Abstract
Although the transporter-like protein Patched (Ptc) is genetically implicated in reception of the extracellular Hedgehog (Hh) protein signal, a clear definition of the Hh receptor is complicated by the existence of additional Hh-binding proteins and, in Drosophila, by the lack of physical evidence for direct binding of Hh to Ptc. Here we show that activity of Ihog (Interference hedgehog), or of its close relative Boi (Brother of Ihog), is absolutely required for Hh biological response and for sequestration of the Hh protein to limit long-range signaling. We demonstrate that Ihog interacts directly with Ptc, is required for presentation of Ptc on the cell surface, and that Ihog and Ptc are both required for high-affinity Hh binding. On the basis of their joint roles in ligand binding, signal transduction, and receptor trafficking, we conclude that Ihog and Ptc together constitute the Drosophila Hh receptor.
Keywords: Brother of Ihog, Hedgehog receptor, Hedgehog signaling, Ihog, Patched
The Hedgehog (Hh) signaling pathway organizes pattern formation in a variety of embryonic tissues ranging from insect segments to the vertebrate limb and neural tube (Nüsslein-Volhard and Wieschaus 1980; Chiang et al. 1996; for review, see Jessell 2000; McMahon et al. 2003). The Hh pathway also functions post-embryonically in homeostatic processes such as tissue maintenance and regeneration by acting on tissue stem or progenitor cells (Nystul and Spradling 2006; Mandal et al. 2007; Takashima et al. 2008; Zhao et al. 2009). Hh pathway dysfunction thus can lead to embryonic pattern disruptions, such as segmentation defects in Drosophila (for review, see Ingham and McMahon 2001) or holoprosencephaly (for review, see Muenke and Beachy 2000) and other birth defects in humans; post-embryonic dysfunction can result in proliferative disorders such as the growth of malignant tumors (for review, see Varjosalo and Taipale 2008) or tissue degeneration (Lavine et al. 2008).
The secreted Hh protein heading this signaling pathway is produced as a precursor that undergoes cleavage and cholesterol modification in an autoprocessing reaction, followed by further covalent addition of palmitate (for review, see Mann and Beachy 2004). The mature, dually lipid-modified protein signal (HhNp) is then released from cells by an active process that requires Dispatched (Burke et al. 1999; Ma et al. 2002) and involves other protein and lipoprotein components (Han et al. 2004; Glise et al. 2005; Gorfinkiel et al. 2005; Hollway et al. 2006; Kawakami et al. 2005; Panáková et al. 2005; Woods and Talbot 2005). Upon release, typically from a localized source, the Hh protein then elicits concentration-dependent cellular differentiation or proliferation responses from cells in surrounding tissues and structures.
The Hh receptor has several unusual features, the most striking of which may be a separation of its Hh-sensing function from signal transmission to the cell's interior. The latter function (signal transmission) is mediated by Smoothened (Smo), a seven-transmembrane protein that acts via an intracellular signal cascade to activate the latent cytoplasmic transcription factor Ci (Cubitus interruptus) in Drosophila and the homologous Gli proteins in vertebrates. Smo is not involved, however, in direct binding and sensing of the extracellular Hh signal, which instead appears to involve the transporter-like protein, Patched (Ptc), which contains 12 transmembrane segments. In the absence of Hh, Ptc indirectly inhibits Smo, possibly via transport of a small molecule intermediate (Taipale et al. 2002). In the presence of Hh, Ptc inhibition of Smo is blocked, and pathway activation by Hh is functionally equivalent to loss of Ptc (for review, see Lum and Beachy 2004).
A role for Ptc in sensing the Hh protein is consistent with genetic analysis (Ingham et al. 1991; Sampedro and Guerrero 1991), and studies in mammals suggest that Ptc interacts directly with the Hh protein (Marigo et al. 1996; Stone et al. 1996; Fuse et al. 1999). However, several other mammalian Hh-binding proteins that contribute to biological activity of the pathway have been identified (Chuang and McMahon 1999; Okada et al. 2006; Tenzen et al. 2006; Yao et al. 2006; Zhang et al. 2006; Allen et al. 2007; Martinelli and Fan 2007), thus complicating the simple conclusion that Ptc is the binding component of the Hh receptor. Genetic studies in Drosophila implicate Ptc in a second function beyond Smo regulation; namely, the sequestration of Hh protein within the imaginal disc epithelium to limit its long-range signaling ability (Chen and Struhl 1996). This function might most simply be accounted for by Hh binding, but no direct interaction of Hh protein with Drosophila Ptc has been demonstrated.
More recent studies in Drosophila cultured cells suggest that high-affinity Hh binding and transcriptional response require expression of not only Ptc, but also Ihog (Interference hedgehog) (Yao et al. 2006). Ihog is a type I single-span transmembrane protein with four extracellular Ig domains, two extracellular fibronectin type III (FNIII) domains, and a cytoplasmic domain unrelated to sequences of known structure or function. Biochemical and structural studies have shown that Fn1, the first FNIII domain, directly contacts HhN (McLellan et al. 2006; Yao et al. 2006). Fn1 alone, however, is insufficient for high-affinity binding of Hh, either alone or in synergy with Ptc, and the physical basis for interaction between Ihog and Ptc is unknown. In addition, ihog mutant phenotypes in embryos and imaginal discs are mild (Yao et al. 2006), possibly due to functionally overlapping expression of a related Drosophila protein, Boi (Brother of Ihog), that in cultured cells can functionally substitute for Ihog. Curiously, although the mammalian members of the Ihog family, Cdo and Boc, both contribute to aspects of Hh signaling (Okada et al. 2006; Tenzen et al. 2006; Yao et al. 2006; Zhang et al. 2006), they bind to mammalian Hh proteins via a nonorthologous FNIII repeat (Tenzen et al. 2006; Yao et al. 2006; McLellan et al. 2008).
To further define the nature of the Hh receptor and elucidate the mechanistic roles of Ihog proteins in Hh receptor function, we focus here on the Drosophila ihog and boi genes and their protein products. We demonstrate by genetic analysis that maternal and zygotic loss of ihog and boi function produces severe defects in Hh target gene expression and segmental patterning in embryos. We further demonstrate that Ihog or Boi protein activity is required for all Hh-dependent target gene expression and patterning functions in the wing imaginal disc, and for sequestration of Hh protein to limit long-range signaling. We demonstrate biochemically that the Fn2 domains of Ihog/Boi interact physically with Ptc, and that this domain is required for presentation of Ptc on the cell surface. Both Fn1 and Fn2 domains are required for formation of a high-affinity multimolecular complex of Ihog with Ptc and HhN. On the basis of these studies, we conclude that Ihog and Ptc together constitute the Hh receptor in Drosophila.
Results
Mutation of the ihog locus
Although ptc function is known to be required genetically for sequestration of the Hh signal (Chen and Struhl 1996) and Ptc protein has been suggested to bind to Hh (Lu et al. 2006), these studies did not incorporate analysis of the more recently identified Hh-binding proteins, Ihog and Boi (Yao et al. 2006). In addition, previous genetic analysis revealed only mild or intermediate phenotypes of ihog mutations in embryos and imaginal discs, and did not account for the overlapping expression and function of boi, for which no mutant allele existed (Yao et al. 2006).
To definitively examine the role of Ihog family proteins in Hh pathway function, we designed and introduced precise DNA lesions into the Drosophila germline (Gong and Golic 2003) that inactivate ihog and boi function. The ihog mutation was designed to replace a 2781-base-pair (bp) contiguous genomic segment containing the full ihog coding sequence by the mini white (w+) marker (Fig. 1A), thus resulting in a clean ihog mutation that leaves adjacent genes intact (Figs. 1A; Supplemental Fig. S1). Introduction of the ihog DNA lesion into the Drosophila germline was confirmed by RT–PCR analysis (Supplemental Fig. S2A), and the ihog mutant allele was found to be homozygous viable and fertile, unlike previously described deletion alleles that also disrupt function of a vital gene ∼200 bp to the 3′ side of ihog (Yao et al. 2006).
