Abstract
Theoretical models predict that selfish DNA elements require host sex to persist in a population. Therefore, a transposon that induces sex would strongly favor its own spread. We demonstrate that a protein homologous to transposases, called α3, was essential for mating type switch in Kluyveromyces lactis. Mutational analysis showed that amino acids conserved among transposases were essential for its function. During switching, sequences in the 5′ and 3′ flanking regions of the α3 gene were joined, forming a DNA circle, showing that α3 mobilized from the genome. The sequences encompassing the α3 gene circle junctions in the mating type α (MATα) locus were essential for switching from MATα to MATa, suggesting that α3 mobilization was a coupled event. Switching also required a DNA-binding protein, Mating type switch 1 (Mts1), whose binding sites in MATα were important. Expression of Mts1 was repressed in MATa/MATα diploids and by nutrients, limiting switching to haploids in low-nutrient conditions. A hairpin-capped DNA double-strand break (DSB) was observed in the MATa locus in mre11 mutant strains, indicating that mating type switch was induced by MAT-specific DSBs. This study provides empirical evidence for selfish DNA promoting host sexual reproduction by mediating mating type switch.
Keywords: Mating type switch, transposable element, gene conversion, DNA double-strand break, sexual reproduction
Transposable elements (TEs) have had a profound effect on the shaping of modern genomes. In humans, close to half of the genome can be traced to TEs (Lander et al. 2001). TEs can be broadly divided into retrotransposons, which mobilize through an RNA intermediate, and DNA transposons, which mobilize through a “cut-and-paste” or “copy-and-paste” mechanism (Curcio and Derbyshire 2003). According to the parasitic DNA hypothesis, TEs are selfish DNA elements that persist in a population as a function of their ability to self-replicate (Doolittle and Sapienza 1980). The hypothesis that TEs constitute “junk” DNA that do not contribute to host fitness has been challenged (Feschotte and Pritham 2007). One example is the process of V(D)J recombination (Jones and Gellert 2004), during which the Rag1 and Rag2 proteins generate DNA double-strand breaks (DSBs) at recombination signal sequences (McBlane et al. 1995; Ramsden and Gellert 1995). RAG1 is closely related in sequence to Transib elements, a group of TEs found in different invertebrates (Kapitonov and Jurka 2005). Another example is the transposase-derived transcription factors FHY3 and FAR1, which regulate light signaling in Arabidopsis (Lin et al. 2007). It was suggested that FHY3 and FAR1 acquired the DNA-binding and transcriptional activation potential of transposases derived from a mutator-like TE.
Apart from V(D)J recombination, only a handful of examples are known in which programmed DSBs are induced during mitotic growth. Mating type switch in Saccharomyces cerevisiae provides such an example (Herskowitz et al. 1992; Haber 1998). Sexual differentiation in S. cerevisiae involves three cell types: the a-haploid and α-haploid cell types, and the a/α-diploid cell type (Herskowitz 1988). In haploids, the mating type is determined by the allele present in the mating type (MAT) locus. Additional copies of a and α information reside at the transcriptionally silent cryptic mating type loci. A DSB induced in MAT by the HO endonuclease facilitates mating type switch through a directional gene conversion between MAT and one of the cryptic mating type loci. Mating type switch is regulated through HO expression, which only occurs in haploid cells during the G1 phase of the cell cycle. HO-mediated switching is restricted to the Saccharomyces sensu stricto lineage and close relatives such as Candida glabrata, Kluyveromyces delphensis, and Saccharomyces castellii (Butler et al. 2004).
Interestingly, MAT loci from different ascomycetes usually contain a conserved set of regulatory genes (MATa1, MATα1, and MATα2), but also genes that are not shared between yeasts. As an example, the high-mobility group transcription factors Mata2/Mtla2 are present in Kluyveromyces lactis and Candida albicans, where they are important for a-cell identity (Tsong et al. 2006). However, the Mata2 gene has been lost in the Saccharomyces sensu stricto group, where a-cell identity is the default state. In addition, the C. albicans MTL loci contains genes encoding poly(A) polymerases, oxysterol-binding proteins, and phosphatidylinositol kinases (Hull and Johnson 1999), seemingly without functions in mating or determination of cell type identity. The basidiomycete Cryptococcus neoformans has MAT loci spanning >100 kb and encoding >20 genes (Lengeler et al. 2002), including mating pheromones and pheromone receptors. This acquisition of new genes in the MAT loci was suggested to resemble the evolution of sex chromosomes (Fraser et al. 2004).
K. lactis contains a gene in the MATα locus, called MATα3, that has not been found in other yeast species and whose function was unknown. In this study, we present evidence that the α3 protein is an ancient transposase with an essential role in mating type switching from MATα to MATa. During switching, the MATα3 gene (and HMLα3) is mobilized from the genome, forming a circular intermediate, which is similar to mobilization of some TEs (Curcio and Derbyshire 2003). We propose that adaptive evolution of a TE resulted in a novel mechanism for mating type switch in K. lactis, providing the first example of a transposon adapted to facilitate sexual reproduction.
Results
The α3 gene shares similarities with transposase genes
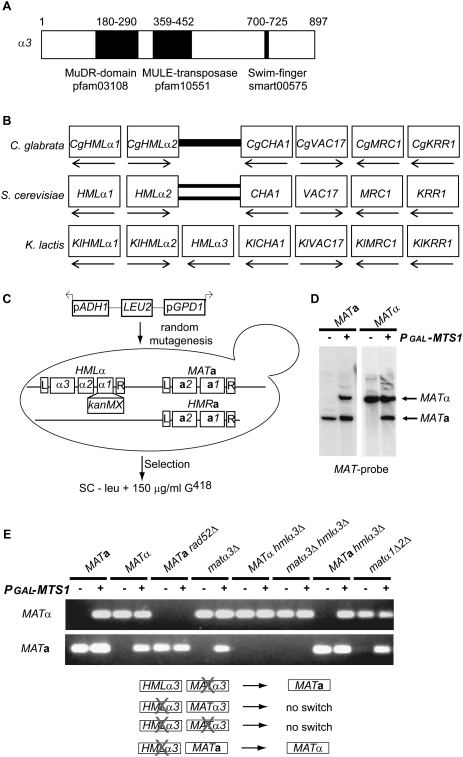
We explored mating type switching in K. lactis, a yeast species lacking a functional HO gene (Fabre et al. 2005). K. lactis contains a duplicated gene in the HMLα and MATα loci, α3, that has not been found in other yeast species. Position-specific iterated (PSI) BLAST (Altschul et al. 1997) searches revealed significant homology between α3 and transposases of the mutator-like (MULE) family (Fig. 1A) from plants, but also uncharacterized ORFs from other organisms (Supplemental Material). The analysis revealed the presence of three different protein domains constituting signature motifs of MULE transposases in α3 (Juretic et al. 2005; Babu et al. 2006). We compared the HMLα locus from K. lactis with the corresponding loci in other ascomycetes using the yeast gene order browser (Byrne and Wolfe 2005). Compared with the HMLα loci in S. cerevisiae and C. glabrata, the genes surrounding HMLα3, as well as their transcriptional orientation, were conserved. However, only K. lactis contained the HMLα3 gene (Fig. 1B). This result, together with the homology with transposases, suggested that the HMLα3 gene originated from a horizontal gene transfer of a TE.
Figure 1.
