Abstract
Study Objectives:
Genetic manipulation of cAMP-dependent protein kinase A (PKA) in Drosophila has implicated an important role for PKA in sleep/wake state regulation. Here, we characterize the role of this signaling pathway in the regulation of sleep using electroencephalographic (EEG) and electromyographic (EMG) recordings in R(AB) transgenic mice that express a dominant negative form of the regulatory subunit of PKA in neurons within cortex and hippocampus. Previous studies have revealed that these mutant mice have reduced PKA activity that results in the impairment of hippocampus-dependent long-term memory and long-lasting forms of hippocampal synaptic plasticity.
Design:
PKA assays, in situ hybridization, immunoblots, and sleep studies were performed in R(AB) transgenic mice and wild-type control mice.
Measurements and Results:
We have found that R(AB) transgenic mice have reduced PKA activity within cortex and reduced Ser845 phosphorylation of the glutamate receptor subunit GluR1. R(AB) transgenic mice exhibit non-rapid eye movement (NREM) sleep fragmentation and increased amounts of rapid eye movement (REM) sleep relative to wild-type mice. Further, R(AB) transgenic mice have more delta power but less sigma power during NREM sleep relative to wild-type mice. After sleep deprivation, the amounts of NREM and REM sleep were comparable between wild-type and R(AB) transgenic mice. However, the homeostatic rebound of sigma power in R(AB) transgenic mice was reduced.
Conclusions:
Alterations in cortical synaptic receptors, impairments in sleep continuity, and alterations in sleep oscillations in R(AB) mice imply that PKA is involved not only in synaptic plasticity and memory storage but also in the regulation of sleep/wake states.
Citation:
Hellman K; Hernandez P; Park A; Abel T. Genetic evidence for a role for protein kinase a in the maintenance of sleep and thalamocortical oscillations. SLEEP 2010;33(1):19-28.
Keywords: Protein kinase A, spindle oscillations, delta, REM sleep, NREM sleep, sleep maintenance, mouse, sleep fragmentation
IDENTIFYING THE MOLECULAR MECHANISMS RESPONSIBLE FOR THE INDUCTION AND MAINTENANCE OF SLEEP IS AN IMPORTANT CLINICAL OBJECTIVE. TO this end, it is important to understand the molecular mechanisms underlying the oscillations in neural activity that accompany specific sleep states. Much work has focused on defining the roles of specific neurotransmitter systems in sleep/wake regulation1–3; however, the role of intracellular signaling pathways in sleep/wake regulation has been less explored. Pharmacologic studies have implicated the cAMP/protein kinase A (PKA) pathway in the regulation of wakefulness and rapid eye movement (REM) sleep. For example, rats have relatively higher levels of cAMP in the cortex, hippocampus, hypothalamus, and the pons-medulla during wakefulness but lower levels during non-rapid eye movement (NREM) sleep and REM sleep.4 Further, treatments that increase cAMP activity (e.g., administration of the phosphodiesterase inhibitor rolipram in rats5 and mutations in Drosophila that increase cAMP6) promote wakefulness. Conversely, decreased cAMP activity has been shown to increase rest in Drosophila,6,7 and a mutation in CREB, a target of PKA, increases sleep in mice.8 In human sleep, the relevance of this pathway was established by a genome-wide examination of sleep traits showing that a single nucleotide polymorphism in the intron region of PDE4D, a cAMP-specific phosphodiesterase, had the most significant association with sleepinesss.9
Genetic approaches are a valuable tool for identifying the role of intracellular signaling pathways in sleep/wake regulation because of their cell type and regional specificity. Although transgenic mice have been used to investigate specific signaling pathways in learning and memory, their use in the study of sleep/wake state regulation has been limited. R(AB) transgenic mice express a dominant negative form of the RIα regulatory subunit of PKA in neurons within the hippocampus and other forebrain regions10 and exhibit impairments in hippocampus-dependent long-term memory, synaptic plasticity, and place cell stability.10–14 Here, we examine electroencephalographic (EEG) and electromyographic (EMG) recordings of transgenic R(AB) and wild-type mice to identify the role of PKA in sleep/wake regulation and sleep oscillations.
MATERIALS AND METHODS
Animals
R(AB) transgenic mice10 were backcrossed in the hemizygous state to C57BL/6J mice for 11 to 13 generations and were bred in our colony under standard conditions. Wild-type mice used in these studies were littermates of R(AB) transgenic mice. Food and water were provided ad libitum. Mice were maintained on a 12-hour light/12-hour dark cycle with lights on at 0700. Both female and male mice were used in a balanced fashion, and we did not observe an effect of sex in any of our experiments. For genotyping, tail DNA was analyzed by Southern blot analysis using a transgene-specific probe.10 All animal care and experiments were carried out in accordance with the National Institute of Health guidelines and were fully approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Surgery
Adult mice (12-24 weeks of age, n = 10 wild-type, n = 13 R(AB) transgenic) were implanted with EEG and EMG electrodes under isoflurane anesthesia. An electric drill was used to thin the skull near the place of electrode implantation. EEG electrodes were implanted 1 mm bilateral to midline and 1.3 mm posterior to bregma on each side. A reference EEG electrode was placed 1.5 mm posterior to lambda over the cerebellum.15 Electrodes were held in place with glass ionomer resin (Ketac-cem, 3M, Maplewood, MN).16 Electrodes consisted of silver ball electrodes (Φ = 0.33 mm) insulated with Teflon, soldered to gold socket contacts (Plastics One, Roanoke, VA) and pushed into a 6-pin plug (363 plug, Plastics One), which was then attached to a fully rotating commutator (SLC6, Plastics One).17 All recordings were analyzed with the right parietal EEG electrode and referenced to the cerebellum electrode. In our experience, alterations in behavioral states are most evident using this electrode arrangement. Mice were allowed 2 weeks recovery from surgery before recording.
Data Acquisition
EEG signals were filtered at 0.3 to 60 Hz (1/2 amplitude, 6 dB/octave), and EMG signals were filtered at 1 to 100 Hz with 12A5 amplifiers (Astro-Med, West Warwick, RI) and sampled at 256 Hz with 12-bit resolution (similar to Graves et al.8). Data acquisition and visual scoring of EEG/EMG recordings were performed with custom software that is available for Microsoft Windows from the authors.
Following 24 hours of baseline recording, 9 wild-type mice and 12 R(AB) transgenic mice were sleep deprived by gentle handling for 6 consecutive hours starting at 07:00. Sleep deprivation by brief gentle tactile stimulation (stroking head vibrissae and back) of the individual mouse was performed whenever sleep was observed in the EEG/EMG.8,17 Although some brief sleep occurs during this period of deprivation, there was no difference in the amount of sleep obtained between the groups during the sleep-deprivation period: wild-type: 33 ± 5 minutes; R(AB): 27 ± 4 minutes p = 0.41). This technique has been reported to be less stressful than other methods of sleep deprivation.18–21 The effects of sleep deprivation were examined in 2-hour windows and compared with baseline data from the same time points.
Data Analysis
Behavioral states were scored in 4-second epochs during the 24-hour baseline period, the 6-hour sleep-deprivation period, and the 18-hour recovery period. Observers blind to experimental group and condition visually scored EEG and EMG recordings (wakefulness, NREM sleep, REM sleep) according to previously established criteria,8,22 which were similar to human criteria.23 During NREM sleep, K-complexes, spindles, and delta activity were present along with reduced EMG tone. During REM sleep, there was a further reduction in EMG tone, and theta oscillations dominated the EEG signals. Wakefulness was characterized by bursts of high-frequency EEG activity (30+ Hz) and EMG activity.
To measure changes in sleep microstructure, we tabulated the amount of time spent continuously in each sleep state over the entire baseline period and performed a survival analysis, a robust statistical technique for examining sleep fragmentation.24 Survival analysis was performed using Statview (version 5.0.1, SAS Institute, 1999, Cary, NC).
