Abstract
The ruminant conceptus undergoes a period of elongation that is required for maternal recognition of pregnancy, prior to attaching to the endometrium. The purpose of these studies was to investigate the role of proline-rich 15 (PRR15) in the sheep conceptus by examining mRNA expression, protein localization, and the effect of PRR15 mRNA degradation. Conceptuses were collected on Days 11, 13, 15, 16, 17, 21, and 30 after mating. Quantitative RT-PCR showed expression of PRR15 mRNA corresponded with the process of trophoblast elongation, with peak expression occurring on Days 15 and 16. A recombinant ovine PRR15 was generated and used to create polyclonal antibodies against PRR15. Immunohistochemistry of a Day 15 conceptus indicated that PRR15 was localized predominantly in the nucleus of the trophectoderm and extraembryonic primitive endoderm. To test whether PRR15 was required during early conceptus development, RNA interference was employed. Blastocysts collected on Day 8 after mating were infected with a lentivirus expressing a short-hairpin RNA (shRNA) that targeted PRR15 mRNA for degradation, an shRNA containing a three-nucleotide mismatch to PRR15 mRNA, or a lentivirus expressing no shRNA. After infection, blastocysts were transferred into recipient ewes and collected back on Day 15 of gestation. Although the majority of the control and mismatched shRNA-treated conceptuses elongated and survived to Day 15, none of the embryos treated with the lentivirus expressing shRNA against PRR15 mRNA elongated, and most died. In conclusion, expression of PRR15 mRNA occurred during a narrow window of conceptus development, and degradation of PRR15 mRNA led to conceptus demise or abnormal development.
Keywords: conceptus, implantation, placenta, trophoblast
Proline-rich 15 (PRR15) is highly expressed during a narrow window of conceptus development, and targeted degradation of PRR15 mRNA leads to conceptus demise.
INTRODUCTION
In all ungulate mammals, including ruminants, the trophoblast of the conceptus goes through a period of rapid elongation immediately prior to uterine attachment [1, 2]. Little is known about the cellular mechanisms involved in conceptus elongation in ruminants, partly because of the difficulty in the in vitro culture of elongated conceptuses [1, 3]. In all domestic ruminants, embryo mortality is most prevalent during this elongation phase of embryonic development, whereas later losses during the fetal period are only estimated at 5% [4, 5]. For a ruminant embryo to survive and establish itself in the uterus, it must avoid rejection by the maternal immune system and secrete factors that enable maintenance of the corpus luteum (CL). This is especially important during the “critical period” (Days 13–15 in sheep and Days 15–17 in cattle), during which the majority of embryo mortality occurs [6]. The predominant regulatory factor during this critical period in ruminants is interferon-tau (IFNT). First identified as ovine trophoblast protein-1, IFNT prevents luteolysis, allowing for maternal recognition of pregnancy in ruminants [7, 8]. However, many other factors, including growth factors, cell-adhesion molecules, cytokines, extracellular matrix metalloproteinases, hormones, and transcription factors, are also produced by the conceptus, enabling it to establish itself and grow in the uterus [9].
The gene that is the focus of these studies was first identified as G90 in the gastrointestinal tract of the mouse [10] and during early embryonic development in various tissues of the mouse [11]. Later, this same gene was independently identified by our laboratory [12] by RNA differential display in trophoblast tissue during critical periods of conceptus development in cattle and was called periattachment factor (PF). Both G90 and PF are now identified as proline-rich 15 (PRR15). PRR15 mRNA in the bovine conceptus was at greater concentrations on Day 17.5 than on Day 15.5 of pregnancy [12]. Further screening of PRR15 mRNA by Northern blot hybridization resulted in detection in a Day 25 cDNA library but no detection in Day 30 to Day 36 bovine trophoblasts. In a panel of seven adult bovine tissues, PRR15 mRNA was only detected faintly in the kidney [12]. Sequence analysis of PRR15 indicated an open reading frame (ORF) encoding a 126-amino acid protein. The inferred amino acid sequence predicted four putative protein kinase C (PKC) phosphorylation sites, two casein kinase II phosphorylation sites, as well as a nuclear targeting sequence [12], but no signal sequence for secretion. In silico analysis identified homologues of PRR15 in the human, baboon, chimpanzee, mouse, dog, and rat genomes. Interestingly, all six phosphorylation sites as well as the nuclear targeting sequence are identical between the cow and human sequences. A number of expressed sequence tags (ESTs) were also identified, including those derived from human placenta, colon, kidney, and stomach, as well as the mouse lung, neonatal eye, mammary gland, and brain. Further work confirmed the presence of PRR15 mRNA in the ovine conceptus on Day 15 and faintly in the adult sheep lung and kidney [13]. The time period of PRR15 expression [12] in trophoblast tissue corresponds with critical events during gestation, such as maternal recognition of pregnancy and the beginning of a close apposition of the embryonic trophoblast to the uterine endometrium.
A common method for beginning to elucidate the function of a novel gene is through reverse genetics and the creation of a knockout animal; however, this approach is difficult and inefficient in species other than mice [14]. RNA interference (RNAi), an evolutionarily conserved mechanism first described in the worm Caenorhabditis elegans, is a mechanism by which double-stranded RNA (dsRNA) activates sequence-specific gene silencing [15, 16]. In mammalian cells, dsRNAs can be presented in many forms; however, those longer than 30 nucleotides lead to interferon-mediated antiviral responses and nonspecific downregulation of gene expression. For this reason, either the injection of synthetic small interfering RNAs (siRNAs) 21–22 nucleotides long that mimic the products of Dicer, or vectors that express a short-hairpin RNA (shRNA) often are used to induce gene silencing. Because they are cleaved into siRNA within the cell, it is thought that shRNAs may be more efficient in targeting genes for silencing than in vitro-produced siRNA [17]. The use of lentiviruses to create transgenic or knockout animals with relatively high efficiency has been reported in mice and other species, particularly when compared to other methods of generating transgenic animals [14]. Recently, two separate groups have shown that lentiviral transduction of mouse blastocysts was not deleterious to blastocyst survival and resulted in trophoblast-specific transgenesis in all transduced embryos [18, 19].
The aim of these studies was to determine the precise timing of PRR15 mRNA expression during sheep conceptus development, to localize PRR15 in the conceptus at the time of peak mRNA expression and, finally, to use RNAi to study the development of the ovine conceptus after targeted degradation of PRR15 mRNA.
MATERIALS AND METHODS
Conceptus Tissue Collection
All procedures with animals were approved by the Colorado State University Institutional Animal Care and Use Committee. For analysis of PRR15 mRNA concentration during early gestation and localization of PRR15, conceptuses were collected from mature crossbred ewes on Days 11, 13, 15, 16, 17, 21, and 30 of gestation (n = 4 to 6 conceptuses per day). For collection of Day 11 and Day 13 conceptus tissue, ewes were superovulated by twice-daily i.m. injections for 4 days of follicle-stimulating hormone (FSH; 30, 30, 20, 20, 10, 10, 10, and 10 mg of Folltropin; Bioniche, Belleville, ON, Canada) beginning on Day 7 of the estrous cycle. Prostaglandin F2α (PGF2α; Lutalyse; Pfizer, New York, NY) was administered in two doses (5 mg, i.m., per dose) given 4 h apart beginning at the time of the sixth FSH injection. Conceptuses on Days 15 through 30 of gestation were collected from ewes synchronized by two injections (10 mg, i.m.) of PGF2α given 14 days apart. After observation of standing estrus with a vasectomized ram, all ewes were bred by one of two intact rams. Pregnant ewes were sedated using sodium pentobarbital (20 mg/kg, i.v.), a complete hysterectomy was performed, and the ewes were euthanized (90 mg/kg sodium pentobarbital, i.v.). Conceptuses were flushed from the uterus using Dulbecco modified Eagle medium/Nutrient Mixture F-12 Ham (DMEM/F12; Sigma, St. Louis, MO) supplemented with 0.25% Fraction V BSA (Sigma) warmed to 38°C. All recovered conceptuses were placed into 1.5-ml Eppendorf tubes and briefly centrifuged before removal of any liquid, and then they were frozen at −80°C until total cellular RNA (tcRNA) isolation. For Day 21 and Day 30 conceptuses, the fetus was separated from the trophoblast before freezing.