The α3 gene encoded a putative transposase essential for mating type switch. (A) Schematic drawing of the α3 ORF, indicating the relative positions of protein domains. (B) A synteny view of the HMLα loci from C. glabrata, S. cerevisiae, and K. lactis. Boxes denote genes, and boxes in the same column are orthologs. The gene names for the S. cerevisiae genes are indicated, as well as the transcriptional orientation of the genes (arrows). A thick bar indicates a lack of an ortholog. The double bars indicate lack of an ortholog and an inversion relative to the other genes. (C) Schematic representation of the genetic selection. SAY130 (hmlα1Δ∷kanMX) was mutagenized with pRS405-pApG. The genes within the MATa, HMRa, and HMLα loci are depicted, including the shared flanking repeats L and R. Derepression of hmlα1Δ∷kanMX was selected on plates lacking leucine, containing 150 μg/mL G418. (D) DNA blot analysis of BamHI-digested DNA from SAY45 (MATa) and SAY119 (MATα) containing pGAL-MTS1 (+) or plasmid alone (−), hybridized with a MAT-specific probe. The MATa and MATα bands are indicated. (E, top panel) PCR analysis of mating type switch in SAY45 (MATa), SAY119 (MATα), SAY 507 (MATa rad52Δ), SAY 121 (matα3Δ), SAY120 (hmlα3Δ), SAY126 (matα3Δ hmlα3Δ), SAY122 (MATa hmlα3Δ), and SAY688 (matα1Δα2Δ). Genomic DNA from these strains containing either the pGAL-MTS1 plasmid (+) or the empty vector (−) were subjected to MATa and MATα-specific PCR, and the resulting products were resolved on agarose gels. (Bottom panel) Schematic drawing of the α3 mutant strains indicating if they switched mating type.
Isolation of strains overexpressing Mating type switch 1 (Mts1) in a selection for defective silencing
We hypothesized that α3 was involved in mating type switch in K. lactis, but since our laboratory strains switched mating type very rarely, we had to develop an assay to test this notion. This was accomplished by using a genetic selection. This selection (aimed at identifying genes involved in transcriptional silencing) used a strain containing a silenced hmlα1Δ∷kanMX gene. The strain was mutagenized with linear DNA, taking advantage of the high illegitimate recombination frequency displayed by K. lactis (Kegel et al. 2006), selecting for G418 resistance (Fig. 1C; see the Materials and Methods for details). We used a plasmid containing a selectable marker (LEU2) flanked by strong promoters (GPD1 and ADH1) oriented in opposite directions. By linearizing the plasmid in between the two promoters prior to transformation, the insertion of the plasmid into the genome by illegitimate recombination was expected to cause strong outward-directed transcription. Among the G418-resistant strains recovered were three independent isolates that contained insertions in the same intergenic region (see the Materials and Methods). These insertions resulted in overexpression of the gene adjacent to the insertion (data not shown). This gene was the ortholog of S. cerevisiae RME1, a transcriptional regulator. The similarity between the predicted amino acid sequence of this protein and Rme1 was confined to the C-terminal domain containing three zinc finger motifs (Supplemental Fig. S1). We named this gene MTS1 since overexpression of this gene induced efficient switching in K. lactis (Fig. 1D). This result indicated that the retrieval of the Mts1-overexpressing mutants in the genetic selection was due to a physical translocation of the silenced kanMX gene into the expressed MAT locus, rather than an effect on transcriptional silencing.
HMLα3 was necessary for MATα-to-MATa switching
To investigate whether the α3 gene controlled mating type switch, we assayed switching in a set of K. lactis strains that harbored deletions in the HMLα3 and/or MATα3 genes. The strains contained a plasmid carrying MTS1 under the control of a strong GAL1 promoter to induce switching, or the empty plasmid as control. Switching events were detected by PCR amplification of the MATa or MATα loci (see the Materials and Methods). This assay worked reliably, as wild-type MATa or MATα strains showed Mts1-dependent switching (Fig. 1E), but a MATa rad52 mutant strain did not, since Rad52 is essential for gene conversion (Malone and Esposito 1980). The α1 and α2 proteins are expressed only from the MATα locus and were not required for mating type switch, since a mutation that removed most of these ORFs, including their common promoter region, switched mating type. We tested switching in two single-mutant strains (matα3Δ HMLα3 and MATα3 hmlα3Δ), as well as in two strains completely lacking α3 coding sequences (MATa hmlα3Δ and matα3Δ hmlα3Δ). Both the MATα and the matα3Δ strains containing the hmlα3Δ mutation did not switch mating type. However, the matα3Δ HMLα3 and MATa hmlα3Δ strains switched mating type (Fig. 1E). This showed that HMLα3 was required for switching from MATα to MATa, but not from MATa to MATα. As HMLα3 and MATα3 encode identical proteins, the unique dependence on HMLα3 was surprising, and was confirmed on a DNA blot (Supplemental Fig. S2). Importantly, a plasmid-borne GFP-α3 fusion gene could complement the switching defect of a MATα3 hmlα3Δ strain (Supplemental Fig. S2).
Mts1 binds to both the MATa and MATα loci
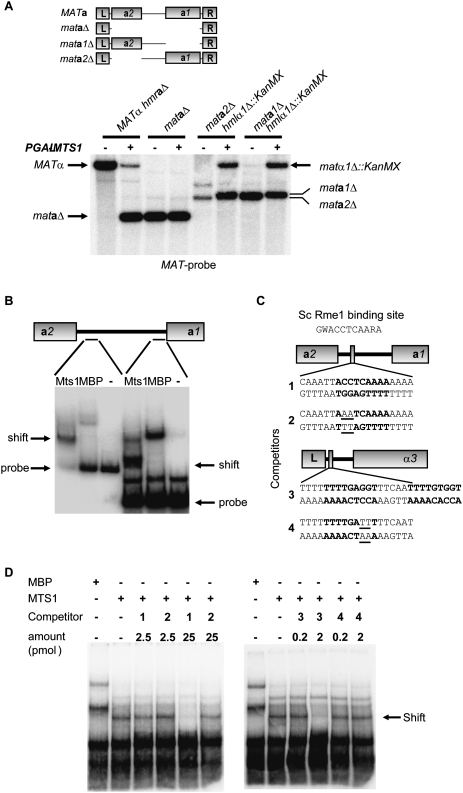
To begin exploring cis-acting sequences in the MATa locus that were necessary for switching, we generated three mutant strains. The mataΔ strain had the entire locus deleted except for the flanking sequences called L (left) and R (right). The L and R sequences are shared between all mating type loci and presumably are used as blocks of homology to resolve recombination intermediates. The genes encoded by the MATa locus were deleted individually in two separate strains, generating mata1Δ and mata2Δ strains, respectively. Overexpression of MTS1 in these strains demonstrated that the mataΔ strain did not switch mating type, whereas the mata1Δ and mata2Δ strains did (Fig. 2A). This indicated that the intergenic sequence in between MATa1 and MATa2 was essential for switching, but that the a1 and a2 gene products were dispensable.
Figure 2.
Identification of Mts1-binding sites in both the MATa and MATα loci. (A, top panel) Schematic drawing showing the deletions generated in the MATa locus. (Bottom panel) DNA blot analysis of BamHI-digested DNA from SAY726 (MATα hmraΔ), SAY727 (mataΔ), SAY990 (mata2Δ), and SAY1003 (mata1Δ) containing pGAL-MTS1 (+) or plasmid alone (−). A MAT-specific probe was used, and the identity of the bands is shown. (B) EMSA using probes from the MATa1–MATa2 intergenic region. Either recombinant Mts1, maltose-binding protein (MBP), or no protein (−) was added to the probes prior to electrophoresis. The location of the free probes and Mts1-dependent mobility shifts are indicated. (C) Drawing of the location of the Mts1-binding sites in the MATa and MATα loci. The DNA sequence of the 19-bp duplexes (1–4) used as competitors in D is shown. Note the second potential binding site at MATα (competitor 3), which was not included on the competing fragment. (Top) The Rme1-binding site in S. cerevisiae is shown. (W) A or T; (R) A or G. (D) EMSA using the probe on the left in B, but also including competitor duplexes with the sequences shown in C. Competitors used and their concentrations are indicated above the lanes.
