Abstract
Genome rearrangements are often associated with genome instability observed in cancer and other pathological disorders. Different types of repeat elements are common in genomes and are prone to instability. S-phase checkpoints, recombination, and telomere maintenance pathways have been implicated in suppressing chromosome rearrangements, but little is known about the molecular mechanisms and the chromosome intermediates generating such genome-wide instability. In the December 15, 2009, issue of Genes & Development, two studies by Paek and colleagues (2861–2875) and Mizuno and colleagues (pp. 2876–2886), demonstrate that nearby inverted repeats in budding and fission yeasts recombine spontaneously and frequently to form dicentric and acentric chromosomes. The recombination mechanism underlying this phenomenon does not appear to require double-strand break formation, and is likely caused by a replication mechanism involving template switching.
Keywords: Inverted repeat, breakage–fusion–bridge cycle, genome instability, template switch, recombination
Maintaining genome stability is crucial for normal cell growth, as revealed by the fact that many cancers and numerous genetic diseases are associated with genome rearrangements (Kolodner et al. 2002). The molecular mechanisms underlying such large-scale chromosome changes, often referred to as gross chromosomal rearrangements (GCRs), are complex. Genetic studies, conducted mostly in the budding yeast Saccharomyces cerevisiae, have helped to identify numerous genes and pathways suppressing such alterations. Many of these have established functions in monitoring the integrity of replication forks (S-phase checkpoints) or in mediating genetic exchanges by means of homologous recombination (HR), an important genome integrity mechanism promoting the repair of double-strand breaks (DSBs) or single-strand gaps formed during replication or arising from processing DNA-damaging lesions. At-risk DNA elements have been identified in the genome: They contain different types of repeat sequences that are prone to instability and influence the rate of genome rearrangements (Gordenin and Resnick 1998).
Several lines of evidence led to the common view that GCR formation is initiated by replication dysfunction. Accordingly, mutations in genes encoding DNA replication proteins or S-phase checkpoint components, or causing dysregulation of replication origins result in genome instability and increased GCR rates (Chen and Kolodner 1999; Myung et al. 2001; Lengronne and Schwob 2002). The DNA structures arising during DNA replication that are processed to yield genome rearrangements are not yet clear, but DSBs have been proposed to mediate many of them. In the December 15, 2009, issue of Genes & Development, two studies by Paek et al. (2009) and Mizuno et al. (2009) uncover novel replication-based mechanisms that likely do not involve a DSB intermediate, and that operate at nearby inverted repeats to generate dicentric chromosome intermediates that, upon segregation, lead to breakage and further chromosome rearrangements.
Inverted repeats, palindromes, and dicentric chromosome formation
Repetitive elements are abundant in genomes, and frequently serve as substrates for genome rearrangements (Batzer and Deininger 2002). The biological basis for this extends beyond the high density of these elements and is explained by their ability to disrupt DNA replication and essential repair processes. For instance, simple repeat sequences located within a short distance of each other are prone to replication errors, leading to small deletion and duplication mutations. Repetitive DNA sequences also induce recombination. Not surprisingly, therefore, low-copy repeats (LCRs) were found to commonly flank rearranged genomic segments associated with recurrent genomic rearrangements found in genomic disorders (Stankiewicz and Lupski 2002). Such rearrangements are most often mediated by nonallelic HR (NAHR), a mechanism of ectopic HR between highly homologous but nonallelic LCR substrates (Stankiewicz and Lupski 2002). While recombination between direct repeats results in deletion and/or duplication of the genomic segment flanked by the LCRs, recombination between inverted repeats can invert the intervening sequence (Inoue and Lupski 2002).
Inverted repeats can be separated by a few base pairs to many kilobases of DNA. Palindromes—a specific type of inverted repeat separated by only very few base pairs—are poorly tolerated in Escherichia coli cells and are underrepresented in the S. cerevisiae and human genomes (Lobachev et al. 2000; Stenger et al. 2001), presumably due to their tendency to form hairpin and cruciform structures (Lobachev et al. 2002; Lemoine et al. 2005), which could be recognized and cleaved by a nuclease (Leach and Stahl 1983) or could affect or slow down DNA replication (Ahmed and Podemski 1998). Studies in budding yeast have shown that palindromes can rearrange to form acentric or dicentric chromosomes (Narayanan et al. 2006; Voineagu et al. 2008), a finding confirmed in fission yeast by a recent study (Mizuno et al. 2009). Acentric and dicentric chromosomes are unstable chromosome intermediates: Acentric palindromic chromosomes have been proposed to be precursors for extrachromosomal elements. The two centromeres of a dicentric chromosome are pulled apart at anaphase to separate nuclei, causing breakage of the dicentric and the production of new ends that fuse to form a new dicentric chromosome, thus establishing a cycle—the breakage–fusion–bridge cycle (BFB)—that is believed to play a major part in gene amplification in cancer (Albrecht et al. 2000; Narayanan et al. 2006; Tanaka and Yao 2009).
