Abstract
MacroH2A1 is a histone variant that is enriched on the inactive X chromosome (Xi) in mammals and is postulated to play an important, but unknown, role in the repression of gene expression. Here we show that, although macroH2A1 marks repressed autosomal chromatin, it positively regulates transcription when located in the transcribed regions of a subset of its target genes. We used chromatin immunoprecipitation (ChIP) coupled with tiling microarrays (ChIP–chip) to determine the genomic localization of macroH2A1 in IMR90 human primary lung fibroblasts and MCF-7 breast cancer cells. The patterns of macroH2A1 deposition are largely similar across the autosomes of both cell lines. Our studies revealed a genomic localization pattern unique among histone variants; namely, the occupation by macroH2A1 of large chromatin domains (>500 kb in some cases) that contain repressive chromatin marks (e.g., histone H3 Lys 27 trimethylation). The boundaries of macroH2A1-containing domains tend to occur in promoter-proximal regions. Not all promoters, however, serve as macroH2A1 boundaries; many macroH2A1-containing chromatin domains invade the transcribed regions of genes whose products play key roles in development and cell–cell signaling. Surprisingly, the expression of a subset of these genes is positively regulated by macroH2A1. MacroH2A1 also plays a role in augmenting signal-regulated transcription, specifically for genes responsive to serum starvation. Collectively, our results document an unexpected role for macroH2A1 in the escape from heterochromatin-associated silencing and the enhancement of autosomal gene transcription.
Keywords: MacroH2A1, histone variant, transcription, ChIP–chip
The incorporation of histone variants into nucleosomes specifies functional differences between chromatin domains. For example, histones H3.3, H2AX, and CENP-A (an H3 variant) replace their canonical histone counterparts at sites of active transcription, DNA double-strand breaks, and centromeres, respectively (Sarma and Reinberg 2005). MacroH2A1 is a histone H2A variant named for its large (∼30-kDa) C-terminal “macro” domain (Pehrson and Fried 1992). MacroH2A1 is enriched on the inactive X chromosome (Xi) in mammals, and has been postulated to play an important, but unknown, role in the repression of gene expression associated with X inactivation (Ladurner 2003). Importantly, macroH2A1 has also been found in a variety of contexts on autosomes (Zhang et al. 2005; Agelopoulos and Thanos 2006; Changolkar and Pehrson 2006; Choo et al. 2006; Buschbeck et al. 2009; Mietton et al. 2009). Although recent studies have suggested a role for macroH2A1 in autosomal gene regulation (Zhang et al. 2005; Agelopoulos and Thanos 2006; Changolkar and Pehrson 2006; Choo et al. 2006; Changolkar et al. 2007, 2008; Bernstein et al. 2008; Buschbeck et al. 2009), many questions about the role of macroH2A1 in the regulation of autosomal genes remain. For example, does macroH2A1 generally mark repressive heterochromatin on autosomes, as it does on the inactive X chromosome? Additionally, does macroH2A1 play a functional role in the regulation of the genes with which it associates?
The N-terminal histone region of macroH2A1 is only 64% identical to histone H2A. Nevertheless, the structure of homotypic and heterotypic macroH2A-containing nucleosomes is largely similar to nonvariant histone nucleosomes (Chakravarthy et al. 2005). While more salt-stable, macroH2A1-containing nucleosomes organize the same amount of DNA (147 base pairs [bp]) as their canonical counterparts (Abbott et al. 2004). There are, however, several features that distinguish macroH2A1-containing chromatin from typical chromatin. MacroH2A1-containing chromatin assembled in vitro is more repressive to transcription than canonical chromatin, specifically blocking transcriptional initiation, not elongation (Doyen et al. 2006). A subset of DNA-binding transcription factors is unable to recognize their binding elements when they are incorporated into macroH2A1-containing chromatin (Angelov et al. 2003; Agelopoulos and Thanos 2006). MacroH2A1-containing nucleosomes are refractory to both the histone acetylation activity of p300 (Doyen et al. 2006) and the nucleosome remodeling activity of the SWI/SNF complex (Angelov et al. 2003). The reduced ability of SWI/SNF to remodel macroH2A1-containing nucleosome appears to be due to a preference of the chromatin remodeler to bind to canonical nucleosomes (Chang et al. 2008). However, certain histone chaperones, such as nucleolin, can stimulate the ability of SWI/SNF to remodel macroH2A1-containing nucleosomes (Angelov et al. 2006).
MacroH2A1 is the founding member of a superfamily of macrodomain-containing proteins that have been identified from bacteria to humans (Pehrson and Fuji 1998). The macrodomain of macroH2A provides a unique platform for the recruitment of protein factors to chromatin, and a handful of such factors have been identified (e.g., PARP-1, HDAC1) (Chakravarthy et al. 2005; Hernandez-Munoz et al. 2005; Ouararhni et al. 2006; Nusinow et al. 2007). Several macrodomains, including the macrodomain of one of the splice variants of macroH2A1 (macroH2A1.1), have been shown to bind NAD+ metabolites, including poly(ADP-ribose), ADP-ribose and O-acetyl-ADP-ribose, with high affinity (Kustatscher et al. 2005; Ahel et al. 2009; Gottschalk et al. 2009; Kraus 2009; Timinszky et al. 2009). While the function of the ligand binding to macrodomains is not completely understood, it suggests that macroH2A1-containing chromatin (at least for macroH2A1.1) has a novel mechanism to regulate chromatin function through this covalently linked ligand-binding domain whose activities may be regulated by the local concentrations of specific ligands. To our knowledge, differential functions of macroH2A1.1 and macroH2A1.2 in the regulation of gene expression have not yet been reported. It is important to note, however, that while macroH2A1.2 is ubiquitously expressed, the expression of the macroH2A1.1 splice variant seems to be restricted to differentiated cells (Pehrson et al. 1997). Additionally, macroH2A1.1 expression is down-regulated in several cancers with poor prognosis (Sporn et al. 2009).
While suggested to play a role in X inactivation, macroH2A1 is expressed equally in both males and females (Rasmussen et al. 1999) and is highly conserved across all vertebrates (Pehrson and Fuji 1998), while X inactivation occurs only in female mammals. Therefore, macroH2A1 likely plays a more general role in the regulation of gene expression than previously appreciated. Consistent with a broader role in gene regulation, macroH2A1 has also been found enriched on the inactive allele of imprinted genes (Choo et al. 2006) and senescence-associated heterochromatic foci (Zhang et al. 2005). Additionally, macroH2A1 is required for the transcriptional silencing of endogenous murine leukemia viruses found in the mouse genome (Changolkar et al. 2008). Most recently, macroH2A was also found at a variety of developmentally regulated genes in a pluripotent teratocarcinoma cell line (Buschbeck et al. 2009). While most of the current literature has focused on the role of macroH2A1 in the repression of gene expression, recent evidence suggests that subpopulations of macroH2A1 may play a very different role in the cell. For example, macroH2A1 phosphorylated on S137 is excluded from the transcriptionally inert inactive X chromosome (Bernstein et al. 2008), suggesting that transcriptional repression may not be the only function of this histone variant.
In the studies described herein, we used chromatin immunoprecipitation (ChIP) coupled with genomic tiling microarrays (i.e., ChIP–chip) to determine the localization of macroH2A1 in the human genome. We found a genomic localization pattern unique among histone variants; namely, the occupation by macroH2A1 of large chromatin domains (>500 kb in some cases). Our results show that, although macroH2A1 marks repressed autosomal chromatin, it positively regulates transcription when located in the transcribed regions of several target genes. Collectively, our results document an unexpected role for macroH2A1 in enhancing the transcription of a subset of autosomal genes.