This result prompted us to screen the 855-base-pair (bp)-long MATa1 and MATa2 intergenic region for Mts1-binding sites. Seven DNA probes spanning this region were tested for binding to recombinant Mts1 using an electrophoretic mobility shift assay (EMSA) (Supplemental Fig. S3). Two of the probes showed specific mobility shifts (Fig. 2B). Using the DNA fragment showing the most robust mobility shift and competitor DNA, we pinpointed one Mts1-binding site to 111–129 bp upstream of the start codon of MATa2 (Fig. 2C). Examination of this 19-bp sequence revealed the presence of a close match to the previously described S. cerevisiae Rme1-binding site (Shimizu et al. 1998). Three similar sites were found within the other probe originating from the MATa1 and MATa2 intergenic region that showed a mobility shift. Interestingly, two closely juxtaposed copies of a very similar sequence were also found upstream of the MATα3 gene close to the L repeat (Fig. 2C). We used 19-bp duplexes corresponding to the site upstream of MATa2 and one of the MATα sites as competitors for the Mts1 mobility shift. Only 2 pmol of a sequence originating from the α3 upstream region abolished the mobility shift (Fig. 2D). For the sequence from the a2 upstream region, 25 pmol were required for efficient competition. In both cases, we used duplexes with 2 bp changed, compared with the genomic sequence as controls. Both competed very inefficiently for the Mts1 mobility shift (Fig. 2D).
Deletion of Mts1-binding sites reduced switching from MATα to MATa
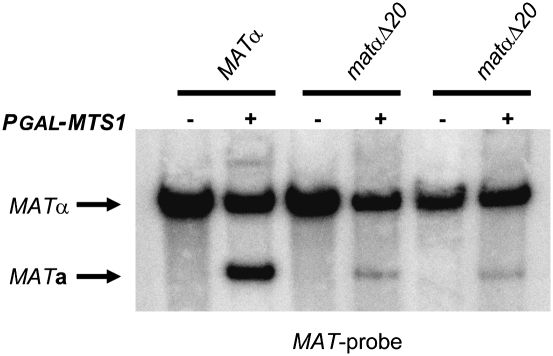
Next, we investigated whether the Mts1-binding sites in the MATα locus were important for switching. We introduced a 20-bp deletion close to the L repeat in the MATα locus (matαΔ20), which deleted the two closely juxtaposed putative Mts1-binding sites shown in Figure 2C. Two independent isolates were transformed with either pGAL-MTS1 plasmid or empty vector, and the mating type switch was scored by genomic DNA blot. The strains harboring the deletion displayed a severe reduction in switching (Fig. 3). Compared with the wild type with a MATa/MATα ratio of 0.38, two isolates of the isogenic mutant strains had ratios of 0.13 and 0.08, respectively. Hence, deletion of these Mts1-binding sites reduced switching by a factor of 3–5. Deleting only one of the Mts1-binding sites in MATα had no effect on switching (data not shown). We concluded that Mts1 had a direct role in mating type switch by binding to distinct binding sites in the MATα locus. It should be pointed out that the defective switching observed in the matαΔ20 strain cannot be a consequence of a decreased expression of the MATα3 gene, since a matα3Δ strain switched mating type with normal efficiency (Supplemental Fig. S2).
Figure 3.
Mts1-binding sites in the MATα locus were important for efficient mating type switch. DNA blot analysis using BamHI-digested DNA from SAY509 (MATα), SAY1377, and 1378 (both matαΔ20, deleting the two putative Mts1-binding sites shown in Fig. 2C), containing pGAL-MTS1 (+) or plasmid alone (−). A MAT-specific probe was used, and the identity of the bands is shown.
MTS1 expression was regulated by nutrients and cell type
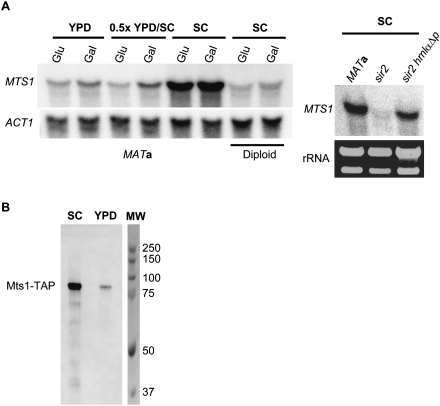
It was observed previously that switch of the mating type in K. lactis did not occur until cells were transferred to malt extract (ME) medium (Herman and Roman 1966), a nutrient-limited medium. To determine if the transcription of MTS1 was different in nutrient-rich (YPD) and synthetic (SC) medium, we measured transcript levels from MATa wild-type cells grown in SC, YPD, and 0.5× SC/YPD using either glucose or galactose as a carbon source. The MTS1 transcript was approximately sixfold more abundant in SC compared with YPD or 0.5× SC/YPD (Fig. 4A), but the carbon source had only minor effects. Since mixing rich medium with synthetic medium (0.5× SC/YPD) led to low expression levels, the rich medium appeared to have a dominant repressing effect.
Figure 4.
Mts1 expression was regulated by nutrients and cell type. (A, left panel) RNA blot analysis using total RNA from SAY45 and SAY45 × SAY119 (diploid) as indicated below the blot. Cells were grown in YPD, SC, or 0.5× YPD/SC using either glucose (glu) or galactose (gal) as a carbon source. The blot was hybridized with an MTS1-specific probe, stripped, and rehybridized with an ACT1-specific probe as a loading control. (Right panel) RNA blot analysis using total RNA from SAY45, SAY102 (sir2Δ), and SAY189 (sir2Δ hmlαΔp). Cells were grown in SC medium and the blot was hybridized with an MTS1-specific probe. As loading control, rRNA bands from the same strains are shown. (B) Protein blot analysis of total proteins (0.1 A600) from SAY988 (MTS1-TAP) grown in either SC or YPD. Anti-TAP (Invitrogen) was used as the primary antibody. Molecular weight markers (MW) are shown on the right.
The fact that MTS1 transcription was also repressed in a MATa/MATα diploid grown in SC medium (Fig. 4A) indicated that an additional repression mechanism was controlling MTS1 expression. To distinguish between ploidy effects and the possibility that MTS1 was repressed by the diploid-specific a1/α2 repressor, we investigated MTS1 transcription in a MATa sir2 and a MATa sir2 hmlαΔp background. In a MATa sir2 strain, MTS1 transcription was repressed. This repression was alleviated by the hmlαΔp mutation that abolishes transcription of the HMLα1 and HMLα2 genes (Fig. 4A, right panel). Therefore, MTS1 transcription was regulated by cell type, likely through the a1/α2 repressor.
A strain containing a MTS1-TAP fusion gene was generated, and we measured the steady-state levels of Mts1-TAP in cells grown in YPD and SC using an anti-TAP antibody. Consistent with the RNA blot, we found sevenfold higher levels of Mts1-TAP in cells grown in SC (Fig. 4B) compared with YPD.
