and further establish the importance of VRK1 during cell proliferation in the testes.
MATERIALS AND METHODS
Animals
C57B/6 mice were purchased from Charles River Laboratories. All mice were maintained in the Medical College of Wisconsin animal colony under specific pathogen-free conditions in accordance with National Institutes of Health and institutional guidelines.
Generation and Evaluation of Vrk1 Chimeric Mice
The RRR178 and XH078 clones of mouse sv129/ola embryonic stem (ES) cells in which a β-galactosidase/neoR (βgeo) gene-trap cassette had been inserted into the Vrk1 locus were obtained from the Mutant Mouse Regional Resource Center, University of California at San Francisco. (For the positioning of these insertions within the Vrk1 locus, see Figs. 1A and 3A.) The RRR178 locus has the official symbol of Vrk1Gt(RRR178)Byg and hereafter will be referred to as GT3. The XH078 locus has the official symbol of Vrk1Gt(X078)Byg and hereafter will be referred to as GT12. The Medical College of Wisconsin Transgenic Core Facility was employed to generate chimeric mice by injection of these ES cells into C57BL/6 blastocysts, which were subsequently implanted into pseudopregnant female CD1 mice. Male chimeras were mated to C57BL/6 females; agouti Aw pups born from these matings were further characterized to monitor germline transmission of the gene-trapped locus. Inheritance of the gene-trapped GT12 locus could be verified by assaying tail tips for β-galactosidase (β-gal) activity (Xgal staining). Because the VRK1-GT3βgeo fusion protein was not active in this assay, mice carrying the GT3 locus were genotyped by Southern blot and PCR as described later.
Genotyping
Genomic DNA from targeted ES cells or from mouse tail tips was harvested by immersion of the cells or tissue in lysis buffer (4 M urea, 0.5% Sarkosyl [Sigma], 10 mM cyclohexane diamine tetraacetic acid, 0.1 M Tris [pH 8], 0.2 M NaCl, and 1 mg/ml of proteinase K) followed by isopropanol precipitation. DNA was analyzed using standard Southern blot protocols employing probes corresponding to exons 2–6 or 11–15, as shown in Supplemental Figures S1 and S2 (all Supplemental Data are available online at www.biolreprod.org). Further Southern blot analysis of the GT12 locus was performed using probes 1 and 2, which map within intron 12 of the Vrk1 gene or βgeo, respectively. Probe 1 was amplified from genomic DNA using primers Ex12ScreenUP (5′-TAGCTGCCTGCGGGGTGTCG-3′) and Ex12ScreenDN (TGGGAGGCAGAGGCAAATGAATC-3′); probe 2 was amplified from genomic DNA using primers βgeoUP1 (5′-TATACGAAGTTATCGCAGATCTGGAC-3′) and βgeoDN1 (5′-AAATGTGAGCGAGTAACAACCCGTC-3′).
PCR Screen
The precise site of the gene-trap insertion in the Vrk1 GT3 mice was determined by sequence analysis of a PCR product amplified from genomic DNA with the Ex3ScreenUP primer (5′-TCTGCCTGATGTTGTGATGAG-3′), which anneals to sequences within Vrk1 intron 3, and the βgeoDN2 primer (5′-TGGCCTGTCCCTCTCACCTTC-3′), which anneals to sequences within the βgeo gene-trap cassette. The site of the GT12 insertion was determined similarly, using the Ex12UP (5′-GACATGGAGTGCTCAGACAC-3′) primer, which anneals within intron 12 of Vrk1, and the βgeoDN2 primer. Subsequent genotyping of Vrk1 GT3 mouse tail tip DNA was performed using the primers Ex3UP (5′-ACTGATGGGGTCTGTGTTTC-3′) and Ex3DN (5′-GGAGTTACTTCTTGGCGCTG-3′), which span the insertion site, and the βgeo-specific primers βgeoUP1 and βgeoDN1.
Xgal Staining of Frozen Sections
Adult testes were fixed for 1.5 h in 4% paraformaldehyde and saturated stepwise in PBS with 10%, 30%, and then 50% sucrose. Tissues were then immersed in OCT medium, frozen in liquid nitrogen, and cut in 10-μm sections that were mounted on slides for storage at −20°C. Slides were warmed to room temperature and then fixed in 0.2% paraformaldehyde for 10 min on ice. They were then washed briefly in wash buffer (0.1 M sodium phosphate [pH 7.4], 0.02% NP40, 0.01% deoxycholate, and 2 mM MgCl2) and overlayed with β-gal stain (wash buffer plus 1 mg/ml of Xgal, 5 mM potassium ferrocyanide, and 5 mM potassium ferricyanide). Testis sections shown were stained for 3 h in a humid chamber at 37°C.
Immunohistochemistry
Tissues were harvested and fixed overnight in zinc formalin, embedded in paraffin, and cut into 4-μm sections. Sections were dewaxed in Citrisolv (Fisher) and rehydrated stepwise through decreasing concentrations of ethanol. Antigen retrieval was performed in 10 mM citric acid buffer (pH 6.0) for 10 min in a 95°C water bath. Primary antibody incubations using the mouse anti-PLZF (sc-28319), mouse anti-PCNA (anti-proliferating cell nuclear antigen; sc-56), or goat anti-c-Kit (sc-1494) were performed overnight (Santa Cruz Biotechnology). The rat GCNA (germ cell-specific nuclear antigen) monoclonal antibody (kindly provided by Dr. George Enders, University of Kansas Medical Center, Kansas City, KS) was used at a 1:100 dilution in an overnight incubation. When mouse primary antibodies were employed, the biotin-conjugated secondary antibody included in the M.O.M. Immunodetection Kit (Vector Laboratories) was used. For the GCNA staining, a biotin-conjugated goat anti-rat secondary antibody (559286; BD Biosciences) was used. For the c-Kit staining, a biotin-conjugated rabbit anti-goat secondary antibody (BA-5000; Vector Laboratories) was used. Staining was then visualized following treatment with Elite ABC Reagent (Vector Laboratories) and incubation with diaminobenzidine reagent. All images shown are representative of sections observed from at least two mice; quantitation was done by counting stained cells within each tubule (n value shown) of a random field.
Semiquantitative RT-PCR
Tissues were harvested from wild-type (+/+) and Vrk1 GT3/GT3 mice of the ages indicated. Total RNA was isolated using RNeasy Plus spin columns (Qiagen). Spleen samples were subjected to an additional on-column DNase treatment. RT-PCR was performed on 2 μg of total RNA using Superscript III Reverse Transcriptase (Invitrogen) and random primers according to the manufacturer's specifications. Equivalent cDNA volumes were used as template in semiquantitative PCR reactions containing [α-32P]dATP (1.6 μCi/nmol). Reactions were performed using the following Vrk1-specific primers: upstream: US1, 5′-GAAGTTCGTACTACGCTGTGGACC-3′, and US2, 5′-CACTTTCACAACACAGGGCG-3′; downstream: DS1, 5′-GACGCGCCCTGTGTTGTGAAA-3′, and DS2, 5′-GTACCGATAAGCAAGGCCATAGTCTA-3′. Amplification of β-actin sequences was performed using primers β-Actin AS, 5′-CTAGAAGCATTGCGGTGGACGATGG-3′, and β-Actin S, 5′-TGACGGGGTCACCCACACTGTGCCC-3′. The upstream primers amplify sequences found in both wild-type and gene-trapped transcripts, whereas the downstream primers only amplify sequences found in full-length Vrk1 mRNAs. The latter primers provide an estimate of gene-trap “splice-around” activity in the GT3 mice. The number of amplification cycles for Vrk1 and β-actin genes was chosen to fall into the exponential phases of amplification (25 and 27 cycles, respectively). The level of amplified products was quantitated by PhosphorImager analysis using ImageQuant software. For each tissue, a ratio of the resulting PCR products, (wtD/GT3D)/(wtU/GT3U), was calculated to assess the reduction of full-length mRNA in the GT3 mice versus the wild-type mice. For amplification of Kit ligand transcript, the primers used were KLUS, 5′-CCCTGAAGACTCGGGCCTA-3′, and KLDS, 5′-CAATTACAAGCGAAATGAGAGCC-3′.
Immunoblot Analysis
Freshly harvested tissues were lysed in ice-cold extraction buffer (20 mM Tris [pH 7.4], 100 mM NaCl, 1% TX-100, 0.05% SDS, and 2 mM MgCl2) containing 1 μg/ml of pepstatin, 1 μg/ml of leupeptin, 1 mM PMSF, 1 mM NaF, 5 nM calyculin A, and 25 U/ml of Benzonase (Sigma) for 15 min. NaCl was then added to a final concentration of 500 mM, and incubation was continued for 15 min, after which the sample was clarified by sedimentation at 14 000 × g for 10 min. The concentration of total protein in the clarified lysates was determined using the BCA Protein Assay Kit (Thermo Scientific).