Results
MacroH2A1 is deposited in large domains distributed similarly throughout the genomes of both IMR90 and MCF-7 cells
While macroH2A1 has been associated with the repressive chromatin of the inactive X chromosome, a broader understanding of the role macroH2A1 plays in autosomal gene regulation is less clear. In order to better understand the localization and function of macroH2A1 in the human genome, we performed ChIP–chip in both the primary human female lung fibroblasts cell line IMR90 and the human female breast cancer cell line MCF-7. Of the three macroH2A isoforms in humans (e.g., macroH2A1.1, macroH2A1.2, and macroH2A2), macroH2A1.2 is by far the predominant form in MCF-7 cells, while both macroH2A1.1 and macroH2A1.2 are similarly expressed in IMR90 cells (Supplemental Fig. S1A,B). MacroH2A2, on the other hand, is not expressed in either cell line.
Using an antibody that recognizes both macroH2A1.1 and macroH2A1.2 (Supplemental Fig. S1C), we performed ChIP–chip experiments in both IMR90 and MCF-7 cells using microarray platforms representing either 244 Mb or 41 Mb of genomic DNA covering a total of 21,567 and 2138 transcription start sites (TSSs), respectively. The results of the ChIP–chip experiments showed excellent specificity, were highly reproducible, and had low false discovery rates (FDRs). The specificity of the macroH2A1 chip signal was demonstrated by a dramatic reduction in ChIP signal (i.e., DNA recovery) upon shRNA-mediated depletion of macroH2A1 (Supplemental Fig. S2A). The reproducibility of the ChIP–chip data was demonstrated by a strong correlation between the window averages from the separate ChIP–chip replicates (Supplemental Fig. S2B). ChIP-quantitative PCR (qPCR) confirmations demonstrated a low FDR for both macroH2A1-bound (FDR < 0.042) and macroH2A1-unbound (FDR < 0.042) regions (Supplemental Fig. S2C). Furthermore, using ectopic expression of Flag-tagged macroH2A1.1 and macroH2A1.2 coupled with ChIP-qPCR, we found that both macroH2A1 isoforms localize to the same genomic regions in all cases tested (data not shown).
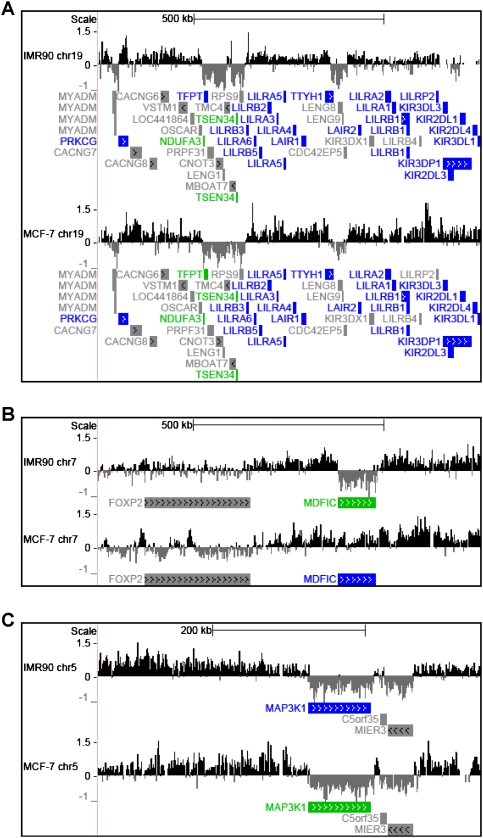
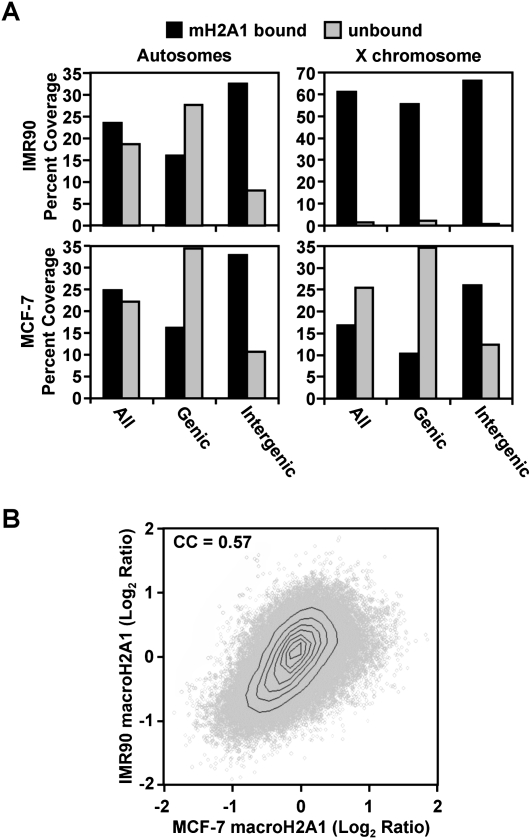
In our initial analysis, we plotted the data for each of the ENCODE regions from both the IMR90- and MCF-7-derived ChIP–chip experiments. We observed that macroH2A1 is incorporated throughout the genome in large chromatin domains that can span >500 kb (Fig. 1; Supplemental Fig. S3). When comparing the deposition of macroH2A1 in IMR90 and MCF-7 cells, we observed many areas with striking similarities, as well as some less frequent differences (see Fig. 1B, for example). Overall, macroH2A1-bound regions occupy nearly a quarter of autosomal DNA in both IMR90 and MCF-7 cells (Fig. 2A). Interestingly, in both IMR90 and MCF-7 cells, macroH2A1 occupies a larger fraction of intergenic regions compared with regions within the bodies of genes (33% and 16%, respectively). Overall, these observations suggested to us that macroH2A1 is a more general feature of chromatin than previously appreciated and may play a role in regulating autosomal gene expression.
Figure 1.
MacroH2A1 is incorporated into chromatin in large domains. University of California at Santa Cruz genome browser-generated (http://genome.ucsc.edu) histogram showing the log2 ratios of macroH2A1 ChIP over input for both IMR90 and MCF-7 cells. The location and orientation of RefSeq genes are depicted below each track. Gene expression in each cell line is color-coded. Expressed genes are colored green, unexpressed genes are colored blue, and genes with ambiguous expression are colored gray. (A) A 1-Mb span of chromosome 19 (ENCODE region ENm007). (B) A 1-Mb span of chromosome 7 (ENCODE region ENm012). (C) A 500-kb span of chromosome 5 (ENCODE region ENr221).
Figure 2.
In addition to the inactive X chromosome, macroH2A1 occupies a large proportion of autosomal chromatin. (A) Histogram showing the percent base coverage of macroH2A1-bound and macroH2A1-unbound regions in either an IMR90 or MCF-7 cell subset for genic or intergenic regions on either autosomes or the X chromosome. (B) Scatter plot of the macroH2A1 ChIP–chip log2 ratios from MCF-7 cells versus IMR90 cells for the 94,966 windows occupied by at least six probes on both array platforms. (CC) Spearman correlation coefficient of the data presented in the scatter plot (P < 10−300).
Two factors likely to contribute to the pattern of macroH2A1-bound and macroH2A1-unbound regions observed in the ChIP–chip analysis are (1) differential incorporation of macroH2A1 into nucleosomes and (2) differential nucleosome density. To determine which of these contributes to the pattern of macroH2A1 genomic occupancy, we determined the average level of histone H3 density in macroH2A1-bound and macroH2A1-unbound regions in MCF-7 cells (Supplemental Fig. S4). Generally, there is no significant difference between the level of H3 found in macroH2A1-bound and macroH2A1-unbound regions. Additionally, macroH2A1 is poorly correlated with H3 in MCF-7 cells (correlation coefficient = 0.033). Overall, these results indicate that the observed patterns of macroH2A1 occupancy generally arise from differences in the location of macroH2A1 incorporation into nucleosomes, rather than differences in nucleosome occupancy.