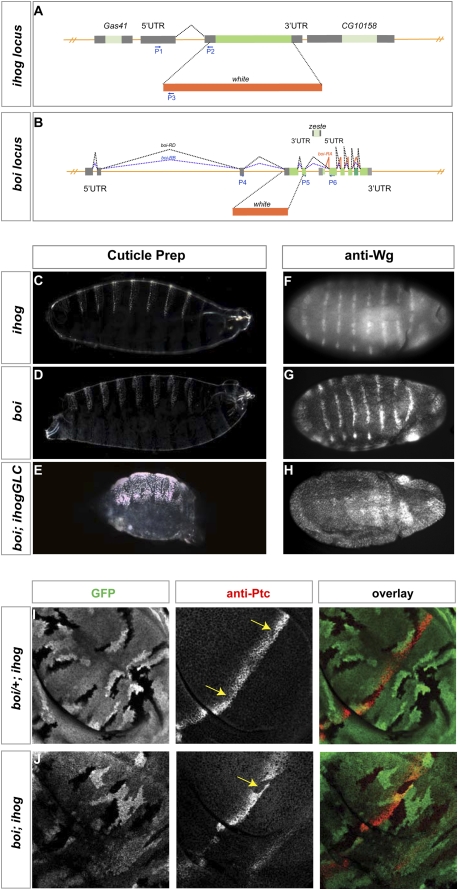
Figure 1.
ihog/boi mutations disrupt embryonic patterning and Hh signal transduction in the wing imaginal disc. (A) Generation of ihog mutation by gene targeting. A genomic region within the ihog locus was replaced by the white marker gene, as shown. Primers used in RT–PCR (Supplemental Fig. S3) are represented as blue arrows. (B) Generation of boi mutation by gene targeting. Splicing patterns of three predicted boi transcripts (boi-RA, red; boi-RB, blue; boi-RD, black) are indicated. The boi-RA transcript initiates from an internal initiation site, thus defining a sixth exon unique to boi-RA. The exon present in Boi-RD, but excluded from Boi-RB, is a darker green. A genomic region was replaced by the white marker gene as shown. Primers used in RT–PCR (Supplemental Fig. S3) are located within exons and are shown as blue arrows. Note that the gene zeste (light green) is transcribed from the opposite strand. (C–E) Cuticle preparations of embryos lacking maternal and zygotic ihog and boi either alone (C,D) or in combination (E). (F–H) Wingless protein expression in stage 10 embryos lacking maternal and zygotic ihog and boi either alone (F,G) or in combination (H). (I) Reduction of Ptc expression (red) in ihog homozygous boi heterozygous mutant clones (boi/+; ihog), marked by loss of GFP expression (yellow arrows). (J) Loss of Ptc expression (red) in boi; ihog homozygous mutant clones, marked by loss of GFP expression (yellow arrow).
Mutation of the boi locus
The boi gene comprises three predicted transcripts derived from a 26-kb genomic region (Figs. 1B; Supplemental Fig. S3). Two of these predicted transcripts encode proteins (Boi-RB and Boi-RD) that, like Ihog, contain four extracellular Ig domains and two FNIII domains, with a single transmembrane domain and a cytoplasmic domain lacking homology with Ihog or with any other domain of known function. The larger Boi-RD protein is predicted to contain 112 internal amino acid residues within its cytoplasmic domain that are absent from Boi-RB, due to exclusion of an internal exon in the Boi-RB transcript; our cDNA clone contains coding sequences like those in Boi-RB. The third predicted transcript is initiated from an internal location, and its encoded protein (Boi-RA) therefore lacks the signal sequence and the four Ig domains, and thus would be unlikely to be secreted. A coding sequence like Boi-RA appears to be expressed in cl-8 Drosophila cultured cells, but is not functional in Hh signaling (Yao et al. 2006).
As coding sequences for Boi-RD and Boi-RB are distributed among multiple exons that span another gene, zeste, that is transcribed from the opposite strand, we selected for replacement by the (w+) marker a 1586-bp region of contiguous genomic sequence that includes the entire fourth exon and a portion of the fifth (Fig. 1B). This replacement does not affect the zeste gene but results in deletion of the AUG initiation codon, the signal sequence, and the four Ig domains from the coding sequence of Boi-RD and Boi-RB (Supplemental Fig. S3). If any protein could be translated from a residual transcript from this modified locus, it would lack a signal sequence and therefore should not be secreted.
Introduction of the boi DNA lesion into the Drosophila germline was confirmed by RT–PCR analysis (Supplemental Fig. S2B), and the boi mutant allele was found to be homozygous viable and fertile. Animals bred to homozygosity for both boi and ihog mutations, however, die in early larval stages (data not shown). Of note, the level of Ihog protein expression increased significantly in boi homozygotes as compared with wild type (Supplemental Fig. S1), suggesting that loss of function for one gene may be compensated by increase in expression of the other, at least in stocks maintained as homozygotes.
Roles of Ihog proteins in embryonic patterning
As maternal expression is known to contribute to ihog function (Yao et al. 2006), we generated ihog maternal germline clones in a boi homozygous mutant background, and thus were able to examine embryos lacking maternal as well as zygotic ihog and boi function. These individuals derive from crosses between boi; ihogGLC females and boi; ihog/+ males, and hereafter are referred to as boi; ihogGLC. Cuticle preparations from these embryos revealed a fully penetrant phenotype in which naked cuticle normally present in the posterior of each segment fails to develop, resulting in a continuous lawn of poorly patterned denticles (Fig. 1E). This cuticle phenotype greatly resembles that of embryos homozygous for hh null alleles (Lee et al. 1992), and is far more severe than that of embryos lacking maternal and zygotic ihog or boi alone (Fig. 1C,D). We also assessed the effects of ihog and boi loss on segmentally repeated expression of the wingless (wg) gene, which depends on Hh signaling for its maintenance (for review, see DiNardo et al. 1994). We similarly noted a severe effect on Wg protein expression in boi; ihogGLC individuals (Fig. 1H), and only a mild effect, if any, on Wg expression in embryos lacking maternal and zygotic ihog or boi alone (Fig. 1F,G). Although maternal loss was required for full expression of the boi; ihog loss-of-function phenotype, such loss did not produce any phenotype in the presence of a paternally provided wild-type allele of ihog. Maternal boi and ihog function is thus neither essential nor sufficient for normal development.
Ihog proteins essential for Hh transduction and target gene expression
As loss of Wg expression and naked cuticle in each segment of the developing embryo can be caused by effects other than simple loss of Hh signaling, we further assessed the function of ihog/boi by examining expression of Hh target genes in the wing imaginal disc, the precursor of the adult wing. Hh expression in the disc is restricted to cells of the posterior compartment, which lack expression of Ci and are not responsive to the Hh signal; target gene expression consequently is limited to a stripe of adjacent cells within the anterior compartment. As the ptc gene not only encodes a Hh receptor component but also is a transcriptional target for Hh pathway activity, its expression serves as an indicator of Hh response and occurs at its highest level in a stripe of anterior cells immediately adjacent to the compartment boundary (Fig. 1I). We generated clones of cells homozygous for ihog in a boi mutant background by FLP-mediated recombination (see the Materials and Methods), and found that loss of both ihog and boi caused a complete loss of Ptc expression (Fig. 1J). Loss of ihog function in a heterozygous boi mutant background, in contrast, only caused partially decreased expression of ptc (Fig. 1I), consistent with previous findings of a partial loss of Hh response in ihog homozygous mutant clones (Yao et al. 2006).
Smo protein levels normally increase in A cells near the anterior/posterior (A/P) compartment boundary in response to Hh signaling (Denef et al. 2000). We noted, however, that Smo levels were not higher in clones of cells lacking ihog/boi function (Supplemental Fig. S4), indicating that Ihog/Boi activity is required for the Hh-dependent stabilization of Smo. This confirms the role of Ihog/Boi in Hh transduction upstream of Smo.
Ihog proteins sequester Hh and limit its range of action
A previously reported aspect of Ptc protein function is the sequestration of Hh protein, thus limiting distal action of the Hh signal (Chen and Struhl 1996). As Ihog/Boi were unknown and therefore were not tested at the time of this analysis, we assessed their contribution to this function by examining the effects of large clones positioned between the P-cell source of Hh protein and A cells that are capable of responding. The normal expression of ptc in wild-type discs is restricted to a stripe of five to 10 cells immediately adjacent to the Hh-expressing P cells. A similar pattern, albeit with some reduction in level of expression, was noted in boi/+; ihog clones with reduced but not absent expression of the Ihog/Boi proteins (Fig. 2A). In contrast, and as noted above, all expression of ptc was lost within A-cell clones lacking both Ihog and Boi (boi; ihog); expression of ptc, however, occurred in cells just outside and to the anterior of such clones, even at distances from the Hh-expressing P cells that greatly exceed the normal five- to 10-cell range of ptc expression (Fig. 2B, see arrow).
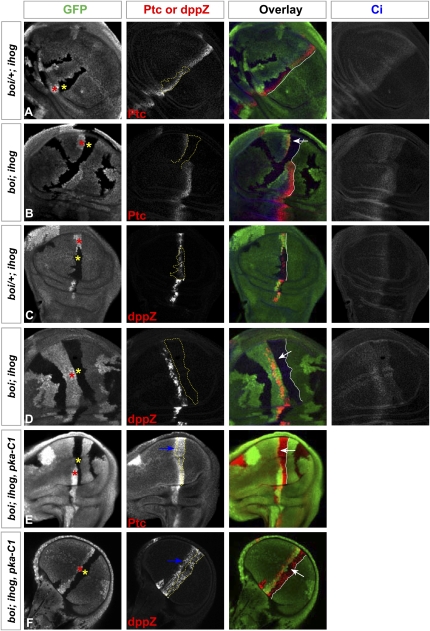
Figure 2.
ihog/boi mutations prevent sequestration of the Hh signal. Each set of panels shows a wing imaginal disc immunostained for GFP (green), Ptc or dppZ (red), and Ci (blue, marking cells of A compartment origin in A–D). Homozygous ihog mutant clones (yellow asterisk) are marked by the loss of GFP expression, and their ihog+/ihog+ sister clones (red asterisk) are marked by elevated levels of GFP expression. (A,C) Homozygous ihog and heterozygous boi mutant clones (boi/+; ihog) in the A compartment behave as wild-type in that only cells a short distance from the Hh-secreting posterior cells (lacking Ci expression) express Ptc (A) and dppZ (C). No Ptc or dppZ expression was detected anterior to the mutant clones. (B,D) Homozygous boi; ihog mutant clones (boi; ihog) originating in the A compartment (as judged by expression of Ci and by position of the ihog+/ihog+ sister clone) fail to express Ptc (B) or dppZ (D). However, Ptc and dppZ expression is noted in thin strips of cells immediately anterior to the borders of these boi; ihog mutant clones, abnormally far from the posterior cell source of the Hh signal, and indicating that sequestration of the Hh signal in the boi; ihog cells failed. White arrows indicate abnormally far-ranging Hh action across mutant clones lacking both Ihog and Boi to induce Ptc or dppZ. Although Ci levels in boi; ihog mutant clones at the compartment boundary are lower than in surrounding cells, these Ci levels are slightly higher than in cells far to the anterior. (E,F) Homozygous boi; ihog; pka-C1 mutant clones originating in the A compartment autonomously express Ptc (E) and dppZ (F), but still fail to sequester the Hh signal. Note expression of Ptc or dppZ in thin strips of cells (blue arrows) immediately anterior to the border (yellow lines) of these boi; ihog; pka-C1 mutant clones, indicating that elevated Ptc expression alone is not sufficient to sequester the Hh signal, and that Ihog family proteins are absolutely required to limit the range of Hh activity. White lines indicate the border of Hh-secreting posterior cells.