In the December 15, 2009, issue of Genes & Development, Paek et al. (2009) found that, in budding yeast, naturally occurring or synthetic inverted repeats separated by ∼1.3–5.5 kb of DNA and having as few as ∼20 base pairs (bp) of homology can fuse to form acentric or dicentric chromosomes (Paek et al. 2009). This nearby inverted repeat fusion phenomenon has been reported previously in bacteria and in fission yeast (Bi and Liu 1996; Albrecht et al. 2000). Since nearby inverted repeats separated by a few kilobases of DNA are unlikely to form cruciform structures as do palindromes, it is most probable that the mechanisms leading to formation of dicentric and acentric chromosomes in these situations are also different. The recent studies by Paek et al. (2009) and Mizuno et al. (2009) characterized the factors influencing the formation of acentric and dicentric chromosome intermediates at nearby inverted repeats or palindromic loci in budding and fission yeast, respectively.
DSBs as important intermediates leading to genome rearrangements
Previous genetic studies aimed at uncovering the factors and pathways that keep the genome rearrangements at very low levels suggested that the majority of rearrangement events are the result of DSB repair processes (Kolodner et al. 2002).
Studies in budding yeast have shown that ∼2% of the DSBs induced by ionizing radiation that fall within repetitive elements give rise to NAHR-mediated chromosomal aberrations reshaping the genome, where the two ends formed by a single DSB at repetitive sequences act independently in strand invasion reactions involving the sister chromatid or the homologous chromosomes (Argueso et al. 2008).
Many of the rearrangements occurring in a genome have been proposed to be associated with ongoing replication. Supportive of the idea that replication stress can be an important source of breaks leading to DNA structural changes, previous studies that characterized the mechanisms of GCR increase in S-phase checkpoint mutants in budding yeast have found that all of the genome rearrangements in these mutants involved the deletion of a chromosome end and de novo addition of a new telomere (Myung et al. 2001). Based on other findings, a model has emerged that suggests that replication forks are prone to stall at repetitive elements (Voineagu et al. 2008, 2009) and collapse in checkpoint mutants (Lopes et al. 2001; Cha and Kleckner 2002), thus generating DNA breaks (Cha and Kleckner 2002) that often become substrates for telomerase, or can be engaged in break-induced replication (BIR), a HR pathway proposed to restart collapsed forks or repair single DSBs (McEachern and Haber 2006).
Although BIR is normally faithful, if the broken end invades a homolog instead of a sister molecule, it can lead to loss of heterozygozity; however, if it involves homologous sequences at a different chromosomal position, then translocation, duplication, or deletion can result. BIR could also lead to complex chromosomal rearrangements if it involves multiple rounds of strand invasion, DNA synthesis, and dissociation, especially if these events occurred within repetitive regions (Narayanan et al. 2006; Smith et al. 2007). Similar to other HR events, BIR is normally dependent on homology and HR factors (McEachern and Haber 2006). However, a new mechanism of BIR involving microhomology-mediated strand invasion has been proposed to account for those complex rearrangements without substantial homologous change (Payen et al. 2008; Hastings et al. 2009). This model proposes that, following resection, the 3′-end ssDNA derived from the DSB of a collapsed fork can anneal with any ssDNA template with which it shares microhomology and which is located in physical proximity, in a HR protein-independent manner (Hastings et al. 2009).
Formation of dicentric chromosomes has also been proposed to occur through several mechanisms involving the formation of a DSB intermediate that could then be processed in several alternative ways (Lobachev et al. 2007). Using different approaches and different organisms, the recent studies by Mizuno et al. (2009) and Paek et al. (2009) come to an astonishingly similar conclusion: Nearby inverted repeats can fuse to form acentric and dicentric chromosomes in a replication-dependent manner, but without involving a DSB as an intermediate (Mizuno et al. 2009; Paek et al. 2009). The factors implicated in this rearrangement and the main arguments against a DSB intermediate are described below.