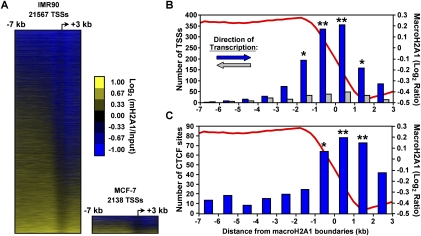
In addition to occupying genic and intergenic regions to a similar extent, macroH2A1 patterns are also generally conserved between the two cell lines. There is a highly significant correlation between corresponding genomic locations in the two data sets (Figs. 1, 2B). This is also observed in heat maps depicting macroH2A1 enrichment in IMR90 and MCF-7 cells, which are ordered for the overall level of macroH2A1 present near TSSs in IMR90 cells (Fig. 3A; Supplemental Fig. S5). While the overall patterns of macroH2A1 deposition are highly conserved between IMR90 and MCF-7 cells, there are some striking differences, most notably on the X chromosome. MacroH2A1 is most widely known for its enrichment on the inactive X chromosome. While IMR90 cells maintain an inactive X chromosome, MCF-7 cells, like many breast cancer cells, have lost this form of epigenetic repression (Sirchia et al. 2005). While macroH2A1 levels are generally high along the X chromosome in IMR90 cells, much less macroH2A1 binding is observed on the X chromosome in MCF-7 cells (Fig. 2A; Supplemental Fig. S3).
Figure 3.
TSSs and CTCF-binding sites are enriched near macroH2A1-containing domain boundaries. (A) Heat map showing macroH2A1 ChIP–chip data for all 21,567 or 2138 promoters from the IMR90 and MCF-7 data, respectively (from −7 kb to +3 kb relative to the TSS), ordered for average macroH2A1 intensity. (B) Histogram representing the locations of TSSs relative to macroH2A1 domain boundaries in IMR90 cells. The bars are color-coded to indicate the direction of gene transcription relative to the orientation of the macroH2A1 boundary. Asterisks denote significant bars with P-value < 10−8 (*) or P-value < 10−67 (**). (C) Histogram representing the locations of CTCF-binding sites relative to macroH2A1 domain boundaries in IMR90 cells. Asterisks denote significant bars with P-value = 10−7 (*) or P-value < 10−12 (**).
MacroH2A1-containing domain boundaries occur near TSSs
The disparity in the distribution of macroH2A1 between intergenic and intragenic regions led us to examine the boundaries between macroH2A1-bound and macroH2A1-unbound regions and their relationship to the TSSs of genes. A macroH2A1 boundary was defined as the midpoint between the edges of adjacent macroH2A1-bound and macroH2A1-unbound regions. Boundaries were considered only if all windows between the macroH2A1-bound and macroH2A1-unbound regions were well tiled (i.e., had only limited repeat masking). Using these criteria, we identified 1,754 macroH2A1 chromatin domain boundaries in IMR90 cells and 255 boundaries in MCF-7 cells. We observed that annotated TSSs are highly enriched within 2 kb of macroH2A1 boundaries (Fig. 3B; Supplemental Fig. S6). Overall, 68% of macroH2A1 boundaries identified in IMR90 cells have a TSS within 2 kb. Notably, when the boundaries are oriented with the macroH2A1-bound side to the left, as shown in Figure 3B (Supplemental Fig. S6), the TSSs of genes transcribed to the right are specifically and significantly enriched. The enrichment of TSSs at macroH2A1 domain boundaries can be observed clearly when the ChIP–chip data are expressed as a heat map anchored by the TSS of each gene on the array (Fig. 3A). Together, these observations suggest that some genes have evolved a mechanism to prevent macroH2A1 from invading their transcribed regions, perhaps in a manner analogous to insulators.
The observation that a subset of TSS might act as macroH2A1 insulators led us to hypothesize that a known insulator protein, such as CTCF, might be involved in establishing the macroH2A1 domain boundaries. In order to explore this possibility, we made use of previously published ChIP–chip data on the genomic localization of CTCF in IMR90 cells across the genome (Kim et al. 2007). Out of nearly 14,000 CTCF-occupied sites identified in IMR90 cells, 3992 are represented on the tiling microarray used for the IMR90 cells in our studies. By calculating the distance of each macroH2A1 boundary to the nearest CTCF-binding site, we found that CTCF is statistically enriched at macroH2A1 boundaries (Fig. 3C). In all, 240 (∼14%) macroH2A1 boundaries are located within 2 kb of a CTCF-binding site. While CTCF occupies a significant fraction of macroH2A1 boundaries, the majority of macroH2A1 boundaries are not marked by this insulator protein, indicating that CTCF-independent mechanisms also play a role in determining where macroH2A1 boundaries occur.
MacroH2A1 marks genes that are involved in signaling and development
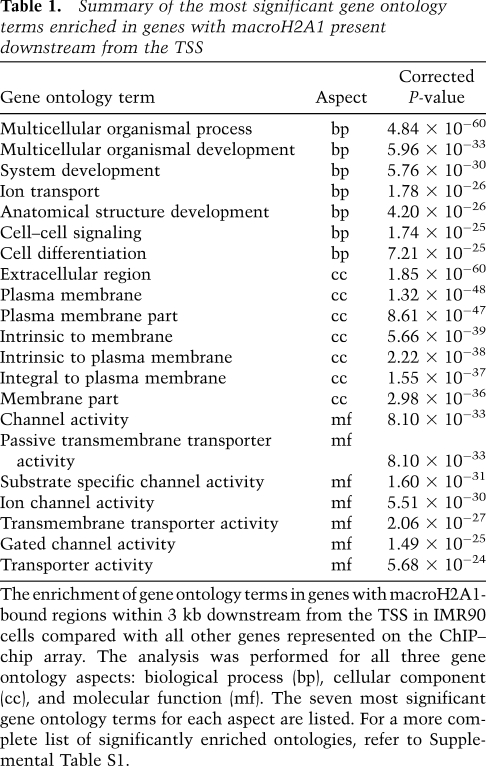
We hypothesized that the presence of macroH2A1 downstream from the TSS might specify distinct sets of functionally related genes. To test this possibility, we determined the ontological gene categories that were enriched in autosomal genes containing macroH2A1 (Table 1; Supplemental Table S1). Genes with macroH2A1 present in the transcribed region encode proteins that (1) play roles in developmental processes and/or (2) function as signaling molecules that act at or are exported from the cell surface (i.e., cytokines, growth factors, ion channels, and receptors). These associations are consistent with previous work (Buschbeck et al. 2009), and suggest that macroH2A1 may play a role in regulating developmental and cell–cell signaling processes.
Table 1.
Summary of the most significant gene ontology terms enriched in genes with macroH2A1 present downstream from the TSS
The enrichment of gene ontology terms in genes with macroH2A1-bound regions within 3 kb downstream from the TSS in IMR90 cells compared with all other genes represented on the ChIP–chip array. The analysis was performed for all three gene ontology aspects: biological process (bp), cellular component (cc), and molecular function (mf). The seven most significant gene ontology terms for each aspect are listed. For a more complete list of significantly enriched ontologies, refer to Supplemental Table S1.