Expression of the dpp target of Hh signaling occurs in a stripe of cells near the A/P boundary, although this expression is first seen a few cells distant from the boundary and extends beyond the normal range of Ptc expression. A similar pattern of dpp expression, indicated by a dpp-lacZ reporter, also was noted in large clones of boi/+; ihog cells with a reduced dosage of Ihog/Boi proteins (Fig. 2C). In clones lacking all Ihog/Boi proteins, dpp expression was completely abolished, but dpp was expressed in cells to the anterior of such clones, again at distances from the Hh-expressing P cells that greatly exceeded the normal range of dpp expression (Fig. 2D, see arrow). These results suggest that Ihog/Boi proteins play a role in sequestration of the Hh signal and limitation of its action on distal cells.
In clones lacking Ihog/Boi, the Ptc protein is not expressed, and the loss of sequestration noted could be due to loss of Ptc (Figs. 1J, 2B). To determine whether Ihog/Boi proteins are required for sequestration in the presence of Ptc, we generated large clones of cells lacking not only Ihog/Boi, but also the catalytic subunit of cAMP-dependent protein kinase (PKA-C1). The loss of PKA-C1 expression causes transcriptional activation of Hh pathway targets through stabilizing effects on Ci, thus resulting in expression of endogenous Ptc (Jiang and Struhl 1995; Lepage et al. 1995; Li et al. 1995; Pan and Rubin 1995; Strutt et al. 1995); as demonstrated previously, loss of PKA-C1 restores sequestration of Hh in clones lacking Smo function (Chen and Struhl 1996). In striking contrast, however, we noted that Ptc expression occurred on the anterior side of clones lacking Ihog/Boi, at an abnormally large distance from the Hh-expressing posterior cells (Fig. 2E, see arrow). Similarly, anterior dpp expression also occurred at distances from the Hh-expressing P cells that greatly exceed the normal range of dpp expression (Fig. 2F, see arrow). These results confirm that Ihog/Boi expression is absolutely required for sequestration of Hh to limit its range of action, and that expression of the Ptc protein alone is not sufficient for this sequestration activity.
Ihog/Boi is required for Hh response, not Hh release
Our results thus far demonstrate a requirement for the activity of Ihog/Boi in various aspects of Hh response, including segmental patterning, activation of specific Hh pathway targets, and sequestration of Hh protein to limit distal signaling. We did not, however, examine a potential role for Ihog/Boi in Hh-producing cells. In order to address this possibility, we removed Ihog/Boi function in cells that are activated simultaneously by MARCM (Lee and Luo 1999) for ectopic Hh expression. When such Hh-expressing anterior clones were examined, we noted that endogenous Ptc protein expression was induced within boi/+; ihog clones (Fig. 3A,B), but not within boi; ihog double-mutant clones (Fig. 3C,D). This result confirms the cell-autonomous requirement for Ihog/Boi in Hh response noted at the compartment boundary (see above), and further demonstrates that this requirement applies even when cells are exposed to high concentrations of Hh protein such as those produced by GAL4-driven expression within the MARCM clone.
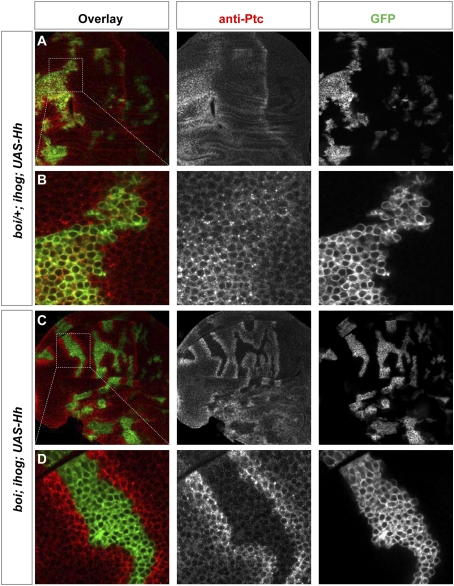
Figure 3.
Ihog/Boi is required for Hh response, not export. Wing discs are immunostained for GFP (green) and Ptc (red). ihog homozygous mutant clones also express active GAL4 (from ActP-Gal4, activated by the MARCM method and indicated by expression of mCD8GFP) and have been induced in boi heterozygous (boi/+; ihog in A,B) or boi hemizygous (boi; ihog in C,D) larvae; cells in these clones simultaneously express a UAS-Hh transgene. Note that in the A compartment Hh is expressed only in GFP-positive cells. (A,B) Ptc expression was induced both inside and outside of the Hh-expressing ihog mutant clone carried by boi heterozygous animals (boi/+; ihog; ActP-Gal4 > UAS-Hh). (C,D) Ptc expression was induced only in cells surrounding the Hh-expressing ihog mutant clones carried by boi hemizygous animals (boi; ihog; ActP–Gal4 > UAS–Hh). boi; ihog mutant cells are thus not capable of responding to the Hh signal, but do produce and release an active Hh signal that stimulates pathway activation in surrounding cells.
We also examined Hh response in cells outside the clones lacking Ihog/Boi function and noted that Ptc expression extends beyond the boundaries of Hh-expressing clones at a range and intensity at least as great as that observed at the A/P compartment boundary (Fig. 3C,D). We also noted that large clones of cells lacking Ihog/Boi within the posterior compartment did not decrease the level or range of endogenous Ptc expression in adjacent anterior cells (Supplemental Fig. S5). From these observations, we conclude that loss of Ihog/Boi function in Hh-expressing cells does not decrease the range or potency of Hh action on adjacent cells, and that Ihog/Boi function is not required for the export of Hh protein from cells producing it.
Ihog interacts physically with Ptc via Fn2
The genetic requirement for Ihog/Boi and Ptc in all aspects of Hh response, particularly in activities such as the sequestration of the Hh ligand (see above), raises the question of whether the genetic collaboration of Ihog/Boi and Ptc may depend on a physical interaction. We addressed this question by immunoprecipitation of wild-type or variant forms of these proteins from transiently transfected Drosophila S2R+ cultured cells. We found that Ptc coprecipitated with Ihog (Fig. 4B), and that this coprecipitation was not affected by deletion of the Fn1 domain, previously shown to interact directly with HhN (McLellan et al. 2006; Yao et al. 2006). Deletion of Fn2 or mutations of particular Fn2 surface residues (Supplemental Fig. S6), in contrast, disrupted or reduced Ptc coprecipitation (Fig. 4B). HhN added to the external medium was also coprecipitated with Ihog from transiently transfected cells, but, unlike the Ptc interaction, this association was dependent on Fn1 and not Fn2 (Fig. 4C). Neither the interaction with Ptc nor Hh was affected by deletion of the Ihog cytoplasmic domain.
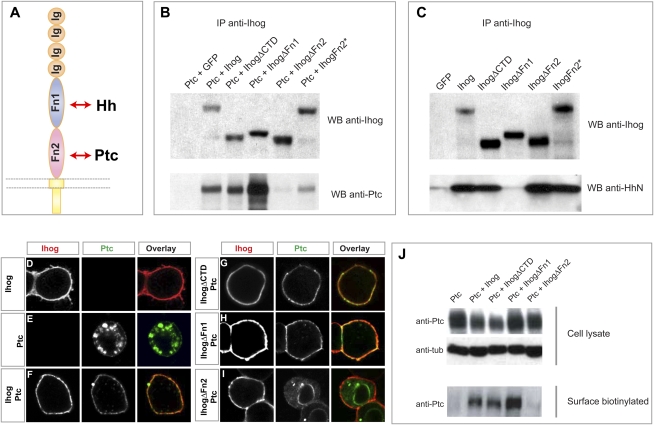
Figure 4.