To analyze spectral properties during various sleep/wake states, fast Fourier transform (FFT) analysis was performed using custom software over all scored data except 1 wild-type mouse and 1 R(AB) transgenic mouse because they displayed evidence of electrical artifact contamination. The FFT was performed with 0.25-Hz resolution (0.25-128 Hz) using a moving window throughout the 24-hour period. Signals were calibrated with 100-μV pulses. In addition to calculating spectral properties from absolute spectra, absolute spectra were normalized to compensate for differences in signal strength and spectral leakage (a property of FFTs that causes bands to spread and blur25 ). A frequency-independent normalization procedure was performed (similar to the procedure used by Borbély et al.26 that characterized increased delta power after sleep deprivation) by calculating the ratio of the power of an individual frequency band within a specific state over that band's contribution to power over all states. Importantly, this technique differs from spectral density normalization, which is confounded when an increase in a single frequency range results in a compensatory decrease in another frequency. The absolute (not normalized) and frequency-independent normalized spectra were calculated and plotted during wakefulness, NREM sleep, and REM for each genotype. Analysis of variance was used to identify frequency ranges that were different between genotypes.
To detect spindles in the EEG recordings during NREM sleep, we modified an existing algorithm used for detecting sharp waves in the hippocampus.27 The resultant algorithm is similar to that used to detect spindles in rats.28 This algorithm examined a continuous FFT of the EEG signal and quantified EEG events that had a 50% minimum increase in 10- to 14-Hz activity lasting for at least 500 milliseconds. This algorithm was based on the definition of human spindles, which are transient 10- to 14-Hz events lasting at least 0.5 seconds.23 The presence of spindles was calculated by this method during the entire 24-hour baseline period during NREM sleep only followed by visual verification. Notably, this algorithm was successful at correctly identifying more than 90% of visually identified spindles in all mice except for 2 wild-type mice and 3 R(AB) transgenic mice that contained electrocardiographic and/or respiratory artifacts in their recordings. In subsequent analyses, spindle amplitude was normalized to compensate for differences in impedance by dividing the absolute power by the mean 4- to 30-Hz power for each individual mouse. It was necessary to exclude the 1- to 4-Hz frequency range because R(AB) mice had increased power in the 1- to 4-Hz range that would mask sensitivity to spindle detection in the 10- to 14-Hz frequency band. The number of spindles, mean duration of spindles, mean maximum spindle amplitude, and mean integrated spindle amplitude (time × amplitude) were determined for each mouse. Differences were compared between genotypes and examined with analysis of variance.
In Situ Hybridization
Two wild-type and 4 R(AB) transgenic mouse brains were dissected and rapidly frozen in Tissue-Tek embedding medium (EMS, Hatfield, PA). Twenty-micrometer sections were fixed and hybridized, as has been previously described,10 to an [γ-35S] dATP-labeled transgene-specific oligonucleotide 5'-CAGGATCCGCTTGGGCTGCAGTTGGACCT-3', that hybridizes to sequences present in the 5' untranslated leader within the transgenic transcript. Slides were exposed for 25 days to Kodak Biomax MR autoradiographic film.
PKA Assay
Cortex was dissected into ice-cold artificial cerebrospinal fluid10 and homogenized in 200 μL of extraction buffer29 (n = 8 wild-type, n = 8 R(AB) transgenic mice). The extract was diluted to a protein concentration of 2 mg/mL30 and kept at 4°C until use. PKA activity was then assayed in duplicate as previously described.31 Upon the addition of cAMP (5 mM) and/or PKI (40 mg/mL; Peninsula Labs), PKA activity (pmol [γ-32P] ATP·min-1·mg-1 protein) was normalized to background activity measured without the addition of substrate.
GluR1 Phosphorylation Assay
To examine downstream effects of reduced PKA activity, we assayed cortical extracts by Western blotting for total and phospho-GluR1.32 Wild-type and R(AB) (4- to 6-month-old) mice were removed from their homecage and killed by cervical dislocation between ZT hours 4 and 6 (n = 5/group). Cortical tissue was rapidly dissected on ice and frozen promptly at −80°C. Tissue was then homogenized in RIPA buffer, with protease and phosphatase inhibitors added fresh. Homogenates were spun at 13,000 rpm at 4°C for 20 minutes. Supernatants were then processed by Western blot analysis for pSer845 and total GluR1. Total protein per microliter of supernatant was determined by Bradford assay, and mouse beta tubulin was used as a loading control. GluR1 and beta tubulin antibodies were obtained from Millipore (1:2000) and Sigma (1:50,000), respectively.
Statistical Analysis
Analysis of variance, correlations, and posthoc tests were performed in Statview.
RESULTS
PKA Activity Is Reduced in the Cortex of R(AB) Transgenic Mice
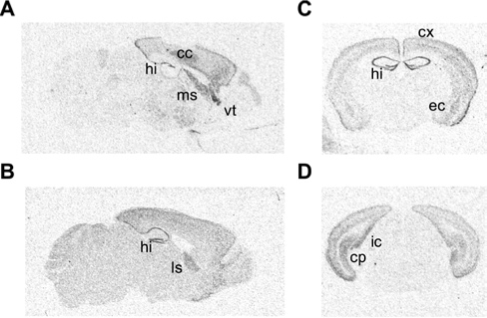
To begin our examination of the role of PKA in sleep/wake regulation, we used in situ hybridization to determine the location of the anatomic expression pattern of the R(AB) transgene (Figure 1). Consistent with the expression pattern of other CaMKIIα driven transgenes, R(AB) was expressed in the hippocampus, cortex, and amygdala but was absent in the pons, medulla, and hypothalamus—regions known to regulate sleep.33, 10, 34
Figure 1.
In situ hybridization showing expression of the R(AB) transgene. A, An autoradiograph of a sagittal section near the midline. Transgenic mRNA expression is observed in the hippocampus (hi), cortex (cx), and lateral septum (ls). B, A sagittal section approximately 0.6 mm lateral from the midline. C, A coronal section approximately 1.2 mm behind the bregma, showing expression in the basolateral amygdala (bla), hippocampus, cortex (cx), and lateral septum (ls) but not in the thalamus or hypothalamus. D, A coronal section 3.8 mm posterior to bregma, revealing staining in the cortex, presubiculim (prs), and caudate putamen (cp) but not in the pontine region.
Previous studies have demonstrated reduced PKA activity in R(AB) transgenic mice; however, these assays were performed using hippocampal extracts.10 Therefore, we conducted an additional PKA activity assay to determine whether the R(AB) transgene also decreased PKA activity in the cortex. As expected, we found that PKA activity in cortex was significantly reduced after cAMP stimulation (basal: wild-type: 150 ± 42 pmol·min-1·mg-1, R(AB) transgenic mice: 100 ± 29 pmol·min-1·mg-1; F1,14 = 1.06, p = 0.34; cAMP stimulated: wild-type: 3510 ± 84 pmol·min-1·mg-1, R(AB) transgenic mice: 3240 ± 69 pmol·min-1·mg-1; F1,14 = 2.8, p < 0.05). Thus, R(AB) transgenic mice also exhibit reduced PKA activity in the cortex, consistent with the pattern of expression of the transgene.
PKA-Dependent Phosphorylation of GluR1 Is Reduced in the Cortex of R(AB) Transgenic Mice
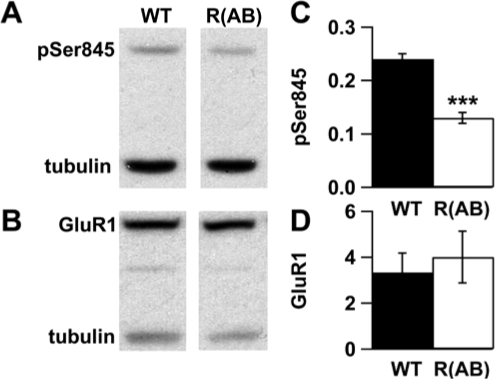
Phosphorylation of the GluR1 subunit of AMPA-type glutamate receptors at Ser845 is known to be a PKA-dependent process.32 Thus, to verify that reduced PKA activity also affects the functionality of downstream targets of PKA, we examined levels of phosphorylated GluR1 within the cortex (Figure 2). Phosphorylation of the GluR1 subunit of AMPA-type glutamate receptors was reduced by 50% in R(AB) mice compared with wild-type controls (p < 0.001). Significant differences were not observed in the levels of total Glur1 (p = 0.65) or the beta tubulin loading control (p = 0.75). Thus, reduced PKA activity within cortex of R(AB) transgenic mice also leads to a decrease in the PKA-dependent phosphorylation of a synaptic plasticity-related molecule without altering its expression.
Figure 2.