MacroH2A1 colocalizes with histone H3K27 trimethylation
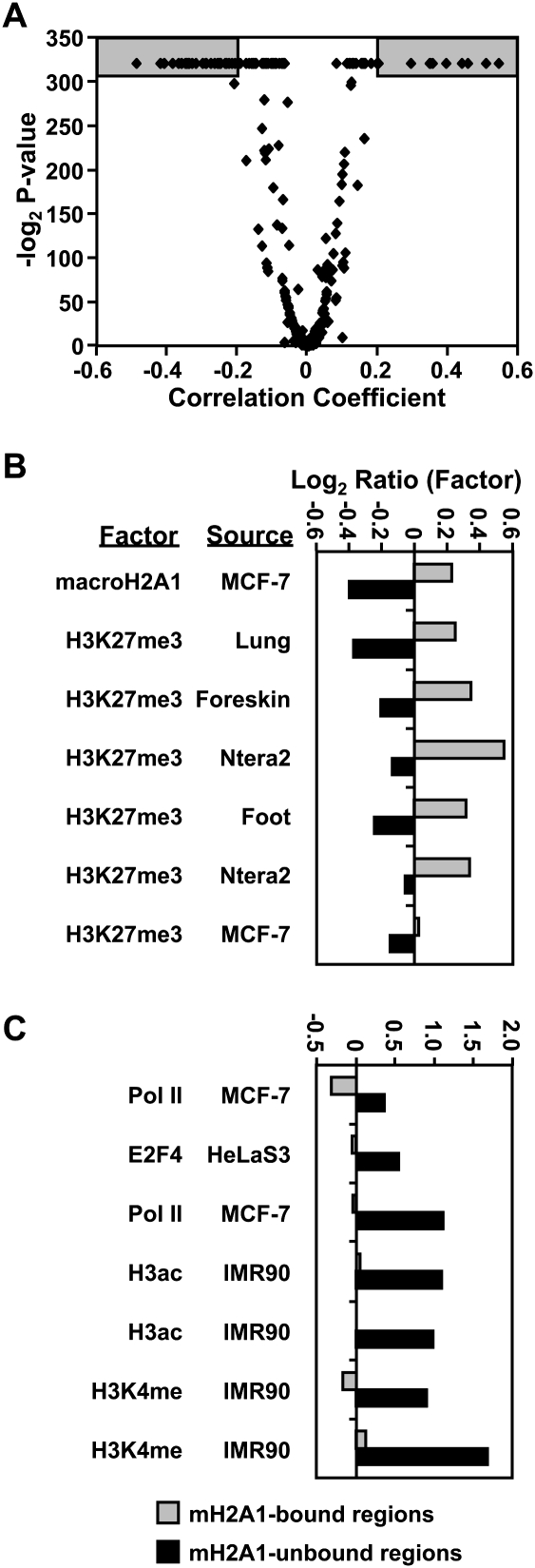
In order to gain a better understanding of the functional characteristics of macroH2A1-containing chromatin, we examined the relationship of macroH2A1 in IMR90 cells with a wide variety of histone modifications and chromatin- and transcription-related factors from 367 ChIP–chip experiments deposited previously into the NCBI Gene Expression Omnibus (GEO) repository. We calculated the Spearman correlation coefficient and associated P-value of the macroH2A1 ChIP–chip data from IMR90 cells with each of the existing ChIP–chip data sets (Fig. 4A; Supplemental Table S2). Not surprisingly, of all of the ChIP–chip data sets included in the analysis, the macroH2A1 data from IMR90 cells correlate most strongly with the macroH2A1 data from MCF-7 cells (correlation coefficient of 0.55), highlighting the similar patterns of macroH2A1 deposition between these two cell types. Interestingly, the next six data sets most positively correlated with the pattern of macroH2A1 in IMR90 cells are all H3K27 trimethylation (Fig. 4B). MacroH2A1 was also found to be negatively correlated with a variety of positive marks for transcription, including RNA polymerase II, histone H3K4 methylation, and histone H3 acetylation (Fig. 4C). Similar observations were made with the MCF-7 cell macroH2A1 ChIP–chip data (Supplemental Fig S7; Supplemental Table S3). The observation that macroH2A1-containing chromatin is enriched for a facultative heterochromatin mark and depleted for marks of active transcription is generally consistent with previous results, suggesting a role for this histone variant in transcriptional repression.
Figure 4.
MacroH2A1 binding positively correlates with heterochromatic chromatin marks and negatively correlates with active chromatin marks. (A) Volcano plot of Spearman's correlation coefficient for the macroH2A1 ChIP–chip data from IMR90 cells with each of 367 ChIP–chip data sets versus the corresponding significance score (−log2 P-value). (B) Average log2 ratios in macroH2A1-bound and unbound regions for the seven most positive correlations. (C) Same as in B for the seven most negative correlations.
MacroH2A1 levels downstream from TSS are negatively correlated with gene expression
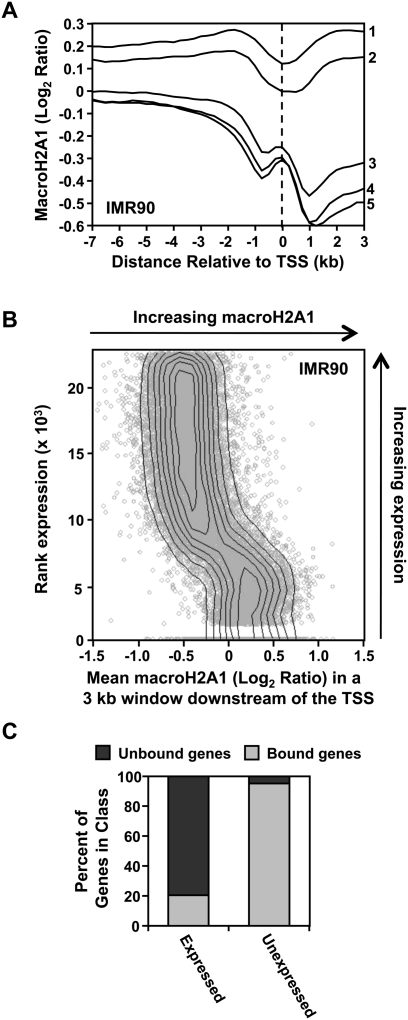
To further explore the relationship between macroH2A1 and gene expression on autosomes, we used genomic run-on (i.e., GRO-seq) expression data from IMR90 cells to determine the average pattern of macroH2A1 deposition at genes separated into different levels of expression (GRO-seq is a more sensitive and direct measurement of transcriptional activity across the genome compared with expression microarrays) (Fig. 5A; Core et al. 2008). This analysis clearly indicates that the level of macroH2A1 downstream from the TSS specifies transcriptional outcomes. For example, genes in the bottom pentile of expression had, on average, the highest levels of macroH2A1 downstream from the TSS (line 1 in Fig. 5A). The general negative correlation between macroH2A1 occupancy of a gene and expression is further illustrated by plotting the average level of macroH2A1 in the proximal 3 kb downstream from each TSS against the rank level of expression (Fig. 5B). As expected from these data, macroH2A1 generally marks the genes with the lowest levels of expression. Consistent with this, the gene set with high levels of macroH2A1 downstream from the TSS is enriched for unexpressed genes (84-fold enrichment, P < 10−300) (Fig. 5C). Similar results for both IMR90 and MCF-7 cells were obtained using available Affymetrix expression microarray data (Supplemental Fig. S8), suggesting similar functions of macroH2A1 in gene regulation in the two cell lines. In this regard, many of the differences in macroH2A1 occupancy in the transcribed regions of genes between IMR90 and MCF-7 cells correspond to differences in gene expression (Supplemental Fig. S9).
Figure 5.