tcRNA Isolation and Quantitative Real-Time PCR
Total cellular RNA was isolated from individual Day 11 to Day 30 ovine conceptuses and Day 21 and Day 30 fetuses using RNeasy kits (Qiagen, Valencia, CA) according to the manufacturer's protocol. Quantity and integrity of RNA were confirmed by the absorbance ratio at 260:280 nm and by electrophoresis on a 1.2% formaldehyde denaturing gel and visualization of the 18S and 28S ribosomal subunits. Samples were stored at −80°C until use. cDNA was generated from approximately 2 μg of tcRNA by RT at 55°C for 50 min using oligo(dT) primers (Superscript III; Invitrogen, Carlsbad, CA). Each cDNA sample was treated with 5 units of RNase H (New England Biolabs, Ipswich, MA) for 20 min at 37°C. To control for variances in the efficiency of the RT reaction, all cDNA was quantified by spectrophotometry based on absorbance ratio at 260:280 nm, and an equal amount of cDNA was used for each sample in the quantitative RT-PCR (qRT-PCR) reaction.
Primers for ovine PRR15 (PRR15) were designed to amplify an intron-spanning, 500-bp product based on the bovine PRR15 (PRR15) cDNA sequence [12]. The PRR15 primers used were: forward, 5′-TAG CTG GAC TGC AGC GAT TT-3′, and reverse, 5′-GAC ACT GGG GTG CTG ATT CT-3′. The primers and Taqman probe sequences for ovine IFNT (IFNT) are able to detect all isoforms of IFNT [20]. Both ovine PRR15 and IFNT mRNA were normalized to ovine GAPDH (GAPDH) mRNA levels after determining no differences existed in ovine GAPDH mRNA concentrations between any day of development (Fisher least significant difference, P > 0.10). The ovine GAPDH forward primer, 5′-GAT TGT CAG CAA TGC CTC CT-3′, and GAPDH reverse primer, 5′-GGT CAT AAG TCC CTC CAC GA-3′, amplified a 94-bp product. For analysis of PRR15 mRNA in the human BeWo choriocarcinoma cell line, primers were designed to amplify a 228-bp, intron-spanning cDNA for human PRR15 (PRR15). The human PRR15 primers used were: forward, 5′-CCA GAA GCC TGA TCT CTC CA-3′, and reverse, 5′-CCC TTT CTC CAC GTG GTC T-3′. For qRT-PCR analysis, human PRR15 mRNA concentrations were normalized to human ribosomal protein S15 (RPS15) mRNA concentrations. The human RPS15 primers amplified a 361-bp product and were: forward, 5′-TTC CGC AAG TTC ACC TAC C-3′, and reverse, 5′-CGG GCC GGC CAT GCT TTA CG-3′. Before qRT-PCR was performed, all primers were tested by amplification of a PCR product that was cloned into pPCR-Script Amp SK(+) vector (Stratagene, La Jolla, CA) and sequenced (Colorado State University Macromolecular Resources) to verify identity. An annealing temperature of 60°C was used for all genes.
For qRT-PCR analysis, a standard curve was created for each gene from 1 × 102 to 1 × 10−6 pg using a PCR product amplified from the sequenced plasmid for each gene. Amplification was detected using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) for ovine PRR15, human PRR15, human RPS15, and ovine GAPDH, and iQ Supermix for ovine IFNT on a Biorad iCycler (Bio-Rad) with the addition of 200 nM forward and reverse primers for each gene. Because of the high guanine and cytosine content of the ovine PRR15 transcript (67%), betaine (N,N,N-trimethyl glycine; Sigma) at 1 M final concentration and Pyrococcus furiosus (Pfu) polymerase (Pfu Turbo Hotstart; Stratagene) at 0.15 units were added per 25-μl qRT-PCR reaction. Samples were amplified by 45 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min. For all genes except ovine IFNT, a melt-curve analysis was conducted by increasing the temperature 0.5°C at 10-sec intervals from 55°C to 100°C. All standards were run in duplicate, and cDNA from each conceptus was run in triplicate. Product specificity was confirmed by observation of a single melt curve for each gene and by electrophoresis and visualization of all qRT-PCR products on a 2% agarose gel. The starting quantity (picograms) of the gene of interest was normalized to the starting quantity (picograms) of ovine GAPDH mRNA.
Recombinant Ovine PRR15 Production
Primers were designed to amplify the entire ORF of ovine PRR15. The primer sequences were: forward, 5′-GCA CAC GTG ACT TCG ACA AA-3′, and reverse, 5′-AAT GGG AAG AGA GTC CTG GAC-3′. Ovine PRR15 was cloned into pENTR/TEV/D-TOPO (Invitrogen), which contains a Tobacco Etch Virus (TEV) protease site on the 5′ end of the PRR15 cDNA, and then was transferred to the glutathione S-transferase (GST) fusion vector, pDEST15 (Invitrogen), by site-specific recombination using LR clonase II (Invitrogen). The resulting vector, pDEST/GST-TEV-ovine PRR15, was transformed into chemically competent BL21(D3) bacteria by standard techniques. A culture of BL21(D3) bacteria was grown in LB-ampicillin (50 μg/ml) at 37°C until the culture grew to an optical density at 260 nm of 0.5. Isopropyl β-d-thiogalactopyranoside (1 mM) was added, and the culture was incubated for 5 h. The bacteria were pelleted and then lysed with 10 ml of B-PER (Pierce, Rockford, IL) and then centrifuged at 27 000 × g for 15 min at 4°C. The supernatant from the centrifuged bacterial lysate was allowed to flow through a glutathione affinity column. The glutathione affinity column was washed with Pierce Wash Buffer 1, followed by Wash Buffer 2 (Pierce 78200). A “cleavage mixture” of 2.5 ml of Wash Buffer 2, 25 μl of AcTEV protease (Invitrogen), and 22 μl of 0.1 M dithiothreitol was mixed and added to the glutathione affinity column, and the column was placed at 4°C overnight. The following day, the column was placed at room temperature for 30 min and then washed twice with 2.5 ml of Wash Buffer to collect the column eluate. The eluate was dialyzed against 1 L of ammonium acetate buffer (50 mM, pH 5.5) for 2 h at 4°C. A chromatography column containing S-Sepharose (Sigma) was equilibrated with 30 ml of ammonium acetate start buffer (50 mM, pH 5.5). The dialyzed protein sample was then added to the S-Sepharose column. The column was washed with start buffer, followed by successive 10-ml fractions of elution buffer (50 mM ammonium acetate, pH 5.5) containing increasing concentrations of NaCl (300 mM, 325 mM, and 350 mM). Fractions were collected upon addition of the 10 ml of each elution buffer. The 280-nm absorbance of each fraction was determined, and an absorbance curve was plotted. Two absorbance peaks were observed: one in the first five fractions and another larger peak in fractions 9–16. The samples of the second absorbance peak were pooled and concentrated by processing in Centricon YM-10 centrifugal filter units (Millipore, Billerica, MA) at 5000 × g for 1 h. A concentrated sample was electrophoresed on a polyacrylamide gel and stained with Coomassie. Only a single band at the expected molecular weight was observed on the Coomassie-stained gel. A sample of the recombinant oPRR15 (roPRR15) was also sequenced (Colorado State University Macromolecular Resources) to confirm its identity.
The recombinant protein was then sent to Covance Immuno Technologies (Denver, PA) for polyclonal antibody production. Two New Zealand White rabbits were injected with 125 μg of roPRR15 with Freund incomplete adjuvant at 21-day intervals repeated six times. Preimmune serum was collected from each rabbit prior to any injections with roPRR15, and antisera were collected after the fifth and sixth injections. The antisera (CSU-αoPRR15–146) and preimmune sera (CSU-oPRR15 PB) collected from the rabbits were used for immunohistochemistry. The antisera was used in Western immunoblot analysis of the recombinant protein, which detected a single band at the correct size. Detection could effectively be abolished by preabsorbing CSU-αoPRR15–146 with roPRR15 (data not shown).