α3 mobilized from the genome in response to Mts1 overexpression
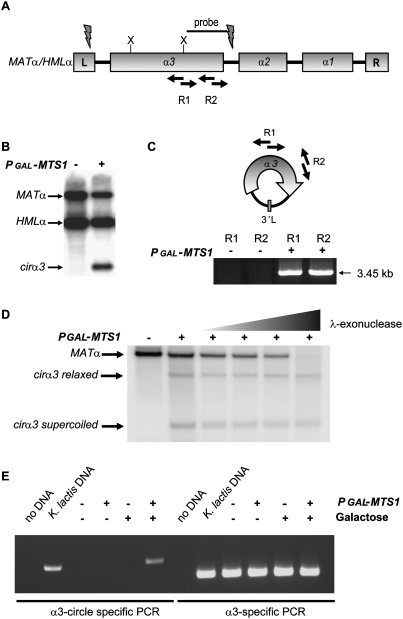
Next, we explored if the α3 gene had other features in common with TEs. Figure 5A schematically shows the MATα/HMLα loci, displaying the three ORFs as well as the flanking sequences L and R. DNA blotting of MATα strains overexpressing Mts1 revealed a band (Fig. 5B) that hybridized to an α3-specific probe (Fig. 5A), which could not be explained as a linear DNA fragment based on the positions of restriction enzyme sites. We hypothesized that this band could represent a circular DNA molecule. To test this, we performed inverse PCR on genomic DNA from strains overexpressing Mts1 using two different primer pairs that could only render a product if a circular DNA molecule was present (Fig. 5C). A PCR product was readily observed. DNA sequencing revealed that the band corresponded to an α3 gene fragment in which the 5′ and 3′ ends were fused. Thus, the band observed on the DNA blot (Fig. 5B) corresponds to a head-to-tail fusion of the α3 gene (the band is missing an internal XbaI fragment in this XbaI digest). To confirm the presence of a circular form of α3, we treated BamHI-digested genomic DNA (there are no BamHI sites in the circle) with λ-exonuclease, which specifically degrades linear DNA but not circles or nicked circles. The results showed that a linear MATα fragment was sensitive to the exonuclease, but a putative supercoiled and relaxed form of the α3 circle was resistant (Fig. 5D). Thus, α3 retained the ability to excise from the genome in a manner similar to many TEs. It should be pointed out that, on DNA blots, the hybridization signal from the novel band was stronger in MATα strains than in MATa strains (data not shown). Hence, most of the signal originated from the MATα locus, although some of it originated from the HMLα locus. To learn more about the formation of the α3, circle we tested whether it could be induced ectopically in S. cerevisiae. A plasmid containing the entire K. lactis HMLα locus was introduced into a wild-type S. cerevisiae strain. Then we introduced either plasmid alone or the pGAL-MTS1 vector. Circle-specific PCR showed that the α3 circle was formed in S. cerevisiae in an MTS1-dependent manner (Fig. 5E). Thus, the only K. lactis-specific loci required for circle formation were MTS1 and HMLα. Notably, circularization is a feature of several TEs (Curcio and Derbyshire 2003) and may be a characteristic of the mutator elements in maize (Sundaresan and Freeling 1987), but is almost unheard of for nontransposable genes.
Figure 5.
The α3 gene mobilized from the genome. (A) Schematic drawing of the MATα/HMLα loci. The position of the probe used in B, the positions of the PCR primers used in C, and XbaI sites are indicated. The flashes indicate where the α3 gene circle is joined. (B) DNA blot analysis of XbaI-digested DNA from SAY119 (MATα) containing pGAL-MTS1 (+) or plasmid alone (−) hybridized with the probe indicated in A. The MATα, HMLα, and cirα3 bands are indicated. (C, top panel) Inverse PCR strategy used to reveal the nature of the cirα3 band. (Bottom panel) Genomic DNA from strain SAY119 containing pGAL-MTS1 (+) or plasmid alone was subjected to inverse PCR using the primer pairs R1 and R2, followed by agarose gel electrophoresis. (D) DNA blot analysis of BamHI-digested DNA from SAY119 (MATα), containing vector alone (−) or pGAL-MTS1 (+). Lanes 3–6 were treated with increasing concentrations of λ-exonuclease (0.02, 0.1, 0.5, and 2.5 U, respectively). The identity of the bands hybridizing to the α3-specific probe is shown beside the blots. (E) S. cerevisiae strain SAY73 containing K. lactis HMLα (M plasmid) was subjected to α3 circle-specific PCR and α3 gene-specific PCR, followed by agarose gel electrophoresis, as indicated. In addition, either pRS414 without insert (−) or pRS414-pGAL-MTS1 (+) was introduced and grown in the presence of glucose (−) or galactose (+), as indicated. As controls, no DNA lanes and K. lactis DNA lanes were included.
Mutations in the DDE motif and SWIM finger of α3 abolished switching
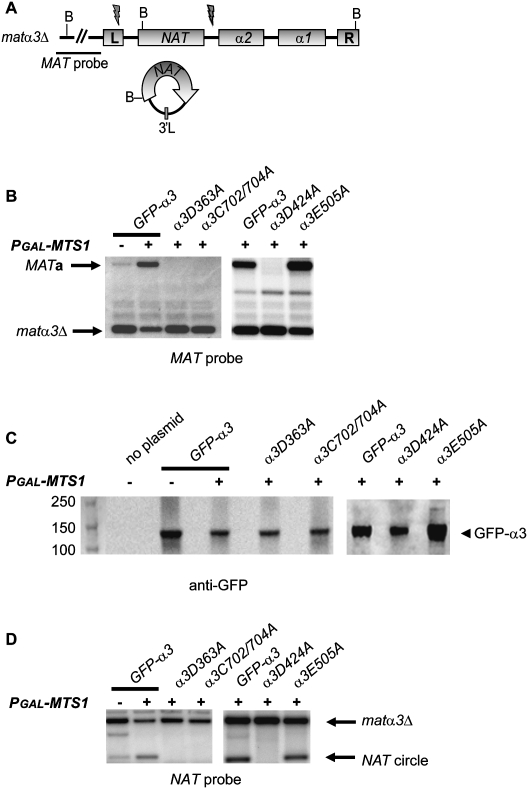
To confirm that α3 indeed was required for switching, we generated matα3Δ∷NAT hmlα3Δ∷NAT alleles (Fig. 6A) that were complete null alleles flush from the start codon to the stop codon. Then, we introduced a plasmid expressing a GFP-α3 fusion gene controlled by the GPD1 promoter. A pGAL-MTS1 plasmid was introduced simultaneously, and we tested whether the strains switched mating type. The results in Figure 6B show that the GFP-α3 fusion gene could complement the switching defect of the matα3Δ hmlα3Δ strain. In the absence of pGAL-MTS1, we observed low-level background switching due to the endogenous MTS1 gene, which is induced in the synthetic growth medium used in this experiment (Fig. 4). To begin characterizing which features of α3 were important for function, we generated mutations in GFP-α3. Multiple sequence alignments with α3 and known mutator transposases are difficult to assemble due to low sequence similarity. However, comparing α3 with the plant transposase-derived protein FAR1 by PSI-BLAST (Supplemental Fig. S4; Lin et al. 2007) predicted that D363 and D424 were part of the catalytic DDE motif. The DDE motif is required for coordination of metal ions in many transposases (Kulkosky et al. 1992; Haren et al. 1999). The position of the “E” residue was ambiguous, but the glutamate in position 505 of α3 aligned with a glutamate in FAR1. We therefore generated α3D363A, α3D424A, and α3E505A alleles. The other allele tested exchanged two of the cysteines in the SWIM finger for alanine residues (α3C702/704A). The only mutant α3 protein that complemented the switching defect was α3E505A (Fig. 6B). By performing protein blots using an anti-GFP antiserum, we showed that all fusion proteins were present at similar steady-state levels (Fig. 6C). D363, D424, and the SWIM finger of α3 were therefore essential for the mating type switch from MATα to MATa. To investigate whether the formation of the α3 gene circle had the same molecular requirements as the mating type switch, we stripped the blots used in Figure 6B and reprobed with a NAT probe. If the formation of the circle required the α3 protein, then we expected to generate a NAT-containing circle with α3 flanking sequences only when a functional α3 allele was present. The results were completely consistent with this notion (Fig. 6D). Thus, the α3 protein was required for excising the α3 gene from the genome.
Figure 6.
Mutations in the DDE motif and the SWIM finger of α3 abolished mating type switch. (A) Schematic drawing of the matα3Δ∷NAT locus with the location of BamHI sites shown. The location of the MAT probe is indicated. Flashes indicate where the α3 gene circle is joined, and below is a drawing of the resulting NAT-containing circle. (B) DNA blot analysis of BamHI-digested genomic DNA from strain SAY1356 (matα3Δ hmlα3Δ) containing a plasmid with a GFP-α3 gene fusion and mutant derivatives, as indicated. The strains contained either pGAL-MTS1 (+) or the empty vector (−). A MAT-specific probe was used, and the MATa and matα3Δ bands are indicated. (C) Shown is a protein blot using an α-GFP antibody against GFP-α3. Total protein (0.1 A600) prepared from cells expressing wild-type or mutant GFP-α3, as indicated, was subjected to electrophoresis and blotting. A strain lacking the GFP-α3 plasmid shows that the antibody was specific. Molecular weight markers in kilodaltons are shown on the left. (D) The same blots as in B, but hybridized with a NAT-specific probe. The probe detects both a matα3Δ∷NAT/hmlα3Δ∷NAT-specific band and a NAT circle with α3 flanking sequences, as indicated.