One-hundred micrograms of each lysate were subjected to immunoblot analysis with the following antibodies: a rabbit polyclonal antiserum raised against a peptide corresponding to amino acids 3–18 of mouse VRK1 (Bethyl), rabbit α-VRK1 (Sigma), mouse α-PCNA (Santa Cruz Biotechnology), or rabbit α-histone H3 phosphoSer10 (Upstate). Quantitation of the immunoreactive signals was performed using an AlphaImager documentation system and FluorChem 8900 software.
RESULTS
VRK1 Expression Within Testis Is Limited to Sertoli Cells and Spermatogonia
To characterize the role of VRK1 in vivo, we obtained two gene-trapped lines of murine ES cells from Baygenomics (now available from the International Gene Trap Consortium) with the goal of producing gene-targeted mice. These ES lines each contain a βgeo insertion, the location of which has been mapped by 5′ rapid amplification of cDNA ends (RACE) analysis to different introns of the murine Vrk1 locus on mouse chromosome 12 (Fig. 1; see also Fig. 3). The splice acceptor site at the 5′ end of the βgeo sequences and the poly-A addition site at the 3′ end lead to the expression of a fusion mRNA that initiates with the first exon of Vrk1 and terminates after the βgeo sequences. The first ES cell line we employed (ID: XH078) contains an insertion within intron 12 of Vrk1; this locus will be referred to as GT12. The VRK1-βgeo fusion protein that would be expressed from this locus lacks only 10–54 (depending on the isoform [12]) of the 441 amino acids found in the full-length VRK1 protein. This fusion protein retains the kinase domain and the nuclear localization signal and, thus, likely mimics the wild-type protein in both function and intracellular localization.
FIG. 1.
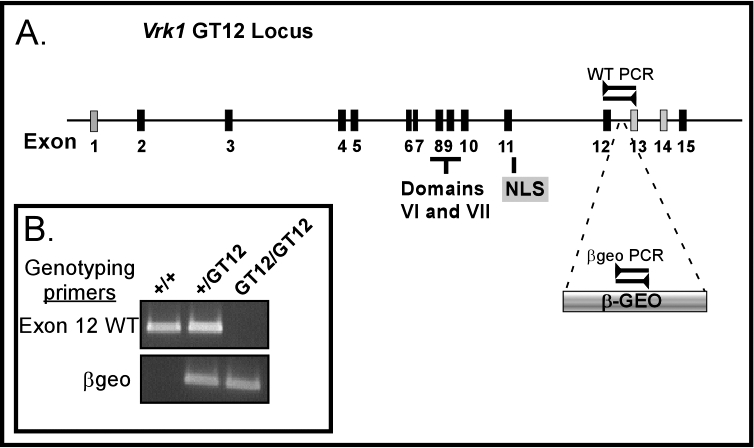
Genotypic analysis of the murine Vrk1 locus; confirmation of gene-trap insertion within intron 12. A) Schematic representation of the gene-trapped GT12 Vrk1 locus is shown, with numbered boxes representing the exons. Exon 1 is not translated, exons 13 and 14 are excluded from some splice variants, and exons 2–12 and 15 are present in all isoforms. Exons 8 and 9 encode key catalytic residues (kinase domains VI and VII) and exon 11 the nuclear localization signal (NLS), as noted. The position of the GT12 βgeo insertion is shown. PCR products used to distinguish the wild-type (WT) locus from one containing the β-Geo insertion are indicated. B) PCR genotyping of tail tip DNA. WT primers span the site of the GT12 insertion (as shown in the above schematic, A) and only amplify a product from the WT Vrk1 allele. Amplification of a product with the βgeo primers, which lie within the gene-trap insertion, confirms the presence of a gene-trapped allele.
Germline-competent chimeric mice were generated following blastocyst injection with this line of ES cells, and we established colonies of heterozygous +/GT12 mice. These mice had a normal appearance and were fertile. Initial genotyping of these mice was accomplished by Southern blot analysis, which verified the insertion of tandem, back-to-back, βgeo insertions within intron 12 (Supplemental Fig. S1). Subsequently, we mapped the precise location of the 5′ end of the βgeo insertion to +312 of intron 12, which allowed us to use PCR to rapidly and accurately genotype subsequent generations of mice. Two primer pairs were used; primer pair WT (wild type) spans the insertion site and, under our experimental conditions, only yields an amplified product in wild-type or heterozygous mice (Fig. 1B, top). The βgeo primer pair amplifies a region of the gene-trap insert (Fig. 1B, bottom). Genotypic analysis of the F2 progeny from +/GT12 × +/GT12 matings yielded +/+, +/GT12, and GT12/GT12 pups in the predicted mendelian ratios. Moreover, the size, behavior, overall health, and life span of the GT12/GT12 mice did not differ from those of their wild-type littermates.
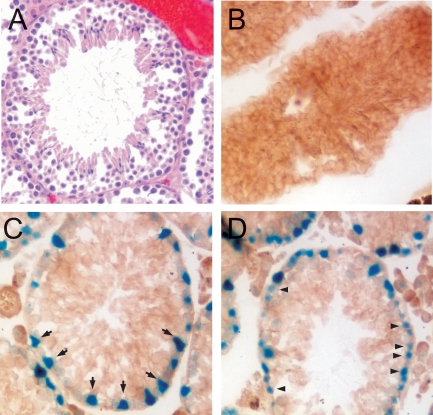
During initial examination of the VRK1-βgeo expression profile by Xgal staining, we found that intact testes harvested from GT12/GT12 mouse stained strongly blue, whereas wild-type testes exhibited no staining (data not shown). This confirms and extends previous studies indicating that murine Vrk1 mRNA is highly expressed in the testis, suggesting that VRK1 may have a role in murine gametogenesis [10]. As expected from the wild-type phenotype of the GT12/GT12 mice, the seminiferous tubules within these testes showed a normal histological organization and contained the full complement of characteristic cell types (Fig. 2A). Frozen sections of the +/+ and GT12/GT12 testes were stained with Xgal to reveal which cell types expressed the VRK1-βgeo fusion protein. No staining was observed in the +/+ samples (Fig. 2B). As seen in two representative GT12/GT12 tubules (Fig. 2, C and D), the Xgal staining was restricted to cells in the basal layer. This basal layer contains the spermatogonial stem cells (type A) and the type B spermatogonia to which they give rise. Also present are the large Sertoli cells, which give both mechanical support and signaling cues to most, if not all, cell types within the tubule. The Sertoli cell nuclei often adopt a somewhat triangular shape, whereas spermatogonia have significantly smaller, round nuclei. These distinctive nuclear morphologies allowed us to determine that both Sertoli cells (Fig. 2C, arrows) and spermatogonia (Fig. 2D, arrowheads) stained positive with Xgal, indicating that VRK1 is expressed in these cell types. We did not observe expression in meiotic cells, spermatids, or sperm. We further confirmed the identity of the stained cells by counterstaining the Xgal-treated sections with DAPI (data not shown), taking advantage of the distinctive nuclear staining of each of these cell types. (Sertoli cells have large, euchromatic nuclei that stain poorly with DAPI except for their distinctive nucleoli. Spermatogonia have smaller, round nuclei with multiple heterochromatic regions.)
FIG. 2.
VRK1 is expressed in the basal layer of the seminiferous tubule in both Sertoli cells and spermatogonia. A) Representative tubule within a GT12/GT12 testis, following formalin fixation and H&E staining, is shown. The GT12/GT12 tubules exhibit wild-type morphology and cellular content. B–D) Frozen sections of testes dissected from 11-wk-old mice were assayed in situ for β-gal activity. A wild-type tubule (B) and GT12/GT12 tubules (C and D). Arrows shown in C point to some of the β-gal+ Sertoli cells. Arrowheads shown in D highlight some of the β-gal+ spermatogonia. Original magnification ×10.