Ser845 phosphorylation of GluR1 is reduced in cortex of R(AB) mice. Representative immunoblots using cortical extracts from a wild-type (WT) and R(AB) transgenic mouse probed for A, phospho-GluR1 (pSer845) and B, total GluR1. C and D, Quantification of immunoblots indicated a significant decrease in pSer845 levels in R(AB) transgenic mice compared to WT mice, whereas levels of total GluR1 were not different. Data are expressed as the mean ratio of pSer845 or GluR1/tubulin ± SEM. ***p < 0.001.
R(AB) Transgenic Mice Have More REM Sleep
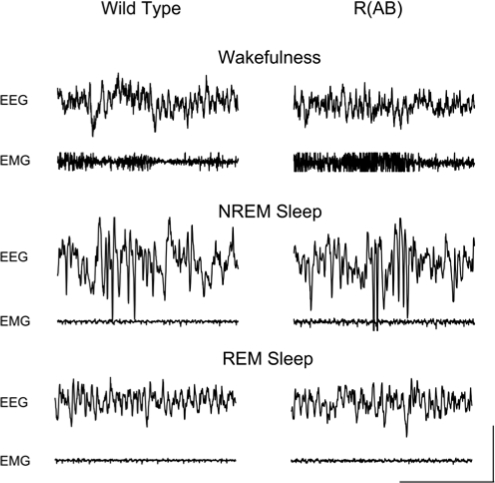
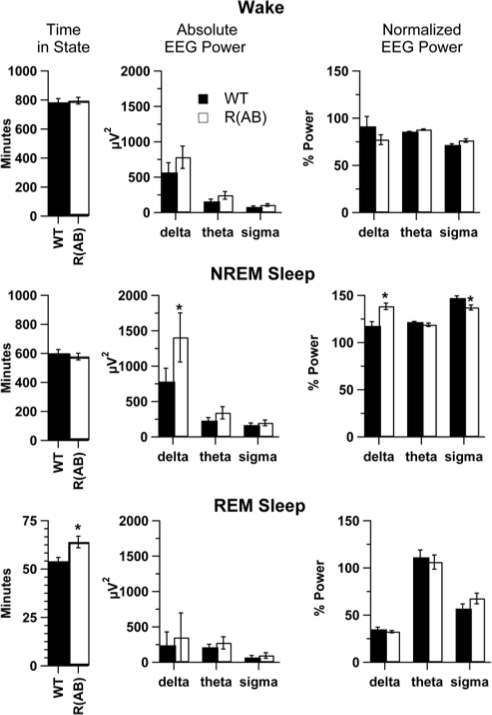
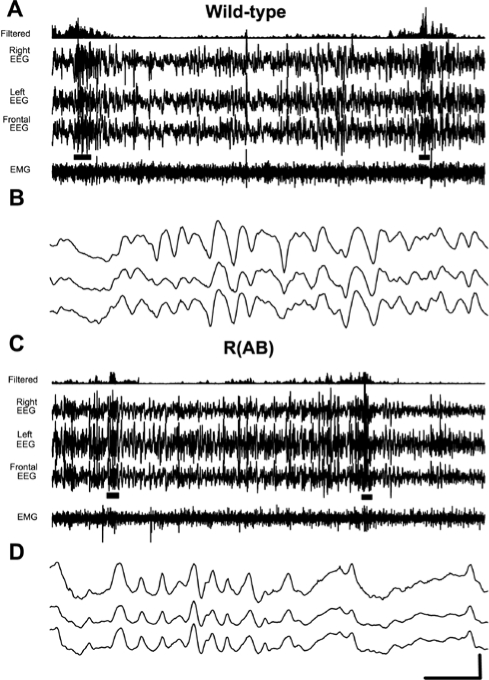
We examined sleep in wild-type (n = 10) and R(AB) transgenic (n = 13) mice with EEG and EMG recordings. There were no obvious differences in the signals upon visual inspection (Figure 3). Wild-type mice had amounts of NREM sleep and REM sleep (Figure 4, left panels) comparable to that of previously published studies of sleep in C57BL/6J mice,17,35 the genetic background on which the R(AB) transgene is maintained. However, R(AB) mice had 19% more REM sleep than wild-type mice (wild-type mice: 54 ± 2 minutes, R(AB) transgenic mice: 64 ± 3 minutes; F1,21 = 6.75, p < 0.05), which translates into a significant increase in the proportion of REM to total sleep time (wild-type mice: 8.3% ± 0.5%, R(AB) transgenic mice: 10% ± 0.5%; F1,21 = 6.0, p < 0.05). There was no significant difference in the total time spent in NREM sleep (wild-type mice: 601 ± 27 minutes, R(AB) transgenic mice: 578 ± 23 minutes; F1,21 = 0.5, p = 0.51) or in total time spent in wakefulness (wild-type mice: 784 ± 26 minutes, R(AB) transgenic mice: 795 ± 23 minutes; F1,21 = 0.1, p = 0.74).
Figure 3.
Sample traces of electroencephalographic (EEG) and electromyographic (EMG) during wakefulness, non-rapid eye movement (NREM), and rapid eye movement (REM) sleep in wild-type and R(AB) transgenic mice. Four-second epochs are shown from a single mouse of each genotype during each of the 3 states. Wakefulness was characterized by bursts of high-frequency EEG activity (30+ Hz) and high-amplitude EMG activity. During NREM sleep, delta activity was present along with reduced EMG tone. During REM sleep, there was a further reduction in EMG tone, and theta oscillations dominated the EEG signals. REM sleep was characterized by predominance of theta activity in the EEG signal. Scale bar shows 2 seconds and 100 μV.
Figure 4.
R(AB) transgenic mice have more rapid eye movement (REM) sleep, increased delta power, and reduced sigma power. The left panels show the average time spent in wakefulness, non-rapid eye movement (NREM) sleep, and REM sleep during the baseline period. The middle panels indicate the total power within the delta (1-4 Hz), theta (6- to 8-Hz), and sigma (10- to 14-Hz) power, as determined by a fast Fourier transform of absolute electroencephalographic (EEG) power over a 24-hour period within each state. R(AB) mice have significantly more delta power than wild-type mice during NREM sleep The right column shows normalized EEG power (see Methods for details). R(AB) mice have significantly more delta power and less sigma power than wild-type mice during NREM sleep. *p < 0.05. Data are expressed as the mean ± SEM. EMG refers to electromyography.
R(AB) Transgenic Mice Have Increased Delta and Decreased Sigma Activity During NREM Sleep
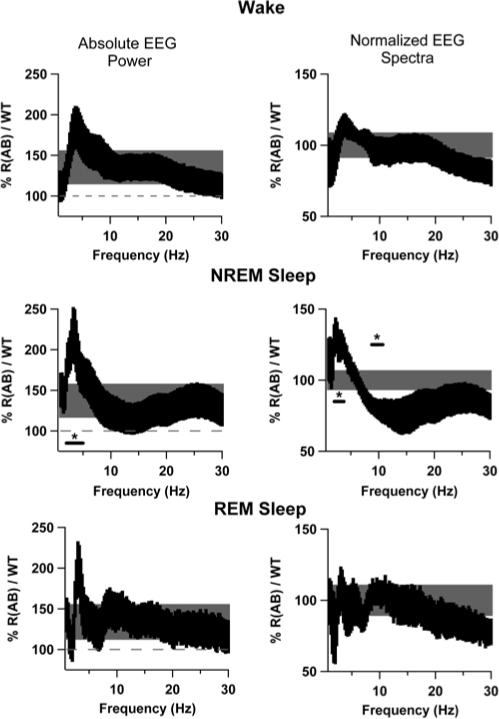
We performed a spectral analysis on recordings from each behavioral state using a FFT over the 24-hour baseline period to identify alterations in sleep oscillations caused by genetic inhibition of PKA (Figures 4 and 5). To examine which frequency bands were most affected, we examined the difference in absolute EEG spectra (Figure 4, middle panels). Overall, R(AB) transgenic mice had more delta, theta, and sigma power than wild-type mice within their EEG signals (Figure 4), but only delta power during NREM sleep was significantly higher compared with wild-type mice in the analysis of absolute EEG spectra (p < 0.05). To compensate for differences produced by alterations in signal strength and spectral leakage,25 frequency-independent normalization was applied (Figures 4 and 5, right panels). EEG spectra were normalized within individual bands to determine the relative contribution of each state toward the power associated with each spectral region. Normalization also demonstrated that R(AB) mice have increased delta (1- to 4-Hz) power during NREM sleep, compared with wild-type mice (p < 0.05). Additionally, normalization unmasked a decrease in sigma (10- to 14-Hz) power during NREM sleep (p < 0.05), likely obscured in the absolute EEG spectra by spectral leakage.25 Examination of the entire spectrum revealed that alterations specifically occurred between 2- to 3.75-Hz and 8.75- to 10.75-Hz in R(AB) mice during NREM sleep (Figure 5). Significant differences in EEG spectra were only observed during NREM sleep and only in the delta and sigma frequency bands.