MacroH2A1 levels downstream from the TSS, while negatively correlated with expression, are not an absolute marker of silent genes. (A) Average macroH2A1 ChIP–chip profiles from IMR90 cells of genes in expression pentiles ranked from least to most expressed based on GRO-seq data. (B) Scatter plot of the average level of macroH2A1 found in the first 3 kb of a gene versus the rank level of gene expression based on GRO-seq data. (C) Histogram depicting the percent of expressed and unexpressed genes containing macroH2A1 downstream from the TSS.
Although we observed a negative correlation between macroH2A1 occupancy and the levels of expression, correlation alone does not prove causation. Using GRO-seq data, we found that ∼12% of transcriptionally active autosomal genes in IMR90 cells contain macroH2A1 downstream from the TSS (Fig. 5C); similar trends were observed for IMR90 and MCF-7 cells using Affymetrix expression microarray data (Supplemental Fig. S8). Thus, while there is a strong negative correlation between macroH2A1 occupancy and gene expression, not all of the genes marked by this histone variant are repressed. Therefore, macroH2A1 is not an absolute determinant of transcriptional repression.
When present downstream from the TSS, macroH2A1 positively regulates the expression of a subset of genes
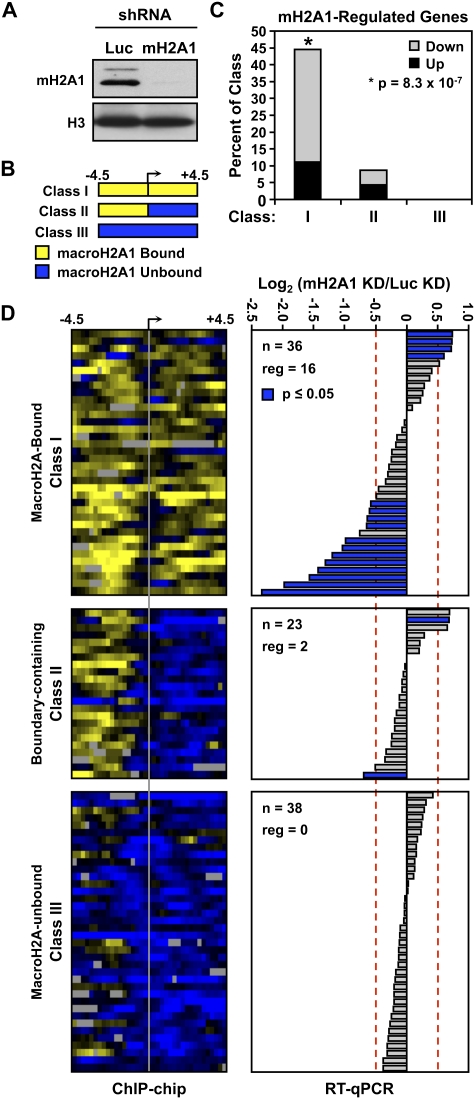
Since a subset of macroH2A1-containing genes escape repression, we re-evaluated the role of macroH2A1 in gene expression. To do so, we generated MCF-7 cell lines stably expressing shRNAs targeting macroH2A1 or luciferase (as a control) (Fig. 6A; Supplemental Fig. S10). RNA isolated from each cell line was used to determine the effect of macroH2A1 knockdown on the expression of a subset of genes chosen without prior knowledge of their macroH2A1-bound status. A gene was considered to be macroH2A1-regulated if the absolute log2 fold change (macroH2A1 knockdown over the luciferase knockdown) was >0.5 and the P-value was <0.05 (two-tailed paired Student's t-test). Of the 97 genes tested, 18 (∼19%) were regulated by macroH2A1 knockdown (Fig. 6C,D). The genes were then split into three classes based on whether they were bound by macroH2A1 both upstream of and downstream from the TSS (class I), were found near a macroH2A1 boundary (class II), or did not contain macroH2A1 either upstream of or downstream from the TSS (class III) (Fig. 6B; Supplemental Table S4). Importantly, class I genes were significantly and specifically enriched for regulation by macroH2A1, with nearly half (∼44%) of the tested genes in this class affected by macroH2A1 knockdown, compared with <3% of the genes that are not bound by macroH2A1 in the transcribed region (Fig. 6C). Furthermore, 75% of the regulated genes were down-regulated upon macroH2A1 knockdown, indicating an unexpected positive role for macroH2A1 in the regulation of these target genes. From this data, we conclude that, although the localization of macroH2A1 downstream from the TSS generally marks repressed domains of autosomal chromatin, macroH2A1 actually protects a subset of its target genes from silencing.
Figure 6.
Depletion of macroH2A1 reveals a positive role for macroH2A1 in the expression of class I genes. (A) Western blot for macroH2A1 in MCF-7 cells with stably integrated expression vectors for shRNAs targeting luciferase (Luc) or macroH2A1 (mH2A). (B) Schematic representation of the three observed macroH2A1-binding patterns relative to the TSSs. (C) Histogram depicting the percent of genes tested from each class that pass both the fold change (i.e., log2 0.5) and P-value (≤0.05) thresholds. The asterisk denotes significant enrichment of regulated genes in class I based on the P-value from a Fisher exact test (P = 8.3 × 10−7). The gray and black shading represents the proportions of genes that are down-regulated or up-regulated in the macroH2A1 knockdown cell line, respectively. (D, left) Heat map depicting the macroH2A1-binding patterns of all genes tested by RT-qPCR for regulation by macroH2A1 knockdown separated into the three macroH2A1-based gene classes. (Right) Log2 fold change in expression upon macroH2A1 knockdown (macroH2A1/Luc) for each of the genes shown in the heat map to the left. The dotted lines depict the log2 fold change threshold of 0.5. Blue bars indicate a P-value of 0.05 or less. (mH2A1) MacroH2A1; (KD) knockdown.
MacroH2A1 affects signal-regulated transcription
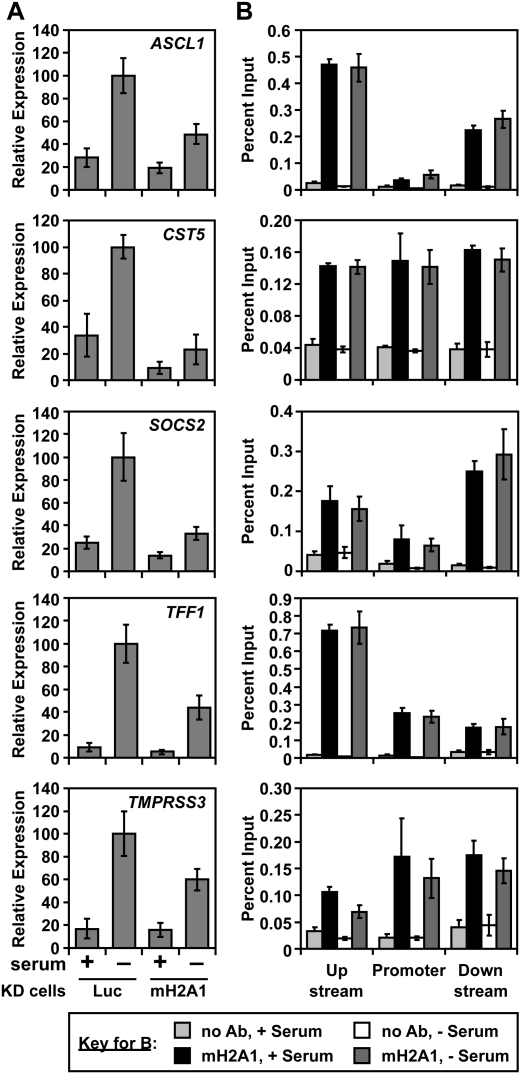
The data described above indicate that macroH2A1 can play a positive role in gene expression at genes where it is bound in the transcribed region. These studies were conducted under normal growth conditions and represent changes in steady-state levels of transcription. To further explore the positive role of macroH2A1 in gene expression, we determined whether macroH2A1 might contribute to the signal-regulated transcription of its target genes. As a model, we used several genes with macroH2A1 bound downstream from the TSS that are induced upon serum starvation in MCF-7 cells (i.e., ASCL1, CST5, SOCS2, TFF1, and TMPRSS3) (Fig. 7A). We hypothesized that this mechanism of activation is likely due to the cessation of signaling cascades initiated by growth factors at the cell surface that actively represses the expression of these genes. Indeed, treatment of MCF-7 cells with various kinase inhibitors stimulates the expression of these serum starvation-induced target genes (Supplemental Fig. S11). Interestingly, induction of these genes by serum starvation is inhibited by knockdown of macroH2A1 (Fig. 7A). Furthermore, induction of these genes by serum starvation does not lead to changes in macroH2A1 deposition upstream of the TSS, at the promoter, or in the transcribed region (Fig. 7B), indicating that removal of this histone variant is not required for the serum starvation-induced transcriptional activation of these genes. Together, these results illustrate that macroH2A1 can play a positive role in the transcription of signal-regulated target genes.