GT3 Locus Is a Hypomorphic Allele that Leads to Reduced Expression of VRK1
The second ES cell line that we obtained from Baygenomics contains a βgeo insertion in intron 3 of VRK1 (ID: RRR178); this allele will be referred to henceforth as GT3. The GT3 locus thus expresses a protein containing only the N-terminal 72 amino acids of VRK1 fused to βgeo. Thus, in contrast to the nearly full-length VRK1 in the GT12 mice, the GT3 VRK1 protein would be highly truncated and catalytically inert, because it is missing all of the kinase domain except the Walker A and B boxes and is also missing the C-terminal region containing the nuclear localization signal (Fig. 3). These ES cells were also used to produce transgenic mice, which were subsequently bred to yield +/GT3 and GT3/GT3 progeny. As with the GT12 mice, initial genotyping of these mice was accomplished by Southern blot analysis, which verified the site of βgeo insertion (Supplemental Fig. S2). Subsequently, we used RACE analysis to map the precise location of the 5′ end of the βgeo insertion to +6174 of intron 3, which allowed us to genotype mice by PCR using primer pairs specific for the wild-type and βgeo locus (Fig. 3, A and B).
FIG. 3.
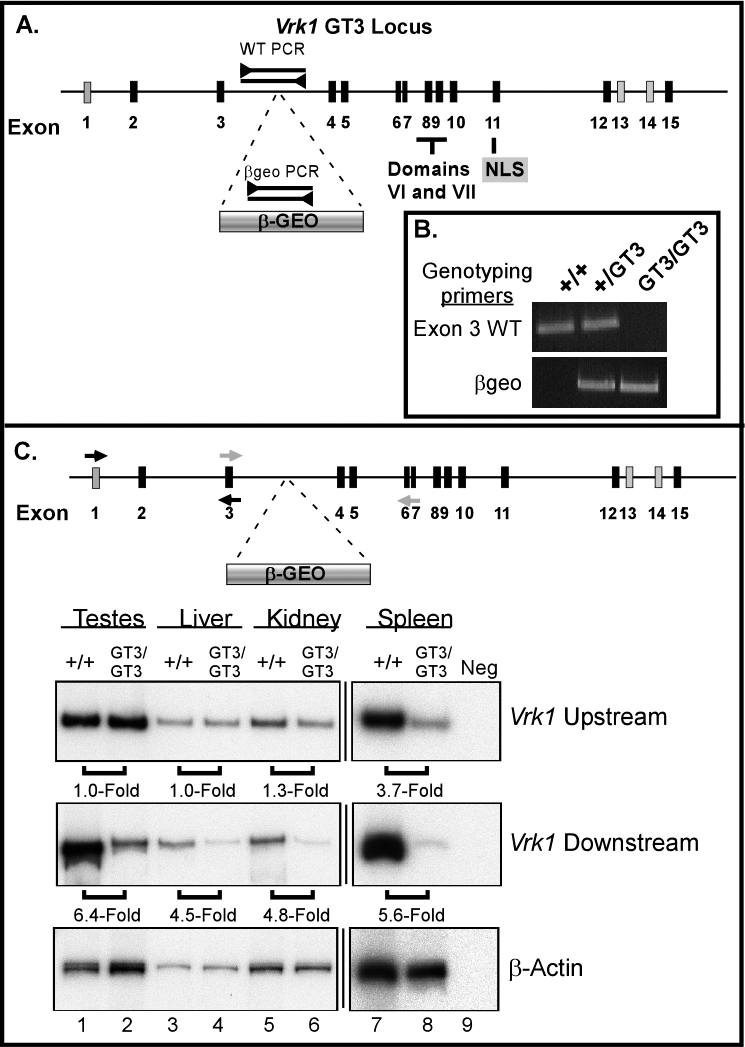
Genotypic and expression analysis of the GT3 Vrk1 locus. A) Schematic representation of the gene-trapped GT3 Vrk1 locus. The murine Vrk1 locus is shown, with numbered boxes representing the exons as described for Figure 1. The position of the GT3 βgeo insertion is shown. NLS, nuclear localization signal. B) Genotyping PCR. Wild-type (WT) primers span the site of the GT3 insertion (as shown above, A), only amplifying a product when a wild-type VRK1 allele is present. βgeo primers confirm the presence of a gene-trapped allele. C) RT-PCR. Tissues were harvested from 16-wk-old +/+ and Vrk1 GT3/GT3 mice. Vrk1-specific cDNA sequences (as well as a β-actin control) were amplified by semiquantitative RT-PCR using the primers shown in the schematic. The upstream primers (black arrows) amplify sequences found in both full-length and gene-trapped transcripts. The relative level of total Vrk1 mRNA in +/+ versus GT3/GT3 tissues was calculated; the fold-difference is shown beneath the appropriate lanes. The downstream primers (gray arrows) amplify sequences found only in full-length mRNAs and, hence, provide an estimate of the frequency with which the gene trap is spliced around in the GT3/GT3 mice. The fold-difference in the level of the full-length Vrk1 mRNA in +/+ versus GT3/GT3 tissues (shown beneath middle panel) was calculated after normalizing to the total Vrk1 mRNA levels found in each tissue.
Genotypic analysis of the F2 progeny from +/GT3 × +/GT3 matings yielded +/+, +/GT3, and GT3/GT3 pups in the predicted mendelian ratios; this conclusion was supported by chi-square analysis. Moreover, the size and life span of the GT3/GT3 mice did not differ from their wild-type littermates. These observations were somewhat surprising, because VRK is essential in both D. melanogaster and C. elegans [15, 16]. Mice encode two catalytically active VRK enzymes; therefore, redundancy between the functions of VRK1 and VRK2 might render the loss of VRK1 less detrimental. RT-PCR analysis of RNA extracted from liver, kidney, and testis indicated that VRK2 expression was not elevated in GT3/GT3 mice (not shown); nevertheless, endogenous levels of VRK2 might be sufficient to overcome the loss of VRK1.
Another explanation for the viability of GT3/GT3 mice was suggested by the observation that in many cases, gene-trapped mice retain some expression of the wild-type protein as a result of incomplete splicing to the βgeo insertion. In essence, the gene-trapped allele is often hypomorphic rather than null [21]. To examine the level to which GT3/GT3 mice express full-length VRK1, we performed semiquantitative RT-PCR of RNA extracted from multiple tissues. Two primer sets were employed. The Vrk1 upstream primer set (shown by black arrows in Fig. 3B) spans exons 1–3 and, thus, should amplify the same product from both the trapped Vrk1-βgeo transcript and any full-length Vrk1 transcript that is produced. In contrast, the Vrk1 downstream primer set (shown by gray arrows in Fig. 3B) spans exons 3–7 of Vrk1; because these exons are not present in the Vrk1-βgeo transcript, these primers will only yield an amplified product from full-length mRNA generated by “splicing around” the trap. Primers specific to β-actin were also used as an internal control.
Previous studies have indicated that Vrk1 transcript is ubiquitously expressed [22], and indeed, the upstream primers yielded a Vrk1 amplicon in all samples tested (Fig. 3B, top). However, the level of Vrk1 expression appeared to vary between the different tissues harvested from wild-type mice. Highly proliferative tissues, such as the testis and spleen, contained more Vrk1 mRNA than nonproliferative tissues, such as the liver and kidney (Fig. 3B, top, compare lanes 1 and 7 to lanes 3 and 5). The fact that Vrk1 exhibits increased expression in highly proliferative tissues was intriguing in light of its previously attributed roles in mitosis and meiosis in D. melanogaster, C. elegans, and tissue culture cells [15, 16, 23].
For most tissues, no difference was found in the upstream amplicon levels between the +/+ and GT3/GT3 samples (Fig. 3, top, compare lanes 1 and 2, lanes 3 and 4, and lanes 5 and 6), indicating that expression of the Vrk1 mRNA is unchanged by the βgeo insertion. In contrast, we consistently observed an approximately 3.7-fold decrease in amplicon levels in the spleen samples harvested from GT3/GT3 mice. This finding suggests that either transcription of the Vrk1 gene is inhibited by the βgeo insertion in the spleen or, more likely, that the cell types that express Vrk1 are partially depleted from the GT3/GT3 spleen.
The RT-PCR analyses performed with the Vrk1 downstream primers (Fig. 3C, middle) allowed us to quantitate the decrement in full-length Vrk1 mRNA in the GT3/GT3 tissues. After normalizing to the signal obtained with the upstream primers (total transcript levels), we calculated a deficiency in full-length Vrk1 mRNA that ranged from 4.5- to 6.4-fold (80–85% decrease) in the various adult tissues. Parallel studies were also performed using tissues from 4-wk-old mice. The amount of full-length VRK1 expressed in GT3/GT3 mice of this age was also significantly reduced compared to +/+ mice (Supplemental Fig. S3). These RT-PCR data indicate the GT3 allele is, indeed, hypomorphic and that GT3/GT3 mice retain approximately 15–20% of the wild-type levels of full-length Vrk1 mRNA in their tissues. This expression profile was also observed at the protein level, as shown by immunoblot analysis (Supplemental Fig. S4). (The absolute levels of full-length Vrk1 mRNA are lowest in the GT3/GT3 spleen [20-fold reduction relative to +/+]. As described earlier, we believe that the observed loss of total Vrk1 transcripts in this tissue likely results from a depletion of Vrk1-expressing cells in the GT3/GT3 spleen.)