Immunohistochemistry
Day 15 conceptuses recovered from pregnant ewes were fixed in 4% paraformaldehyde in PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.3) for 1 h and then placed into 70% ethanol overnight at 4°C before paraffin embedding. Six-micrometer sections were cut and placed onto Superfrost/Plus slides (Fisher Scientific, Waltham, MA) and dried overnight. Slides were then deparaffinized and were rehydrated through a graded ethanol series (100%, 95%, 70%, and 50%). An antigen-retrieval step was performed by incubating slides in a 10 mM sodium citrate/0.05% Tween 20 (Sigma) solution for 30 min at 85°C. Sections were then bathed in 3% hydrogen peroxide for 1 h to quench any endogenous peroxidase activity. Staining was performed using the Vectastain Elite ABC kit according to the manufacturer's protocol (Vector Laboratories, Burlingame, CA). The antisera and preimmune serum were diluted in PBS supplemented with 1.5% (v/v) normal goat serum at a 1:10 000 dilution. Peroxidase activity was visualized using 4% 3,3′diaminobenzidine solution for 90 sec before washing in water and dehydrating through a graded ethanol series. For morphological comparisons, a serial section was also stained with hematoxylin and eosin using standard procedures.
The human choriocarcinoma BeWo cell line was also stained for presence of PRR15. Cells were grown in F12-K media (Mediatech, Herndon, VA) in chamber slides for 24–48 h prior to staining. Cells were then washed twice with PBS and fixed with 4% paraformaldehyde in PBS for 5 min, followed by two more washes in PBS for 3 min each. Cells were then stained with antisera or preimmune sera as described above.
Construction of Lentiviral Vectors
Lentiviral infection of embryos was used to stably integrate and express shRNA targeting the ovine PRR15 mRNA sequence. The lentiviral vector pLL3.7 [21] containing the RNA polymerase III promoter U6, upstream of the multiple cloning site for introduction of shRNA cassettes, as well as a cytomegalovirus promoter upstream of enhanced green fluorescent protein (EGFP) was used. Oligonucleotides were designed to contain (from 5′ to 3′) a 5′Xho overhang, ovine PRR15 target antisense sequence, a TTCAAGAGA loop sequence, ovine PRR15 target sense sequence, six thymidine residues for PolIII termination, and a blunt 3′ end. Oligonucleotides were ordered with 5′ phosphates and then PAGE purified (Invitrogen). Oligonucleotides were diluted in water to 60 pmol/μl. One microliter each of complementary oligonucleotides was annealed in 48 μl of annealing buffer (100 mM potassium acetate; 30 mM HEPES-KOH, pH 7.4; and 2 mM magnesium acetate). The annealing mixture was incubated at 95°C for 4 min and 70°C for 10 min, and then cooled to 4°C. The annealed oligonucleotides were ligated into pLL3.7 that was digested with XhoI/HpaI, and they were treated with 10 units of calf intestinal phosphatase (New England Biolabs). The resulting vectors were sequenced (Colorado State University Macromolecular Resources) to verify correct insertion and sequence identity.
Lentiviral particles were generated in 293FT cells (Invitrogen) grown in high-glucose DMEM medium supplemented with 10% fetal calf serum in an incubator at 37°C and 5% CO2 to approximately 2 × 106 cells on the day of transfection. For each 10-cm dish, serum-free DMEM was mixed with one of the pLL3.7 lentiviral constructs (3.92 μg), as well as the packaging vectors pRΔ8.74 (2.96 μg; gag/pol elements) and pMD2.G (1.12 μg; env elements) in a total volume of 300 μl. The transfection agent Polyfect (80 μl; Qiagen) was added, as well as 1 ml of complete DMEM medium. This transfection mixture was added to 293FT cells with 7 ml of complete DMEM medium. After overnight incubation, the medium was aspirated off and discarded, and fresh medium was added. The medium was then collected at the end of 24 and 48 h of incubation with the 293FT cells. Next, the lentivirus supernatant was ultracentrifuged over a 30% sucrose cushion at 47 000 × g for 2 h at 16°C. The resulting pellets were then resuspended in Hepes-buffered chemical-defined medium for late-stage embryos (HCDM-2) [22], aliquoted, and stored at −80°C for future use. Frozen aliquots of concentrated virus were thawed and 10-fold serially diluted. Serial dilutions of virus were added to six-well plates of HEK293 cells at ∼90% confluency. The numbers of GFP-expressing cells within each well were counted, and the titer was calculated based on the volume of the initial aliquot present in the well from which GFP-expressing cells were counted.
Prior to in vivo studies with ovine embryos, three shRNA sequences were tested for their efficacy in degrading ovine PRR15 mRNA in vitro using the psiCHECK vector (psiCHECK-2; Promega, Madison, WI). With the psiCHECK vector, the oPRR15 ORF was inserted into the multiple cloning region downstream (3′) of the Renilla luciferase gene stop codon. This vector was cotransfected into 293T and Cos-7 cells along with the pLL3.7 vectors containing various shRNA cassettes. One shRNA sequence (pLL3.7-shRNA3) produced 80% knockdown of luciferase expression and was chosen for the in vivo studies. An empty vector expressing no shRNA but producing EGFP was used as a control (pLL3.7). As a second control, a lentiviral vector was generated containing an shRNA targeting the same sequence as pLL3.7-shRNA3, except for 3-bp mismatches with the ovine PRR15 mRNA (pLL3.7-MM) sequence. The shRNA produced by this vector (pLL3.7-MM) is homologous to the human PRR15 mRNA sequence. This vector was then tested on the BeWo cells. Lentiviral particles were generated in 293FT cells and added to BeWo cells in a 1:1 ratio with F12-K medium (Mediatech). The BeWo cells were treated with either pLL3.7-shRNA3, pLL3.7, or pLL3.7-MM. After 48 h of treatment, tcRNA was harvested from the cells using the RNeasy mini kit (Qiagen), and cDNA was generated and analyzed by qRT-PCR as described previously. This was repeated twice, with a new virus being produced in 293FT cells each time.
Embryo Collection and Transfer
Embryo donor ewes were superovulated by twice-daily i.m. injections of FSH for 4 consecutive days (48, 48, 36, 36, 24, 24, 20, and 20 mg; Folltropin). At the time of the fifth FSH injection, PGF2α was administered in two doses (5 mg, i.m., per dose; Lutalyse) given 4 h apart. At the time of the sixth FSH injection, equine chorionic gonadotropin (150 units, s.c.) was administered, and gonadotropin-releasing hormone (50 μg, i.m.; OvaCyst; Vedco, St. Joseph, MO) was administered at the time of the eighth FSH injection. Recipient ewes were given two doses of PGF2α (5 mg, i.m., per dose) 4 h apart beginning at the time the donor ewes received the fourth FSH injection. Standing estrus in all ewes was observed by using a vasectomized ram. After observation of standing estrus, all donor ewes were bred by one of two intact rams. For embryo collection, pregnant ewes were sedated using sodium pentobarbital (20 mg/kg, i.v.), a complete hysterectomy was performed, and the ewes were euthanized (90 mg/kg sodium pentobarbital, i.v.). Conceptuses were flushed from the uterus using modified Dulbecco PBS (m-PBS) [23] supplemented with 0.1% w/v Fraction V BSA warmed to 38°C.
Collected embryos were washed in HCDM-2. Because viruses are unable to cross the zona pellucida, embryos were recovered on Day 8, a time when most are hatched. Recovered embryos that had not already hatched out of the zona pellucida in vivo were mechanically hatched at this time with a microblade or 26-gauge needle. For virus infection, three hatched blastocysts were incubated in 100-μl drops overlaid with mineral oil. Each drop contained 60 μl of chemically defined medium for late-stage embryos (CDM-2) [22], 30 μl of concentrated lentivirus (1 × 107 TU/ml), and 10 μl of polybrene (8 ng/ml; Sigma). Drops containing embryos were cultured in 5% CO2/5% O2 /90% N2 at 37°C for approximately 6 h prior to transfer. Embryos to be transferred were recovered in the morning and transferred the same day in the afternoon. Immediately prior to transfer, embryos were washed in HCDM-2 three times. Embryos from any individual donor ewe were divided among multiple treatments, and multiple treatment groups were transferred on any given day. Only recipient ewes that had come into estrus within 24 h of the donor ewes received embryos.