Cell surface presentation of Ptc is dependent on Ihog. (A) A schematic diagram of the Ihog protein. (B) Drosophila S2R+ cells coexpressing Ptc with wild-type or variant Ihog proteins were lysed, Ihog proteins were immunoprecipitated with anti-Ihog antibody and Protein G beads, and proteins were detected by immunoblotting for Ihog (top panel) or Ptc (bottom panel). Ihog Fn2 but not Fn1 is critical for Ptc binding. Note that two surface residues on the Fn2 domain are mutated in IhogFn2* (see Supplemental Fig. S6). (C) Drosophila S2R+ cells expressing wild-type or variant Ihog proteins were incubated with HhN-conditioned medium, cells were lysed, Ihog proteins were immunoprecipitated with anti-Ihog antibody and Protein G beads, and proteins were detected by immunoblotting for Ihog (top panel) or Hh (bottom panel). Ihog Fn1 but not Fn2 is critical for HhN binding. (D–I) Confocal microscope images showing localization of Ihog (red) and Ptc (green) proteins. Drosophila S2R+ cells were transfected for expression of Ptc and/or Ihog, as indicated. Ihog protein was detected mainly on the cell surface (D), whereas Ptc was localized mainly in intracellular vesicles with a trace of cell surface expression (E). Surface localization of Ptc increased with coexpression of Ihog (F), IhogΔCTD (G), or IhogΔFn1 (H), but not with IhogΔFn2 (I). (J) S2R+ cells transfected for expression of Ptc and Ihog variants were labeled by surface biotinylation. Immunoblots in the top panels show 5% of the biotinylated cell lysate, and the bottom panel shows the biotin-labeled proteins recovered by Streptavidin-agarose beads. Coexpression with Ihog and IhogΔCTD increased levels of Ptc expressed on the surface.
Ihog mediates surface presentation of Ptc
Given that Ptc and Ihog interact physically, we wondered whether they influence each other's localization. To examine these issues, we used S2R+ cultured cells and focused initially on detection of surface proteins by using antibodies that recognize extracellular epitopes and a protocol that omits detergent permeabilization (Fig. 4D–I), or, in other experiments, through use of surface biotinylation to specifically mark and monitor behavior of surface proteins (Fig. 4J).
Immunostaining of cells transfected for expression of Ihog protein confirmed its predominant localization on the cell surface (Fig. 4D). Ptc protein expressed in S2R+ cells, in contrast, is localized primarily in intracellular vesicles, albeit with a trace of surface expression. The predominantly intracellular staining in S2R+ cells reflects the intracellular localization of the great bulk of Ptc protein and indicates some degree of cell permeabilization despite the absence of detergent, possibly due to the fixation step in the protocol. Coexpression with Ihog dramatically shifted the apparent localization of Ptc to the cell surface (Fig. 4F). This Ihog-mediated shift in Ptc localization was not affected by deletion of the C-terminal domain of Ihog (IhogΔCTD) (Fig. 4G), but was dramatically reduced by deletion of Fn2 (Fig. 4I), the Ihog domain that mediates interaction with Ptc. Ihog protein lacking the Hh-interacting Fn1 domain retained the ability to shift Ptc localization to the surface (Fig. 4H). Given this dramatic effect of Ihog on Ptc surface localization, we speculate that the trace levels of Ptc on the surface of cells not transfected for Ihog expression (Fig. 4E) may be due to endogenous expression of Ihog/Boi in this cell line (Supplemental Figs. S1, S7; Yao et al. 2006).
We further examined the trafficking of Ptc using a cell-impermeable biotinylation agent to mark and monitor the surface proteins of transfected S2R+ cells. We noted that, although only trace amounts of Ptc protein expressed alone could be surface-biotinylated, the level of surface-labeled Ptc proteins was increased dramatically by coexpression with Ihog, IhogΔCTD, or IhogΔFn1, but not by IhogΔFn2 (Fig. 4J). These results confirm our conclusions based on immunostaining that Ihog mediates surface expression of Ptc in a manner dependent on the interaction between Ihog and Ptc, mediated by Ihog Fn2.
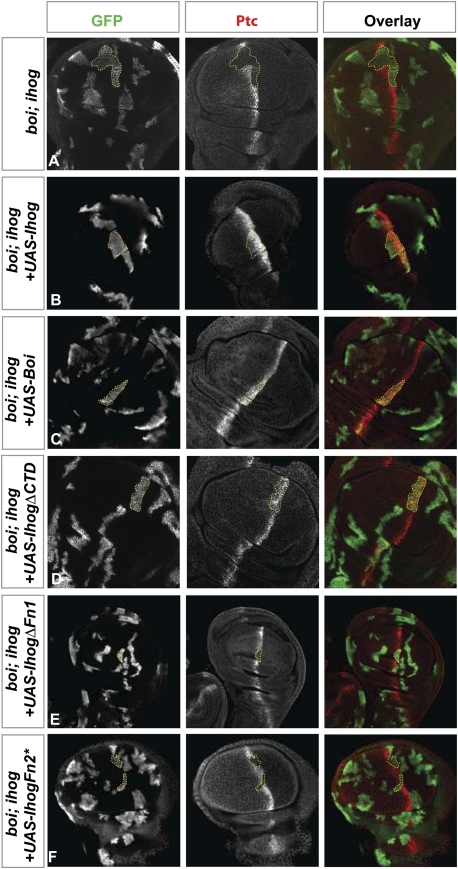
To confirm the importance of these interactions in vivo, we tested the ability of Ihog variants to function in Hh signaling in the wing imaginal disc. We noted that either wild-type Ihog or Boi are capable of rescuing Hh response in clones of cells lacking boi and ihog functions (Fig. 5A–C). The C-terminal domain of Ihog was dispensable for this rescuing activity (Fig. 5D). In striking contrast, Ihog expression constructs lacking Fn1 or Fn2 (Fig. 5E,F, respectively) completely failed to rescue Hh response, indicating that both of these domains are critical for Ihog function in vivo.
Figure 5.
Sequence requirements for function of Ihog proteins in Hh transduction. Each set of panels shows a wing disc immunostained for GFP (green) and Ptc (red). The yellow outlines indicate marked clones in the A compartment near the A/P boundary, where Ptc normally is highly expressed. (A) Loss of Ptc expression in boi hemizygous larvae containing MARCM clones (indicated by expression of mCD8GFP) that lack ihog function (boi; ihog). (B–F) Hemizygous boi larvae with MARCM clones (indicated by expression of mCD8GFP) lacking ihog function (boi; ihog) and specifically expressing wild-type Ihog (B), wild-type Boi (C), or various altered Ihog proteins as indicated (D–F). Note that Ptc expression is rescued by IhogΔCTD, but not by variants with altered or missing Fn1 or Fn2 domains. Note that two surface residues on Fn2 domain are mutated in IhogFn2* (see Supplemental Fig. S6).
Surface presentation of Ptc is insufficient for high-affinity Hh binding
We noted previously in cultured cell-based experiments that Ihog and Ptc act synergistically to mediate binding of HhN (Yao et al. 2006). Given our observation that Ihog expression increases surface localization of Ptc, we considered the possibility that synergistic binding could be due simply to Ihog enhancement of Ptc surface presentation. This possibility is especially relevant in light of the previously reported findings that Ptc proteins lacking C-terminal cytotail residues 1181–1286 (Ptc1180) or 1131–1286 (Ptc1130) are more prominently localized on the cell surface (Johnson et al. 2000; Zhu et al. 2003; Lu et al. 2006), and that S2 cells expressing these truncated forms of Ptc are able to retain HhN (Lu et al. 2006).
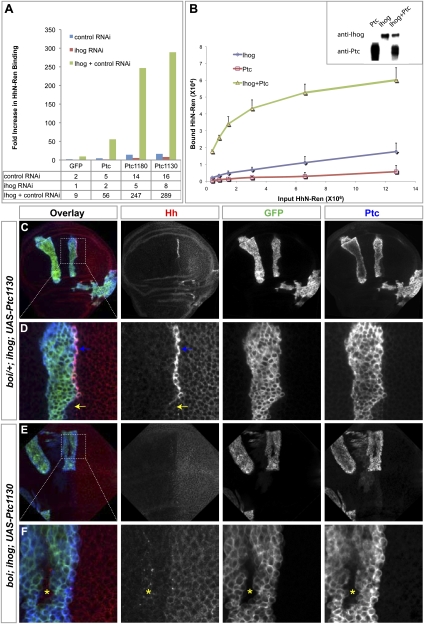
Indeed, we also observed immunostaining of HhN on the surface of S2 cells transfected for expression of Ptc1130 and incubated at 4°C with HhN (Supplemental Fig. S8B). Immunostaining for HhN also was noted on the surface of cells expressing Ptc, albeit at lower levels (Supplemental Fig. S8A). For both Ptc and Ptc1130, however, the amount of HhN immunostaining dramatically increased upon cotransfection with Ihog (Supplemental Fig. S8C,D). To measure this effect, we made use of HhN-Ren, a quantifiable form of HhN fused to the Renilla luciferase enzyme. Consistent with the immunostaining results, we noted that coexpression of Ihog with full-length Ptc, Ptc1180, or Ptc1130 dramatically increased the binding of HhN-Ren by 30-fold or more as compared with cells cotransfected with ihog dsRNA (Fig. 6A). Given the greater surface localization of Ptc1180 and Ptc1130 (Supplemental Fig. S8B; Lu et al. 2006), these results together suggest that the Ihog-mediated increase in HhN binding is not solely dependent on increased surface localization of Ptc.