Figure 5.
Spectral plots indicating increased delta power and reduced sigma power in R(AB) transgenic mice. The left panels are plots within each state of the ratio of R(AB) to wild-type (WT) absolute electroencephalographic (EEG) power expressed as a percentage. The black shaded area represents the ratio of R(AB) and WT mice with 0.25-Hz frequency resolution, thereby indicating alterations within specific frequency regions. The height of the black shaded area includes ± 1 standard error. Because R(AB) mice have more total power across the entire spectrum, we have included a reference band shown in grey, indicating the average total power change across the entire spectrum to help identify specific regions that are significantly increased. Similarly, the height of the grey shaded area includes ± 1 standard error of total mean power change. The underlined area designates a significant increase in 2- to 5-Hz power in R(AB) mice (p < 0.05). To compensate for difference in signal strength, EEG signals were normalized as a function of the total power within each frequency band and are shown in the right column of panels. R(AB) mice had a relative increase in 2.5- to 3.75-Hz power (p < 0.05) and a relative decrease in 8.75- to 10.75-Hz power (p < 0.05).
R(AB) Transgenic Mice Exhibit Fragmented Sleep
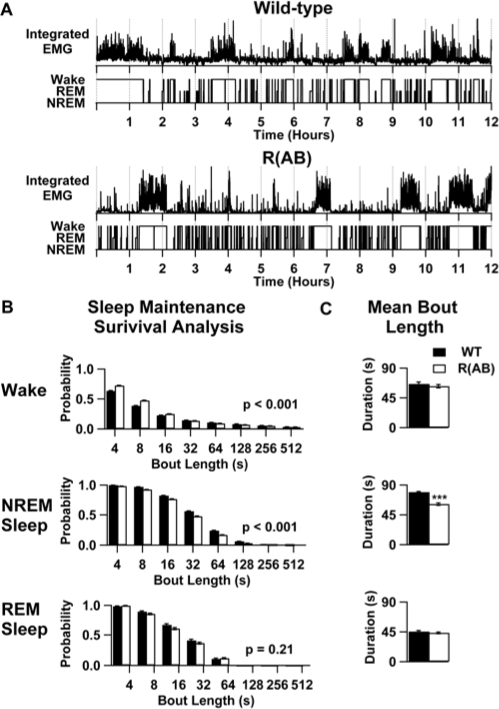
To identify alterations in sleep microstructure, we examined the distribution of sleep/wake bouts in wild-type and R(AB) transgenic mice during the 24-hour baseline period. Examination of individual hypnograms revealed that R(AB) mice transition in and out of NREM sleep more frequently (Figure 6). Survival analysis revealed that R(AB) mice had an increased probability of having shorter sleep bouts, punctuated by brief (4- to 16-s) arousals (Figure 6). Indeed, R(AB) mice had 30% shorter NREM sleep-bout lengths (p < 0.001) over the baseline period, relative to wild-type mice. In contrast, R(AB) mice had 12% more REM bouts than wild-type mice (p = 0.16). Although not statistically significant, this increase can be extrapolated to produce a phenotype of 7.5 minutes of increased REM sleep. Because REM sleep-bout length in R(AB) mice was only increased by 4% (p = 0.63), it is likely that the increased time R(AB) mice spend in REM (described above) is due to an increase in the number of bouts rather than bout duration. Indeed, we found a correlation between the amount of time spent in REM sleep and the number of REM entries in both wild-type and R(AB) transgenic mice during the 24-hour baseline period (r2 = 0.71, p < 0.0001), further suggesting that the phenotype of increased REM sleep may be the result of increased entry into REM sleep.
Figure 6.
R(AB) transgenic mice have fragmented non-rapid eye movement (NREM) sleep. A: Examples of sleep fragmentation during the light period. Integrated electromyography (EMG) power for each epoch in a wild-type (WT) mouse and an R(AB) transgenic mouse during the light period. Beneath the integrated EMG records are the state hypnograms The increased NREM sleep-wake and wake-NREM sleep transitions in R(AB) transgenic mice can be observed by the increased frequency of EMG events and transitions in the hypnogram in R(AB) transgenic mice. B: The cumulative probability of maintenance of each state was determined by survival analysis over the 24-h baseline period. These graphs depict the probability of maintaining a specific state in each genotype. P values are shown for the significance in difference for the survival distribution between genotypes. C: panels show mean bout length within each state. Data are expressed as the mean ± SEM. ***p < 0.001. REM refers to rapid eye movement sleep.
R(AB) Transgenic Mice Show Reduced Spindle Amplitude During NREM Sleep
To elucidate the cause of the reduction in sigma power in R(AB) mice, we analyzed spindle activity in both groups. Alterations in the sigma frequency range are thought to be caused by alterations in spindles.36 Furthermore, spindle activity in mice can be altered by the administration of cholinergic agonists, resulting in reduced sigma power.37 In light of these findings, we calculated the number of spindles over the baseline period and determined their mean duration, maximum amplitude, and integrated amplitude. Overall, spindles in both groups of mice were similar to those observed in humans in frequency (10- to14-Hz), density (2 per 30 seconds), and amplitude (20 μV) (Figure 7). In some cases, mouse spindles were as long as human spindles (0.5-2.0 seconds) but were typically shorter in duration (0.5 −1.0 seconds in total length or 0.5-1.0 seconds shorter). The maximum amplitude and integrated amplitude of spindles in R(AB) mice were 8% smaller than those of wild-type mice (p < 0.05; F1,14 = 2.6; and p < 0.05; F1,14 = 2.6, respectively). No change was seen in the number (wild-type: 2248 ± 146, R(AB): 2420 ± 195; F1,14 = 0.8, p = 0.42 ) or duration of spindles (wild-type: 587 ± 15 ms, R(AB): 582 ± 15 ms; F1,14 = 1.2, p = 0.24) during the baseline period. These results show that the reduced sigma power in R(AB) mice is likely caused by a reduction in maximum spindle amplitude and integrated amplitude.
Figure 7.
Examples of spindles from wild-type and R(AB) transgenic mice. A and C show 1 minute of non-rapid eye movement (NREM) sleep activity recorded from 3 different electroencephalographic (EEG) leads and 1 electromyographic (EMG) lead in wild-type and R(AB) mice, respectively. EEG activity from the right EEG lead was filtered using an inverse fast Fourier transform (10- to 14-Hz), integrated, and displayed above the right EEG trace. Spindles underlined in black beneath the EEG signals in A and C are enlarged in B and D, respectively. Note that the spindles are preceded by K-complexes in B and D. The horizontal scale bar indicates 7.5 seconds for traces A and C and 0.25 seconds for traces B and D. The vertical scale bar indicates 100 μV for all traces.
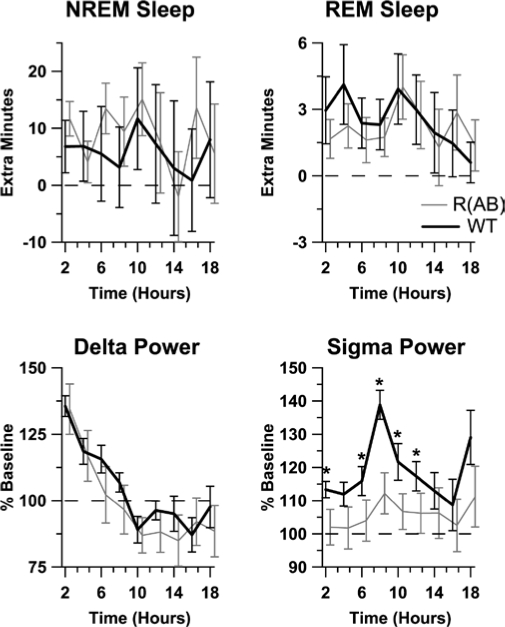
Homeostatic Changes in Sigma Power After Sleep Deprivation Are Reduced in R(AB) Transgenic Mice
To determine the effect of the genetic inhibition of PKA on sleep homeostasis, we characterized sleep rebound in mice subjected to 6 hours of sleep deprivation starting at ZT hour 0. Figure 8 (top panels) show that both wild-type and R(AB) transgenic mice demonstrated significant NREM sleep and REM sleep rebound during the recovery period (p values < 0.05). No significant differences in the time spent in NREM or REM sleep were found between the 2 groups (p values > 0.05).