Transfer was performed using a fine, fire-polished glass pipette inserted through a puncture wound into the uterine horn ipsilateral to the CL. Three embryos were transferred into each recipient. Each recipient was given flunixin meglumine immediately prior to surgery and again 12 h after surgery (75 mg, i.m.; VedaGesic; Vedco). Recipient ewes were sedated using sodium pentobarbital (20 mg/kg, i.v.). An endotracheal tube was inserted, and recipients were maintained on 2 L/min O2 and 1%–2% isoflurane (Altane; Minrad Inc., Bethlehem, PA) during the transfer procedure. After transfer, recipient ewes were maintained on an ad libitum diet of alfalfa hay and monitored daily for 7 days after transfer. On Day 7 after transfer (Day 15 of gestation), the recipient ewes were euthanized as described previously, and conceptuses were recovered. After flushing, both uterine horns were cut longitudinally to visually inspect for any elongated conceptuses that may have attached to the endometrium. Total cellular RNA from lentiviral-treated conceptuses was isolated and cDNA generated as previously described.
Recovered embryos were analyzed for EGFP expression to ensure lentiviral infection had occurred using the following EGFP primers: forward, 5′-TCT TCT TCA AGG ACG ACG GCA ACT-3′, and reverse, 5′-TGT GGC GGA TCT TGA AGT TCA CCT-3′. With the same cDNA from lentiviral-infected embryos that was used for qRT-PCR analysis, EGFP was amplified by 40 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec. A portion of the PCR product was electrophoresed on 2% agarose gel, and a portion of the PCR product was sequenced (Colorado State University Macromolecular Resources) to verify specificity.
Statistical Analysis
All data were analyzed using SAS software (SAS Institute, Cary, NC). For all qRT-PCR data, ovine GAPDH-normalized ovine PRR15 and ovine IFNT mRNA concentrations were subjected to ANOVA using the PROC GLM procedure, with regression contrasts performed to further characterize the expression pattern. Comparisons between days of gestation, tissue types, and lentiviral constructs were made using Tukey honestly significant difference test. Due to heterogeneity of variance, qRT-PCR data for oPRR15 mRNA across different days of gestation and data for lentiviral-treated embryos were log-transformed. Ovine IFNT mRNA data and PRR15 mRNA data (Days 21 and 30 trophoblast and fetus) were not log-transformed because with these data, heterogeneity of variance did not occur. The percent of embryos recovered was analyzed by χ2.
RESULTS
Conceptus qRT-PCR During Early Gestation
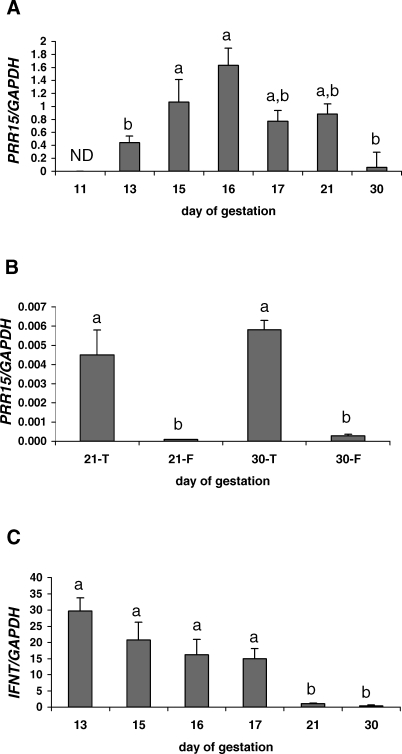
Quantitative RT-PCR was performed in sheep to accurately define the temporal pattern of gene expression during early conceptus development. Although the housekeeping gene, ovine GAPDH, mRNA was detected in Day 11 conceptuses, no PRR15 mRNA was detected at this stage of gestation. This observation was based on the lack of an amplification curve in the qRT-PCR reaction, as well as the lack of a band after electrophoresis of the qRT-PCR products on a 2% agarose gel. PRR15 mRNA exhibited a quadratic expression pattern (P < 0.05) from Day 13 to Day 30 of gestation, with peak mRNA levels detected on Day 16 (Fig. 1A). Expression of PRR15 mRNA on Days 15 and 16 was significantly greater than on Day 13 or Day 30 (P < 0.05). A comparison of PRR15 mRNA concentration between the fetal and trophoblast tissue of Day 21 and Day 30 conceptuses was also performed. Ovine PRR15 mRNA concentrations were greater (P < 0.05) in trophoblast tissue than fetal tissue on Days 21 and 30 (Fig. 1B). There were no significant differences in PRR15 mRNA concentrations between Day 21 and Day 30 for either the trophoblast or fetal tissue.
FIG. 1.
Quantitative RT-PCR of ovine PRR15 (A and B) and IFNT (C) mRNA concentration normalized to GAPDH mRNA in ovine conceptuses. A) Log-transformed PRR15 mRNA concentrations exhibited a quadratic expression pattern (P < 0.05). ND, no PRR15 mRNA amplification detected at this day. B) PRR15 mRNA concentrations in ovine trophoblasts (T) and fetuses (F). C) IFNT mRNA exhibited a linear expression pattern (P < 0.01). Data represent least squares means ± SEM. Bars with different lowercase letters above the bar differ (P < 0.05).
Because little is known about ovine PRR15 or its function, we compared the mRNA concentrations of PRR15 to those of a gene of known importance during early pregnancy, IFNT. From Day 13 to Day 30, IFNT mRNA exhibited a linear decrease in expression (P < 0.01; Fig. 1C). The concentrations of IFNT mRNA on Day 21 and Day 30 were lower than any other day of gestation studied (P < 0.05).
Immunohistochemistry of Conceptuses
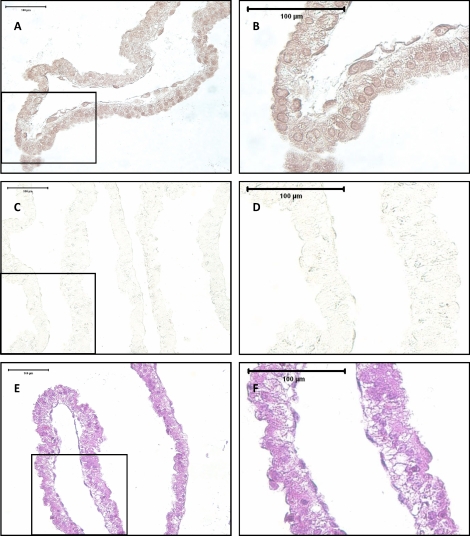
The peak concentration of ovine PRR15 mRNA as detected by qRT-PCR was on Days 15 and 16 of gestation. For this reason, Day 15 conceptus tissue was fixed and paraffin embedded in preparation for immunohistochemistry. Immunohistochemistry using an ovine PRR15-specific antibody on Day 15 conceptus tissue showed staining throughout the length of the conceptus (Fig. 2A). Staining appeared to be predominantly nuclear, with some lighter staining in the cytoplasm (Fig. 2B). PRR15 was detected in the trophectoderm, as well as the extraembryonic primitive endoderm of the conceptus. No staining was detected in sections stained with preimmune serum (Fig. 2, C and D).
FIG. 2.
Immunohistochemistry for PRR15 in serial sections of a paraffin-embedded Day 15 ovine conceptus. A) Conceptus stained using rabbit anti-PRR15 antibody. B) Magnification of a portion of A. C) Staining with preimmune serum from the same rabbit used to generate an antibody against PRR15. D) Magnification of a portion of C. E) Hematoxylin and eosin staining. F) Magnification of a portion of E.