Figure 6.
Presentation of Ptc on the cell surface is insufficient for high-affinity Hh binding. (A) S2R+ cells transfected for expression of Ptc, Ptc1180, or Ptc1130 together with control dsRNA showed mild increase in binding of HhN-Ren (Renilla luciferase-tagged form of HhN) as compared with cells expressing GFP. Cells expressing Ptc, Ptc1180, or Ptc1130 together with dsRNA against ihog showed significantly reduced binding to HhN-Ren, whereas cells expressing Ptc, Ptc1180, or Ptc1130 together with Ihog and control dsRNA showed dramatically enhanced binding to HhN-Ren. Note that the fold increase in HhN-Ren binding was calculated by normalizing to cells transfected for control expression of GFP together with dsRNA against ihog. Similar to full-length Ptc protein, constitutively surface-localized Ptc1180 and Ptc1130 (Supplemental Fig. S8; Lu et al. 2006) also require Ihog for high-affinity HhN binding. (B) Cell-free binding of HhN to membrane vesicles containing Ihog, Ptc, or the two combined. HhN-Ren binding to vesicles was enhanced dramatically with vesicles from Hi5 cells expressing both Ihog and Ptc. The inset shows levels of Ihog and Ptc present in vesicles from cells expressing either protein alone or in combination. (C–F) Each set of panels shows a wing disc immunostained for GFP (green), Hh (red), and Ptc (blue). MARCM clones (indicated by expression of mCD8GFP) lacking ihog and induced in boi heterozygous (boi/+; ihog in C,D) or boi hemizygous (boi; ihog in E,F) larvae simultaneously express a UAS-Ptc1130 transgene driven by ActP-Gal4. (C,D) HhNp protein accumulated dramatically in boi/+; ihog clones expressing Ptc1130. Note that high-level HhNp accumulation occurred only in the cells that immediately abut Hh-expressing P cells (blue arrows), but not in other boundary cells outside the clone that lack expression of Ptc1130 (yellow arrows). We also noted such accumulations at the boundaries of Ptc1130-expressing clones within the posterior compartment. (E,F) No HhN accumulated in boi; ihog clones expressing Ptc1130. Note that HhN traveled through the Ptc1130-expressing boi; ihog mutant clones and accumulated in wild-type cells immediately anterior to the mutant clone (yellow asterisks).
To remove surface localization as a variable in Ptc access to and binding of extracellular HhN, we developed an assay in which membranes from heterologous cells were isolated and tested for their ability to bind and coprecipitate HhN-Ren. We found that binding of HhN-Ren depended on coexpression of Ihog and Ptc, as dramatically higher binding was observed for combined expression even with lower levels of Ihog and Ptc proteins (Fig. 6B). As membrane vesicles in these cell-free extracts are not shielded from exposure to HhN-Ren, we conclude that Ihog contributes directly to binding of HhN by Ptc, in addition to its effect on Ptc surface localization.
Ptc expression alone is insufficient for sequestration of the Hh ligand in vivo, and this activity requires expression of Ihog/Boi (see above). As wild-type Ptc is localized mainly inside cells, we tested the ability of surface-localized Ptc1130 to sequester Hh ligand in the absence of Ihog/Boi function. For this purpose, we examined wing disc clones with MARCM-activated expression of Ptc1130 for their ability to sequester Hh ligand when occurring adjacent to Hh-expressing posterior cells. We found that HhNp accumulated dramatically in boi/+; ihog clones expressing Ptc1130 (Fig. 6C,D, blue arrows); this accumulation occurred in the cells that immediately abut Hh-expressing posterior cells, but not in other boundary cells outside the clones that lack expression of Ptc1130 (Fig. 6D, yellow arrows). The accumulation of HhNp is thus dependent on expression of Ptc1130; this accumulation is also dependent on expression of Ihog/Boi, as no accumulation is seen in clones expressing Ptc1130 but lacking Ihog/Boi function (boi; ihog) (Fig. 6E,F). Binding of Hh by Ptc in imaginal discs or cultured cells thus requires Ihog/Boi, even if Ptc surface localization is increased by deletion of its C-terminal cytoplasmic tail.
Ihog, Ptc, and HhN form a complex
We noted enhanced binding of HhN in imaginal discs, cultured cells, and membrane preparations in a manner dependent on expression of both Ihog/Boi and Ptc; this effect could be due to formation of a complex incorporating favorable energetic interactions between all three of the components: HhN, Ihog, and Ptc. Indeed, we already established pairwise interactions between Ihog and Ptc (Fig. 4B) and between Ihog and HhN (Fig. 4C; McLellan et al. 2006; Yao et al. 2006). Alternatively, the high-affinity complex might only involve HhN and Ptc, with the role of Ihog being to help recruit HhN to the membrane.
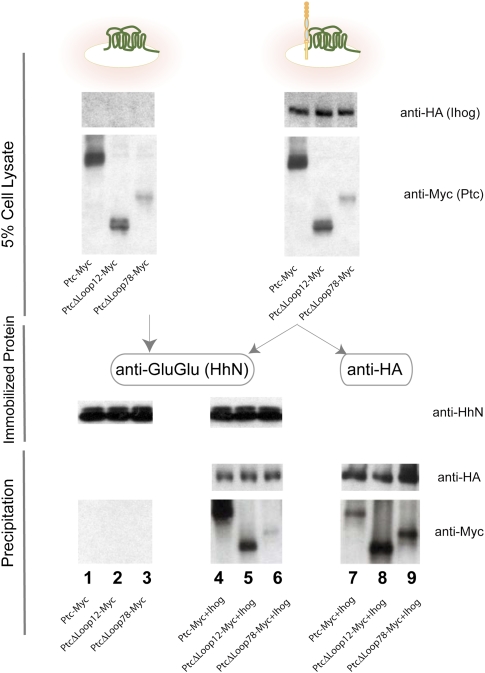
To investigate the possibility of an interaction involving all three components, we used the GluGlu monoclonal antibody attached to Sepharose beads to purify a form of GluGlu epitope-tagged HhN (Supplemental Fig. S9), and incubated this immobilized HhN with detergent-solubilized extracts from S2R+ cells expressing Ptc. Despite testing many buffer and salt conditions and a variety of detergent types and concentrations, we did not observe a HhN/Ptc interaction (Fig. 7, lane 1; data not shown). Knowing that Ihog and Ptc in vesicles bind HhN at much higher affinity than either alone (Fig. 6B), we produced detergent extracts from cells coexpressing Ptc and Ihog, and in this setting indeed observed precipitation of Ptc by HhN (Fig. 7, lane 4). Of course, given the already established interactions between HhN and Ihog, and between Ihog and Ptc, it is possible that Ptc is precipitated indirectly by interacting with Ihog that is bound to immobilized HhN. We therefore compared the amount of Ptc detected from these same extracts when Ihog is precipitated via its HA epitope tag in the absence of HhN protein (Fig. 7, lane 7). We noted that, despite a higher quantity of Ihog precipitated, significantly less Ptc was detected (Fig. 7, cf. lanes 7 and 4), indicating that the presence of HhN enriches Ptc in the complex. Similar results were noted for Boi, HhN, and Ptc (Supplemental Fig. S10). Together with our previous observations, these results indicate that Ptc and HhN contribute favorable energetic interactions to a complex that also benefits from the previously identified interactions between Ptc/Ihog and Ihog/HhN (see above). We note that Ptc variants lacking either of its two large extracellular loops are able to interact with Ihog (Fig. 7, lanes 8,9) as well as or better than wild-type Ptc (Fig. 7, lane 7), but that these deleted variants are impaired in their ability to be enriched by HhN in the presence of Ihog (Fig. 7, cf. lanes 5 and 8, lanes 6 and 9). The Ptc extracellular loops thus appear to be involved in formation of a complex containing Ptc, Ihog, and HhN. Fn1 and Fn2 from Ihog or Boi also appear to be involved in formation of this complex, as deletion of either one similarly impairs the ability of Ihog or Boi to participate in this complex (Supplemental Fig. S10).
Figure 7.
A complex of Ihog, Ptc, and HhN. Immobilized HhN preferentially binds detergent-solubilized Ptc in the presence of Ihog. GluGlu-tagged HhN protein was purified using Sepharose beads containing the anti-GluGlu monoclonal antibody, and these beads were incubated with detergent-solubilized extracts from S2R+ cells expressing Ptc variants (lanes 1–3), or Ptc variants coexpressed with HA-tagged Ihog (lanes 4–6). Precipitation of Ptc by Sepharose HhN-GluGlu beads depended critically on the presence of Ihog, and was much reduced by deletion of the large extracellular Ptc loops (Δloop1,2 or Δloop7,8). (Lane 7) In comparison, precipitation of Ihog with anti-HA beads precipitated significantly less Ptc.
