Abstract
Following cessation of growth, yeast cells remain viable in a non-dividing state for a period of time known as the chronological life span (CLS). Autophagy is a degradative process responsible for amino acid recycling in response to nitrogen starvation and amino acid limitation. We have investigated the role of autophagy during chronological aging of yeast grown in glucose minimal media containing different supplemental essential and non-essential amino acids. Deletion of ATG1 or ATG7, both of which are required for autophagy, reduces CLS, whereas deletion of ATG11, which is required for selective targeting of cellular components to the vacuole for degradation, does not reduce CLS. The non-essential amino acids isoleucine and valine, and the essential amino acid leucine, extend CLS in autophagy-deficient as well as autophagy-competent yeast. This extension is suppressed by constitutive expression of GCN4, which encodes a transcriptional regulator of general amino acid control (GAAC). Consistent with this, GCN4 expression is reduced by isoleucine and valine. Furthermore, elimination of the leucine requirement extends CLS and prevents the effects of constitutive expression of GCN4. Interestingly, deletion of LEU3, a GAAC target gene encoding a transcriptional regulator of branched side chain amino acid synthesis, dramatically increases CLS in the absence of amino acid supplements. In general, this indicates that activation of GAAC reduces CLS whereas suppression of GAAC extends CLS in minimal medium. These findings demonstrate important roles for autophagy and amino acid homeostasis in determining CLS in yeast.
Keywords: aging, autophagy, amino acid homeostasis, Saccharomyces cerevisiae
Introduction
The yeast Saccharomyces cerevisiae has been used as a model eukaryote to study the aging process for 50 years (Mortimer and Johnson, 1959). Similar to cells in metazoans, yeast cells undergo replicative and chronological aging. Replicative life span is defined as the number of divisions a cell undergoes before becoming senescent. Chronological life span (CLS) is defined as the length of time that a non-dividing cell remains viable. Viability is usually defined as the capacity to resume mitosis, which is a biologically meaningful measure of how well cells survive in a non-dividing state. In yeast, the CLS may range from days to months, depending on the genetic background and growth conditions. Like in other organisms, chronological aging in yeast results from a series of time- and metabolism-dependent changes that result in reduced viability, and is distinguished from loss of viability due to the secondary effects of deleterious mutations or adverse environmental factors (e.g., temperature extremes, toxic substances). Cellular processes that forestall such changes promote chronological longevity.
In free-living organisms such as yeast, the main factor that gives rise to a non-dividing state is nutrient scarcity, which triggers a shift from a growth-based metabolism to a survival-based metabolism. Arguably, yeast in the wild spend much of their time in a non-dividing, quiescent state known as stationary phase. Importantly, stationary phase is not simply a cessation of cell division. During stationary phase, yeast cells actively respond to changes in nutrient availability by undertaking a series of metabolic and cell biological changes that enhance cell survival, including reinforcement of the cell wall, accumulation of storage carbohydrates (e.g., glycogen and trehalose), and increased stress resistance, all of which are orchestrated by specific programs of gene expression (reviewed in Gray et al., 2004; Herman, 2002). Thus, yeast in stationary phase remain metabolically and biosynthetically active, albeit at reduced levels, and remain responsive to environmental signals. In general, stationary phase yeast share similarities in metabolism and gene expression with non-dividing differentiated (G0 phase) mammalian cells that undergo chronological aging (Allen et al., 2006).
Most studies of CLS in yeast have been carried out on cells grown in glucose-containing media, such as non-defined rich (YPD) medium and synthetic complete (SC) medium. Although routinely used for these purposes, these media are not well suited to the study of cellular pathways that respond to nutrient scarcity. Nitrogen sources are both abundant and multiple in rich media and pathways that respond to nitrogen limitation are not fully activated. In their natural environment, yeast encounter nutrient limitations different from those associated with growth in rich media in the laboratory. Yeast in the wild are prototrophs that synthesize most of their metabolites from simple carbon and nitrogen sources, inorganic salts, and trace elements. Carbon and nitrogen frequently become limiting under these conditions in the environment. Considering this, we view synthetic minimal media as more closely approximating conditions in the wild insofar as nutrient availability is concerned.
One of the main cellular pathways that responds to nitrogen limitation is autophagy. Autophagy is both inducible and constitutively active at a low level in most cells, and functions to degrade cellular components to recycle amino acids and other metabolites. Multiple autophagic pathways are known in yeast (Hung et al., 2004; Kvam and Goldfarb, 2007; Nair and Klionsky, 2005). Macroautophagy is the process by which a double membrane autophagosome is formed around an organelle or region of cytoplasm. The autophagosome outer membrane fuses with the membrane of the vacuole, degradation takes place in the vacuolar interior, and nutrients recycle across the vacuolar membrane to the cytoplasm (Yang et al., 2006). For the sake of simplicity in this report, we will use the term autophagy to refer to macroautophagy. Autophagy is generally considered to be a non-specific process, but specialized autophagic pathways are known to handle specific targets. Autophagy is the primary mechanism for the turnover of damaged or superfluous organelles, such as mitochondria and peroxisomes (Dunn et al., 2005; Kissova et al., 2007; Kundu and Thompson, 2005). In yeast, autophagy is upregulated by, and required for survival during, nitrogen starvation. In the laboratory, autophagy is routinely induced by transferring rapidly growing yeast from YPD rich medium to starvation medium containing glucose, but lacking ammonium sulfate and essential amino acids.