The sequences at the MATα3 circle junction were required for switching from MATα to MATa
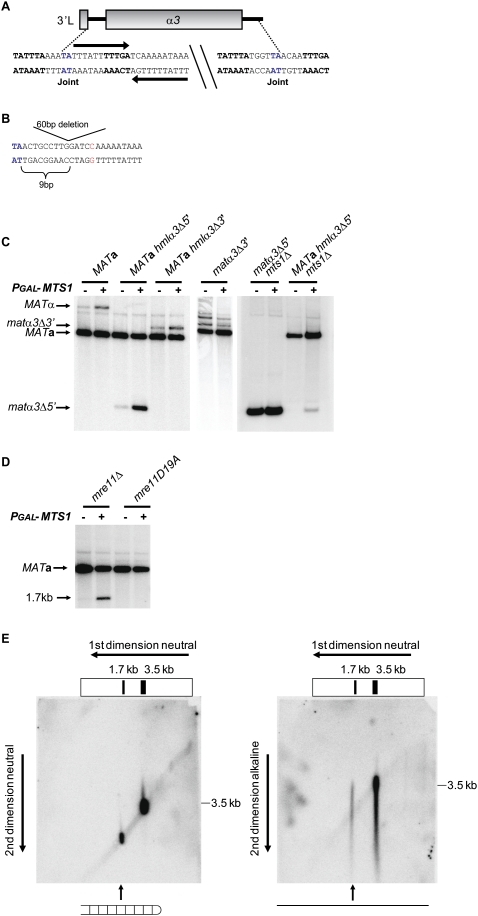
Next, we investigated the role of the DNA sequences encompassing the circle junctions, both for mating type switching and for formation of the α3 gene circle. Sequencing the α3 circle junction revealed that the fusion took place at a TA sequence, the 5′ junction resided within the L repeat, and the 3′ junction was in the α3–α2 intergenic region (Fig. 7A). The 5′ junction sequence had an 11-bp inverted repeat immediately downstream from the TA sequence. In addition, there were other sequences shared between the 5′ and 3′ junctions (see Fig. 7A). First, we investigated the importance of these sequences at HMLα by generating strains with deletions of the 5′ junction (hmlα3Δ5′) or the 3′ junction (hmlα3Δ3′). Overexpressing Mts1 in a MATa hmlα3Δ3′ strain did not result in α3 circle formation, showing that the sequences at the 3′ junction were essential. Strikingly, deletion of the 5′ junction resulted in a novel circle junction. This novel junction also occurred at a TA sequence, and the distance with respect to the α3 ORF was maintained (Fig. 7B). Hence, the sequence spanning the 5′ circle junction in wild-type strains was not essential for excising the HMLα3 gene.
Figure 7.
Switching from MATα to MATa required the sequences spanning the MATα3 circle junctions and formation of DNA hairpins in the MATa locus. (A) Schematic drawing of the DNA sequences spanning the α3 circle junctions. The TA sequence in blue indicates the fusion point, bases in bold indicate sequences shared between the 5′ and 3′ circle junctions, and arrows show a 22-bp inverted repeat. (B) A 60-bp deletion (5′ junction sequence) and a 26-bp deletion (3′ junction sequence) were introduced in HMLα, resulting in either a novel circle junction or no circle formation, respectively. Shown is the novel circle junction generated in the hmlα3Δ5′ mutant. Note the 9-bp spacing from the start of the deletion to a novel fusion point, which was the same spacing as in the wild-type junction. For cloning purposes, a C (red) was introduced in the 60-bp deletion of the 5′ junction sequence. (C) DNA blot analysis using BamHI-digested DNA from SAY45 (MATa), SAY875 (MATa hmlα3Δ5′), SAY876 (MATa hmlα3Δ3′), SAY1194 (matα3Δ3′), SAY930 (matα3Δ5′ mts1Δ), and SAY931 (MATa hmlα3Δ5′ mts1Δ). The strains contained either pGAL-MTS1 (+) or plasmid alone (−). A MAT-specific probe was used, and the identity of the bands is indicated. (D) DNA blot analysis using BamHI-digested DNA from SAY559 (MATa mre11Δ) and SAY973 (MATa mre11-D19A) containing pGAL-MTS1 (+) or plasmid alone (−). A MAT-specific probe was used, and the identity of the bands hybridizing to the probe is shown beside the blot. (E) Two-dimensional gel electrophoresis followed by hybridization to a MAT-specific probe. (Left panel) Both dimensions were performed under neutral conditions (1× TAE). (Right panel) The first dimension was performed under neutral conditions, and the second dimension was performed under denaturing conditions (30 mM NaOH, 0.1 mM EDTA). The size of a 3.5-kb marker in the second dimension is shown on the left of both panels. Indicated below the panels is the duplex versus ssDNA nature of the hairpin DNA intermediate.
Then, we explored the relationship between circle formation and mating type switching. We investigated switching in MATa hmlα3Δ5′ and MATa hmlα3Δ3′ strains and found that both strains switched their mating type (Fig. 7C).
We noticed low levels of switching in the absence of Mts1 overexpression (Fig. 7C). To test whether the endogenous MTS1 gene mediated this switch, we explored switching in a MATa hmlα3Δ5′ mts1Δ double-mutant strain. Removing the endogenous MTS1 gene abolished background switching, which could be rescued by introduction of the pGAL-MTS1 plasmid. Hence, endogenous levels of MTS1 could mediate mating type switch.
Next, we investigated the importance of the circle junction sequences at the MATα locus. We thus generated matα3Δ5′ mts1Δ and matα3Δ3′ mutant strains. Overexpression of Mts1 in these strains showed that neither of them switched their mating type to MATa (Fig. 7C). Thus, the circle junction sequences were essential for switching when present at MATα. Consequently, for switching from MATα to MATa, generating a MAT-derived normal α3 circle was essential. These observations strongly suggested that, for switching from MATα to MATa, the DNA lesions generated during MATα3 excision were essential switching intermediates.
Formation of a hairpin-capped DSB at MATa
We tested the trans-acting molecular requirements for switching. In both MATa and MATα strains, several genes known to mediate gene conversion were essential for switching (data not shown). A MATa mre11Δ strain did not undergo switching (Fig. 7D), but surprisingly, a novel faster migrating band hybridizing to the MAT-specific probe was observed. We hypothesized that this band represented a switching intermediate that was stabilized in mre11Δ strains. The size of the band (1.7 kb) indicated the presence of a DSB in the MATa1–MATa2 intergenic region. An allele carrying a point mutation, mre11D19A, corresponding to the mre11D16A allele in S. cerevisiae, was generated. Mre11D16A was shown previously to be strongly compromised for Mre11 exonuclease activity (Lewis et al. 2004). The mre11D19A strain neither switched mating type nor generated the intermediate seen in the mre11Δ strain. Hence, the Mre11 exonuclease activity was required for completing the switch, but lack of it was not sufficient for stabilizing the intermediate.