Lentiviral Infection and Embryo Transfer
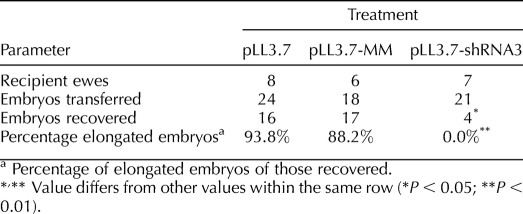
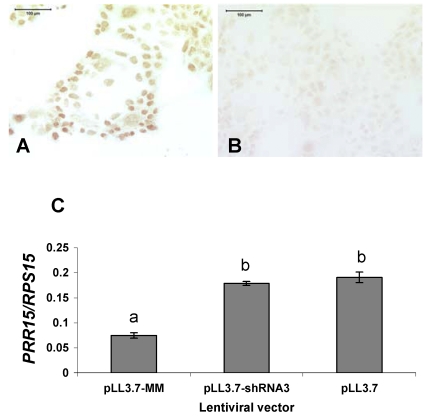
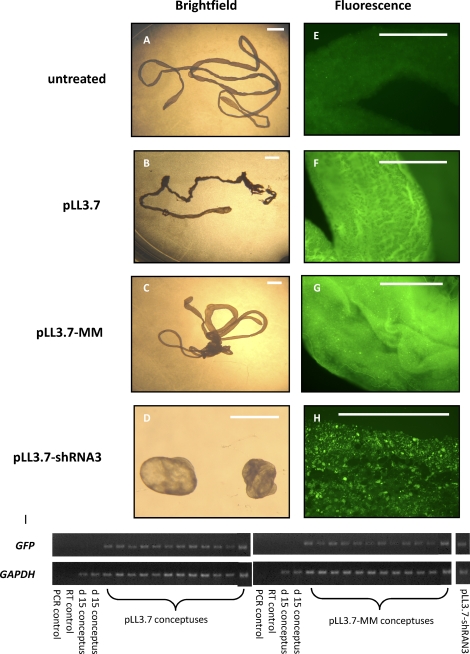
A total of 63 lentiviral-infected embryos were transferred into 21 recipient ewes. The results from each treatment are summarized in Table 1. Although 93.8% of the empty vector (pLL3.7)-treated and 88.2% mismatched shRNA (pLL3.7-MM)-treated conceptuses had elongated by Day 15 of gestation, 0% of the knockdown vector (pLL3.7-shRNA3)-treated conceptuses had elongated by this point. Only 4 of 21 embryos transferred were recovered back for the pLL3.7-shRNA3 treatment, indicating a high degree of embryo mortality in this treatment. These four pLL3.7-shRNA3 embryos were recovered from three separate ewes. Transfer of pLL3.7-treated and pLL3.7-MM-treated embryos resulted in 66.7% and 94.4% recovery rates, respectively. The pLL3.7-MM lentiviral construct targeted the human PRR15 mRNA sequence and was tested on BeWo cells in a 48-h culture. Immunohistochemistry in BeWo cells shows that PRR15 is expressed in, and localizes to, the nucleus, similarly to the ovine conceptus (Fig. 3, A and B). The pLL3.7-MM construct resulted in 63% knockdown in human PRR15 mRNA compared with controls in BeWo cells (Fig. 3C). This was a significant decrease (P < 0.05) in human PRR15 mRNA concentration compared with both pLL3.7-treated and pLL3.7-shRNA3-treated cells. This demonstrates the target specificity of pLL3.7-shRNA3 and that pLL3.7-MM can induce RNAi when the target mRNA is present.
TABLE 1.
Summary of sheep embryo transfer.
FIG. 3.
A) BeWo cells stained with rabbit anti-ovine PRR15 antibody. B) BeWo cells stained with preimmune serum from the same rabbit used to generate an antibody against ovine PRR15. Bars = 100 μm. C) Quantitative RT-PCR of human PRR15 mRNA in BeWo cells treated with lentiviral vectors expressing shRNA against human PRR15 mRNA (pLL3.7-MM) or against ovine PRR15 mRNA (pLL3.7-shRNA3) and an empty vector control (pLL3.7). Data are least squares means ± SEM. Bars with different lowercase letters above the bars differ (P < 0.05).
At the time of embryo collection, a subset of each treatment was assessed for EGFP expression by fluorescence microscopy. A portion of the conceptus was dissected off and placed in 70% ethanol and then placed on a glass slide, and fluorescence was assessed using filters at 480-nm excitation and 535-nm emission. An untreated Day 15 conceptus was used as a control. No EGFP expression was observed in the untreated sample (Fig. 4E), whereas all lentivirus-treated samples showed EGFP fluorescence (Fig. 4, F–H). All conceptus samples to be used for qRT-PCR were analyzed for EGFP transgene expression by PCR. All samples indicated that insertion of the lentiviral vector DNA had occurred (Fig. 4I).
FIG. 4.
Brightfield (A–D) and fluorescence (E–H) microscopy of Day 15 conceptuses treated with one of three lentiviral constructs and an untreated control. Bar = 1 mm. I) The PCR for GFP and GAPDH was performed on cDNA from 12 pLL3.7 conceptuses, 12 pLL3.7-MM conceptuses, a pool of two pLL3.7-shRNA3, and two untreated control conceptuses. No GFP was detected in the PCR control, RT control, or untreated Day 15 control conceptuses. GAPDH mRNA was present in all conceptus samples.
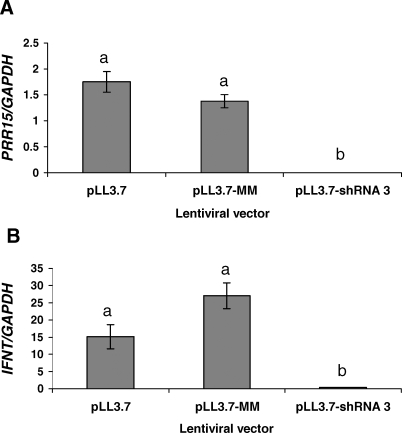
For qRT-PCR analysis, 12 pLL3.7, 12 pLL3.7-MM, and two pLL3.7-shRNA3 conceptuses were used. There was no difference in either ovine PRR15 mRNA or IFNT mRNA between pLL3.7-treated and pLL3.7-MM-treated conceptuses (Fig. 5). For conceptuses treated with pLL3.7-shRNA-3, PRR15 mRNA was undetectable, and IFNT mRNA was reduced (P < 0.05).
FIG. 5.
Quantitative RT-PCR analysis of log-transformed ovine PRR15 (A) and IFNT (B) mRNA in Day 15 conceptuses treated with one of three lentiviral constructs (pLL3.7, pLL3.7-MM, or pLL3.7-shRNA3). Data represent least squares means ± SEM. Bars with different lowercase letters above the bar differ (P < 0.05).
DISCUSSION
This study provides evidence that expression of the gene PRR15 is required for normal conceptus development in sheep. The process of elongation is required for conceptus survival in all ungulate species [1, 2]. Quantitative RT-PCR was used to precisely determine the timing of PRR15 mRNA expression in the sheep conceptus during development from Day 11 to Day 30 of gestation (Fig. 1A). The time point when PRR15 mRNA was first detected (Day 13) roughly coincides in time with when the ovine conceptus undergoes a period of rapid elongation [24, 25]. On Day 11, ovine conceptuses retain a spherical shape, and elongation begins around Day 12 [24, 25]. However, there is variation in the exact time of onset of elongation [1, 2]. In this study, conceptuses recovered on Day 11 of gestation were indeed spherical, and no PRR15 mRNA was detectable at this time, whereas the housekeeping gene, GAPDH, mRNA was present. After PRR15 mRNA was first detected on Day 13, PRR15 mRNA exhibited a quadratic pattern of expression, with peak mRNA concentration detected at Day 16 of gestation before declining on Days 17 through 30. The increase in PRR15 mRNA up to Day 16 correlates well with previous studies that reported an increase in embryo length up to Day 15 and the beginning of a close apposition of the conceptus to the uterus [26, 27]. On Day 16 in sheep, the conceptus begins to adhere to the endometrium, and elongation has stopped [7, 28]. Although there are larger variances in PRR15 mRNA concentration on Day 16, this could be due to an inherent variation in ovine conceptus size [2, 29]. The decline in PRR15 mRNA from Days 17–30 roughly agrees with Northern blot hybridization results obtained in the cow [12, 13]. In the cow, PRR15 mRNA was undetectable from Day 30 to Day 36 but was detectable at Days 21 and 28, indicating a decrease in expression in this species as well. It is interesting to note the abrupt decrease in PRR15 mRNA concentrations in ovine trophoblast occurs within days after the burst of expansion (Fig. 1A). Initial Northern blot hybridization results indicated PRR15 mRNA may be trophoblast specific [13]. However, using qRT-PCR we were able to detect PRR15 mRNA in the sheep fetus, although at a much lower concentration than what was observed in the trophoblast (Fig. 1B). Furthermore, although PRR15 mRNA was greater in trophoblast than fetal tissue on Days 21 and 30, these days represent a significant decrease after the peak PRR15 mRNA concentration on Day 16 (Fig. 1A). Based on these observations, it appears likely that PRR15 may have a functional role in the trophoblast, and not the fetus, during conceptus expansion.