Discussion
Ihog proteins function in all aspects of Hh response
Using the targeted alleles of ihog and boi developed in this study, we provided evidence that Ihog proteins are an essential component required for all biological responses to the Hh signal, including target gene induction and patterning in the embryonic segment and in the wing imaginal disc. The central role of Ihog proteins in Hh response was not noted previously because of the functionally overlapping expression of Ihog and Boi in embryos and imaginal discs, which complicates genetic screens and analysis and accounts for our observation that neither the ihog nor boi targeted alleles are lethal in homozygous form. The cl-8 cells used in our genome-scale RNAi screen (Lum et al. 2003a), in contrast, do not express Boi, and this exposed a critical role for Ihog and facilitated initial discovery of this essential component. In addition to functional overlap, analysis of these functions has been complicated by the required removal of all maternal function for fully penetrant expression of embryonic phenotypes, although maternal expression is neither necessary nor sufficient for Hh response.
Ihog signaling function integrates modular activities of Fn1 and Fn2
The interaction of Ihog Fn2 with Ptc is essential for presentation of wild-type Ptc on the cell surface. We cannot, at present, distinguish between the possibilities that Ihog-mediated surface presentation of Ptc is due to an increased rate of transport to the surface or to an increased duration of residence on the surface. Whatever the mechanism, Fn2 can interact with Ptc in vitro and promote surface presentation of Ptc in cells, even in the absence of Fn1. Similarly, Fn1 alone can interact with HhN in vitro (McLellan et al. 2006), and Fn1 and Fn2 thus have demonstrably independent functions. Neither domain alone, however, suffices for formation of a high-affinity complex, and the presence of both domains is required for Hh signal reception and transduction and participation in signaling in vivo (see Fig. 5).
The role of Ihog proteins in Hh binding
In addition to surface presentation of Ptc (discussed above), our evidence indicates that Ihog proteins also play a direct role in binding the Hh ligand in a multimolecular receptor complex that is critical for transduction. We thus found, as reported previously (Lu et al. 2006), that Hh ligand is bound to the surface of cultured cells expressing a variant of Ptc (Ptc1130) with increased localization on the surface (Supplemental Fig. S8). We also found, with the use of quantitative assays, that endogenous Ihog expressed in these cultured cells contributes critically to binding, and that additional Ihog expression can dramatically enhance binding (Fig. 6A). In addition, expression of Ptc1130 in the wing imaginal disc clearly produces visible accumulation of the Hh protein on what appears to be the surface of anterior cells at the compartment boundary (Fig. 6C,D); this accumulation depends critically on the expression of Ihog/Boi (Fig. 6E,F).
Consistent with the role of Ihog in binding, we also noted a striking contribution of Ihog to binding in membrane vesicle preparations when present in combination with Ptc (Fig. 6B). In addition, we were able to use purified, immobilized HhN and detergent-solubilized extracts containing Ptc and Ihog to demonstrate Ihog-dependent, enhanced precipitation of Ptc (Fig. 7). In these biochemical experiments, we observed that immobilized HhN fails to precipitate detergent-solubilized Ptc alone, but does so in the presence of detergent-solubilized Ihog, and that Ihog alone precipitates Ptc much less efficiently than when HhN is present. This enhancement of Ptc precipitation was dependent on the presence of both the HhN-binding Fn1 domain and the Ptc-binding Fn2 domain of Ihog, consistent with the formation of a multimolecular complex involving HhN, Ptc, and Ihog (Supplemental Fig. S10). Similar results were noted for Boi (Supplemental Fig. S10).
It is interesting to note that we were unable to observe much if any interaction between HhN and Ptc in the absence of Ihog. Formally, it is possible that the interaction of Ptc with HhN is indirect and mediated through enhanced Ihog interaction due to Ptc-induced multimerization or allosteric effects on Ihog. We believe this is unlikely, however, because Ihog is capable of dimerization in the absence of Ptc (McLellan et al. 2006), and because the HhN-interacting surface of Ihog is located on the Fn1 domain, which folds independently and is quite distinct from the Ptc-interacting Fn2 domain, thus making allostery unlikely (Supplemental Fig. S6; McLellan et al. 2006). We thus favor the interpretation that favorable energetic contributions in the multimolecular receptor/ligand complex derive from Ptc–HhN contacts as well as contacts between Ihog–Ptc and Ihog–HhN.
It is important to note that, despite a direct physical interaction of Ihog and Ptc and their mutual contributions to formation and surface presentation of receptor, and to ligand binding, these two pathway components have opposing roles in pathway regulation. Ihog proteins are thus absolutely required for pathway activation in response to Hh ligand, whereas Ptc alone suffices for suppression of Smo activity in the absence of ligands.
Mammalian Ihog proteins
Functional genetic analyses of the mammalian Ihog proteins Cdo and Boc have revealed roles in Hh signaling. Cdo mutant mice thus display mild to intermediate forms of holoprosencephaly, a classic manifestation of Hh signaling deficiency, with the severity of the effect depending on strain background and subject to modifying effects of mutations in other Hh pathway components (Cole and Krauss 2003; Tenzen et al. 2006; Zhang et al. 2006; Allen et al. 2007). Boc mutant mice also show defects in Hh signal-dependent axonal pathfinding by dorsal neurons with ventral commissural projections in the developing neural tube (Okada et al. 2006; Yam et al. 2009). Neither of these mutants displays phenotypes as severe as those seen in the Shh mutant mouse, or in the Smo mutant, which affects all aspects of Hh signaling. It is possible, however, that a systematic analysis of the double mutant Cdo; Boc animals might reveal more severe phenotypes, as we noted here for ihog; boi in Drosophila. In addition, our phenotypic characterization of ihog and boi mutants was not designed to reveal defects in axonal pathfinding functions like that of murine Boc, and the possibility of such a function in Drosophila remains to be explored.
Materials and methods
Constructs
Constructs used in generating transgenic flies were subloned into pW25 (ihog and boi target constructs) or pUAST. Expression constructs of Ihog variants and Ptc variants used in Drosophila cell culture were fused in-frame with a C-terminal HA tag or 3xMyc tag, respectively, and were cloned into pAcSV plasmid. Constructs used in the baculovirus expression system were subcloned into pVL1392 or pVL1393 (BD Biosciences). GluGlu-tagged HhN was generated by inserting the GluGlu tag between Hh codons for amino acids 254 and 255, followed by codons for amino acids 256 and 257 and a stop codon TAA.
Antibodies
Antibodies and dilutions used were mouse anti-Ptc, 1:50 (Capdevila et al. 1994); mouse anti-βgal, 1:100 (Promega, Z378); mouse anti-Smo, 1:50 (Lum et al. 2003b); mouse anti-En 4D9, 1:50 (Patel et al. 1989); mouse anti-Wg 4D4, 1:50 (Brook and Cohen 1996); mouse anti-HA, 1:1000 (HA.11, Covance); mouse anti-β-tubulin E7, 1:5000 (Developmental Studies Hybridoma Bank, developed by M. Klymkowsky); rabbit anti-HhN, 1:1000 (Tabata and Kornberg 1994) for immunostaining; rabbit anti-HhN, 1:1000 for Western blotting (Lee et al. 1992); rabbit anti-Myc, 1:1000 (A-14, Santa Cruz Biotechnologies); rabbit anti GFP, 1:1000 (Invitrogen, A-11122); rat monoclonal anti-Ci 2A1, 1:50 (Motzny and Holmgren 1995); rat anti-Ihog, 1:200 for immunostaining and 1:1000 for immunoblotting (Yao et al. 2006); HRP-conjugated goat anti-mouse (Promega, W4021); HRP-conjugated goat anti-rabbit (Jackson Immuno-Research Laboratory, 111-035-144); and HRP-conjugated goat anti-rat (Jackson Immuno-Research Laboratory, 112-035-167). Fluorophore-conjugated secondary antibodies were from Invitrogen.
Purification and immobilization of HhN
Culture of Sf9 and Hi5 cells and production of recombinant baculovirus were as described (Dukkipati et al. 2006). Hi5 cells used for protein production were infected with baculovirus for expression of HhN-GluGlu. Forty-eight hours after infection, cells were cleared by centrifugation at 1000g and the supernatant was concentrated and incubated with GluGlu monoclonal antibody-affinity matrix (Covance, AFC-115P) overnight at 4°C. Beads were washed and stored at 4°C until use.
Protein immunoprecipitation
Drosophila S2R+, S2, and S2-HhN stable cells were cultured as described (Lum et al. 2003b). Transfection was carried out using FuGENE 6 transfection reagent (Roche). Forty-eight hours after transfection, S2R+ cells were lysed in 0.5% Digitonin (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 40 μM low-molecular-weight Heparin, 1 mM CaCl2, protease inhibitors) for 30 min at room temperature. The lysate was clarified by centrifugation, and proteins were immobilized by binding to anti-HA-affinity matrix (Roche, 11815016001) or GluGlu monoclonal antibody-affinity matrix with prebound HhN for 2 h at room temperature. Beads were washed and proteins were recovered directly in SDS-PAGE sample buffer (for the anti-HA matrix) or by elution with a peptide containing the GluGlu epitope (Anaspec, 62189).
Cell immunostaining
Forty-eight hours after transfection, S2R+ cells were then washed twice with PBS, fixed in 4% formaldehyde (Ted Pella) in PBS, blocked by 1.5% normal goat serum (NGS) in PBS, incubated with primary antibody in PBS containing 1.5% NGS for 1 h at room temperature, washed three times with 0.1% Tween-20/PBS, incubated with secondary antibody, and washed.