Figure 7.
MacroH2A1 modulates signal-regulated transcriptional responses of macroH2A1-containing genes. (A) Relative expression of five macroH2A1-containing, serum starvation-activated genes in luciferase or macroH2A1 knockdown cells either in serum (+) or deprived of serum for 24 h (−). Experiments were scaled to yield a value of 100 for the Lucifierase knockdown serum-starved samples. All differences between serum-deprived Luciferase and macroH2A knockdown samples were significant (P < 0.02). (B) MacroH2A1 ChIP from MCF-7 cells with or without 24 h of serum starvation.
Discussion
Recent genomic localization studies on the role of macroH2A in gene expression have focused on either the inactive X chromosome in mice (Mietton et al. 2009) or promoter-proximal regions of a subset of developmentally regulated autosomal genes in human teratocarcinoma cells (Buschbeck et al. 2009). The data that we present here are consistent with these previously published results, but go beyond these studies to augment our understanding of macroH2A1 function in several important respects. First, we demonstrate that macroH2A1 is present in large domains spread across a quarter of the autosomal genome in both breast cancer cells and primary lung fibroblasts, and that the pattern of deposition is conserved between cell types. Second, we show that the boundaries of the macroH2A1-enriched domains are found near TSSs and CTCF-binding sites. Third, we demonstrate that macroH2A1 colocalizes with the facultative heterochromatin mark, histone H3K27 trimethylation. Fourth, we show that the presence of macroH2A1 downstream from the TSS is associated with transcriptional repression. However, we also show that, while most genes with high levels of macroH2A1 downstream from the TSS are silenced, some macroH2A1-marked transcribed regions are, in fact, active. Surprisingly, macroH2A1 is a positive regulator of a subset of these genes. Finally, we provide additional support for a role of macroH2A1 as a positive effector of gene expression by demonstrating that macroH2A1 augments serum starvation-induced transcriptional responses. Together, these results provide a new view of the localization and function of this unique histone variant in the genome.
First described as a factor that is present on the inactive X chromosome, macroH2A was thereby characterized as a repressive histone mark. Our data and previously published studies confirmed the enrichment macroH2A1 on the inactive X (Mietton et al. 2009). The inactive X is assembled into a facultative heterochromatin typified by the polycomb-repressive complex and repressive histone marks including histone H3K27 trimethylation (Chow and Heard 2009). These factors have roles on both the inactive X chromosome and autosomes, and macroH2A1 is no exception. Just as H3K27 trimethylation is present on autosomes, our work demonstrates that macroH2A1 is also present in large domains of heterochromatin on autosomes that can be hundreds of kilobases in length. In fact, macroH2A1-containing domains are often coincident with H3K27 trimethylation.
Chromatin is organized into functional domains that often end at boundary elements or insulators (Phillips and Corces 2009). Insulators function as a barrier to prevent heterochromatin from silencing genes found in transcriptionally permissive regions. We found that a subset of macroH2A1 domain boundaries could be explained by the presence of binding sites for the well-studied chromatin insulator protein, CTCF. However, not all macroH2A1 boundaries contain binding sites for CTCF, which begs the question: What additional factors can function as macroH2A1 boundaries? Interestingly, we found a subset of promoter-proximal regions can function as macroH2A1 domain boundaries. However, we have, so far, been unable to identify DNA motifs enriched at these promoters using either directed or de novo motif-finding techniques.
Our work and the previous work of others (Buschbeck et al. 2009) support the observation that macroH2A1 levels are inversely correlated with gene expression. While the majority of genes that contain macroH2A1 are silent, a considerable fraction of macroH2A1-containing genes escapes this typical repression and is expressed. How can these outliers be explained? One possible explanation is that, while macroH2A1 is recruited to heterochromatic regions of the genome, it does not play a general role in heterochromatic associated transcriptional repression. Support for this line of reasoning comes from studies of the macroH2A1 knockout mouse, which, besides being viable and fertile, undergoes normal X inactivation (Changolkar et al. 2007). Further support for the idea that macroH2A1 is not generally a repressive chromatin factor comes from the fact that we did not observe a wholesale increase in the expression of genes bound by macroH2A1 when the histone variant was knocked down. Finally, macroH2A1 can also augment transcriptional responses to serum deprivation, demonstrating that macroH2A1 not only enhances basal transcription, but can also boost transcriptional responses to certain stimuli. Far from acting as a functionally repressive chromatin component, our results demonstrate that macroH2A1 is, in fact, a positive regulator of a subset of the genes that contain macroH2A1 in the transcribed region. Furthermore, it suggests that the true role of macroH2A1 is more complicated than previously appreciated. MacroH2A1 is recruited to repressive heterochromatin, but in a significant number of instances, macroH2A1 is required to escape heterochromatic repression.
Future work is required to determine the mechanisms by which macroH2A1 allows its target genes to escape heterochromatic gene silencing. However, it is interesting to hypothesize that the large macrodomain-containing nonhistone region of macroH2A1 plays a role in this process. This domain represents a platform that can (1) recruit chromatin-dependent transcriptional regulators (e.g., HP1β, HDAC1, and PARP-1) (Chakravarthy et al. 2005; Changolkar and Pehrson 2006; Ouararhni et al. 2006; Nusinow et al. 2007), (2) provide a novel platform for covalent modifications (e.g., S137 phosphorylation) (Bernstein et al. 2008), and (3) bind small molecule ligands (e.g., the NAD+ metabolites poly[ADP-ribose], ADP-ribose, and O-acetyl-ADP-ribose, in the case of macroH2A1.1) (Kustatscher et al. 2005; Timinszky et al. 2009). All of these features may locally regulate the function of macroH2A1-containing nucleosomes and thereby affect the expression of a subset of the genes to which it is bound.
Materials and methods
Cell lines and treatments
MCF-7 human breast cancer cells were maintained in MEM supplemented with 5% calf serum. IMR90 human primary lung fibroblast cells were maintained in MEM supplemented with 10% fetal bovine serum. MacroH2A1-depleted MCF-7 cells were generated by retroviral-mediated gene transfer of two shRNA sequences specifically targeting the macroH2A1 mRNA using the pSUPER.retro system (Oligoengine) with puromycin and G418 for drug selection. The targeting sequences used for macroH2A1 were 5′-GCAATGCAGCGAGAGACAACA-3′ and 5′-GCGTGTGTTGTGGTGCTTTAT-3′. The target sequences were chosen using the BLOCK-iT RNAi Designer (Invitrogen; https://rnaidesigner.invitrogen.com/rnaiexpress). Control cells harboring shRNA sequences directed against luciferase (5′-GATATGGGCTGAATACAAA-3′) were generated in parallel.