IFNT is the pregnancy recognition signal in ruminants, secreted from Day 10 to Day 21 in the sheep [30, 31]. For this reason, we also measured ovine IFNT mRNA during conceptus development along with PRR15 mRNA. The linear decrease in IFNT mRNA from Day 13 to Day 30 (Fig. 1C) agrees with previous reports that described a decrease in IFNT mRNA from Day 14 to Day 22 of pregnancy in the sheep using dot blots [31], as well as a peak of IFNT transcription on Day 13 [32]. Ovine IFNT binds to the IFNAR1 and IFNAR2 receptors used by all type I interferons, and these receptors are present on most mammalian cells and are components of signal transduction pathways that induce IFNT-stimulated genes [8, 31, 32]. Because of the increase in ovine IFNT just prior to PRR15 mRNA expression, it is possible that signaling by IFNT or an interferon-stimulated gene could induce PRR15 transcription. However, initial work in our lab detected PRR15 mRNA in the trophoblasts of nonruminant species, such as the horse [13], and cDNAs have been identified in the human placenta, neither of which expresses IFNT; therefore, it is unlikely that IFNT and PRR15 are interacting with each other. Thus, PRR15 likely has a separate but important function during this time period of conceptus development.
Recombinant ovine PRR15 was used to generate polyclonal antibodies against PRR15 in the rabbit. Immunohistochemistry of a Day 15 ovine conceptus (Fig. 2) showed predominantly nuclear staining, confirming initial data generated by amino acid sequence analysis that reported a putative nuclear targeting sequence [12]. Staining was also performed in BeWo cells that were used for testing control shRNA constructs. These cells also showed distinct nuclear staining, indicating similar localization across species in another placental cell type. Staining was also apparent in the cytoplasm in the conceptuses; however, it is unknown how long PRR15 remains in the cytoplasm after translation. There are four putative PKC phosphorylation sites within ovine PRR15, suggesting PKC phosphorylation may be a prerequisite to trafficking into the nucleus. Although we hypothesized that PRR15 staining would be located in the trophectoderm only, it was also apparent in the extraembryonic primitive endoderm. The cells of the extraembryonic primitive endoderm develop from the inner cell mass (ICM) and migrate beneath the trophectoderm, further differentiating into the visceral endoderm beneath the ICM and the parietal endoderm (PE) surrounding the rest of the blastocoele cavity [25]. This PE layer of cells also elongates in ruminant embryos, along with the trophectoderm [7, 25, 33]. In sheep, the PE cells stretch and take on a multinucleated syncytial morphology but retain the morphological characteristics of polarized epithelium [25]. A multinucleated PE has also been observed in elongating bovine and porcine conceptuses [25, 34]. The PE cells have epithelial cell characteristics, such as desmosome-associated tonofilaments, structural polarization, and microvilli [2]. The morphology of the PE could be the result of cellular adaptation to manage the rapid elongation of the trophoblast [25]. Therefore, it is not surprising that ovine PRR15 is found in both the trophectoderm and extraembryonic primitive endoderm of elongating conceptuses if it is involved in transcriptionally regulating trophoblast outgrowth and migration. Furthermore, qRT-PCR that compared ovine PRR15 mRNA in trophoblast to that in fetus on Days 21 and 30 indicated that PRR15 mRNA is present in fetal tissues as well (Fig. 1B), indicating it is not exclusively trophoblast derived.
To begin to elucidate the function of PRR15 during early conceptus development, we used a lentiviral vector that expresses an shRNA directed against the PRR15 mRNA sequence. Knockdown of ovine PRR15 mRNA resulted in embryo mortality by Day 15 of pregnancy, or a complete block of conceptus elongation, whereas a large majority of control embryos elongated (88.2%–93.8%) and were recovered (66.7%–94.4%; Table 1). Because of the normal recovery rates in both sets of control embryos compared with the very low (19%) recovery rate of those treated with pLL3.7-shRNA3, it is clear that treatment with this construct resulted in conceptus demise in the majority of these treated embryos. The exact size of each conceptus recovered was not measured. However, it is known that the rates of elongation and size at any particular day after conception are variable [1, 2, 24]. It is possible that in vitro culture and/or lentiviral treatment may have affected the rate of elongation in conceptuses in this study. This would not be surprising, because lentiviral infection of murine blastocysts can result in some developmental delay [19], and it is well documented that in vitro culture for any time period may delay development [35]. Regardless of any developmental delay that may have occurred due to the effect of culture or lentivirus, it is clear that the vast majority of our control lentiviral-treated embryos were able to survive and elongate by Day 15, whereas reduced PRR15 mRNA resulted in significant conceptus demise.
Under normal in vivo conditions, PRR15 mRNA was undetectable prior (Day 11) to initiation of elongation, and then it increased around the time conceptus elongation begins (Fig. 1A). Recovered conceptuses treated with pLL3.7-shRNA3 had a morphology similar to that of an unelongated Day 11–12 conceptus, and no ovine PRR15 mRNA was detected in these conceptuses (Figs. 4D and 5A). Conceptus elongation is required for expression of IFNT, which allows for maternal recognition of pregnancy and conceptus survival [36, 37]. Therefore, it is possible that failure to elongate resulted in conceptus demise and resorption in embryos treated with pLL3.7-shRNA3. Failure to elongate in conceptuses treated with pLL3.7-shRNA3 may also help explain the reduction in IFNT mRNA observed in the unelongated conceptuses that were recovered (Fig. 5). However, conceptus demise due to factors other than elongation cannot be ruled out, and the high resorption rate of these conceptuses may indicate that conceptus demise occurred prior to the process of elongation. Although peak expression of PRR15 mRNA coincided with the time of trophoblast attachment to the uterus in sheep (Fig. 1A), PRR15 appears to have a functional role earlier than this time, because PRR15 mRNA degradation resulted in few embryos surviving to the time of attachment. The mRNA expression pattern of PRR15 and the results observed after knockdown of PRR15 mRNA suggest that PRR15 is playing a critical role in early development of the ruminant conceptus, but likely not in the attachment process. Our qRT-PCR data indicated that PRR15 mRNA began to decrease as soon as attachment begins (Fig. 1A).
Previous studies have identified global gene expression changes during the elongation period of conceptus growth [28, 38–40]. However, most of these studies identified previously known genes, and none provided evidence of critical function using a knockdown or knockout approach. Suppressive subtractive hybridization was used to identify a highly upregulated novel gene during the elongation process in porcine conceptuses [38]. However, sequence alignment between this gene (GenBank accession no. AC009682) and PRR15 showed no sequence similarity, and no further study on the function of this novel gene has been reported. In contrast to most genes studied during conceptus elongation [41, 42], PRR15 mRNA does not continue to increase throughout development, but peaks on Day 16 before declining markedly through Day 30 in trophoblast tissue. Day 16 corresponds to the beginning of attachment of the trophoblast to the uterus [27, 28]. Perhaps physical attachment to the endometrium and signaling through integrins or extracellular matrix proteins function to reduce transcription of PRR15 mRNA in the ovine trophoblast at this time.