Male and Female GT3/GT3 Mice Are Infertile
As mentioned earlier, matings of +/GT3 with +/GT3 mice, and of +/GT12 with +/GT12 mice, yielded F2 progeny of all predicted genotypes in the correct mendelian ratios. Thus, both gene-trapped alleles support viability in the homozygous state. Matings of GT12/GT12 × GT12/GT12 mice were productive, and we have maintained a colony of GT12/GT12 mice over many generations. Thus, the GT12 allele supports both viability and fertility in the homozygous state.
In contrast, the GT3 allele had quite a different impact. No litters were obtained from multiple matings of GT3/GT3 males with GT3/GT3 females, although copulation plugs were observed and matings of +/GT3 mice continued to be productive over many generations. To determine whether this fertility problem was gender specific, we mated nine GT3/GT3 males to +/+ females and nine GT3/GT3 females to +/+ males. These matings were initiated when the GT3/GT3 mice were 6–10 wk of age and were allowed to continue for 6–8 wk. From the matings set up with the nine GT3/GT3 male mice, we obtained only a single litter; one 6-wk-old GT3/GT3 male fathered a litter but could produce no further litters despite repeated attempts. None of the other eight GT3/GT3 males were fertile. In addition, none of the nine GT3/GT3 females produced any litters when mated to +/+ males. Thus, in the homozygous state, the GT3 allele renders both male and female mice infertile.
VRK1-Deficient Testes Display Progressive Loss of Mitotic and Meiotic Cells
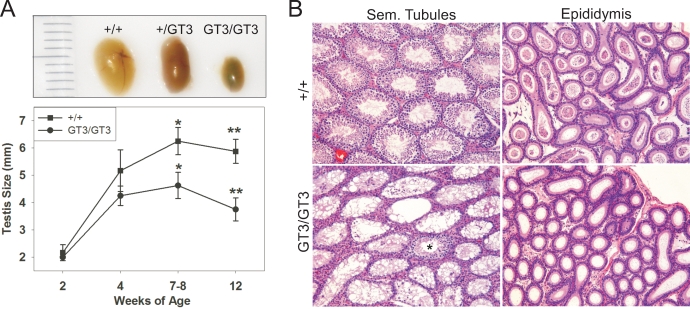
Both male and female GT3/GT3 mice are infertile, but in the present study, we focused on identifying the defect in males. As shown in Figure 4A (top), a gross analysis of the testes of adult +/+, +/GT3, and GT3/GT3 mice revealed a clear decrease in the size of the testis in the GT3/GT3 mice, although the overall body sizes of all mice were comparable. When this analysis was performed at various times of postnatal development, we observed little difference in testis size at 2 or 4 wk of age (Fig. 4A, bottom). However, a few weeks after puberty, the difference in size between the testes of +/+ and GT3/GT3 mice was readily observed and statistically significant (P < 0.03). The cellular basis for this difference in testis size became apparent upon histological examination. While hematoxylin and eosin (H&E)-stained sections of +/+ testes from 11- to 12-wk-old mice showed the expected complement of mitotic spermatogonia and meiotic cells in each tubule, the GT3/GT3 tubules appeared devoid of virtually all cell types with the exception of Sertoli cells (Fig. 4B, left). Furthermore, a stark difference was also seen in the epididymal tubules of these mice. Whereas the lumens of more than 90% of epididymal tubules of +/+ mice contained sperm, none of the tubule lumens in the 12-wk GT3/GT3 epididymis contained sperm.
FIG. 4.
The testes of adult GT3/GT3 mice are reduced in size and display a loss of cellularity. A, top) Photograph illustrating the relative size of testes from 12-wk-old +/+, +/GT3, and GT3/GT3 mice. A, bottom) Testes from mice of the indicated ages were measured lengthwise; data are depicted graphically. Error bars represent standard error between samples; at least five testes were measured for each age of mice. For the samples harvested at 7–8 and 12 wk, a statistically significant difference is found between the sizes of the +/+ and GT3/GT3 testes, as determined by the Mann-Whitney rank sum test. *P < 0.03, **P < 0.001. B) H&E-stained sections of the testis and epididymis taken from 12-wk-old +/+ and GT3/GT3 mice. With the rare exception (one example marked with asterisk), the seminiferous tubules in the GT3/GT3 testis lack most spermatogonia and all meiotic cells. Similarly, the lumens of the GT3/GT3 epididymis are devoid of sperm. Original magnification ×4.
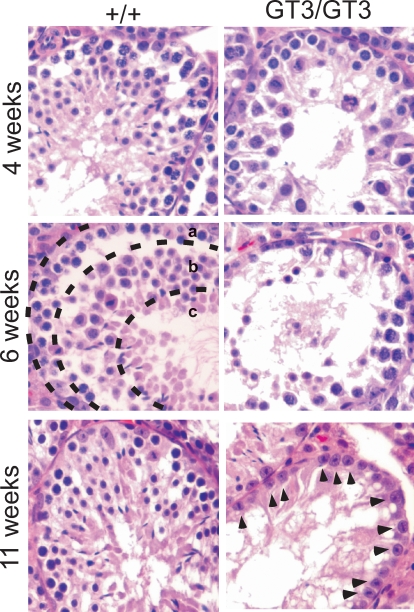
To determine the age of onset for this loss of cellularity, we examined testes from younger mice beginning at 2 wk of age. We found that +/+ and GT3/GT3 testes were indistinguishable at 2 wk (not shown) and 4 wk (Fig. 5) of age, before the onset of puberty. At 6 wk of age, GT3/GT3 tubules displayed a range of phenotypes. Some GT3/GT3 tubules, like their wild-type counterparts, contained spermatogonia, the full complement of cells in meiosis I, and round and elongated spermatids (marked as a, b, and c, respectively, in the +/+ sample shown in Fig. 5). However, many GT3/GT3 tubules displayed a striking loss of cellular density. As illustrated by the image in Figure 5, these tubules showed a clear reduction in the number of meiotic cells and spermatids. Some GT3/GT3 tubules (not shown) exhibited what appeared to be a “Sertoli Cell Only” phenotype even at this early age. Histological examination of epididymal sections from 6-wk-old mice showed that whereas sperm were observed in more than 90% of the lumens of +/+ samples, sperm were only seen in less than 20% of the lumens of GT3/GT3 samples (not shown). GT3/GT3 mice examined between the ages of 7–10 wk displayed an increasingly severe phenotype, with many tubules showing reduced cellularity and an absence of meiotic cells. By 11 wk, the GT3/GT3 tubules appeared to contain only Sertoli cells (Fig. 5, arrowheads), whereas the wild-type testes retained all cell types inherent to spermatogenesis.
FIG. 5.
VRK1-deficient testes show a progressive loss of cellularity and absence of spermatogenesis with increasing postnatal age. Sections of paraffin-embedded testes harvested from +/+ or GT3/GT3 mice at 4, 6, and 11 wk of age were stained with H&E. The image of the 6-wk-old +/+ tubule is subdivided into zones a, b, and c to indicate the location of spermatogonia/Sertoli cells, meiotic cells, and elongated spermatids, respectively. These cell types can also be found in some tubules of the 6-wk-old GT3/GT3 mice, albeit at lower numbers, but are absent in older GT3/GT3 mice. By 11 wk of age, only one basal layer of cells, which is largely comprised of Sertoli cells (arrowheads), remains in the GT3/GT3 sample. Original magnification ×20.