Where indicated, MCF-7, luciferase knockdown, and/or macroH2A1 knockdown cells were serum-starved by washing the cells in PBS and replacing the medium with MEM without serum for 24 h. Where indicated, cells were treated with 100 ng/mL TPA for 3 h (expression analysis) or 1.5 h (ChIP assays).
ChIP
ChIP was performed essentially as described previously (Krishnakumar et al. 2008). The immunoprecipiations were performed from cross-linked MCF-7 and IMR90 cells with antibodies against macroH2A1 (Upstate Biotechnologies, 07-219) or histone H3 (Abcam, ab1791-100), or without antibodies as a control. Antibodies were screened for specificity by immunoblotting MCF-7 acid-extracted cell lysates. For gene-specific ChIP analyses, qPCR was used to determine the enrichment of immunoprecipitated material relative to the input material using gene-specific primer sets to the specified regions. Each ChIP experiment was conducted a minimum of three times with independent chromatin isolates to ensure reproducibility. For the ChIP–chip analyses, two replicates of the immunoprecipitated genomic DNA were blunted, amplified by ligation-mediated PCR (LM-PCR), and used to probe a DNA microarray (Nimblegen).
ChIP–chip
The ChIP–chip sample processing and analysis were done essentially as described previously (Krishnakumar et al. 2008). LM-PCR-amplified immunoprecipitated DNA samples were labeled with Cy5, and the LM-PCR-amplified reference DNA samples were labeled with Cy3. The fluorescently labeled immunoprecipitated and reference samples were combined and used to probe a factory-designed (HX1, Nimblegen) or a custom human oligonucleotide genomic array (Nimblegen). The HX1 ChIP–chip array contained ∼2.2 million probes, which tile all ENCODE regions, as well as ∼20,000 TSSs typically from −7 kb to +3 kb, with a mean probe spacing of 100 bp. We also used a custom-designed array that contained ∼390,000 probes, which tile all ENCODE regions, as well as ∼1500 TSSs typically −25 kb to +5 kb, with a mean probe spacing of 76 bp (Krishnakumar et al. 2008). The macroH2A1 ChIP–chip analyses were run two times to ensure reproducibility. Detailed information about genomic regions included on the custom array and the data from the hybridizations described in this study can be accessed from the NCBI GEO Web site (http://www.ncbi.nlm.nih.gov/geo) using accession numbers GSE9607 (MCF-7) and GSE18633 (IMR90).
Genomic data analyses
The genomic data analyses were performed using the statistical programming language R (R Development Core Team 2006). All data processing scripts are available on request. The log2 ratio data from each array was subjected to lowess normalization (Smyth and Speed 2003). The normalized data were scaled to equivalent sum of squares, and then the between array mean log2 ratio was determined for each probe. An error model was generated using a 1-kb moving window with 250-bp steps in which both the mean probe log2 ratio and P-values were calculated for each window. The P-values were calculated using the nonparametric Wilcoxon signed-rank test. The histograms depicting the genomic location of macroH2A1 in MCF-7 and IMR90 (Fig. 1) were generated using the University of California at Santa Cruz (UCSC) genome browser (Kent et al. 2002). The TSS-anchored heat maps used to visualize the ChIP–chip data (Fig. 3A) were generated with Java Treeview (Saldanha 2004). The contour lines on scatter plots (Fig. 2B) were generated using two-dimensional kernel density estimation (Venables and Ripley 2002).
Identification of macroH2A1-bound and macroH2A1-unbound regions
Significant bound regions were defined as at least three consecutive windows with (1) positive means, (2) at least six probes, and (3) P-values <0.016. Significant unbound regions were defined as at least three consecutive windows with (1) negative means, (2) at least six probes, and (3) P-values <0.016. The use of these selection criteria were justified by a low false positive rate (FPR) as determined by ChIP-qPCR (macroH2A1-bound region, FDR < 4.2%; macroH2A1-unbound region, FDR < 4.2%).
Boundary finding
MacroH2A1 domain boundaries were defined as the regions occurring between the edges of adjacent macroH2A1-bound and macroH2A1-unbound regions. Only boundaries exclusively containing well-tiled windows (at least six probes per window, indicating a lack of extensive repeat masking) were considered. Only under this criterion can the central location of the boundary be determined accurately. One-thousand-seven-hundred-fifty-four and 255 boundaries that satisfied these criteria were identified in IMR90 and MCF-7 cells, respectively.
Boundary to TSS distances
The distance of each boundary to the closest TSS (Fig. 2B) was determined using RefSeq gene annotations downloaded from the UCSC genome browser (Karolchik et al. 2003). A Fisher exact test was used to determine the significance of the patterns observed in the boundary histograms, compared with the background pattern obtained by determining the distance of all windows with at least six probes to the closest TSS, accounting for the fact that the boundary can occur in two possible orientations.
ChIP–chip-based classification of genes
For the ChIP–chip-based classification of genes, the genes on the ChIP–chip array were defined and macroH2A1-bound or macroH2A1-unbound. MacroH2A1-bound genes were defined as those that contained a macroH2A1-bound region within 3 kb downstream from the TSS. MacroH2A1-unbound genes were defined by the presence of windows from a macroH2A1-unbound region within 3 kb downstream from the TSS and the lack of a macroH2A1-bound region.
Expression-based classification of genes
The relative expression levels of genes in IMR90 cells were determined using GRO-seq data described in Core et al. (2008). The genes were then ordered for average expression signals, and the genes in each pentile were collected to determine the average macroH2A1-binding profile for each group (Fig. 5A). Expression microarray data were used to confirm the results of the GRO-seq analysis (Supplemental. Fig S8). For the microarray expression-based categorization of genes, three replicates of previously described MCF-7 expression microarray data (Affymetrix U133A, GEO accession no. GSE9253) were associated with the genes represented on the ChIP–chip array (Krishnakumar et al. 2008). The two replicates of IMR90 expression data used to categorize the IMR90 ChIP data were described previously (Heintzman et al. 2007). In cases where multiple probe sets corresponded to a given gene, all signals from the probe sets were averaged.
Distribution of expressed and unexpressed genes among the ChIP–chip classes
For the GRO-seq-based expression analysis (Fig. 5), genes were labeled expressed or unexpressed according to the methods described in Core et al. (2008). For the microarray-based expression analysis (Supplemental Fig S8), a gene on the ChIP–chip array was marked as unambiguously expressed or unexpressed, and all probe sets from all three expression array replicates corresponding to the gene were flagged unanimously present or absent, respectively. Any genes on the array not meeting these criteria were marked as ambiguous and were removed from the expression-based categorization analysis.
Gene ontology analysis of the ChIP–chip classes
The gene ontology analysis was done using the Generic Gene Ontology Term Finder (http://go.princeton.edu/cgi-bin/GOTermFinder/GOTermFinder) (Boyle et al. 2004). The collection of all genes that contained well-tiled windows with 4.5 kb both upstream of and downstream from the TSS—and that thus had the potential to be placed in one of the three macroH2A-defined gene classes—were used as the background for the analysis. An FDR of 1% was used as the cutoff for significant enrichment.