Factors and mechanisms implicated in the fusion of nearby inverted repeats or palindromes
The studies by Mizuno et al. (2009) and Paek et al. (2009) analyzed the mechanisms through which nearby inverted repeats lead to GCRs and chromosome instability. In one study, Paek et al. (2009) focused their attention on a highly unstable region of the budding yeast chromosome VII containing nearby inverted repeats. They report that those inverted repeats first fuse to form a dicentric chromosome. This is not a site-specific phenomenon, as such fusion events are shown to occur at other inverted repeats present at different sites in the yeast genome. The instability of this locus was shown previously by Weinert's group (Admire et al. 2006 to increase when DNA replication was disrupted, such as in checkpoint mutants where replication forks stalling at the repeats fail to be stabilized). In this recent study, Paek et al. (2009) found that the fusion observed, which led to dicentric chromosome formation, did not require known DSB repair (HR, nonhomologous end-joining [NHEJ], or single-strand annealing [SSA] pathways) or replication fork pathways (post-replication repair [PRR] and BIR factors). Interestingly, the only factor found to promote fusion was Srs2, a helicase that removes Rad51 from ssDNA filaments (Krejci et al. 2003; Veaute et al. 2003), suggesting that regulating Rad51 activity may affect this process.
On the other hand, the experimental system of Mizuno et al. (2009) uses a construct consisting of two replication termination sequences (RTS1) placed in inverted orientation at the ura4+ locus in fission yeast (Lambert et al. 2005), and other modifications of this construct creating a perfect palindrome, a palindrome interrupted by a 14-bp spacer, and other constructs aimed at assessing the effect of the orientation and size of the repeat element (RTS1) or of other factors that may intervene in the replication of this genomic segment (Mizuno et al. 2009). The RTS1 locus is naturally present into the Schizosaccharomyces pombe genome, and ensures efficient mating type switching in fission yeast by imposing replication fork arrest and regulating the direction of replication (Dalgaard and Klar 2001; Codlin and Dalgaard 2003). Fork arrest at RTS1 requires the Rtf1 protein: In its absence, forks no longer arrest at RTS1, which is then replicated normally (Lambert et al. 2005). Thus, the RTS1 sequences used by Mizuno et al. (2009) offer the means to conditionally and efficiently stall replication forks by attenuating rtf1+ transcription. Previous work conducted in Carr's laboratory (Lambert et al. 2005) has shown that, in such an experimental system, in the presence of Rtf1, >95% of the forks arrest at RTS1 and restart in a manner dependent on HR proteins.
Mizuno et al. (2009) found that replication fork arrest within the palindrome causes a high frequency of chromosomal rearrangement in which the inverted repeats fuse to form acentric and dicentric chromosomes. In contrast to the situation described by Paek et al. (2009), the majority of the fusion events are dependent on HR protein functions (Mizuno et al. 2009). If the initial pausing within the RTS1 repeats would generate a DSB intermediate that will then promote fork restart in a HR-dependent manner, then one would predict that those DSB intermediates should become visible after inhibition of the restart events in HR-deficient cells (or both recombination and resection-defective strains). However, strikingly, DSBs are not detectable by pulse field gel analysis of the molecular species arising during this chromosomal rearrangement event (Mizuno et al. 2009). Thus, although the occasional formation of DSBs during arrest at such elements cannot be ruled out, the evidence provided by Mizuno et al. (2009) compellingly suggests that the vast majority of these restart events do not involve DSBs as intermediates.
Replication-mediated mechanisms for fusion of nearby inverted repeats
Based on the observation that such fusion events are stimulated by replication arrest within the repeated sequence or by replication problems, the two recent studies (Mizuno et al. 2009; Paek et al. 2009) propose a replication-based mechanism involving an aberrant switch of templates (therefore called faulty template switch mechanisms) to account for the chromosomal rearrangements observed. These template switch events are likely triggered by fork stalling and exposure of ssDNA gaps at the stalled fork. Previously, to account for complex rearrangements occurring in genome disorders, an aberrant type of template switch mechanism involving template exchanges between different replication forks has been proposed. Following stalling of the lagging replisome—imposed by regions having abundant repetitive elements—there is a switch to a nearby template at another fork located in proximity (Branzei and Foiani 2007; Lee et al. 2007).