Figure 8.
R(AB) transgenic mice have normal homeostatic sleep recovery, but long-term alterations in sigma power were different between genotypes. The increase in non-rapid eye movement (NREM) sleep, rapid eye movement (REM) sleep, delta power, and sigma power relative to baseline after sleep deprivation was tabulated in 2-h bins. Values for R(AB) mice were offset for clarity. Bottom axis indicates hours after sleep deprivation. *p < 0.05. Data are expressed as the mean ± SEM. WT refers to wild-type mice.
Sleep deprivation is known to increase delta and sigma power during NREM sleep, reflecting increased homeostatic drive.38,39 Therefore, we examined alterations in NREM sleep spectra during recovery from 6 hours of sleep deprivation (Figure 8, bottom panels). EEG spectra after sleep deprivation were compared with that measured during the baseline period. Both wild-type and R(AB) mice had increased delta power after sleep deprivation, but wild-types maintained a higher increase in sigma power during NREM sleep, as compared with R(AB) mice (p < 0.05). No effect of genotype was observed on theta power in any state (p values > 0.5) or on delta and sigma power during wakefulness or REM sleep (p values > 0.5). These results suggest that rebound in sigma oscillations after sleep deprivation is reduced in R(AB) transgenic mice.
DISCUSSION
In the present study, we used a genetic approach to identify a potential role for PKA in sleep/wake regulation. Specifically, we examined sleep in R(AB) transgenic mice that express a dominant negative form of the regulatory subunit of PKA within the forebrain, including the cortex, hippocampus, amygdala, and lateral septum. We found that R(AB) transgenic mice have fragmented sleep, reflecting an impairment in sleep continuity. Further, R(AB) transgenic mice have more REM sleep and delta activity during NREM sleep. Conversely, R(AB) mice have less sigma activity during periods of NREM sleep. In response to sleep deprivation, reduced homeostatic increases in EEG spectra were apparent in R(AB) transgenic mice without any significant alteration in the time spent in NREM or REM sleep. This is the first study of the effects of a transgene expressed primarily in the forebrain on sleep/wake regulation. Our results are in agreement with the literature suggesting that the hypothalamus is the main control center for the time spent in sleep or wakefulness1–3 but that local regulatory control of sleep and sleep homeostasis can occur within the cortex.15,40–42 Our results further suggest a role for PKA in REM sleep regulation and in the delta and sigma activity that occur during NREM sleep.
The Role of PKA and CREB in Sleep/Wake Regulation
PKA plays a critical role in the regulation of the transcription factor CREB,43 which itself has been implicated in the maintenance of wakefulness.8 Nevertheless, CREB mice had overall more dramatic alterations in sleep/wake regulation than did the R(AB) transgenic mice examined here, perhaps because the CREB mutant mice lacked CREB from all cells and not from restricted neuronal circuits. Previous studies have demonstrated that CREBαΔ mutant mice have approximately 100 minutes less wakefulness over a 24-hour light/dark cycle.8 Analysis of the sleep microstructure of CREB knockout mice suggests that these mice had difficulty maintaining wakefulness because of reductions in wake bout lengths restricted to the dark period. Interestingly, R(AB) transgenic mice had increased REM sleep, but normal amounts of NREM sleep, when compared with wild-type mice, whereas CREBαΔ knockout mice had no significant increase in the proportion of REM sleep to NREM sleep. These differences may arise due to fact that CREBαΔ knockout mice have globally decreased levels of CREB, whereas expression of the transgene in R(AB) is more restricted. Yet, we still expected to replicate portions of the CREBαΔ phenotype because increased wakefulness produces increased levels of CREB phosphorylation in cortex.44 In fact, in Drosophila, mutations involving PKA have activity phenotypes comparable to CREB mutations.6 Expression of a constitutively active catalytic subunit of PKA outside of the core α/β mushroom body regions increases wakefulness in Drosophila.7 The fact that R(AB) transgenic mice do not phenocopy CREBαΔ mice suggests either that the maintenance of wakefulness by CREB occurs outside of cortex or that proteins other than PKA that act on CREB, such as CaMKIV45 or RSK,46 may be involved. In addition, it is important to note that R(AB) transgenic mice have a different genetic background (C57BL/6J) than the CREBαΔ knockout mice (F1 hybrid of C57BL/6J 129/SvEvTac). Such differences in genetic background might also contribute to disparities in the phenotype of R(AB) transgenic mice and CREBαΔ knockout mice because different mouse strains are known to have different sleep/wake characteristics.17,47
Reductions in PKA Activity in Cortex Can Produce Symptoms of Sleep-Maintenance Insomnia
The sleep fragmentation and changes in EEG spectra in R(AB) are consistent with observations of human sleep-maintenance insomnia. Sleep-maintenance insomnia is a prevalent form of insomnia primarily manifested by sleep fragmentation and non-restorative sleep in the generally healthy population and in diseased populations, particularly those with chronic pain and depression.48–52 Whereas humans have been reported to have 7 ± 2 awakenings per night, sleep-maintenance insomniacs have 19 ± 3 awakenings per night.53 During the light cycle, when mice consolidate their sleep, wild-type mice have 46 ± 6 arousals, whereas R(AB) transgenic mice have 66 ± 7 arousals. In an EEG study, it was found that patients with sleep-maintenance insomnia have increased delta power,53 similar to the observed increase in delta power in R(AB) transgenic mice that we observe here.
Interestingly, a reduction of spindle activity (activity in the human sigma power band 13-to17-Hz) is seen in sleep-maintenance insomnia53 and in diseases that produce sleep fragmentation, including fibromyalgia,54 sleep disordered breathing,55,56 and Creutzfeldt-Jakob disease,57 as well as with aging.58 Spindles also produce long-lasting hypersynchronized inhibition of thalamic and cortical neurons that could effectively shunt arousing sensory inputs.59 In fact, drugs that treat insomnia enhance or increase spindle activity,60–63 suggesting that spindles may serve an important role in sleep maintenance. It will be interesting to examine the effects of hypnotics on R(AB) transgenic mice in future experiments. Further work will also be necessary to validate our spindle-analysis technique, as this represents a novel innovation to better understand alterations in cortical dynamics within R(AB) transgenic mice.
Similar techniques in rats28 and humans64 have tied increased spindle density to mechanisms involved in memory consolidation. Although the role of spindles in memory consolidation remains to be elucidated, R(AB) transgenic mice have impaired memory consolidation, and observations of reduced spindle activity are consistent with this notion.
PKA, Synaptic Plasticity, Memory Storage, and Sleep
Sleep and sleep oscillations are postulated to be important to memory consolidation,65–67 although this belief has been challenged.68–70 Sleep oscillations are hypothesized to mediate the transfer of information acquired during prior wakefulness from the hippocampus to cortex for more permanent storage by repeatedly activating synapses associated with a particular memory representation.71,72 Although it is difficult to test such a hypothesis directly, our results suggest some testable predictions of this hypothesis. Because R(AB) transgenic mice have reduced spindle/sigma activity during sleep, as well as reduced phosphorylation of GluR1 within cortex, we envisage that spindles during sleep could modulate or be modulated by GluR1 phosphorylation. For example, it is known that synaptic plasticity in barrel cortex utilizes a PKA-dependent mechanism of phosphorylation of GluR1,73 and the phosphorylation of GluR1 is modulated by sleep/wake state.74 Furthermore, phosphorylation of GluR1 increases the probability of channel opening32 and receptor incorporation75,76 and thereby increases synaptic currents, which are important for the manifestation of spindle oscillations.77 Work in vitro has supported a role for cAMP, but not PKA, in the modulation of spindle oscillations.78 It is likely that interaction between PKA and sleep oscillation is complex and could involve the participation of other ion channels and transcriptional regulation. Nevertheless, we predict that memory consolidated during sleep oscillations involves GluR1 phosphorylation within cortex, and manipulations involving learning that affect sleep oscillations may be measured, in part, by assessing GluR1 phosphorylation.