Cell surface biotinylation
S2R+ cells were washed with PBS and incubated for 30 min in PBS containing 1 mg/mL Sulfo-NHS-LC-Biotin (Pierce, 21335). The reaction was quenched by washing the cells with 100 mM glycine in PBS three times. Cells were then lysed in 0.5% Digitonin (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, protease inhibitors) for 30 min. The lysate was clarified by centrifugation, and biotinylated protein was recovered by binding to Streptavidin-Sepharose beads (GE Healthcare, 17-5113-01) for 2 h at room temperature. Beads were washed and protein was recovered in SDS-PAGE sample buffer.
Cell-based binding assay
S2R+ cells were transfected for 48 h followed by incubation with conditioned medium containing HhN-Ren (Yao et al. 2006) protein for 1 h at 4°C. Cells then were washed three times with cold PBS and lysed in 250 μL (per well of a six-well plate) of passive lysis buffer (Promega), and 50 μL of lysate were used to measure luciferase activities.
Cell-free binding assay
Hi5 cells were infected with baculovirus for expression of Ihog, Ptc, or Ihog plus Ptc. Cells were harvested 48 h after infection by centrifugation at 800g for 15 min. The cell pellet was resuspended in 3 vol of buffer A (Brown et al. 2002), homogenized with 50 strokes in a tight Dounce homogenizer, and centrifuged at 800g for 10 min at 4°C. The supernatant from this spin was collected to prepare the membrane fraction by centrifugation at 18,000g for 15 min at 4°C. The resulting membrane pellets were resuspended in conditioned medium containing HhN-Ren. After 1 h of incubation at 4°C, the membrane pellets were washed three times with PBS and solubilized in 200 μL of passive lysis buffer (Promega), and 50 μL of lysate were used to measure luciferase activity.
Drosophila strains
Mutant and transgenic strains are described in the following references: pka-C1B3 (Lane and Kalderon 1993), Actin-Gal4 (Ito et al. 1997), hs-FLP (Golic and Lindquist 1989), tubP-Gal80 (Lee and Luo 1999), dpp-lacZ (BS3.0, from Bloomington Stock Center), UAS-mCD8GFP (Lee and Luo 1999), UAS-Hh (Azpiazu et al. 1996),UAS-Ptc and UAS-Ptc1130 (Johnson et al. 2000), and 2Xubi-nGFP FRT40A (Bloomington Stock Center). ORFs encoding full-length or altered Ihog and Boi were cloned into pUAST to generate UAS-Ihog, UAS-Boi, UAS-IhogCTD (1-742), UAS-IhogΔFn1 (delete aa467-577), UAS-IhogFn2* (K653E,Q655E; mutations in Fn2) (see Supplemental Fig. S6). ihog pka-C1B3 FRT40A recombinant chromosome was selected from male offspring of a mating of ihog/pka-C1B3 FRT40A heterozygous females and Sco/Cyo balancer males.
Generating ihog and boi mutations by gene targeting
The ends-out gene targeting strategy (Gong and Golic 2003) was used to generate ihog and boi mutations. DNA fragments flanking the target locus were prepared by PCR of 4- to 5-kb fragments from genomic DNA using two pairs of primers for ihog: 5′-TTAGCGGCCGCAGAGCGAGATAAGCTGGCACAGG-3′/5′-GACGGTACCTGTGCATCCCTACGCCCGATG-3′ and 5′-AATGGGCCGCCGACTGATTCTAGGTGGGGAAACG-3′/5′-GCGCGTACGTCACCTTGTATGAGGTTTCGCCA-3′; and two pairs of primers for boi: 5′-TACGCGGCCGCTCCACTGCCTTATTGGCTGGCAC-3′/5′-GACCGCATGCTCCAAAAACAAGAGCGGCAGAC-3′ and 5′-ATAGGCGCGCCCTCTAACCATTTGACAGGCGAGG-3′/5′-GTCCGTACGTGTGTGTGAGTGTGTGGCTTGG-3′. These fragments were cloned into the vector pW25. Transgenic flies carrying the ihog target construct on the X chromosome or boi target construct on the second chromosome were crossed to flies carrying the hs-FLP recombinase and I-SceI endonuclease, and the progeny were screened for precise gene targeting as described (Gong and Golic 2003) .
We note here that cuticle phenotypes for our new ihog and boi alleles in embryos lacking maternal and zygotic function may differ from those reported previously for ihog due to compensatory gains in expression of one gene that might be adaptive and could occur when stocks are maintained as homozygous mutants for the other gene. Thus, for example, Ihog protein levels are significantly higher in boi mutant homozygotes as compared with wild-type (Supplemental Fig. S1). The homozygous embryos in our current studies derive from homozygous stocks where such compensatory gain might occur, whereas our previous studies used an allele maintained as a heterozygote because of an associated lethal mutation in an adjacent gene (Yao et al. 2006). Such compensatory gain in homozygous stocks could also have influenced the effects of reduced boi/ihog gene dosage in our current wing imaginal disc studies (i.e., loss of all ihog and half of boi function [75% of gene dosage lost] produces a mild reduction of Ptc, similar to loss of ihog only [50% of gene dosage lost] in our previous study).
Genotype of larvae for generating mosaic clones
Germline clones and somatic clones were generated by FRT-FLP recombination (Golic 1991; Chou et al. 1993; Xu and Rubin 1993; Lee and Luo 1999). The genotypes used in our analysis were as follows.
Wing disc clones marked by absence of GFP.
ihog mutant clones in a boi heterozygous background: y w boi hs-FLP/X; ihog FRT40A/ubi-nGFP FRT40A; dppZ/+. ihog mutant clones in a boi homozygous or hemizygous background: y w boi hs-FLP/y w boi or Y; ihog FRT40A/ubi-nGFP FRT40A; dppZ/+. ihog and pka-C1B3 double mutant clones in a boi homozygous or hemizygous background: y w boi hs-FLP/y w boi or Y; ihog pka-C1B3 FRT40A/ubi-nGFP FRT40A; dppZ/+.
Wing disc MARCM clones.
ihog mutant clones in a boi heterozygous background: y w boi hs-FLP UAS-mCD8GFP/X; ihog FRT40A/tubP-GAL80 FRT40A; ActP-Gal4/Tm6B. Positively marked ihog mutant clones in a boi homozygous or hemizygous background: y w boi hs-FLP UAS-mCD8GFP/y w boi or Y; ihog FRT40A/tubP-GAL80 FRT40A; ActP-Gal4/Tm6B. Positively marked ihog mutant clones in a boi homozygous or hemizygous background with specific expression of a gene of interest to be tested for rescue: y w boi hs-FLP UAS-mCD8GFP/y w boi or Y; ihog FRT40A/tubP-GAL80 FRT40A; ActP-Gal4/UAS-gene-of-interest (for specific expression of rescue constructs).
Germline clones.
ihog germline clones in a boi homozygous or hemizygous background: y w boi hs-FLP; ihog FRT40A/P[ovoD1]2La P[ovoD1]2Lb FRT40A X y w boi; ihog FRT40A/Cyo[act-GFP].
RT–PCR analysis
Total RNA was extracted from cultured cells or tissues using RNeasy Mini-Kit (Qiagen). cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad), and PCR reactions (iTaq DNA Polymerase from Bio-Rad) were performed using 5 μL of cDNA as a template.
Image collection and quantification of flurescence intensity
Confocal images were collected via Zeiss LSM 510 and were processed with LSM software and ImageJ.
Acknowledgments
We thank M. Frasch, K.C. Garcia, J. Jiang, T.B. Kornberg, R. Krauss, T. Lee, L. Luo, D. Pan, M. Scott, G. Struhl, T. Tabata, A. Zhu, Developmental Studies Hybridoma Bank, and Bloomington Stock Center for fly strains and reagents; L. Luo for critical review of the manuscript; Ya-Hui Chou, M. Kato, and S. Park for experimental advice; and J.S. McLellan and D.J. Leahy for designing the IhogK653E, Q655E mutant. P.A.B. is an Investigator of the Howard Hughes Medical Institute. N.S. and X.Z. are Damon Runyon Fellows supported by the Damon Runyon Cancer Research Foundation (DRG-1891-05 and DRG-1915-06, respectively).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1870310.
Supplemental material is available at http://www.genesdev.org.