As controls for our in vivo RNAi experiments, we used a lentiviral vector expressing no shRNA (pLL3.7) and a vector expressing an shRNA with a 3-bp mismatch to the ovine PRR15-specific shRNA sequence (pLL3.7-shRNA3). There was no difference in expression of either PRR15 or IFNT mRNA in Day 15 conceptuses treated with either control vector (Fig. 5). Others have reported that mismatches in an shRNA sequence can abolish the silencing effect [17]. Importantly, we have shown that the pLL3.7-MM is able to induce RNA degradation of human PRR15 mRNA, where there are no mismatched nucleotides with the endogenous sequence (Fig. 3). Therefore, the pLL3.7-MM is a control vector that is able to produce an shRNA that is recognized by Dicer and can be complexed to the RISC machinery. In our study, the elongation and survival rate of pLL3.7-MM-treated embryos was not different from those treated with the empty pLL3.7 vector. The survival rate for pLL3.7 and pLL3.7-MM treatments, as assessed by total number of embryos recovered at Day 15, was 66.7% to 94.4%, respectively (Table 1). This is well within the normal range (60%–90%) reported for transfer of nonfrozen blastocyst-stage embryos in sheep [43–45]. Although the recovery rate for pLL3.7-MM embryos was numerically greater than that of pLL3.7 embryos, there was a slightly reduced percentage of embryos recovered from this treatment that were elongated (88.2% vs. 93.8%, respectively), and these unelongated conceptuses would likely not have continued on to produce viable offspring (Table 1). Therefore, if embryo survival is calculated as the percentage of elongated embryos recovered on Day 15, the rates for pLL3.7-treated and pLL3.7-MM-treated embryos are more similar, at 62.5% and 83.3%, respectively. These survival rates also agree with previous studies using lentiviral vector transduction of embryos at various stages in mice, rats, pigs, and cows that have all resulted in a high rate of embryo viability (>70%) [14]. High rates of transgene integration (70%–100%) and expression (64%–100%) using lentiviral-based vectors have been reported in pigs and cows [46, 47]. These studies were done by injection into the perivitelline space of the zygote or unfertilized oocyte. Therefore, only one cell was available for transduction. In our study, we incubated blastocyst-stage embryos with the lentivirus, exposing the entire population of trophoblast cells to the lentivirus. Blastocyst-stage ovine embryos contain 55–140 trophoblast cells [48, 49]. Therefore, even based on a low rate of integration and expression, this likely produces knockdown of PRR15 mRNA efficiently. Indeed, all conceptuses in this study observed by microscopy or by PCR analysis showed that EGFP expression had occurred (Fig. 4). Although we were unable to differentiate trophoblast vs. inner cell mass infection in the sections used for EGFP fluorescence, it is very likely that infection was limited to trophoblasts. Previous studies in mouse embryos reported that lentiviral infection is limited to the trophoblast layer, even after 24 h of culture with lentivirus [18, 19]. These studies showed transgene expression continued to be limited to trophoblast derivates to Embryonic Day 19.5 [18, 19]. Furthermore, the lentiviral vector used in our study was replication deficient, and it would therefore only be able to infect the outermost layer of cells, and after the initial infection it would not produce more viral particles capable of entering the blastoceole cavity and infecting other cells [21]. The number of actual integration sites for the lentiviral vector expression constructs in our experiment is unknown; however, it has been reported that only one copy of an shRNA transgene is sufficient to provide body-wide gene silencing in a mouse [50].
Two recent studies in mice demonstrate that lentiviral infection of blastocyst-stage embryos is an effective approach to study trophoblast gene function [18, 19]. We used a similar approach in ovine embryos to successfully degrade PRR15 mRNA and showed that PRR15 is required for successful ovine conceptus development. In summary, PRR15 mRNA expression coincides with the process of trophoblast elongation, and targeted degradation of PRR15 mRNA results in conceptus demise or retarded development.
Footnotes
Supported by National Research Initiative Competitive Grant 2005-35203-15885 from the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service; by National Institutes of Health grant R01HD043089; and by College of Veterinary Medicine (Colorado State University) and Biomedical Sciences College Research Council grant 1-54421. S.H.P. was supported by National Institutes of Health Reproductive Biology Training Grant T32HD07031.
REFERENCES
- Blomberg L, Hashizume K, Viebahn C.Blastocyst elongation, trophoblastic differentiation, and embryonic pattern formation. Reproduction 2008; 135: 181–195. [DOI] [PubMed] [Google Scholar]
- Betteridge KJ, Flechon JE.The anatomy and physiology of pre-attachment bovine embryos. Theriogenology 1988; 29: 155–187. [Google Scholar]
- Vejlsted M, Du Y, Vajta G, Maddox-Hyttel P.Post-hatching development of the porcine and bovine embryo-defining criteria for expected development in vivo and in vitro. Theriogenology 2006; 65: 153–165. [DOI] [PubMed] [Google Scholar]
- Jonker FH.Fetal death: comparative aspects in large domestic animals. Anim Reprod Sci 2004; 82–83: 415–430. [DOI] [PubMed] [Google Scholar]
- Sreenan JM, Diskin MG.The extent and timing of embryonic mortality in the cow. Sreenan JM, Diskin MG.Embryonic Mortality in Farm Animals Boston:Martinus Nijhoff;1986: 1–9. [Google Scholar]
- Binelli M, Thatcher WW, Mattos R, Baruselli PS.Antiluteolytic strategies to improve fertility in cattle. Theriogenology 2001; 56: 1451–1463. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Johnson GA, Bazer FW, Burghardt RC.Implantation mechanimsms: insights from the sheep. Reproduction 2004; 128: 657–668. [DOI] [PubMed] [Google Scholar]
- Demmers KJ, Derecka K, Flint A.Trophoblast interferon and pregnancy. Reproduction 2001; 121: 41–49. [DOI] [PubMed] [Google Scholar]
- Regnault TRH, Galan HL, Parker TA, Anthony RV.Placental development in normal and compromised pregnancies—a review. Placenta 2002; 16(suppl A):S119–S129. [DOI] [PubMed] [Google Scholar]
- Krause R, Hemberger M, Himmelbauer H, Kalscheuer V, Fundele RH.Identification and characterization of G90, a novel mouse RNA that lacks an extensive open reading frame. Gene 1999; 232: 35–42. [DOI] [PubMed] [Google Scholar]
- Meunier D, Peters T, Luttges A, Curfs J, Fundele R.Preferential expression of the G90 gene in post-mitotic cells during mouse embryonic development. Anat Embryol 2003; 207: 109–117. [DOI] [PubMed] [Google Scholar]
- Glover MD, Seidel GE., JrIncreased messenger RNA for allograft inflammatory factor-1, LERK-5, and a novel gene in 17.5 day relative to 15.5 day bovine embryos. Biol Reprod 2003; 69: 1002–1012. [DOI] [PubMed] [Google Scholar]
- Wright CD. Regulation of placental development. Fort Collins, CO:: Colorado State University;; 2005. Ph.D. Dissertation. [Google Scholar]