The model of Mizuno et al. (2009) resembles this previous one, and the investigators propose that, following fork arrest within the repeat, HR proteins promote the formation of a recombinogenic 3′ end that can invade the opposite repeat located in physical proximity due to the cruciform conformation that the palindromic substrate under study is prone to adopt (Fig. 1A; Mizuno et al. 2009). This creates the formation of a Holliday junction intermediate, the resolution of which results in either fully replicated chromosomes having the original conformation, or acentric and dicentric chromosome formation (Mizuno et al. 2009).
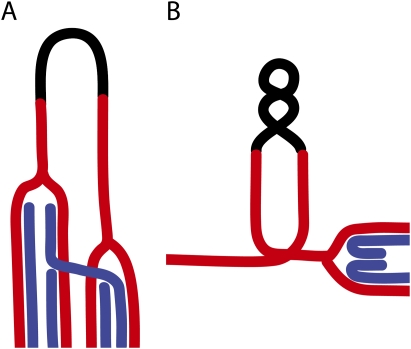
Figure 1.
Template switch events at nearby inverted repeats. Inverted repeats are shown in red, the DNA segment interrupting the repeats are shown in black, and the newly synthesized DNA are shown in blue. In A, the DNA segment containing the inverted repeats (palindrome) adopts a cruciform structure. One replication fork arrests within the repeat and can invade the one of the opposite repeat (Mizuno et al. 2009). In B, the nearby inverted repeats are separated by longer DNA segments, but chromatin looping can bring these elements into physical proximity. Slowing down or fork arrest during replication of the repeat sequence will lead to accumulation of positive supercoil ahead of the replication fork, and this can induce fork regression (Olavarrieta et al. 2002). Following reversal of the four-way junction (Atkinson and McGlynn 2009), one of the newly synthesized strands containing some segment of one of the repeat sequences can reanneal (reinvade) to the wrong repeat sequence, leading to the fusion (joining) of the repeat sequences (Paek et al. 2009).
In the model proposed by Paek et al. (2009), the fork stalled within the repeats fails to be rescued by pathways dependent on HR or PRR factors shown previously to be involved in template switch formation (Branzei et al. 2008; Branzei and Foiani 2009), and undergoes an alternate type of template switch involving fork regression (Fig. 1B; Atkinson and McGlynn 2009). Reversed forks are not common replication intermediates occurring at stalled forks (Sogo et al. 2002), but accumulation of a positive superhelical strain in front of the stalled fork can lead to the formation of four-way junctions or reversed forks (Postow et al. 2001; Olavarrieta et al. 2002). It is possible that nearby inverted repeats are involved in looping events that may form topological barriers, induce fork arrest, and promote regression of the stalled fork (Fig. 1). If, during this fork regression process, the nascent chain pairs with the incorrect, nearby sequence with which it bears some elements of homology (Paek et al. 2009), then when the regressed fork is reversed back, the nascent chain can reanneal or reinvade the wrong sequence, leading to the fusion events observed by Paek et al. (2009) (Fig. 1).
The protein promoting this pairing event is not yet known, as the HR factor Rad52, which has SSA activity, is not required for the fusion (Paek et al. 2009). However, if different pathways contribute to the outcome, the individual role of the distinct factors involved may be difficult to establish genetically. In vitro, Rad51 inhibits the SSA activity of Rad52 (Wu et al. 2008). The observation that Srs2 (in which Rad51 is somewhat up-regulated by failure to disrupt Rad51 filaments) is required for the fusion events observed may mirror a possible role for Rad52 in this process. In the template switch model proposed by Paek et al. (2009), completion of DNA replication following this faulty strand annealing leads to the formation of a dicentric or acentric chromosome.
The models proposed are speculative, but offer a method of investigating the exact mechanisms and factors implicated in such deleterious template exchanges. As large inverted repeats with high sequence homology are abundant in the human genome (Warburton et al. 2004), and dicentric chromosomes increase the genomic instability and likely the cancer risk (Tanaka and Yao 2009), characterizing the replication-associated events and factors implicated in the formation of such chromosome intermediates is important also for understanding the underlying causes of chromosomal rearrangements characteristic of oncogenic transformation.
Acknowledgments
We thank all members of our laboratories for helpful discussions. The work in D.B.’s laboratory is supported by ERC grant 242928 and the Associazione Italiana per la Ricerca sul Cancro. The work in M.F.’s laboratory is supported by grants from Telethon, the Associazione Italiana per la Ricerca sul Cancro, and the European Community.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1884810.