VRK1 Deficiency Results in Progressive Loss of GCNA+ and PCNA+ Spermatogonia
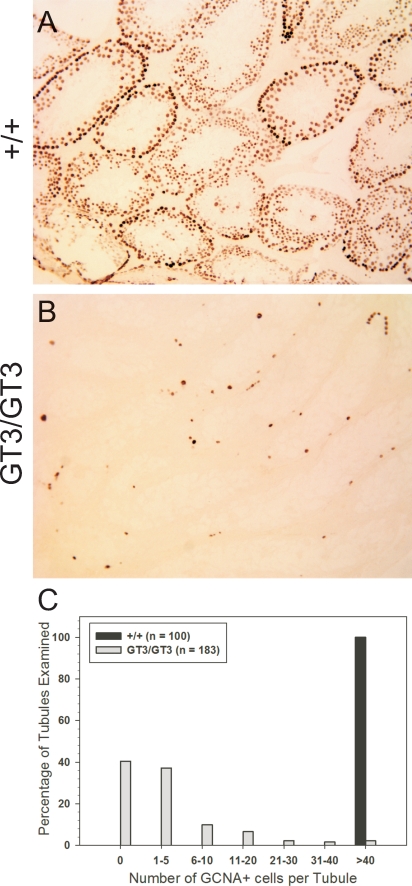
These data indicated a progressive defect in GT3/GT3 mice characterized by a loss of spermatogenesis and a reduction in the complexity of cell types within the seminiferous tubules. We next set out to define more precisely the stage of spermatogenesis that was affected by the VRK1 deficiency. To address this question, we performed immunohistochemical analyses of the seminiferous tubules using antibodies specific for GCNA, PCNA, and KIT. GCNA has been shown to be present in spermatogonial stem cells, mitotic spermatogonia, and cells in the prediplotene stages of meiosis I [24]. In +/+ testes, GCNA staining was strongest in the peripheral spermatogonia but also clearly present in cells undergoing meiosis I (Fig. 6A). In comparison, adult GT3/GT3 testes were largely devoid of GCNA+ tubules (Fig. 6B). Quantitation of the number of GCNA+ cells per tubule in +/+ and GT3/GT3 tubules revealed that approximately 80% of the GT3/GT3 tubules contained less than five cells per tubule, whereas 100% of the +/+ tubules had more than 40 GCNA+ cells per tubule (Fig. 6C). We next examined PCNA staining, which is a more specific marker of proliferating type A and B spermatogonia. Strong PCNA-staining of the rapidly proliferating spermatogonia was seen in the testes harvested from 4-wk-old mice of both the +/+ and GT3/GT3 gentotype (Fig. 7A, top). In adult +/+ mice (age, 14 wk), abundant PCNA+ cells continued to be observed (>40 PCNA+ cells/tubule), substantiating the ongoing proliferation of spermatogonia in these mice. However, PCNA+ cells were largely absent (<5 PCNA+ cells/tubule) from more than 90% of GT3/GT3 tubules (Fig. 7A, bottom, quantitated in Fig. 7B), revealing a severe deficit of proliferating spermatogonia in these VRK1-deficient testes. To further identify the stage at which spermatogenesis is blocked, we also utilized an antibody specific for KIT, a well-characterized marker of early spermatogonial differentiation [25–27]. Whereas KIT+ spermatogonia were clearly present in +/+ testis, adult GT3/GT3 tubules were devoid of any staining (Supplemental Fig. S5), confirming the absence of differentiating cells.
FIG. 6.
Vrk1 deficiency results in a loss of GCNA+ spermatogonia. Sections of paraffin-embedded testes from adult (age, 12 wk) mice were subjected to immunohistochemical analysis using an antibody specific for the germ cell nuclear antigen (GCNA). The GCNA staining was observed as expected in spermatogonia and cells proceeding through meiosis I in +/+ tubules (A), but very few cells in GT3/GT3 tubules (B) stained with the GCNA antibody. The number of GCNA+ cells per tubule was determined after examining many tubules (n value shown) from four testes of mice of each genotype (C). Original magnification ×10.
FIG. 7.
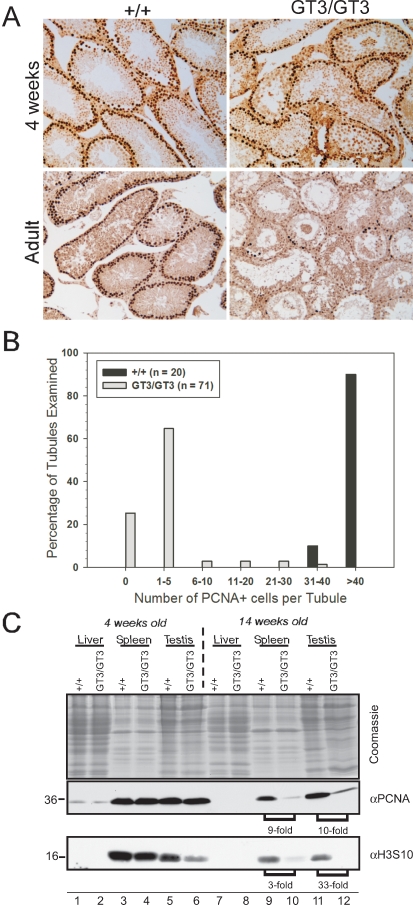
Vrk1 deficiency results in a progressive loss of markers of cellular proliferation. A) Sections of paraffin-embedded testes from 4-wk-old and adult (age, 14 wk) mice were subjected to immunohistochemical analysis using an antibody specific for PCNA. At 4 wk of age, PCNA staining was observed as expected in spermatogonia and early meiotic cells in both +/+ and GT3/GT3 tubules. However, by 14 wk, GT3/GT3 tubules contained very few PCNA+ cells. Original magnification ×10. B) The number of PCNA+ cells per tubule was determined after examining many tubules (n value shown) from four testes of adult mice of each genotype. C) Immunoblot analysis of proliferative markers. Liver, spleen, and testis were harvested from 4- and 14-wk-old mice. Whole-cell lysates were prepared, and 100 μg of protein were subjected to electrophoretic fractionation. Confirmation that the amounts of protein being analyzed were comparable was obtained by Coomassie blue staining (top), and the levels of PCNA and H3pser10 were visualized by immunoblot analysis. The fold-changes in proliferative markers between 14-wk-old +/+ and GT3/GT3 mice are depicted beneath the appropriate bracketed lanes.
To complement these histological analyses of GT3/GT3 tissues, we performed immunoblot analyses of +/+ and GT3/GT3 tissue lysates (Fig. 7C). Because VRK1 has been associated with cell proliferation, and because GT3/GT3 testes have shown a loss of PCNA+ cells, we probed these extracts with antibodies specific for the proliferative marker PCNA and the mitotic marker phosphohistone H3(Ser10). In the samples harvested from the 4-wk-old mice, PCNA levels did not vary between +/+ and GT3/GT3 samples, although a modest, but reproducible, diminution in the level of H3(Ser10) was observed in the GT3/GT3 testis. The results were far more striking in the samples harvested from adult mice. The levels of PCNA in the GT3/GT3 spleen and testis were approximately 10-fold lower than those in the +/+ samples (Fig. 7C, compare lanes 9 and 10 and lanes 11 and 12). These data correlate well with the dramatic loss of PCNA+ cells observed in GT3/GT3 testes by immunohistochemistry (Fig. 7A, bottom). The levels of H3(Ser10) expression were also reduced in GT3/GT3 tissues, being threefold lower in spleen and 33-fold lower in testis. Together, these data provide strong evidence that depletion of VRK1 in the tissues examined severely compromises cellular proliferation.
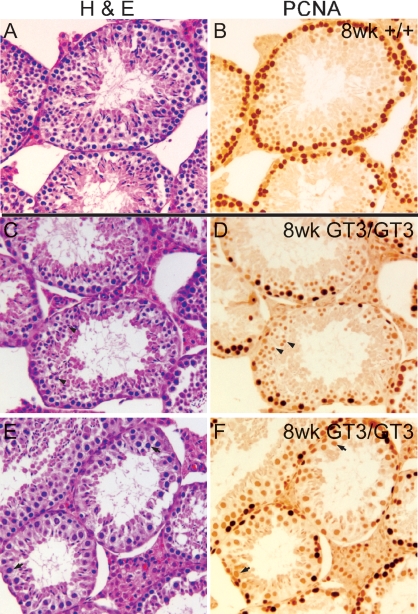
A block in the proliferation and/or differentiation of spermatogonia would explain the absence of meiotic cells and mature sperm in the adult GT3/GT3 mice. To further demonstrate that the loss of spermatogonia is an early defect in VRK1-deficient mice, we examined the tubules of 8-wk-old GT3/GT3 mice, which are at an intermediate stage in tubule degeneration. Many tubules were already devoid of spermatogonia. However, tubules were observed that contained only 10–20 PCNA+ spermatogonia but still contained elongated spermatids, round spermatids (Fig. 8, C and D, arrowheads), and cells in meiosis I (Fig. 8, E and F). Thus, in the tubules that were shown to still be supporting meiosis and spermiogenesis by H&E, a decline in PCNA+ cells was already evident (Fig. 8). These data argue that whereas the initial first wave of spermatogenesis occurs fairly normally in GT3/GT3 mice, a progressive loss of proliferating spermatogonia leads to a subsequent arrest in spermatogenesis and, therefore, to tubule collapse.