Multiple ChIP–chip correlation analysis
Data series containing IMR90 and MCF-7 ChIP–chip data were downloaded from the GEO Web site (http://www.ncbi.nlm.nih.gov/geo) (Barrett et al. 2009). Data series were chosen using two criteria: (1) The genomic data from the chosen series was performed using a two-color platform (i.e., Nimblegen or Agelent) and (2) the data series contained samples generated from IMR90 and MCF-7 cells. In total, 362 ChIP–chip data sets from the following GEO series were included in the analysis: GSE1778, GSE2072, GSE6292, GSE7118, GSE8887, GSE2672, GSE4355, GSE4355, GSE10504, GSE4905, GSE5175, GSE6625, GSE8667, GSE12126, GSE6624, GSE6634, GSE8716, GSE12126, GSE5445, GSE9015, GSE12650, GSE5559, GSE8716, GSE6385, GSE8855, GSE9029, GSE2672, GSE5559, and GSE13051. The data were processed with 1-kb windows identical to that of the macroH2A1 analysis. The Spearman correlation between macroH2A1 and each of the downloaded ChIP–chip data sets was then determined. The score of each correlation was defined as the negative of the log10 of the P-value for the correlation.
Gene-specific expression analysis
Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer's protocols. The RNA was reverse-transcribed and subjected to qPCR using gene-specific primers (sequences available on request). All target gene transcripts were normalized to β-actin. Each experiment was conducted a minimum of three times with independent isolates of total RNA. The P-values were determined using a two-tailed paired Student's t-test. The significance of the enrichment of macroH2A1 functional targets in the macroH2A1-bound class was determined using Fisher's exact test.
Acknowledgments
We thank J. Lis, A. Clark, J. Waterfall, D. Ruhl, M. Kininis, and T. Zhang for critical reading of this manuscript, technical advice, and helpful discussions. This work was supported by grants from the NIH/NIDDK (DK069710), the Cornell University Center for Vertebrate Genomics, and the Endocrine Society to W.L.K.; a post-doctoral fellowship from the American Heart Association (AHA) and from the NIH/NIDDK (DK079847-01) to M.J.G.; and a predoctoral fellowship from the AHA to K.M.F.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1876110.
Supplemental material is available at http://www.genesdev.org.
References
- Abbott DW, Laszczak M, Lewis JD, Su H, Moore SC, Hills M, Dimitrov S, Ausio J. Structural characterization of macroH2A containing chromatin. Biochemistry. 2004;43:1352–1359. doi: 10.1021/bi035859i. [DOI] [PubMed] [Google Scholar]
- Agelopoulos M, Thanos D. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J. 2006;25:4843–4853. doi: 10.1038/sj.emboj.7601364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S, Bouvet P, Dimitrov S. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell. 2003;11:1033–1041. doi: 10.1016/s1097-2765(03)00100-x. [DOI] [PubMed] [Google Scholar]
- Angelov D, Bondarenko VA, Almagro S, Menoni H, Mongelard F, Hans F, Mietton F, Studitsky VM, Hamiche A, Dimitrov S, et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Muratore-Schroeder TL, Diaz RL, Chow JC, Changolkar LN, Shabanowitz J, Heard E, Pehrson JR, Hunt DF, Allis CD. A phosphorylated subpopulation of the histone variant macroH2A1 is excluded from the inactive X chromosome and enriched during mitosis. Proc Natl Acad Sci. 2008;105:1533–1538. doi: 10.1073/pnas.0711632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO:TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschbeck M, Uribesalgo I, Wibowo I, Rue P, Martin D, Gutierrez A, Morey L, Guigo R, Lopez-Schier H, Di Croce L. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol. 2009;16:1074–1079. doi: 10.1038/nsmb.1665. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S, Luger K. Structural characterization of the histone variant macroH2A. Mol Cell Biol. 2005;25:7616–7624. doi: 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EY, Ferreira H, Somers J, Nusinow DA, Owen-Hughes T, Narlikar GJ. MacroH2A allows ATP-dependent chromatin remodeling by SWI/SNF and ACF complexes but specifically reduces recruitment of SWI/SNF. Biochemistry. 2008;47:13726–13732. doi: 10.1021/bi8016944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changolkar LN, Pehrson JR. macroH2A1 histone variants are depleted on active genes but concentrated on the inactive X chromosome. Mol Cell Biol. 2006;26:4410–4420. doi: 10.1128/MCB.02258-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changolkar LN, Costanzi C, Leu NA, Chen D, McLaughlin KJ, Pehrson JR. Developmental changes in histone macroH2A1-mediated gene regulation. Mol Cell Biol. 2007;27:2758–2764. doi: 10.1128/MCB.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changolkar LN, Singh G, Pehrson JR. macroH2A1-dependent silencing of endogenous murine leukemia viruses. Mol Cell Biol. 2008;28:2059–2065. doi: 10.1128/MCB.01362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo JH, Kim JD, Chung JH, Stubbs L, Kim J. Allele-specific deposition of macroH2A1 in imprinting control regions. Hum Mol Genet. 2006;15:717–724. doi: 10.1093/hmg/ddi485. [DOI] [PubMed] [Google Scholar]
- Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyen CM, An W, Angelov D, Bondarenko V, Mietton F, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, Dimitrov S. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol Cell Biol. 2006;26:1156–1164. doi: 10.1128/MCB.26.3.1156-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL. New functions for an ancient domain. Nat Struct Mol Biol. 2009;16:904–907. doi: 10.1038/nsmb0909-904. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- Ladurner AG. Inactivating chromosomes: A macro domain that minimizes transcription. Mol Cell. 2003;12:1–3. doi: 10.1016/s1097-2765(03)00284-3. [DOI] [PubMed] [Google Scholar]
- Mietton F, Sengupta AK, Molla A, Picchi G, Barral S, Heliot L, Grange T, Wutz A, Dimitrov S. Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol Cell Biol. 2009;29:150–156. doi: 10.1128/MCB.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Hernandez-Munoz I, Fazzio TG, Shah GM, Kraus WL, Panning B. Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem. 2007;282:12851–12859. doi: 10.1074/jbc.M610502200. [DOI] [PubMed] [Google Scholar]
- Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, Robin P, Mietton F, Harel-Bellan A, Dimitrov S, Hamiche A. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes & Dev. 2006;20:3324–3336. doi: 10.1101/gad.396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson JR, Fried VA. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- Pehrson JR, Fuji RN. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 1998;26:2837–2842. doi: 10.1093/nar/26.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson JR, Costanzi C, Dharia C. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J Cell Biochem. 1997;65:107–113. doi: 10.1002/(sici)1097-4644(199704)65:1<107::aid-jcb11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2006. [Google Scholar]
- Rasmussen TP, Huang T, Mastrangelo MA, Loring J, Panning B, Jaenisch R. Messenger RNAs encoding mouse histone macroH2A1 isoforms are expressed at similar levels in male and female cells and result from alternative splicing. Nucleic Acids Res. 1999;27:3685–3689. doi: 10.1093/nar/27.18.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Biol. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Sirchia SM, Ramoscelli L, Grati FR, Barbera F, Coradini D, Rossella F, Porta G, Lesma E, Ruggeri A, Radice P, et al. Loss of the inactive X chromosome and replication of the active X in BRCA1-defective and wild-type breast cancer cells. Cancer Res. 2005;65:2139–2146. doi: 10.1158/0008-5472.CAN-04-3465. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Sporn JC, Kustatscher G, Hothorn T, Collado M, Serrano M, Muley T, Schnabel P, Ladurner AG. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene. 2009;28:3423–3428. doi: 10.1038/onc.2009.26. [DOI] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. Modern applied statistics with S. Springer; New York: 2002. [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]