- Park, F. Lentiviral vectors: are they the future of animal transgenesis? Physiol Genomics 2007; 31: 159–173. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC.Potent and specific genetic interference by double-standed RNA in Caenorhabditis elegans. Nature 1998; 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Hannon GJ.RNA interference. Nature 2002; 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS.Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev 2002; 16: 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Ueshin Y, Isotani A, Saito-Fujita T, Nakashima H, Kimura K, Mizoguchi A, Oh-hora M, Mori Y, Ogata M, Oshima R, Okabe M, et al. Complementation of placental defects and embryonic lethality by trophoblast-specific lentiviral gene transfer. Nat Biotech 2007; 25: 233–237. [DOI] [PubMed] [Google Scholar]
- Georgiades P, Cox B, Gertsenstein M, Chawengsaksophak K, Rossant J.Trophoblast specific gene manipulation using lentivirus-based vectors. Biotechniques 2007; 42: 317–324. [DOI] [PubMed] [Google Scholar]
- Michael DD, Alvarez IM, Ocón OM, Powell AM, Talbot NC, Johnson SE, Ealy AD.Fibroblast growth factor-2 is expressed by the bovine uterus and stimulates interferon-τ production in bovine trophectoderm. Endocrinology 2006; 147: 3571–3579. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, Gertler FB, Scott ML, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 2003; 33: 401–406. [DOI] [PubMed] [Google Scholar]
- De La Torre-Sanchez JF, Gardner DK, Preis K, Seidel GE., JrMetabolic regulation of in-vitro-produced bovine embryos. I. Effects of metabolic regulators at different glucose concentrations with embryos produced by semen from different bulls. Reprod Fertil Dev 2006; 18: 585–596. [DOI] [PubMed] [Google Scholar]
- Elsden RP, Seidel GE., Jr . Procedures for Recovery, Bisection, Freezing, and Transfer of Bovine Embryos, 6th ed. Fort Collins, CO:: Colorado State University;; 1995. [Google Scholar]
- Rowson LE, Moor RM.Development of the sheep conceptus during the first fourteen days. J Anat 1966; 100: 777–785. [PMC free article] [PubMed] [Google Scholar]
- Flechon JE, Flechon B, Degrouard J, Guillomot M.Cellular features of the extra-embryonic endoderm during elongation in the ovine conceptus. Genesis 2007; 45: 709–715. [DOI] [PubMed] [Google Scholar]
- Nephew KP, McClure KE, Pope WF.Embryonic migration relative to maternal recognition of pregnancy in sheep. J Anim Sci 1989; 67: 999–1005. [DOI] [PubMed] [Google Scholar]
- Guillomot M.Cellular interactions during implantation in domestic ruminants. J Reprod Fertil 1995; 49(suppl):39–51. [PubMed] [Google Scholar]
- Cammas L, Reinaud P, Dubois O, Bordas N, Germain G, Charpigny G.Identification of differentially regulated genes during elongation and early implantation in the ovine trophoblast using complementary DNA array screening. Biol Reprod 2005; 72: 960–967. [DOI] [PubMed] [Google Scholar]
- Carnegie JA, McCully ME, Robertson HA.The early development of the sheep trophoblast and the involvement of cell death. Am J Anat 1985; 174: 471–488. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Burghardt RC, Johnson GA, Bazer FW.Conceptus signals for establishment and maintenance of pregnancy. Anim Reprod Sci 2004; 82–83: 537–550. [DOI] [PubMed] [Google Scholar]
- Hansen TR, Imakawa K, Polites HG, Marotti KR, Anthony RV, Roberts RM.Interferon of embryonic origin is expressed transiently during early pregnancy in the ewe. J Biol Chem 1988; 263: 12801–12804. [PubMed] [Google Scholar]
- Leaman DW, Cross JC, Roberts RM.Multiple regulatory elements are required to direct trophoblast interferon gene expression in choriocarcinoma cells and trophectoderm. Mol Endocrinol 1994; 8: 456–468. [DOI] [PubMed] [Google Scholar]
- McGeady TA, Quinn PJ, FitzPatrick ES, Ryan MT.Veterinary Embryology Oxford:Blackwell;2006: 17–104. [Google Scholar]
- Geisert RD, Brookbank JW, Roberts RM, Bazer FW.Establishment of pregnancy in the pig II. Cellular remodeling of the porcine blastocyst during elongation on Day 12 of pregnancy. Biol Reprod 1982; 27: 941–955. [DOI] [PubMed] [Google Scholar]
- Rizos D, Clemente M, Bermejo-Alvarez P, de La Fuenta J, Lonergan P, Gutierrez-Adan A.Consequences of in vitro culture conditions on embryo development and quality. Reprod Domest Anim 2008; 43(suppl 4):44–50. [DOI] [PubMed] [Google Scholar]
- Farin CE, Imakawa K, Roberts RM.In situ localization of mRNA for the interferon, ovine trophoblast protein-1, during early embryonic development of the sheep. Mol Endocrinol 1989; 3: 1099–1107. [DOI] [PubMed] [Google Scholar]
- Guillomot M, Michel C, Gaye P, Charlier N, Trojan J, Martal J.Cellular localization of an embryonic interferon, ovine trophoblastin and its mRNA in sheep embryos during early pregnancy. Biol Cell 1990; 68: 205–211. [DOI] [PubMed] [Google Scholar]
- Ross JW, Ashworth MD, Hurst AG, Malayer JR, Geisert RD.Analysis and characterization of differential gene expression during rapid trophoblastic elongation in the pig using suppression subtractive hybridization. Reprod Biol Endocrinol 2003; 1: 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushizawa K, Herath CB, Kaneyama K, Shiojima S, Hirasawa A, Takahashi T, Imai K, Ochiai K, Tokunaga T, Tsunoda Y, Tsujimoto G, Hashizume K.cDNA microarray analysis of bovine embryo gene expression profiles during the pre-implantation period. Reprod Biol Endocrinol 2004; 2: 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg LA, Long EL, Sonstegard TS, Van Tassell CP, Dobrinsky JR, Zuelke KA.Serial analysis of gene expression during elongation of the peri-implantation porcine trophectoderm (conceptus). Physiol Genomics 2005; 20: 188–194. [DOI] [PubMed] [Google Scholar]
- Kliem A, Tetens F, Klonisch T, Grealy M, Fisher B.Epidermal growth factor receptor and ligands in elongating bovine blastocysts. Mol Reprod Dev 1998; 51: 402–412. [DOI] [PubMed] [Google Scholar]
- Dore JJE, Wilkinson JR, Godkin JD.Early gestational expression of transforming growth factor beta isoforms by the ovine placenta. Biol Reprod 1995; 53: 143–152. [DOI] [PubMed] [Google Scholar]
- Cognie Y.State of the art in sheep-goat embryo transfer. Theriogenology 1999; 51: 105–116. [DOI] [PubMed] [Google Scholar]
- Baril G, Traldi AL, Cognié Y, Leboeuf B, Beckers JF, Mermillod P.Successful direct transfer of vitrified sheep embryos. Theriogenology 2001; 56: 299–305. [DOI] [PubMed] [Google Scholar]
- Papadopoulos S, Rizos D, Duffy P, Wade M, Quinn K, Boland MP, Lonergan P.Embryo survival and recipient pregnancy rates after transfer of fresh or vitrified, in vivo or in vitro produced ovine blastocysts. Anim Reprod Sci 2002; 74: 35–44. [DOI] [PubMed] [Google Scholar]
- Hofmann A, Kessler B, Ewerling S, Weppert M, Vogg B, Ludwig H, Stojkovic M, Boelhauve M, Brem G, Wolf E, Pfeifer A.Efficient transgenesis in farm animals by lentiviral vectors. EMBO Rep 2003; 4: 1054–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A, Zakhartchenko V, Weppert M, Sebald H, Wenigerkind H, Brem G, Wolf E, Pfeifer A.Generation of transgenic cattle by lentiviral gene transfer into oocytes. Biol Reprod 2004; 71: 405–409. [DOI] [PubMed] [Google Scholar]
- Wintenberger-Torres S, Flechon JE.Ultrastructural evolution of the trophoblast cells of the pre-implantation sheep blastocyst from day 8 to day 18. J Anat 1974; 118: 143–153. [PMC free article] [PubMed] [Google Scholar]
- Hardy K, Handyside AH, Winston RML.The human blastocyst: cell number, death and allocation during late preimplantation development in vitro. Development 1989; 107: 597–604. [DOI] [PubMed] [Google Scholar]
- Seibler J, Küter-Luks B, Kern H, Streu S, Plum L, Mauer J, Kühn R, Brüning JC, Schwenk F.Single copy shRNA configuration for ubiquitous gene knockdown in mice. Nucleic Acids Res 2005; 33: e67 [DOI] [PMC free article] [PubMed] [Google Scholar]