FIG. 8.
Proliferating spermatogonia are lost first during GT3/GT3 tubule degeneration. Serial sections of testes from 8-wk-old +/+ (A and B) and GT3/GT3 (C–F) mice were stained with H&E (A, C, and E) or subjected to immunohistochemistry using a PCNA-specific antibody (B, D, and F). Those GT3/GT3 tubules containing an intermediate number of PCNA+ cells were examined for the presence of other cell types and were found to contain cells in meiosis I (arrows; bottom), round spermatids (arrowheads; top) and elongated spermatids. Original magnification ×10.
GT3/GT3 Mice Retain ZBTB16+ (PLZF+) Spermatogonia
The loss of GCNA+, PCNA+, and KIT+ spermatogonia in GT3/GT3 mice led us to question whether a deficiency in VRK1 leads to the absence or loss of undifferentiated spermatogonial stem cells.To test this possibility, we examined ZBTB16 (PLZF), which is expressed in type A spermatogonia but is subsequently downregulated upon differentiation of these cells [28, 29]. Thus, ZBTB16 is a marker of spermatogonial stem cells and undifferentiated progenitor cells. As shown in Figure 9, at 4 wk of age ZBTB16-expressing cells were visible within the basal layer of the tubules of both wild-type and GT3/GT3 testes. In adult mice, ZBTB16+ spermatogonia remained present in testes of both genotypes. Quantitation of the number of ZBTB16+ cells at the periphery of each tubule revealed that GT3/GT3 testes have at least as many, and perhaps slightly more, ZBTB16+ cells per tubule as +/+ testes when normalized for the differences in tubule size between the genotypes (4.44 vs. 3.25, respectively; P = 0.02 by Mann-Whitney rank sum test). In sum, these data indicate that whereas active spermatogenesis occurs until puberty in GT3/GT3 mice, the numbers of dividing spermatogonia and meiotic cells drop precipitously in the first few weeks thereafter, resulting in a concomitant loss of more mature spermatids. However, because ZBTB16+ cells are not lost at these time points, the phenotype of VRK1-deficient testes clearly is not caused by a loss per se of the earliest spermatogonia. Rather, it may be that these stem cells cannot differentiate into the more rapidly dividing types of spermatogonia, or that these spermatogonia are unable to proliferate when VRK1 is depleted.
FIG. 9.
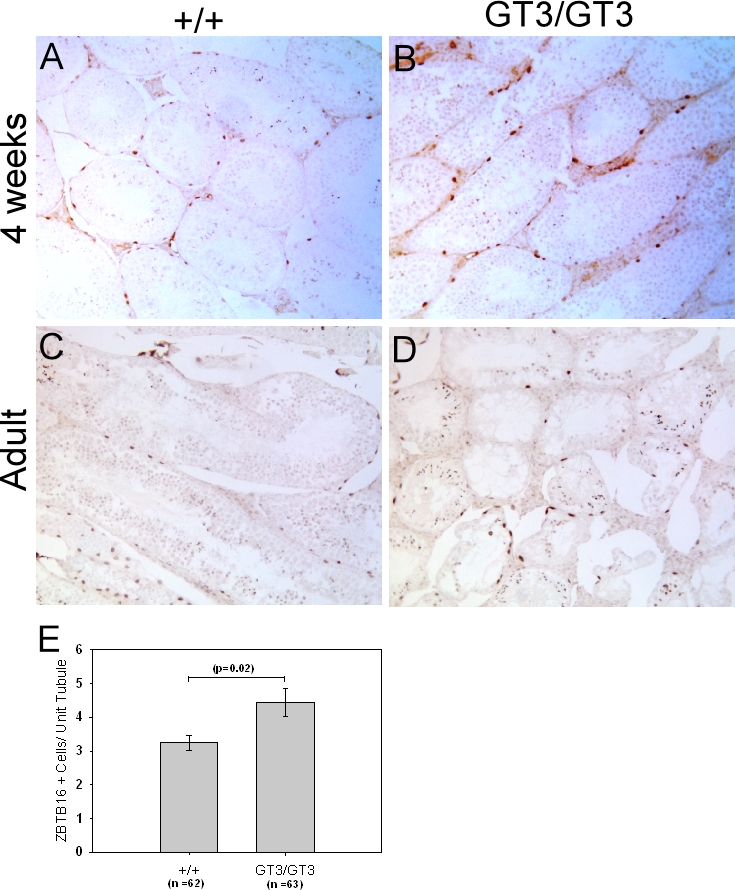
Vrk1-deficient mice retain early PLZF+ (ZBTB16+) spermatogonial stem cells. Sections of paraffin-embedded testes from 4-wk-old and adult (age, 14 wk) mice were subjected to immunohistochemical analysis using an antibody specific for the early spermatogonial marker Zbtb16. A and B) At 4 wks of age, ZBTB16 staining was observed as expected in a subset of peripheral spermatogonia in both +/+ and GT3/GT3 tubules. C and D) At 14 wk, ZBTB16+ cells continue to be observed in +/+ and GT3/GT3 spermatogonia, with some staining of the elongated spermatids as well. E) The number of ZBTB16+ cells per tubule was determined after examining tubules (n value shown) from four testes of adult mice of each genotype. Quantitation represents the number of ZBTB16+ cells per tubule, normalized to the relative area of the tubule being examined. Only ZBTB16+ cells at the basal aspect of the tubule were counted in this analysis. Original magnification ×10.
DISCUSSION
The mammalian vaccinia-related kinases (VRK1, VRK2, and VRK3) form a distinct branch of the casein kinase family. A single VRK homolog is encoded in the genomes of D. melanogaster and C. elegans, and the early developmental arrest that accompanies ablation of its expression indicates that this kinase is essential [15, 30]. With regard to dVRK, also known as NHK-1, the most severe alleles cause an embryonic lethal phenotype, whereas milder, hypomorphic alleles spare viability but cause male and female sterility [15, 31, 32]. Diminished NHK-1 function causes meiotic defects associated with aberrant chromatin modification, spindle formation, chromosome condensation, and karyomere formation. RNAi-mediated depletion of NHK-1 in cultured S2 cells has adverse effects on mitosis, leading to irregular chromosome condensation and to the presence of unaligned and lagging chromosomes [15]. Similar phenotypes have been observed for C. elegans, in which siRNA-mediated depletion of cVRK was shown to cause aberrant meiotic and mitotic spindles as well as altered chromatin organization [30].
In mammals, VRK1 is the ortholog most closely related to the D. melanogaster and C. elegans VRK enzymes, in that it is catalytically active and exhibits a nuclear localization. The present study represents to our knowledge the first genetic analysis of the function of VRK1 in mice. The GT3 allele contains a βgeo gene-trap insertion within intron 3 that should lead to the expression of a highly truncated fusion protein encoding only 72 amino acids of VRK1 fused to βgeo. As is often seen [21], splicing to the gene trap is incomplete, and approximately 15–20% of the Vrk1 transcripts splice around the trap and express full-length VRK1. Thus, the GT3/GT3 mice are, in fact, hypomorphic for VRK1.
Examination of hypomorphic models has been extremely useful for highlighting cell types and pathways that are most sensitive to the loss of a protein while leaving many other tissues unaffected. For example, complete disruption of the LKB1 and PDK1 kinases causes embryonic arrest, but hypomorphic (10–20% of wild-type levels) expression of these kinases is sufficient to support development and allow further study of the kinases [33, 34]. Likewise, our work shows that diminished VRK1 expression has no apparent impact on development, is compatible with viability, and is well tolerated by many tissues and organs. Given the demonstration that VRK is essential in other model organisms, we believe that the hypomorphic nature of the GT3 allele enabled us to avoid an embryonic lethal phenotype and uncover the vital role of VRK1 in spermatogenesis. The fact that the mice are viable and healthy may suggest VRK1 is less important in other tissues, perhaps because of redundancy within the VRK family or with other Ser/Thr protein kinases. The protein most similar to VRK1 is VRK2, which may be able to fulfill some of the roles played by VRK1. Interestingly, a possible role for VRK2 itself in spermatogenesis was suggested by the fact that Pog −/− mice, which lack the Pog (proliferation of germ cells) gene, have a milder infertility defect than gsd (germ-cell deficient) mice, which lack both Pog and the adjacent Vrk2 gene [35].