Based on previous studies that showed a requirement for Mre11 in opening hairpins (Paull and Gellert 1998; Trujillo and Sung 2001), we hypothesized that the intermediate observed in the mre11Δ strain could be a DNA hairpin. To test this notion, we performed two-dimensional gel electrophoresis (Fig. 7E). The first dimension was performed under neutral conditions. Gel slices were cut from the lanes, turned perpendicular, and subjected to second-dimension electrophoresis under either neutral or alkaline conditions. Neutral/neutral electrophoresis generated the expected size pattern of 3.5-kb and 1.7-kb duplexes. When the second dimension was performed under alkaline conditions, the 1.7-kb duplex gave rise to a band that migrated as an ∼3.4-kb, ssDNA molecule (Fig. 7E). The 3.5-kb band gave rise to the expected 3.5-kb single-stranded band on the alkaline blot. This result indicated that the DSB observed in the mre11Δ strain terminated in a DNA hairpin. We also used a probe that hybridized to the distal side of the DSB in the mre11Δ strain, which likewise revealed the presence of a DNA hairpin (data not shown). We concluded that, during MATa to MATα switching, a hairpin-capped DSB was generated in the MATa locus.
Discussion
Evolution of mating type switching in ascomycetes
Here, we demonstrate a novel mechanism for mating type switching in K. lactis, which is induced by a DNA-binding protein (Mts1) and requires a transposase-like protein (α3). Given that K. lactis contains an HO pseudogene (Fabre et al. 2005), it seems likely that an HO-mediated switching mechanism was the ancestral state and that the MTS1/α3-mediated mechanism described here arose later. This prediction is at odds with a previous study (Butler et al. 2004) that suggested that the HO gene arose after S. cerevisiae and K. lactis diverged from each other. At that time, however, the K. lactis HO pseudogene had not been described. We suggest that Mts1 directly induces switching, since two Mts1-binding sites in the MATα locus were important. However, the Mts1 amino acid sequence does not suggest that Mts1 catalyzes DNA cleavage. We speculate that Mts1 may recruit α3 or other factors to the MAT loci to facilitate DNA cleavage. Interestingly, the transcription of Mts1 was repressed by the a/α-cell type and nutrients. The cell type regulation is similar to the regulation of HO transcription in S. cerevisiae, limiting switching to haploid cells. The nutrient regulation of MTS1 explains an early observation (Herman and Roman 1966) that showed that switching in K. lactis only occurred in nutrient-limited media. Recent observations indicate that the RAS/cAMP pathway is responsible for the nutrient regulation of MTS1 transcription (E Barsoum and S Åström, unpubl.). We suggest that this is an adaptation to the fact that the diplophase is unstable in K. lactis. If a clone of haploid cells is deprived of nutrients, some cells in the population must switch mating type prior to mating, meiosis, and spore formation. The induction of MTS1 transcription in nutrient-limited conditions may facilitate this development.
We obtained preliminary data indicating that other yeast species may share a switching mechanism with K. lactis. We explored the closest relatives to K. lactis, collectively known as clade 11 (Kurtzman 2003; Lachance 2007), for the presence of α3-like sequences. Because Kluyveromyces dobzhanskii contained an α3-like gene whose size was altered (possibly circularized) in response to K. lactis Mts1 overexpression, it was likely to use a similar mechanism (data not shown). An α3 probe also hybridized to DNA from Kluyveromyces wickerhamii and Kluyveromyces marxianus. Hence, several other ascomycetes may have a similar mating type switching mechanism.
The role of α3 in mating type switch
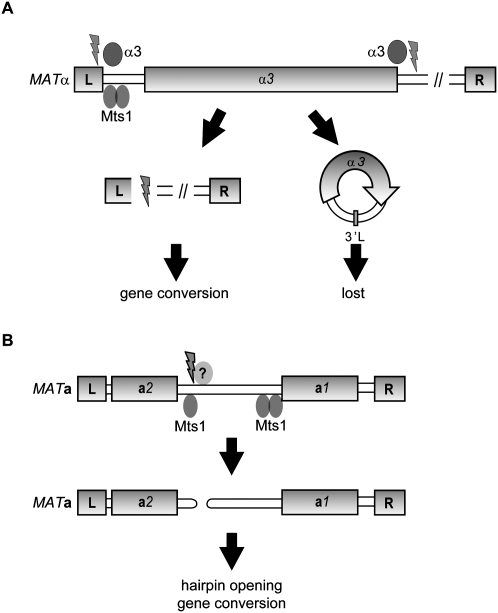
According to a theoretical model by Hickey (1982), spread of TEs in a population depends on sexual reproduction. Definitive empirical evidence for this model is difficult to obtain, but asexuals and self-fertilizing sexuals appear to contain fewer TEs in their genomes than obligate sexually reproducing organisms (Wright and Finnegan 2001). One prediction of this model is that a TE that caused the host to reproduce sexually should be successfully transmitted. Therefore, once established at the MATα/HMLα loci, a TE that reintroduced a complete sexual cycle to its host would favor its own spread through the population. The α3 gene described in this study fits this model. We suggest a two-step model for MATα-to-MATa switching in K. lactis, in which Mts1 binds to sites in the MATα locus and facilitates α3-dependent excision of the MATα3 gene (Fig. 8). We assume that the α3 gene circles are passively lost in subsequent cell cycles. Supporting this notion, we never observed α3 transposition or detected α3 remnants elsewhere in the K. lactis genome. The observation that the α3 gene circle junctions were important for switching from MATα to MATa strongly indicates that the DNA lesions generated during MATα3 mobilization were essential switching intermediates. We propose that the α3 gene circle junctions are required for proper binding/activity of α3. The resulting DNA lesions are then channeled into a gene conversion pathway in which the HMRa sequence is copied into the MAT locus. Consistent with this view, switching in K. lactis requires the recombination proteins Rad51 and Rad52 (Fig. 1E; data not shown).
Figure 8.
Model for mating-type switch in K. lactis. (A) In MATα cells, binding of Mts1 to two sites close to the L repeat is important to induce switching. The α3 transposase-like protein presumably acts at sites flanking the MATα3 gene, resulting in excision of an α3 gene circle. The circles are lost in subsequent cell cycles, as they lack an origin of replication. The resulting DNA lesions are channeled into a gene conversion pathway, in which the repetitive L and R sequences, present also at the HMRa locus, act as blocks of homology to resolve the recombination intermediates. (B) In MATa cells, binding of Mts1 to several sites in the MATa1– MATa2 intergenic region induces switching. An unknown protein generates a hairpin-capped DSB. The hairpin is opened in an Mre11-dependent manner, and the DSB induces a gene conversion using the HMLα locus as donor sequence.
It is intriguing that HMLα3 is essential for mating type switch, but MATα3 is not. That HMLα3 escapes silencing is obvious, which was expected since we observed previously that both MATα3 and HMLα3 are expressed (Åström et al. 2000). We hypothesize that the MATα3 gene is not expressed during switching. This idea is based on the observation that expression of GFP-α3 from an ectopic promoter can complement the switching defect of a MATα hmlα3Δ strain. Alternatively, if α3 is unstable and has a late role in the switching process, then this may explain the dependence on HMLα3.