References
- Allen BL, Tenzen T, McMahon AP. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes & Dev. 2007;21:1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu N, Lawrence PA, Vincent JP, Frasch M. Segmentation and specification of the Drosophila mesoderm. Genes & Dev. 1996;10:3183–3194. doi: 10.1101/gad.10.24.3183. [DOI] [PubMed] [Google Scholar]
- Brook WJ, Cohen SM. Antagonistic interactions between wingless and decapentaplegic responsible for dorsal–ventral pattern in the Drosophila Leg. Science. 1996;273:1373–1377. doi: 10.1126/science.273.5280.1373. [DOI] [PubMed] [Google Scholar]
- Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- Burke R, Nellen D, Bellotto M, Hafen E, Senti KA, Dickson BJ, Basler K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Pariente F, Sampedro J, Alonso JL, Guerrero I. Subcellular localization of the segment polarity protein patched suggests an interaction with the wingless reception complex in Drosophila embryos. Development. 1994;120:987–998. doi: 10.1242/dev.120.4.987. [DOI] [PubMed] [Google Scholar]
- Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- Cole F, Krauss RS. Microform holoprosencephaly in mice that lack the Ig superfamily member Cdon. Curr Biol. 2003;13:411–415. doi: 10.1016/s0960-9822(03)00088-5. [DOI] [PubMed] [Google Scholar]
- Denef N, Neubüser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- DiNardo S, Heemskerk J, Dougan S, O'Farrell PH. The making of a maggot: Patterning the Drosophila embryonic epidermis. Curr Opin Genet Dev. 1994;4:529–534. doi: 10.1016/0959-437x(94)90068-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukkipati A, Vaclavikova J, Waghray D, Garcia KC. In vitro reconstitution and preparative purification of complexes between the chemokine receptor CXCR4 and its ligands SDF-1α, gp120-CD4 and AMD3100. Protein Expr Purif. 2006;50:203–214. doi: 10.1016/j.pep.2006.07.016. [DOI] [PubMed] [Google Scholar]
- Fuse N, Maiti T, Wang B, Porter JA, Hall TM, Leahy DJ, Beachy PA. Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc Natl Acad Sci. 1999;96:10992–10999. doi: 10.1073/pnas.96.20.10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glise B, Miller CA, Crozatier M, Halbisen MA, Wise S, Olson DJ, Vincent A, Blair SS. Shifted, the Drosophila ortholog of Wnt inhibitory factor-1, controls the distribution and movement of Hedgehog. Dev Cell. 2005;8:255–266. doi: 10.1016/j.devcel.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Golic KG. Site-specific recombination between homologous chromosomes in Drosophila. Science. 1991;252:958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N, Sierra J, Callejo A, Ibañez C, Guerrero I. The Drosophila ortholog of the human Wnt inhibitor factor Shifted controls the diffusion of lipid-modified Hedgehog. Dev Cell. 2005;8:241–253. doi: 10.1016/j.devcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Han C, Belenkaya TY, Wang B, Lin X. Drosophila glypicans control the cell-to-cell movement of Hedgehog by a dynamin-independent process. Development. 2004;131:601–611. doi: 10.1242/dev.00958. [DOI] [PubMed] [Google Scholar]
- Hollway GE, Maule J, Gautier P, Evans TM, Keenan DG, Lohs C, Fischer D, Wicking C, Currie PD. Scube2 mediates Hedgehog signalling in the zebrafish embryo. Dev Biol. 2006;294:104–118. doi: 10.1016/j.ydbio.2006.02.032. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes & Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Taylor AM, Nakano Y. Role of the Drosophila patched gene in positional signalling. Nature. 1991;353:184–187. doi: 10.1038/353184a0. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: Inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Protein kinase A and hedgehog signaling in Drosophila limb development. Cell. 1995;80:563–572. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Milenkovic L, Scott MP. In vivo functions of the patched protein requirement of the C terminus for target gene inactivation but not hedgehog sequestration. Mol Cell. 2000;6:467–478. doi: 10.1016/s1097-2765(00)00045-9. [DOI] [PubMed] [Google Scholar]
- Kawakami A, Nojima Y, Toyoda A, Takahoko M, Satoh M, Tanaka H, Wada H, Masai I, Terasaki H, Sakaki Y, et al. The zebrafish-secreted matrix protein you/scube2 is implicated in long-range regulation of hedgehog signaling. Curr Biol. 2005;15:480–488. doi: 10.1016/j.cub.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Lane ME, Kalderon D. Genetic investigation of cAMP-dependent protein kinase function in Drosophila development. Genes & Dev. 1993;7:1229–1243. doi: 10.1101/gad.7.7a.1229. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, Kovacs A, Ornitz DM. Hedgehog signaling is critical for maintenance of the adult coronary vasculature in mice. J Clin Invest. 2008;118:2404–2414. doi: 10.1172/JCI34561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- Lepage T, Cohen SM, Diaz-Benjumea FJ, Parkhurst SM. Signal transduction by cAMP-dependent protein kinase A in Drosophila limb patterning. Nature. 1995;373:711–715. doi: 10.1038/373711a0. [DOI] [PubMed] [Google Scholar]
- Li W, Ohlmeyer JT, Lane ME, Kalderon D. Function of protein kinase A in hedgehog signal transduction and Drosophila imaginal disc development. Cell. 1995;80:553–562. doi: 10.1016/0092-8674(95)90509-x. [DOI] [PubMed] [Google Scholar]
- Lu X, Liu S, Kornberg TB. The C-terminal tail of the Hedgehog receptor Patched regulates both localization and turnover. Genes & Dev. 2006;20:2539–2551. doi: 10.1101/gad.1461306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Beachy PA. The Hedgehog response network: Sensors, switches, and routers. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003a;299:2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, Weis-Garcia F, Gong R, Wang B, Beachy PA. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell. 2003b;12:1261–1274. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- Ma Y, Erkner A, Gong R, Yao S, Taipale J, Basler K, Beachy PA. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell. 2002;111:63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- Mandal L, Martinez-Agosto J, Evans C, Hartenstein V, Banerjee U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446:320–324. doi: 10.1038/nature05585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RK, Beachy PA. Novel lipid modifications of secreted protein signals. Annu Rev Biochem. 2004;73:891–923. doi: 10.1146/annurev.biochem.73.011303.073933. [DOI] [PubMed] [Google Scholar]
- Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- Martinelli DC, Fan CM. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes & Dev. 2007;21:1231–1243. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Yao S, Zheng X, Geisbrecht BV, Ghirlando R, Beachy PA, Leahy DJ. Structure of a heparin-dependent complex of Hedgehog and Ihog. Proc Natl Acad Sci. 2006;103:17208–17213. doi: 10.1073/pnas.0606738103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Zheng X, Hauk G, Ghirlando R, Beachy PA, Leahy DJ. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature. 2008;455:979–983. doi: 10.1038/nature07358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- Motzny CK, Holmgren R. The Drosophila cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech Dev. 1995;52:137–150. doi: 10.1016/0925-4773(95)00397-j. [DOI] [PubMed] [Google Scholar]
- Muenke M, Beachy PA. Genetics of ventral forebrain development and holoprosencephaly. Curr Opin Genet Dev. 2000;10:262–269. doi: 10.1016/s0959-437x(00)00084-8. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Nystul T, Spradling A. Breaking out of the mold: Diversity within adult stem cells and their niches. Curr Opin Genet Dev. 2006;16:463–468. doi: 10.1016/j.gde.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Okada A, Charron F, Morin S, Shin DS, Wong K, Fabre PJ, Tessier-Lavigne M, McConnell SK. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature. 2006;444:369–373. doi: 10.1038/nature05246. [DOI] [PubMed] [Google Scholar]
- Pan D, Rubin GM. cAMP-dependent protein kinase and hedgehog act antagonistically in regulating decapentaplegic transcription in Drosophila imaginal discs. Cell. 1995;80:543–552. doi: 10.1016/0092-8674(95)90508-1. [DOI] [PubMed] [Google Scholar]
- Panáková D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- Patel NH, Martin-Blanco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Guerrero I. Unrestricted expression of the Drosophila gene patched allows a normal segment polarity. Nature. 1991;353:187–190. doi: 10.1038/353187a0. [DOI] [PubMed] [Google Scholar]
- Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- Strutt DI, Wiersdorff V, Mlodzik M. Regulation of furrow progression in the Drosophila eye by cAMP-dependent protein kinase A. Nature. 1995;373:705–709. doi: 10.1038/373705a0. [DOI] [PubMed] [Google Scholar]
- Tabata T, Kornberg TB. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell. 1994;76:89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- Takashima S, Mkrtchyan M, Younossi-Hartenstein A, Merriam JR, Hartenstein V. The behaviour of Drosophila adult hindgut stem cells is controlled by Wnt and Hh signalling. Nature. 2008;454:651–655. doi: 10.1038/nature07156. [DOI] [PubMed] [Google Scholar]
- Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, McMahon AP. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell. 2006;10:647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Taipale J. Hedgehog: Functions and mechanisms. Genes & Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- Woods IG, Talbot WS. The you gene encodes an EGF-CUB protein essential for Hedgehog signaling in zebrafish. PLoS Biol. 2005;3:476–487. doi: 10.1371/journal.pbio.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yam PT, Langlois SD, Morin S, Charron F. Sonic hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron. 2009;62:349–362. doi: 10.1016/j.neuron.2009.03.022. [DOI] [PubMed] [Google Scholar]
- Yao S, Lum L, Beachy P. The ihog cell-surface proteins bind Hedgehog and mediate pathway activation. Cell. 2006;125:343–357. doi: 10.1016/j.cell.2006.02.040. [DOI] [PubMed] [Google Scholar]
- Zhang W, Kang JS, Cole F, Yi MJ, Krauss RS. Cdo functions at multiple points in the Sonic Hedgehog pathway, and Cdo-deficient mice accurately model human holoprosencephaly. Dev Cell. 2006;10:657–665. doi: 10.1016/j.devcel.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Zhao C, Chen A, Jamieson C, Fereshteh M, Abrahamsson A, Blum J, Kwon H, Kim J, Chute J, Rizzieri D, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AJ, Zheng L, Suyama K, Scott MP. Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes & Dev. 2003;17:1240–1252. doi: 10.1101/gad.1080803. [DOI] [PMC free article] [PubMed] [Google Scholar]