Our observation that VRK1 was expressed in Sertoli cells and spermatogonia extends an initial report of human VRK1 expression within seminiferous tubules [36]. Our expression data, combined with the infertility of GT3/GT3 mice, led us to examine the impact of VRK1 deficiency on spermatogenesis. Gross examination of GT3/GT3 testes indicated that in postpubertal mice, these testes were significantly smaller than those of their +/+ counterparts. Subsequent histological analysis of these testes was extremely informative, because the cell types responsible for spermatogenesis can be readily observed in each seminiferous tubule. Briefly, undifferentiated (type A) spermatogonial stem cells are found at the periphery of the tubule along with the Sertoli cells. Spermatogonial stem cells are capable of both self-renewal and differentiation; expression of the transcription factor ZBTB16 is a defining characteristic of these cells [28, 29]. Differentiation produces type B spermatogonia; these cells express markers such as KIT and PCNA and form the population that will undergo meiosis. As the early meiotic cells known as primary spermatocytes become secondary spermatocytes and then spermatids, they move continually closer to the lumen of the tubule. By determining which cell types were absent in the GT3/GT3 testes, we sought to pinpoint the steps at which VRK1 expression was critical. In young males 2–6 wk of age, little difference could be observed between the +/+ and GT3/GT3 testes. Both +/+ and GT3/GT3 mice possessed all cell types expected to be present at a particular time in development, leading us to conclude that VRK1 deficiency does not impede the first postnatal wave of spermatogenesis that occurs during this time. However, during the next few weeks, the GT3/GT3 mice exhibit a progressive loss of GCNA+, PCNA+, and KIT+ premeiotic cells. As a result, all later stages of meiotic cells were also progressively depleted. However, the pool of undifferentiated ZBTB16+ cells remained constant over this time period. Thus, VRK1 is not necessary to maintain the earliest spermatogonial lineage, unlike the transcriptional repressor ZBTB16 [28, 29]. Therefore, we postulate that VRK1 is needed either for the differentiation of type A spermatogonial stem cells into type B spermatogonia, the ongoing proliferation of type B cells required for the homeostasis of the precursor population, or both. In this regard, the phenotype of VRK1-deficient mice is similar to that of mice lacking either the Sohlh-1 or -2 transcriptional regulators; the testes of mice lacking these proteins retain ZBTB16+ cells but lack other differentiated cell types [37, 38]. However, whereas the expression of Sohlh-1 or -2 is limited to spermatogonia, VRK1 appears to be expressed both in Sertoli cells and in the basal layer of spermatogonia. This expression profile indicates that the loss of the differentiating pool of spermatogonia in VRK1-deficient testes might be the result of a cell-autonomous role for VRK1 in spermatogonial differentiation and/or proliferation. Alternatively, wild-type levels of VRK1 expression in the supporting Sertoli cells might be needed for them to provide the requisite signals to the spermatogonia. Such a requirement has been found for the Sertoli-specific factors ERM and Kit ligand (KITL), the absence of which can lead to a “Sertoli Cell Only” phenotype in the seminiferous tubules of adult mice [39–41]. Distinguishing between these two possibilities is an important goal for future studies. However, semiquantitative RT-PCR analysis revealed no significant difference in the expression of KL between +/+ and GT3/GT3 testis (Supplemental Fig. S6), suggesting that the VRK1-deficient Sertoli cells retain the ability to express at least this critical paracrine factor.
On a molecular level, the impact of VRK1 depletion likely is multifactorial. The VRK1/BAF pathway has been examined in mammalian tissue culture and has been documented via genetic analysis of model organisms [16, 23]. VRK1-mediated phosphorylation of BAF inhibits its ability to bind DNA and interact with protein partners. Thus, dynamic phosphorylation of BAF likely is a positive regulator of nuclear envelope breakdown and a negative regulator of nuclear envelope reformation and of BAF-mediated recruitment of chromatin to the nuclear periphery. Indeed, in C. elegans, VRK1 depletion has been shown to reduce BAF phosphorylation and to cause defects in nuclear envelope reformation at the end of mitosis. Aberrant nuclear envelope structures were also observed upon expression of a BAF mutant that exhibited reduced phosphorylation in comparison to the wild-type protein, providing correlative evidence that the VRK1-BAF pathway is causing the observed phenotype. In D. melanogaster, comparable sterility phenotypes are seen upon depletion of VRK1 or expression of a nonphosphorylatable variant of BAF [23]. In both cases, formation of the meiotic karyosome is impeded by aberrant recruitment of the chromosomes to the nuclear periphery. It seems highly likely that the VRK/BAF axis will be evolutionarily conserved, and that future studies will confirm VRK1-mediated phosphorylation of BAF plays important roles in mitosis and meiosis by regulating nuclear envelope dynamics.
Murine VRK1 may exert a pro-proliferative role by phosphorylating other substrates in mitosis or, indeed, during distinct phases of the cell cycle. Phosphorylation of histone H3 has been reported; this phosphorylation would be likely to affect chromosome condensation during mitosis. VRK1 has recently been shown to interact with RAN and with polo-like kinase 3 [42, 43]. RAN not only is involved in nucleocytoplasmic transport but also plays an important role in mitotic spindle assembly and chromosome segregation [44]; PLK3 is implicated in a signaling cascade that affects microtubule dynamics [45] as well as Golgi fragmentation [46]. VRK1 has also been reported to phosphorylate and activate the transcription factor CREB, which stimulates the expression of cyclin D, an important mediator of the G1/S transition. This pathway may also be important in the pro-proliferative role of VRK1.
Although the present study is focused solely on the male infertility defect in the hypomorphic Vrk1 mice, it is also intriguing that VRK1 is highly expressed in the spleen and that the proliferative markers PCNA and phosphohistone H3(Ser10) are greatly reduced in the spleens of VRK1-deficient mice. These data suggest that other proliferative pathways, such as erythropoiesis, which occurs in the murine spleen, might be impaired in the VRK1-deficient mice. Moreover, our studies indicate that female mice homozygous for the hypomorphic Vrk1 allele are also infertile. It will be of great interest to determine the basis for this infertility. The studies described herein indicate that VRK1 plays essential and evolutionarily conserved roles in gametogenesis and fertility in worms, flies, and mammals. The VRK1-deficient line of mice that we have established will provide a fruitful system with which to examine the important contributions of this kinase to cell proliferation and gametogenesis.
Supplementary Material
Acknowledgments
We thank Stephen Duncan for his advice and guidance throughout this project, Lynn Gruman for assistance with the histological analyses, and Kathy Boyle for helpful comments during the preparation of the manuscript.
Footnotes
Supported, in part, by funds to P.T. from the Advancing a Healthier Wisconsin initiative at the Medical College of Wisconsin. M.S.W. was supported, in part, by a postdoctoral fellowship from the Region V “Great Lakes” RCE (NIH award 1-U54-AI-057153).