Similarities between mating type switch and V(D)J recombination
During V(D)J recombination, hairpin intermediates are formed at the end of coding segments (Roth et al. 1992). The resolution of these hairpins, followed by joining of two coding segments, generates the recombined immunoglobulin gene. The Rag1 and Rag2 proteins are responsible for generating the hairpins during V(D)J recombination. In K. lactis, we observe a hairpin-capped DSB in a MATa mre11Δ strain, but not in a wild-type strain or a MATα mre11Δ strain. The in vivo observation of hairpins during V(D)J recombination was made in thymocytes from SCID mice (Roth et al. 1992), which are compromised in nonhomologous end-joining (NHEJ). Hence, in both cases, the presence of hairpins appears to be transient, and defects in DNA repair proteins stabilize them. The inability of the mre11Δ strain to process hairpins is consistent with observations that the MRX(N) [Mre11/Rad50/Xrs2(Nbs1)] complex from both yeast and humans exhibits manganese-dependent cleavage of hairpin structures in vitro (Paull and Gellert 1998; Trujillo and Sung 2001). It was shown previously in S. cerevisiae that MRX and Sae2 process hairpin intermediates, thereby preventing genome rearrangements (Lobachev et al. 2002). In this study, the hairpins were formed at long inverted repeats (IR) consisting of human Alu sequences, and the IRs were in fact responsible for DSB induction. The longest IR in the MATa1–MATa2 intergenic region was only 7 bp. We therefore suggest that hairpin formation in the MATa1–MATa2 intergenic region is independent of long IRs similar to the transesterification reaction performed by Rag1/2 (Roth et al. 1992; McBlane et al. 1995). The hairpin intermediate was not stabilized by an N-terminal phosphoesterase mutation (mre11D19A), suggesting that the resulting protein was able to process the hairpin. The corresponding mutant in S. cerevisiae (mre11D16A) produces a protein with a severe defect in the exonuclease activity of Mre11 (Lewis et al. 2004). A K. lactis mre11D19A mutant strain is methyl methanesulfonate (MMS)-sensitive, similar to an S. cerevisiae mre11D16A mutant strain, suggesting that the K. lactis mre11D19A allele is also defective in exonuclease activity. We surmise that Mre11 exonuclease activity is not important for opening the hairpin, but that another function of Mre11 performs this task. Alternatively, another nuclease, such as Sae2, performs the actual hairpin opening, as suggested in S. cerevisiae (Lengsfeld et al. 2007). It should be noted, however, that Mre11D19A could not complete the switching process from MATa to MATα, indicating that Mre11 exonuclease activity performs an important function during the process.
Mating type switch from MATα to MATa requires the HMLα3 gene, but switching from MATa to MATα did not. Therefore, in the MATa locus, DNA lesions must arise independently of α3 (Fig. 8). We hypothesize that the enzymes that generate lesions in MATa form hairpins, which are stabilized in the absence of Mre11. As the only known enzymes that generate hairpins after DNA cleavage are transposases (Zhou et al. 2004) or their derivatives (Roth et al. 1992), the unidentified enzymes that generate lesions in MATa may be related to transposases.
We did not observe DSBs in the MAT locus during switching in wild-type K. lactis strains. In addition, alkaline gels did not reveal any single-stranded lesions in the MAT region (data not shown). The GAL1–10 promoter used to drive Mts1 expression is leaky in glucose-containing medium in K. lactis. The result is that switching is not synchronously induced, which may provide an explanation for the lack of an observed DSB. It is also feasible that the DNA lesions are very transient. The lesions may be introduced in a synaptic complex (Polard and Chandler 1995), a protein–DNA assembly in which the donor and acceptor sites are correctly aligned, as shown for V(D)J recombination and a number of transposons (Curcio and Derbyshire 2003). Cutting the acceptor sites is almost immediately followed by strand invasion of the donor, limiting the time a DNA lesion can be observed. In models incorporating a synaptic complex, the sequences of both the donor and acceptor loci contribute to successful transposition. However, during switching in K. lactis, we never observed a mutation in the donor sequences that reduce switching. In the MATα hmraΔ strain, for example, in which the entire HMRa locus was deleted except for the L and R sequences, mataΔ strains were generated efficiently upon Mts1 overexpression. Based on these findings, we find it unlikely that synaptic complexes are essential intermediates during switching in K. lactis.
Finally, preliminary evidence suggests that switching is directional in K. lactis (Supplemental Fig. S2). Whether or not the mechanism is similar to S. cerevisiae remains unknown, but will be the subject of future studies.
In summary, we show that an element similar to TEs directs mating type switch in K. lactis. Our observations are consistent with the idea that this TE was fixed in the K. lactis genome by aiding host sexuality.
Materials and methods
Yeast strains
The yeast strains used in this study are listed in Supplemental Table S1. Gene disruptions/deletions/fusions relied on homologous recombination using either a one-step (Kegel et al. 2006) or a two-step (Orr-Weaver et al. 1981) method. The one-step gene disruption procedure used kanMX or NAT PCR fragments amplified from pFA6a-KANMX (Bahler et al. 1998) or pAG25 (Goldstein and McCusker 1999). Genomic manipulations were confirmed by DNA blots or genomic PCR. Sequences of oligonucleotides used are available on request. SAY1346 (hmlα3Δ∷NAT) and SAY1348 (hmlα3Δ∷kanMX) were generated by transforming the appropriate PCR fragment into SAY572 (nej1Δ∷LEU2), selecting for nourseothricin (90 μg/mL; CloNat) or Geneticin (200 μg/mL) resistance. Strains SAY1351 (MATa hmlα3Δ∷NAT) and SAY1446 (MATα hmlα3Δ∷kanMX) were obtained by crossing SAY1346 and SAY1348 with a MATα strain. Strains SAY726 (hmraΔ∷NAT) and SAY727 (mataΔ∷NAT), in which the entire sequence between the L and R repeats were deleted at HMRa and MATa, respectively, were generated by the one-step procedure. Generation of SAY990 (mata2Δ∷NAT) and SAY1003 (mata1Δ∷NAT) used the same procedure, but in these strains only the MATa1 or MATa2 ORFs (from the start codon to the stop codon) were deleted. SAY973 (mre11D19A) was generated as follows: Plasmid pJ18 was linearized with BamHI and introduced into CK213 by selecting for uracil prototrophs. The URA3 gene was popped out on 5-fluoro-orotic acid (5-FOA), and the segregants were screened for the mre11D19A mutation using genomic PCR, followed by DNA sequencing. SAY1351 was transformed with pPMB35 (pGAL-MTS1), and the plasmid was selected against on 5-FOA. Segregants in which a mating type switch event occurred were identified by PCR. SAY1356 (hmlα3Δ∷NAT matα3Δ∷NAT) was recovered among the segregants. SAY875 (hmlα3Δ∷5′junction) and SAY876 (hmlα3Δ∷3′junction) were generated by transforming MunI-linearized p648 or PacI-linearized p650 into CK213-4C, respectively, selecting for Uracil prototrophs and a pop-out procedure. SAY1194 (matα3Δ∷3′junction) was obtained through the introduction of pGAL-MTS1 into SAY876, as described above. SAY754 (mts1Δ∷LEU2) was generated by introducing a BamHI–PstI DNA fragment from pCXJ10-mts1Δ∷LEU2 into AKY125 (lig4Δ∷kanMX), selection for Leu+ isolates, and a cross with CK213-4C. SAY930 (matα3Δ∷5′junction mts1Δ∷LEU2) and SAY931 (hmlα3Δ∷5′junction mts1Δ∷LEU2) were generated by crossing SAY875 with SAY754 (mts1Δ∷LEU2) containing pGAL-MTS1. SAY688 (matα1α2Δ∷LEU2) was generated by transforming SAY119 with PacI-linearized p392, selecting for uracil prototrophs, followed by the pop-out procedure. Strain SAY988 (MTS1-TAP), in which a TAP tag was fused to the C terminus of Mts1, was generated as described (Puig et al. 2001) by transforming strain SAY572 (nej1Δ∷LEU2) with the appropriate PCR fragment. SAY1377 and SAY1378 (matαΔ20), carrying deletions of the two Mts1-binding sites residing close to the L repeat in the MATα locus, were generated by transforming SAY509 (MATα nej1Δ∷LEU2) with HpaI-linearized pEB85, followed by a pop-out procedure.