References
- Admire A, Shanks L, Danzl N, Wang M, Weier U, Stevens W, Hunt E, Weinert T. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes & Dev. 2006;20:159–173. doi: 10.1101/gad.1392506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A, Podemski L. Observations on template switching during DNA replication through long inverted repeats. Gene. 1998;223:187–194. doi: 10.1016/s0378-1119(98)00159-0. [DOI] [PubMed] [Google Scholar]
- Albrecht EB, Hunyady AB, Stark GR, Patterson TE. Mechanisms of sod2 gene amplification in Schizosaccharomyces pombe. Mol Biol Cell. 2000;11:873–886. doi: 10.1091/mbc.11.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso JL, Westmoreland J, Mieczkowski PA, Gawel M, Petes TD, Resnick MA. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci. 2008;105:11845–11850. doi: 10.1073/pnas.0804529105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Bi X, Liu LF. DNA rearrangement mediated by inverted repeats. Proc Natl Acad Sci. 1996;93:819–823. doi: 10.1073/pnas.93.2.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Template switching: From replication fork repair to genome rearrangements. Cell. 2007;131:1228–1230. doi: 10.1016/j.cell.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair (Amst) 2009;8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Codlin S, Dalgaard JZ. Complex mechanism of site-specific DNA replication termination in fission yeast. EMBO J. 2003;22:3431–3440. doi: 10.1093/emboj/cdg330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes & Dev. 2001;15:2060–2068. doi: 10.1101/gad.200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordenin DA, Resnick MA. Yeast ARMs (DNA at-risk motifs) can reveal sources of genome instability. Mutat Res. 1998;400:45–58. doi: 10.1016/s0027-5107(98)00047-5. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet. 2002;3:199–242. doi: 10.1146/annurev.genom.3.032802.120023. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Leach DR, Stahl FW. Viability of λ phages carrying a perfect palindrome in the absence of recombination nucleases. Nature. 1983;305:448–451. doi: 10.1038/305448a0. [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CMB, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Schwob E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1) Mol Cell. 2002;9:1067–1078. doi: 10.1016/s1097-2765(02)00513-0. [DOI] [PubMed] [Google Scholar]
- Lobachev KS, Stenger JE, Kozyreva OG, Jurka J, Gordenin DA, Resnick MA. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- Lobachev KS, Rattray A, Narayanan V. Hairpin- and cruciform-mediated chromosome breakage: Causes and consequences in eukaryotic cells. Front Biosci. 2007;12:4208–4220. doi: 10.2741/2381. [DOI] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- Mizuno KI, Lambert S, Baldacci G, Murray JM, Carr AM. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes & Dev. 2009;23:2876–2886. doi: 10.1101/gad.1863009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell. 2006;125:1283–1296. doi: 10.1016/j.cell.2006.04.042. [DOI] [PubMed] [Google Scholar]
- Olavarrieta L, Martinez-Robles ML, Sogo JM, Stasiak A, Hernandez P, Krimer DB, Schvartzman JB. Supercoiling, knotting and replication fork reversal in partially replicated plasmids. Nucleic Acids Res. 2002;30:656–666. doi: 10.1093/nar/30.3.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek AL, Kaochar S, Jones H, Elezaby A, Shanks L, Weinert T. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes & Dev. 2009;23:2861–2875. doi: 10.1101/gad.1862709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: Conformations at the fork. Proc Natl Acad Sci. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev. 2002;12:312–319. doi: 10.1016/s0959-437x(02)00304-0. [DOI] [PubMed] [Google Scholar]
- Stenger JE, Lobachev KS, Gordenin D, Darden TA, Jurka J, Resnick MA. Biased distribution of inverted and direct Alus in the human genome: Implications for insertion, exclusion, and genome stability. Genome Res. 2001;11:12–27. doi: 10.1101/gr.158801. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yao MC. Palindromic gene amplification–an evolutionarily conserved role for DNA inverted repeats in the genome. Nat Rev Cancer. 2009;9:216–224. doi: 10.1038/nrc2591. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: Interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci. 2008;105:9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, Mirkin SM. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat Struct Mol Biol. 2009;16:226–228. doi: 10.1038/nsmb.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Inverted repeat structure of the human genome: The X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14:1861–1869. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC. Rad51 protein controls Rad52-mediated DNA annealing. J Biol Chem. 2008;283:14883–14892. doi: 10.1074/jbc.M801097200. [DOI] [PMC free article] [PubMed] [Google Scholar]