Although the notion of sleep oscillations playing a role in memory consolidation requires further experimental evidence, there is evidence that repeated disruption of sleep causes memory impairments. Sleep fragmentation by enforced intermittent treadmill activity in rats is sufficient to produce deficits in the Morris water maze and hippocampal long-term potentiation.79 Even repeated interruption of sleep by noise is sufficient to impair spatial long-term memory in rats.80 Epidemiologic studies investigating the chronic interruption of sleep by noise shows that long-term memory processes are affected.81–84 The most adverse form of sleep fragmentation in humans, obstructive sleep apnea, is also known to impair long-term memory.85–87 Also, patients with developmental disorders and accompanying cognitive impairments display sleep fragmentation.88–90 Therefore, reducing sleep fragmentation by enhancing sleep may be a relevant therapeutic target for the treatment of some developmental disorders. Because PKA activity is reduced in some developmental disorders91 and mutations in the cAMP/PKA pathway are involved in the etiology of others,92 further investigation of the role of the cAMP/PKA pathway in sleep/wake regulation may prove to be efficacious in the treatment of developmental disorders associated with learning and memory impairments and alterations in sleep.
DISCLOSURE STATEMENT
This was not an industry supported study. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
The authors thank Alan Young for performing the PKA assays. The authors thank Jacqueline Cater, Laurel Graves, Larry Sanford, Matthew Scharf, Allan Pack, Steven Thomas and Sigrid Veasey for valuable advice. This work was supported by grants from NIA (AG-18199), NIMH (MH-60244), NHLBI (HL-60287), the Whitehall Foundation, the John Merck Foundation and the David and Lucile Packard Foundation (T.A.); NIMH (MH-64329) (K.H.); NSF (0706858) (P.H).
REFERENCES
- 1.Hobson JA, Pace-Schott EF. The cognitive neuroscience of sleep: neuronal systems, consciousness and learning. Nat Rev Neurosci. 2002;3:679–93. doi: 10.1038/nrn915. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, Greco MA. Sleep circuitry and the hypnotic mechanism of GABAA drugs. J Clin Sleep Med. 2006;2:S19–26. [PubMed] [Google Scholar]
- 3.McCarley RW. Neurobiology of REM and NREM sleep. Sleep Med. 2007;8:302–30. doi: 10.1016/j.sleep.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Ogasahara S, Taguchi Y, Wada H. Changes in the levels of cyclic nucleotides in rat brain during the sleep-wakefulness cycle. Brain Res. 1981;213:163–71. doi: 10.1016/0006-8993(81)91256-7. [DOI] [PubMed] [Google Scholar]
- 5.Lelkes Z, Alfoldi P, Erdos A, Benedek G. Rolipram, an antidepressant that increases the availability of cAMP, transiently enhances wakefulness in rats. Pharmacol Biochem Behav. 1998;60:835–9. doi: 10.1016/s0091-3057(98)00038-0. [DOI] [PubMed] [Google Scholar]
- 6.Hendricks JC, Williams JA, Panckeri K, et al. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–15. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 7.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–60. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 8.Graves LA, Hellman K, Veasey S, Blendy JA, Pack AI, Abel T. Genetic evidence for a role of CREB in sustained cortical arousal. J Neurophysiol. 2003;2:1152–9. doi: 10.1152/jn.00882.2002. [DOI] [PubMed] [Google Scholar]
- 9.Gottlieb DJ, O'Connor GT, Wilk JB. Genome-wide association of sleep and circadian phenotypes. BMC Med Genet. 2007;8(Suppl 1):S9. doi: 10.1186/1471-2350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abel T, Nguyen PV, Barad M, Deuel TA, Kandel ER, Bourtchouladze R. Genetic demonstration of a role for PKA in the late phase of LTP and in hippocampus-based long-term memory. Cell. 1997;88:615–26. doi: 10.1016/s0092-8674(00)81904-2. [DOI] [PubMed] [Google Scholar]
- 11.Rotenberg A, Abel T, Hawkins RD, Kandel ER, Muller RU. Parallel instabilities of long-term potentiation, place cells, and learning caused by decreased protein kinase A activity. J Neurosci. 2000;20:8096–102. doi: 10.1523/JNEUROSCI.20-21-08096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Small SA, Wu EX, Bartsch D, et al. Imaging physiologic dysfunction of individual hippocampal subregions in humans and genetically modified mice. Neuron. 2000;28:653–64. doi: 10.1016/s0896-6273(00)00144-6. [DOI] [PubMed] [Google Scholar]
- 13.Woo NH, Abel T, Nguyen PV. Genetic and pharmacological demonstration of a role for cyclic AMP-dependent protein kinase-mediated suppression of protein phosphatases in gating the expression of late LTP. Eur J Neurosci. 2002;16:1871–6. doi: 10.1046/j.1460-9568.2002.02260.x. [DOI] [PubMed] [Google Scholar]
- 14.Gelinas JN, Tenorio G, Lemon N, Abel T, Nguyen PV. Beta-adrenergic receptor activation during distinct patterns of stimulation critically modulates the PKA-dependence of LTP in the mouse hippocampus. Learn Mem. 2008;15:281–9. doi: 10.1101/lm.829208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber R, Deboer T, Tobler I. Topography of EEG dynamics after sleep deprivation in mice. J Neurophysiol. 2000;84:1888–93. doi: 10.1152/jn.2000.84.4.1888. [DOI] [PubMed] [Google Scholar]
- 16.Chemelli RM, Willie JT, Sinton CM, et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell. 1999;98:437–51. doi: 10.1016/s0092-8674(00)81973-x. [DOI] [PubMed] [Google Scholar]
- 17.Veasey SC, Valladares O, Fenik P, et al. An automated system for recording and analysis of sleep in mice. Sleep. 2000;23:1025–40. [PubMed] [Google Scholar]
- 18.Fenzl T, Romanowski CP, Flachskamm C, et al. Fully automated sleep deprivation in mice as a tool in sleep research. J Neurosci Methods. 2007;166:229–35. doi: 10.1016/j.jneumeth.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Meerlo P, Easton A, Bergmann BM, Turek FW. Restraint increases prolactin and REM sleep in C57BL/6J mice but not in BALB/cJ mice. Am J Physiol Regul Integr Comp Physiol. 2001;281:R846–54. doi: 10.1152/ajpregu.2001.281.3.R846. [DOI] [PubMed] [Google Scholar]
- 20.Youngblood BD, Zhou J, Smagin GN, Ryan DH, Harris RB. Sleep deprivation by the “flower pot” technique and spatial reference memory. Physiol Behav. 1997;61:249–56. doi: 10.1016/s0031-9384(96)00363-0. [DOI] [PubMed] [Google Scholar]
- 21.Campbell IG, Guinan MJ, Horowitz JM. Sleep deprivation impairs long-term potentiation in rat hippocampal slices. J Neurophysiol. 2002;88:1073–6. doi: 10.1152/jn.2002.88.2.1073. [DOI] [PubMed] [Google Scholar]
- 22.Ouyang M, Hellman K, Abel T, Thomas SA. Adrenergic signaling plays a critical role in the maintenance of waking and in the regulation of REM sleep. J Neurophysiol. 2004;92:2071–82. doi: 10.1152/jn.00226.2004. [DOI] [PubMed] [Google Scholar]
- 23.Rechtschaffen A, Kales A. A manual of standard terminology, techniques and scoring system for sleep staging human subjects. Bethesda, MD: Public Health Service; 1968. [Google Scholar]
- 24.Norman RG, Scott MA, Ayappa I, Walsleben JA, Rapoport DM. Sleep continuity measured by survival curve analysis. Sleep. 2006;29:1625–31. doi: 10.1093/sleep/29.12.1625. [DOI] [PubMed] [Google Scholar]
- 25.Muthuswamy J, Thakor NV. Spectral analysis methods for neurological signals. J Neurosci Methods. 1998;83:1–14. doi: 10.1016/s0165-0270(98)00065-x. [DOI] [PubMed] [Google Scholar]