REFERENCES
- Oatley JM, Brinster RL.Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol 2008; 24: 263–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcia R, Lopez-Borges S, Vega FM, Lazo PA.Kinetic properties of p53 phosphorylation by the human vaccinia-related kinase 1. Arch Biochem Biophys 2002; 399: 1–5. [DOI] [PubMed] [Google Scholar]
- Lopez-Borges S, Lazo PA.The human vaccinia-related kinase 1 (VRK1) phosphorylates threonine-18 within the mdm-2 binding site of the p53 tumor suppressor protein. Oncogene 2000; 19: 3656–3664. [DOI] [PubMed] [Google Scholar]
- Santos CR, Rodriguez-Pinilla M, Vega FM, Rodriguez-Peralto JL, Blanco S, Sevilla A, Valbuena A, Hernandez T, van Wijnen AJ, Li F, de Alava E, Sanchez-Cespedes M, Lazo PA.VRK1 signaling pathway in the context of the proliferation phenotype in head and neck squamous cell carcinoma. Mol Cancer Res 2006; 4: 177–185. [DOI] [PubMed] [Google Scholar]
- Sevilla A, Santos CR, Barcia R, Vega FM, Lazo PA.c-Jun phosphorylation by the human vaccinia-related kinase 1 (VRK1) and its cooperation with the N-terminal kinase of c-Jun (JNK). Oncogene 2004; 23: 8950–8958. [DOI] [PubMed] [Google Scholar]
- Sevilla A, Santos CR, Vega FM, Lazo PA.Human vaccinia-related kinase 1 (VRK1) activates the ATF2 transcriptional activity by novel phosphorylation on Thr-73 and Ser-62 and cooperates with JNK. J Biol Chem 2004; 279: 27458–27465. [DOI] [PubMed] [Google Scholar]
- Valbuena A, Suarez-Gauthier A, Lopez-Rios F, Lopez-Encuentra A, Blanco S, Fernandez PL, Sanchez-Cespedes M, Lazo PA.Alteration of the VRK1-p53 autoregulatory loop in human lung carcinomas. Lung Cancer 2007; 58: 303–309. [DOI] [PubMed] [Google Scholar]
- Vega FM, Sevilla A, Lazo PA.p53 stabilization and accumulation induced by human vaccinia-related kinase 1. Mol Cell Biol 2004; 24: 10366–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KJ, Patrick DR, Bissell MJ, Fournier MV.Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS ONE 2008; 3: e2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezu J, Oku A, Jones MH, Shimane M.Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with structural similarity to vaccinia virus B1R kinase. Genomics 1997; 45: 327–331. [DOI] [PubMed] [Google Scholar]
- Vega FM, Gonzalo P, Gaspar ML, Lazo PA.Expression of the VRK (vaccinia-related kinase) gene family of p53 regulators in murine hematopoietic development. FEBS Lett 2003; 544: 176–180. [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Traktman P.Characterization of three paralogous members of the mammalian vaccinia related kinase family. J Biol Chem 2004; 279: 7934–7946. [DOI] [PubMed] [Google Scholar]
- Kang TH, Kim KT.Negative regulation of ERK activity by VRK3-mediated activation of VHR phosphatase. Nat Cell Biol 2006; 8: 863–869. [DOI] [PubMed] [Google Scholar]
- Kang TH, Kim KT.VRK3-mediated inactivation of ERK signaling in adult and embryonic rodent tissues. Biochim Biophys Acta 2008; 1783: 49–58. [DOI] [PubMed] [Google Scholar]
- Cullen CF, Brittle AL, Ito T, Ohkura H.The conserved kinase NHK-1 is essential for mitotic progression and unifying acentrosomal meiotic spindles in Drosophila melanogaster. J Cell Biol 2005; 171: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanacz M, Klerkx EP, Galy V, Santarella R, Lopez-Iglesias C, Askjaer P, Mattaj IW.Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in postmitotic nuclear envelope assembly. EMBO J 2007; 26: 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klerkx EP, Lazo PA, Askjaer P.Emerging biological functions of the vaccinia-related kinase (VRK) family. Histol Histopathol 2009; 24: 749–759. [DOI] [PubMed] [Google Scholar]
- Kang TH, Park DY, Choi YH, Kim KJ, Yoon HS, Kim KT.Mitotic histone H3 phosphorylation by vaccinia-related kinase 1 in mammalian cells. Mol Cell Biol 2007; 27: 8533–8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TH, Park DY, Kim W, Kim KT.VRK1 phosphorylates CREB and mediates CCND1 expression. J Cell Sci 2008; 121: 3035–3041. [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Wiebe MS, Traktman P.The vaccinia-related kinases phosphorylate the N′ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol Biol Cell 2006; 17: 2451–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss AK, Thomas T, Gruss P.Efficiency assessment of the gene trap approach. Dev Dyn 1998; 212: 171–180. [DOI] [PubMed] [Google Scholar]
- Zelko I, Kobayashi R, Honkakoski P, Negishi M.Molecular cloning and characterization of a novel nuclear protein kinase in mice. Arch Biochem Biophys 1998; 352: 31–36. [DOI] [PubMed] [Google Scholar]
- Lancaster OM, Cullen CF, Ohkura H.NHK-1 phosphorylates BAF to allow karyosome formation in the Drosophila oocyte nucleus. J Cell Biol 2007; 179: 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders GC, May JJ.Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Dev Biol 1994; 163: 331–340. [DOI] [PubMed] [Google Scholar]
- Yoshinaga K, Nishikawa S, Ogawa M, Hayashi S, Kunisada T, Fujimoto T, Nishikawa S.Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development 1991; 113: 689–699. [DOI] [PubMed] [Google Scholar]
- Ohta H, Tohda A, Nishimune Y.Proliferation and differentiation of spermatogonial stem cells in the w/wv mutant mouse testis. Biol Reprod 2003; 69: 1815–1821. [DOI] [PubMed] [Google Scholar]
- Schrans-Stassen BH, van de Kant HJ, de Rooij DG, van Pelt AM.Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology 1999; 140: 5894–5900. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE.Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 2004; 36: 647–652. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP.Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 2004; 36: 653–659. [DOI] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Morton DG, Gunsalus KC, Reinke V, Kim SK, Kemphues KJ.Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr Biol 2002; 12: 1959–1964. [DOI] [PubMed] [Google Scholar]
- Aihara H, Nakagawa T, Yasui K, Ohta T, Hirose S, Dhomae N, Takio K, Kaneko M, Takeshima Y, Muramatsu M, Ito T.Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo. Genes Dev 2004; 18: 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, Khandan T, Ito T, Orr-Weaver TL.A histone code in meiosis: the histone kinase, NHK-1, is required for proper chromosomal architecture in Drosophila oocytes. Genes Dev 2005; 19: 2571–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, McCarthy A, Smith D, Green KA, Grahame HD, Ashworth A, Alessi DR.Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. EMBO J 2005; 24: 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor MA, Mora A, Ashby PR, Williams MR, Murray-Tait V, Malone L, Prescott AR, Lucocq JM, Alessi DR.Essential role of PDK1 in regulating cell size and development in mice. EMBO J 2002; 21: 3728–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Bishop CE.Late onset of spermatogenesis and gain of fertility in POG-deficient mice indicate that POG is not necessary for the proliferation of spermatogonia. Biol Reprod 2003; 69: 161–168. [DOI] [PubMed] [Google Scholar]
- Valbuena A, Lopez-Sanchez I, Vega FM, Sevilla A, Sanz-Garcia M, Blanco S, Lazo PA.Identification of a dominant epitope in human vaccinia-related kinase 1 (VRK1) and detection of different intracellular subpopulations. Arch Biochem Biophys 2007; 465: 219–226. [DOI] [PubMed] [Google Scholar]
- Hao J, Yamamoto M, Richardson TE, Chapman KM, Denard BS, Hammer RE, Zhao GQ, Hamra FK.Sohlh2 knockout mice are male-sterile because of degeneration of differentiating type A spermatogonia. Stem Cells 2008; 26: 1587–1597. [DOI] [PubMed] [Google Scholar]
- Ballow D, Meistrich ML, Matzuk M, Rajkovic A.Sohlh1 is essential for spermatogonial differentiation. Dev Biol 2006; 294: 161–167. [DOI] [PubMed] [Google Scholar]
- Ohta H, Yomogida K, Dohmae K, Nishimune Y.Regulation of proliferation and differentiation in spermatogonial stem cells: the role of c-kit and its ligand SCF. Development 2000; 127: 2125–2131. [DOI] [PubMed] [Google Scholar]
- Vincent S, Segretain D, Nishikawa S, Nishikawa SI, Sage J, Cuzin F, Rassoulzadegan M.Stage-specific expression of the Kit receptor and its ligand (KL) during male gametogenesis in the mouse: a Kit-KL interaction critical for meiosis. Development 1998; 125: 4585–4593. [DOI] [PubMed] [Google Scholar]
- Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, Zhao GQ, Arber S, Kurpios N, Murphy TL, Cheng AM, Hassell JA, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 2005; 436: 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez I, Sanz-Garcia M, Lazo PA.Plk3 interacts and specifically phosphorylates VRK1 in Ser342, a downstream target in a pathway that induces Golgi fragmentation. Mol Cell Biol 2008; 29: 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Garcia M, Lopez-Sanchez I, Lazo PA.Proteomic identification of nuclear Ran GTPase as an inhibitor of human VRK1 and VRK2 (vaccinia-related kinase) activities. Mol Cell Proteomics 2008; 7: 2199–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HY, Zheng Y.Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev 2004; 18: 512–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Xie S, Chen J, Fukasawa K, Naik U, Traganos F, Darzynkiewicz Z, Jhanwar-Uniyal M, Dai W.Cell cycle arrest and apoptosis induced by human Polo-like kinase 3 is mediated through perturbation of microtubule integrity. Mol Cell Biol 2002; 22: 3450–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Wang Q, Ruan Q, Liu T, Jhanwar-Uniyal M, Guan K, Dai W.MEK1-induced Golgi dynamics during cell cycle progression is partly mediated by Polo-like kinase-3. Oncogene 2004; 23: 3822–3829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.