Genetic selection
The genetic selection in which the MTS1 alleles were isolated was performed as follows: SAY130 (hmlα1Δ∷kanMX) was transformed with pRS405-pApG using the lithium acetate method. Prior to transformation, pRS405-pApG was linearized in between the ADH1 and GPD1 promoters using BamHI. Among 7 × 104 transformed colonies, we isolated 39 stable G418-resistant strains. For allelism tests, the revertants were crossed to SAY119 and tetrad analysis was performed. Cosegregation of G418 resistance and Leu+ indicated that the plasmid insertion was allelic to the mutation that caused G418 resistance, a criterion that 27 isolates satisfied. For determining the exact locations of the pRS405-pApG insertions, we performed inverse PCR. Five micrograms of genomic DNA from the revertant strains were digested with MspI for 1 h at 37°C, heat-inactivated for 20 min at 65°C, and diluted 10-fold in 1× ligase buffer, and 1 U of T4 DNA ligase was added. After ligation, the DNA was precipitated, lyophilized, and resuspended in 20 μL of water. One microliter of this mixture was PCR-amplified using ADH1 primer pairs 549 (5′-CTATCAAGTATAAATAGACCTGC-3′) and 554 (5′-GGAAGGCCGTATACCGTTG-3′) and GPD1 primer pairs 550 (5′-GAGTGAAACTGTTACGTTACC-3′) and 555 (5′-CGAAACTGCCAATACCAAGC-3′), thus generating PCR fragments with DNA flanking the insertion point of pRS405-pApG. The PCR fragments were sequenced (Macrogen), and the sequences were compared with the K. lactis genomic sequence (http://cbi.labri.fr/Genolevures). The MTS1-1, MTS1-2, and MTS1-3 strains had plasmid insertions at −324, −351, and −709 relative to the MTS1 start codon, respectively. Six of the isolated strains had independent insertions of pRS405-pApG in the SIR2 ORF. The remaining 18 isolates will be presented elsewhere.
Plasmids
Cloning was performed using standard methods (Sambrook and Russell 2001). For details, please see the Supplemental Material.
Methods
DNA/RNA/protein blots, DNA/RNA/protein preparations, and transformation of yeast and Escherichia coli followed standard protocols (Schiestl and Gietz 1989; Ausubel 1999; Sambrook and Russell 2001). Yeast media was prepared as described (Burke et al. 2000). EMSAs were performed as described (Sjöstrand et al. 2002) using ∼10 ng of partially purified maltose-binding protein (MBP)/Mts1 in each reaction. The two probes showing a specific mobility shift corresponded to sequences 37–185 bp upstream of the a2 ORF and 1–131 bp upstream of the a1 ORF. Recombinant MBP–Mts1 was partially purified and digested with factor Xa using the procedure recommended by the manufacturer (New England Biolabs). Site-directed mutagenesis was performed according to the protocol outlined in the Stratagene QuickChange procedure. For DNA/RNA blots, 32P-labeled probes were used and quantified when indicated using a PhosphorImager (Fuji). λ-Exonuclease digestion of genomic DNA was performed for 15 min at 37°C using the buffer supplied by the manufacturer (New England Biolabs). For protein blots, the anti-TAP primary antibody (Invitrogen) was diluted 1:500 and the anti-GFP antibody (Clontech) was diluted 1:1000. Mating type-specific PCR used primer sets 193 (5′-CCAATTCAAAATTGCTACCC-3′) and 553 (5′-GTCAAGCGCCTTGTCCAATG-3′) for MATa and 551 (5′-CCTTGATGAACTCAGTCTGAG-3′) and 553 for MATα. Amplifications used standard conditions with an annealing temperature of 55°C.
Acknowledgments
We thank S. Carter, M. Mannervik, and P. Ljungdahl for critical reading of the manuscript, and members of the laboratory for helpful suggestions. This work was supported by grants from the Swedish Research Council and the Swedish Cancer Society to S.U.Å.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.557310.
Supplemental material is available at http://www.genesdev.org.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åström SU, Kegel A, Sjöstrand JO, Rine J. Kluyveromyces lactis Sir2p regulates cation sensitivity and maintains a specialized chromatin structure at the cryptic α-locus. Genetics. 2000;156:81–91. doi: 10.1093/genetics/156.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM. Short protocols in molecular biology: A compendium of methods from current protocols in molecular biology. Wiley; New York: 1999. [Google Scholar]
- Babu MM, Iyer LM, Balaji S, Aravind L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 2006;34:6505–6520. doi: 10.1093/nar/gkl888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in yeast genetics: A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- Butler G, Kenny C, Fagan A, Kurischko C, Gaillardin C, Wolfe KH. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc Natl Acad Sci. 2004;101:1632–1637. doi: 10.1073/pnas.0304170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: Combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ, Derbyshire KM. The outs and ins of transposition: From mu to kangaroo. Nat Rev Mol Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Fabre E, Muller H, Therizols P, Lafontaine I, Dujon B, Fairhead C. Comparative genomics in hemiascomycete yeasts: Evolution of sex, silencing, and subtelomeres. Mol Biol Evol. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2004;2:e384. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Haber JE. Matin type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- Haren L, Ton-Hoang B, Chandler M. Integrating DNA: Transposases and retroviral integrases. Annu Rev Microbiol. 1999;53:245–281. doi: 10.1146/annurev.micro.53.1.245. [DOI] [PubMed] [Google Scholar]
- Herman A, Roman H. Allele specific determinants of homothallism in Saccharomyces lactis. Genetics. 1966;53:727–740. doi: 10.1093/genetics/53.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I, Rine J, Strathern JN. Mating type determination and matin type interconversion in Saccharomyces cerevisiae. In: Jones EW, et al., editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press; New York, NY: 1992. pp. 583–656. [Google Scholar]
- Hickey DA. Selfish DNA: A sexually-transmitted nuclear parasite. Genetics. 1982;101:519–531. doi: 10.1093/genetics/101.3-4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Johnson AD. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science. 1999;285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- Jones JM, Gellert M. The taming of a transposon: V(D)J recombination and the immune system. Immunol Rev. 2004;200:233–248. doi: 10.1111/j.0105-2896.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005;15:1292–1297. doi: 10.1101/gr.4064205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel A, Martinez P, Carter SD, Åström SU. Genome wide distribution of illegitimate recombination events in Kluyveromyces lactis. Nucleic Acids Res. 2006;34:1633–1645. doi: 10.1093/nar/gkl064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 2003;4:233–245. doi: 10.1016/S1567-1356(03)00175-2. [DOI] [PubMed] [Google Scholar]
- Lachance MA. Current status of Kluyveromyces systematics. FEMS Yeast Res. 2007;7:642–645. doi: 10.1111/j.1567-1364.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lengeler KB, Fox DS, Fraser JA, Allen A, Forrester K, Dietrich FS, Heitman J. Matin type locus of Cryptococcus neoformans: A step in the evolution of sex chromosomes. Eukaryot Cell. 2002;1:704–718. doi: 10.1128/EC.1.5.704-718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Storici F, Van Komen S, Calero S, Sung P, Resnick MA. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics. 2004;166:1701–1713. doi: 10.1534/genetics.166.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318:1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- Malone RE, Esposito RE. The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci. 1980;77:503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: A model system for the study of recombination. Proc Natl Acad Sci. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Polard P, Chandler M. An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes & Dev. 1995;9:2846–2858. doi: 10.1101/gad.9.22.2846. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes & Dev. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- Roth DB, Menetski JP, Nakajima PB, Bosma MJ, Gellert M. V(D)J recombination: Broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2001. [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Li W, Covitz PA, Hara M, Shindo H, Mitchell AP. Genomic footprinting of the yeast zinc finger protein Rme1p and its roles in repression of the meiotic activator IME1. Nucleic Acids Res. 1998;26:2329–2336. doi: 10.1093/nar/26.10.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöstrand JO, Kegel A, Åström SU. Functional diversity of silencers in budding yeasts. Eukaryot Cell. 2002;1:548–557. doi: 10.1128/EC.1.4.548-557.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Freeling M. An extrachromosomal form of the Mu transposons of maize. Proc Natl Acad Sci. 1987;84:4924–4928. doi: 10.1073/pnas.84.14.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- Wright S, Finnegan D. Genome evolution: Sex and the transposable element. Curr Biol. 2001;11:R296–R299. doi: 10.1016/S0960-9822(01)00168-3. [DOI] [PubMed] [Google Scholar]
- Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]