- 26.Borbély AA, Baumann F, Brandeis D, Strauch I, Lehmann D. Sleep deprivation: effect on sleep stages and EEG power density in man. Electroencephalogr Clin Neurophysiol. 1981;51:483–95. doi: 10.1016/0013-4694(81)90225-x. [DOI] [PubMed] [Google Scholar]
- 27.Nadasdy Z, Hirase H, Czurko A, Csicsvari J, Buzsaki G. Replay and time compression of recurring spike sequences in the hippocampus. J Neurosci. 1999;19:9497–507. doi: 10.1523/JNEUROSCI.19-21-09497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eschenko O, Molle M, Born J, Sara SJ. Elevated sleep spindle density after learning or after retrieval in rats. J Neurosci. 2006;26:12914–20. doi: 10.1523/JNEUROSCI.3175-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandon EP, Zhuo M, Huang YY, et al. Hippocampal long-term depression and depotentiation are defective in mice carrying a targeted disruption of the gene encoding the RI beta subunit of cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1995;92:8851–5. doi: 10.1073/pnas.92.19.8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Clegg RA, West DW, Aitchison RE. Protein phosphorylation in rat mammary acini and in cytosol preparations in vitro. Phosphorylation of acetyl-CoA carboxylase is unaffected by cyclic AMP. Biochem J. 1987;241:447–54. doi: 10.1042/bj2410447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banke TG, Bowie D, Lee H, Huganir RL, Schousboe A, Traynelis SF. Control of GluR1 AMPA receptor function by cAMP-dependent protein kinase. J Neurosci. 2000;20:89–102. doi: 10.1523/JNEUROSCI.20-01-00089.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benson DL, Isackson PJ, Gall CM, Jones EG. Contrasting patterns in the localization of glutamic acid decarboxylase and Ca2+/calmodulin protein kinase gene expression in the rat central nervous system. Neuroscience. 1992;46:825–49. doi: 10.1016/0306-4522(92)90188-8. [DOI] [PubMed] [Google Scholar]
- 34.Mayford M, Wang J, Kandel ER, O'Dell TJ. CaMKII regulates the frequency-response function of hippocampal synapses for the production of both LTD and LTP. Cell. 1995;81:891–904. doi: 10.1016/0092-8674(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 35.Franken P, Malafosse A, Tafti M. Genetic variation in EEG activity during sleep in inbred mice. Am J Physiol. 1998;275:R1127–37. doi: 10.1152/ajpregu.1998.275.4.R1127. [DOI] [PubMed] [Google Scholar]
- 36.Amizca F, Steriade M. The K-complex: its slow rhythmicity and relation to delta waves. Neurology. 1980;49:952–9. doi: 10.1212/wnl.49.4.952. [DOI] [PubMed] [Google Scholar]
- 37.Douglas CL, Baghdoyan HA, Lydic R. Postsynaptic muscarinic M1 receptors activate prefrontal cortical EEG of C57BL/6J mouse. J Neurophysiol. 2002;88:3003–9. doi: 10.1152/jn.00318.2002. [DOI] [PubMed] [Google Scholar]
- 38.Borbely AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1:195–204. [PubMed] [Google Scholar]
- 39.Dijk DJ, Hayes B, Czeisler CA. Dynamics of electroencephalographic sleep spindles and slow wave activity in men: effect of sleep deprivation. Brain Res. 1993;626:190–9. doi: 10.1016/0006-8993(93)90579-c. [DOI] [PubMed] [Google Scholar]
- 40.Huber R, Esser SK, Ferrarelli F, Massimini M, Peterson MJ, Tononi G. TMS-induced cortical potentiation during wakefulness locally increases slow wave activity during sleep. PLoS ONE. 2007;2:e276. doi: 10.1371/journal.pone.0000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker MP, Stickgold R, Alsop D, Gaab N, Schlaug G. Sleep-dependent motor memory plasticity in the human brain. Neuroscience. 2005;133:911–7. doi: 10.1016/j.neuroscience.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Massimini M, Ferrarelli F, Esser SK, et al. Triggering sleep slow waves by transcranial magnetic stimulation. Proc Natl Acad Sci U S A. 2007;104:8496–501. doi: 10.1073/pnas.0702495104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–23. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 44.Cirelli C, Tononi G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000;885:303–21. doi: 10.1016/s0006-8993(00)03008-0. [DOI] [PubMed] [Google Scholar]
- 45.Matthews RP, Guthrie CR, Wailes LM, Zhao X, Means AR, McKnight GS. Calcium/calmodulin-dependent protein kinase types II and IV differentially regulate CREB-dependent gene expression. Mol Cell Biol. 1994;14:6107–16. doi: 10.1128/mcb.14.9.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xing J, Ginty DD, Greenberg ME. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273:959–63. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 47.Franken P, Malafosse A, Tafti M. Genetic determinants of sleep regulation in inbred mice. Sleep. 1999;22:155–69. [PubMed] [Google Scholar]
- 48.Rosenberg RP. Sleep maintenance insomnia: strengths and weaknesses of current pharmacologic therapies. Ann Clin Psychiatry. 2006;18:49–56. doi: 10.1080/10401230500464711. [DOI] [PubMed] [Google Scholar]
- 49.National Sleep Foundation. Sleep in America Poll. 2002. [Google Scholar]
- 50.Carney CE, Segal ZV, Edinger JD, Krystal AD. A comparison of rates of residual insomnia symptoms following pharmacotherapy or cognitive-behavioral therapy for major depressive disorder. J Clin Psychiatry. 2007;68:254–60. doi: 10.4088/jcp.v68n0211. [DOI] [PubMed] [Google Scholar]
- 51.Khatami R, Zutter D, Siegel A, Mathis J, Donati F, Bassetti CL. Sleep-wake habits and disorders in a series of 100 adult epilepsy patients--a prospective study. Seizure. 2006;15:299–306. doi: 10.1016/j.seizure.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 52.Edinger JD, Wohlgemuth WK, Radtke RA, Marsh GR, Quillian RE. Cognitive behavioral therapy for treatment of chronic primary insomnia: a randomized controlled trial. Jama. 2001;285:1856–64. doi: 10.1001/jama.285.14.1856. [DOI] [PubMed] [Google Scholar]
- 53.Besset A, Villemin E, Tafti M, Billiard M. Homeostatic process and sleep spindles in patients with sleep-maintenance insomnia: effect of partial (21 h) sleep deprivation. Electroencephalogr Clin Neurophysiol. 1998;107:122–32. doi: 10.1016/s0013-4694(98)00048-0. [DOI] [PubMed] [Google Scholar]
- 54.Landis CA, Lentz MJ, Rothermel J, Buchwald D, Shaver JL. Decreased sleep spindles and spindle activity in midlife women with fibromyalgia and pain. Sleep. 2004;27:741–50. doi: 10.1093/sleep/27.4.741. [DOI] [PubMed] [Google Scholar]
- 55.Ondze B, Espa F, Dauvilliers Y, Billiard M, Besset A. Sleep architecture, slow wave activity and sleep spindles in mild sleep disordered breathing. Clin Neurophysiol. 2003;114:867–74. doi: 10.1016/s1388-2457(02)00389-9. [DOI] [PubMed] [Google Scholar]
- 56.Himanen SL, Virkkala J, Huupponen E, Hasan J. Spindle frequency remains slow in sleep apnea patients throughout the night. Sleep Med. 2003;4:361–6. doi: 10.1016/s1389-9457(03)00155-2. [DOI] [PubMed] [Google Scholar]
- 57.Landolt HP, Glatzel M, Blattler T, et al. Sleep-wake disturbances in sporadic Creutzfeldt-Jakob disease. Neurology. 2006;66:1418–24. doi: 10.1212/01.wnl.0000210445.16135.56. [DOI] [PubMed] [Google Scholar]
- 58.Crowley K, Trinder J, Kim Y, Carrington M, Colrain IM. The effects of normal aging on sleep spindle and K-complex production. Clin Neurophysiol. 2002;113:1615–22. doi: 10.1016/s1388-2457(02)00237-7. [DOI] [PubMed] [Google Scholar]
- 59.Glenn LL, Steriade M. Discharge rate and excitability of cortically projecting intralaminar thalamic neurons during waking and sleep states. J Neurosci. 1982;2:1387–404. doi: 10.1523/JNEUROSCI.02-10-01387.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azumi K, Shirakawa S. Characteristics of spindle activity and their use in evaluation of hypnotics. Sleep. 1982;5:95–105. doi: 10.1093/sleep/5.1.95. [DOI] [PubMed] [Google Scholar]
- 61.Hirshkowitz M, Thornby JI, Karacan I. Sleep spindles: pharmacological effects in humans. Sleep. 1982;5:85–94. doi: 10.1093/sleep/5.1.85. [DOI] [PubMed] [Google Scholar]
- 62.Suetsugi M, Mizuki Y, Ushijima I, Kobayashi T, Watanabe Y. The effects of diazepam on sleep spindles: a qualitative and quantitative analysis. Neuropsychobiology. 2001;43:49–53. doi: 10.1159/000054865. [DOI] [PubMed] [Google Scholar]
- 63.Walsh JK, Deacon S, Dijk DJ, Lundahl J. The selective extrasynaptic GABAA agonist, gaboxadol, improves traditional hypnotic efficacy measures and enhances slow wave activity in a model of transient insomnia. Sleep. 2007;30:593–602. doi: 10.1093/sleep/30.5.593. [DOI] [PubMed] [Google Scholar]
- 64.Molle M, Eschenko O, Gais S, Sara SJ, Born J. The influence of learning on sleep slow oscillations and associated spindles and ripples in humans and rats. Eur J Neurosci. 2009;29:1071–81. doi: 10.1111/j.1460-9568.2009.06654.x. [DOI] [PubMed] [Google Scholar]
- 65.Graves L, Pack A, Abel T. Sleep and memory: a molecular perspective. Trends in Neuroscience. 2001;24:237–43. doi: 10.1016/s0166-2236(00)01744-6. [DOI] [PubMed] [Google Scholar]
- 66.Hellman K, Abel T. Molecular mechanisms of memory consolidation. In: Maqet P, Smith C, Stickgold R, editors. Sleep and Brain Plasticity. Oxford, England: Oxford; 2003. [Google Scholar]
- 67.Stickgold R, Hobson J, Fosse R, Fosse M. Sleep, learning, and dreams: off-line memory reprocessing. Science. 2001;294:1052–7. doi: 10.1126/science.1063530. [DOI] [PubMed] [Google Scholar]
- 68.Vertes RP, Siegel JM. Time for the sleep community to take a critical look at the purported role of sleep in memory processing. Sleep. 2005;28:1228–9. doi: 10.1093/sleep/28.10.1228. discussion 30-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vertes RP. Memory consolidation in sleep; dream or reality. Neuron. 2004;44:135–48. doi: 10.1016/j.neuron.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 70.Frank MG, Benington JH. The role of sleep in memory consolidation and brain plasticity: dream or reality? Neuroscientist. 2006;12:477–88. doi: 10.1177/1073858406293552. [DOI] [PubMed] [Google Scholar]
- 71.Buzsaki G. Memory consolidation during sleep: a neurophysiological perspective. J Sleep Res. 1998;7(Suppl 1):17–23. doi: 10.1046/j.1365-2869.7.s1.3.x. [DOI] [PubMed] [Google Scholar]
- 72.Siapas AG, Wilson MA. Coordinated interactions between hippocampal ripples and cortical spindles during slow-wave sleep. Neuron. 1998;21:1123–8. doi: 10.1016/s0896-6273(00)80629-7. [DOI] [PubMed] [Google Scholar]
- 73.Hardingham N, Wright N, Dachtler J, Fox K. Sensory deprivation unmasks a PKA-dependent synaptic plasticity mechanism that operates in parallel with CaMKII. Neuron. 2008;60:861–74. doi: 10.1016/j.neuron.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 74.Vyazovskiy VV, Cirelli C, Pfister-Genskow M, Faraguna U, Tononi G. Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat Neurosci. 2008;11:200–8. doi: 10.1038/nn2035. [DOI] [PubMed] [Google Scholar]
- 75.Esteban JA, Shi SH, Wilson C, Nuriya M, Huganir RL, Malinow R. PKA phosphorylation of AMPA receptor subunits controls synaptic trafficking underlying plasticity. Nat Neurosci. 2003;6:136–43. doi: 10.1038/nn997. [DOI] [PubMed] [Google Scholar]
- 76.Zhu JJ. Activity level-dependent synapse-specific AMPA receptor trafficking regulates transmission kinetics. J Neurosci. 2009;29:6320–35. doi: 10.1523/JNEUROSCI.4630-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lytton WW, Destexhe A, Sejnowski TJ. Control of slow oscillations in the thalamocortical neuron: a computer model. Neuroscience. 1996;70:673–84. doi: 10.1016/s0306-4522(96)83006-5. [DOI] [PubMed] [Google Scholar]
- 78.Luthi A, McCormick DA. Modulation of a pacemaker current through Ca(2+)-induced stimulation of cAMP production. Nat Neurosci. 1999;2:634–41. doi: 10.1038/10189. [DOI] [PubMed] [Google Scholar]
- 79.Tartar JL, Ward CP, McKenna JT, et al. Hippocampal synaptic plasticity and spatial learning are impaired in a rat model of sleep fragmentation. Eur J Neurosci. 2006;23:2739–48. doi: 10.1111/j.1460-9568.2006.04808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rabat A, Bouyer JJ, George O, Le Moal M, Mayo W. Chronic exposure of rats to noise: relationship between long-term memory deficits and slow wave sleep disturbances. Behav Brain Res. 2006;171:303–12. doi: 10.1016/j.bbr.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 81.Hygge S, Evans GW, Bullinger M. A prospective study of some effects of aircraft noise on cognitive performance in schoolchildren. Psychol Sci. 2002;13:469–74. doi: 10.1111/1467-9280.00483. [DOI] [PubMed] [Google Scholar]
- 82.Clark C, Martin R, van Kempen E, et al. Exposure-effect relations between aircraft and road traffic noise exposure at school and reading comprehension: the RANCH project. Am J Epidemiol. 2006;163:27–37. doi: 10.1093/aje/kwj001. [DOI] [PubMed] [Google Scholar]
- 83.Stansfeld SA, Berglund B, Clark C, et al. Aircraft and road traffic noise and children's cognition and health: a cross-national study. Lancet. 2005;365:1942–9. doi: 10.1016/S0140-6736(05)66660-3. [DOI] [PubMed] [Google Scholar]
- 84.Matsui T, Stansfeld S, Haines M, Head J. Children's cognition and aircraft noise exposure at home--the West London Schools Study. Noise Health. 2004;7:49–58. [PubMed] [Google Scholar]
- 85.Salorio CF, White DA, Piccirillo J, Duntley SP, Uhles ML. Learning, memory, and executive control in individuals with obstructive sleep apnea syndrome. J Clin Exp Neuropsychol. 2002;24:93–100. doi: 10.1076/jcen.24.1.93.973. [DOI] [PubMed] [Google Scholar]
- 86.Naegele B, Thouvard V, Pepin JL, et al. Deficits of cognitive executive functions in patients with sleep apnea syndrome. Sleep. 1995;18:43–52. [PubMed] [Google Scholar]
- 87.Bedard MA, Montplaisir J, Richer F, Rouleau I, Malo J. Obstructive sleep apnea syndrome: pathogenesis of neuropsychological deficits. J Clin Exp Neuropsychol. 1991;13:950–64. doi: 10.1080/01688639108405110. [DOI] [PubMed] [Google Scholar]
- 88.Leitner Y, Bloch AM, Sadeh A, et al. Sleep-wake patterns in children with intrauterine growth retardation. J Child Neurol. 2002;17:872–6. doi: 10.1177/08830738020170121901. [DOI] [PubMed] [Google Scholar]
- 89.Lindblom N, Heiskala H, Kaski M, Leinonen L, Laakso ML. Sleep fragmentation in mentally retarded people decreases with increasing daylength in spring. Chronobiol Int. 2002;19:441–59. doi: 10.1081/cbi-120002880. [DOI] [PubMed] [Google Scholar]
- 90.Quine L. Sleep problems in children with mental handicap. J Ment Defic Res. 1991;35:269–90. doi: 10.1111/j.1365-2788.1991.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 91.Kim SH, Nairn AC, Cairns N, Lubec G. J Neural Transm Suppl. 2001. Decreased levels of ARPP-19 and PKA in brains of Down syndrome and Alzheimer's disease; pp. 263–72. [DOI] [PubMed] [Google Scholar]
- 92.Mantovani G, Romoli R, Weber G, et al. Mutational analysis of GNAS1 in patients with pseudohypoparathyroidism: identification of two novel mutations. J Clin Endocrinol Metab. 2000;85:4243–8. doi: 10.1210/jcem.85.11.6986. [DOI] [PubMed] [Google Scholar]