Although autophagy is expected to contribute to chronological longevity in yeast, it has not been widely studied in this context to our knowledge. However, it is clear that autophagy promotes longevity in other species. In C. elegans and D. melanogaster, inactivation of autophagy slows turnover of damaged proteins and reduces life span (Juhasz et al., 2007; Juhasz and Neufeld, 2008; Toth et al., 2008). Elevation of basal levels of autophagy in neuronal tissues in D. melanogaster appears to boost longevity (Simonsen et al., 2008). Autophagy is required for life span extension by calorie restriction in C. elegans (Jia and Levine, 2007), and is elevated in cardiac tissue in rats on a calorie restricted diet (Wohlgemuth et al., 2007), implicating autophagy as a mediator of the life span prolonging effects of calorie restriction. Furthermore, the other major turnover pathway in yeast, the ubiquitin/proteasome turnover pathway, contributes to chronological longevity in yeast (Chen et al., 2004; Chen et al., 2006). Given our understanding of the role of autophagy in cell survival in yeast, we expected that autophagy and amino acid availability would be important factors in influencing chronological longevity in yeast. The studies described below confirm this and demonstrate that autophagy and regulated synthesis of branched side chain amino acids are required for chronological longevity of yeast grown in minimal medium.
Materials and Methods
Yeast strains and plasmids
Yeast strains and plasmids are listed in Table 1. Yeast strains derived from BY4742 (Brachmann et al., 1998) contained KanMX4 marked deletions generated as part of the systematic gene deletion project (Winzeler et al., 1999) and were obtained from Daniel J. Klionsky (University of Michigan) or EUROSCARF. Yeast strains in the W303 background (Bernales et al., 2006; Bernales et al., 2007) were obtained from Sebastian Schuck and Peter Walter (University of California, San Francisco). Plasmids p164 and p238 (Mueller and Hinnebusch, 1986) were obtained from Alan Hinnebusch (National Institute of Child Health and Human Development, NIH). Plasmid pCuGFPAUT7(416) (Kim et al., 2001) was obtained from Daniel J. Klionsky. Plasmid transformants were prepared using lithium acetate (Gietz and Woods, 2002), grown on SD minimal medium, and streaked to single colonies prior to use. Yeast strains YAA1, YAA3, YAA5, and YAA7 were constructed by transforming hoΔ, atg1Δ, atg7Δ, and atg11Δ strains, respectively, with a LEU2-containing PCR product prepared using plasmid pRS315 (Sikorski and Hieter, 1989) as template DNA. Leu+ transformant genomic DNAs were screened by PCR using primers flanking the LEU2-containing PCR product to confirm integration at the LEU2 locus (primer sequences are available upon request).
Table 1 .
Yeast strains and plasmids.
| Strain | Genotype | Reference |
|---|---|---|
| BY4742 | MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Brachmann et al., 1998 |
| WT | BY4742 hoΔ::KanMX4 | Winzeler et al., 1999 |
| atg1Δ | BY4742 atg1Δ::KanMX4 | Winzeler et al., 1999 |
| atg7Δ | BY4742 atg7Δ::KanMX4 | Winzeler et al., 1999 |
| atg11Δ | BY4742 atg11Δ::KanMX4 | Winzeler et al., 1999 |
| leu3Δ | BY4742 leu3Δ::KanMX4 | Winzeler et al., 1999 |
| leu3Δ | BY4742 leu3Δ::KanMX4 | Winzeler et al., 1999 |
| YAA1 | BY4742 LEU2 hoΔ::KanMX4 | This study |
| YAA3 | BY4742 LEU2 atg1Δ::KanMX4 | This study |
| YAA5 | BY4742 LEU2 atg7Δ::KanMX4 | This study |
| YAA7 | BY4742 LEU2 atg11Δ::KanMX4 | This study |
| W303 | MATa ade2-1 can1–100 his3–11,15 leu2–3,112 trp1-1 ura3-1 | Bernales et al., 2006 |
| atg1Δ (W) | W303 atg1Δ::KanMX4 | Bernales et al., 2006 |
| atg7Δ (W) | W303 atg7Δ::KanMX4 | Bernales et al., 2006 |
| atg11Δ (W) | W303 atg11Δ::KanMX4 | Bernales et al., 2006 |
| Plasmid | Relevant functional information | |
| YCp50 | Yeast shuttle vector containing CEN4 and URA3 | Rose et al., 1987 |
| p164 | YCp50 containing GCN4 | Mueller and Hinnebusch, 1986 |
| p238 | YCp50 containing GCN4C constitutive expression allele | Mueller and Hinnebusch, 1986 |
| pCuGFP-AUT7(416) | Yeast shuttle vector pRS416 containing CEN4 and URA3, and encoding a GFP-ATG8 fusion protein under control of the CUP1 promoter. | Kim et al., 2001 |
Microbiological methods
Rich medium (YPD) and synthetic dextrose (SD) minimal medium, both containing 2% glucose, were prepared according to Sherman (Sherman, 2002) (Table 2). Synthetic complete (SC) media containing 2% glucose were prepared according to two commonly used formulae. We refer to Sherman’s formula (Sherman, 2002) as “SC1” and Fink’s formula (Amberg et al., 2005; Styles, 2002) as “SC2” (Table 2). SC1 and SC2 differ in how they are prepared. SC1 is prepared by adding individual supplements to SD after autoclaving. To prepare SC2, a mixture of supplements in powder form is added prior to autoclaving. Filter-sterilized stock solutions of glucose (20% stock) and supplements (prepared as described in reference Sherman, 2002) were added to synthetic base medium. Where indicated, supplements were added to a final concentration equal to three times (3X) the standard amount (Sherman, 2002). G418 sulfate (Mediatech, Inc) was added to a final (active) concentration of 250 µg/ml. Yeast were grown at 30°C. Measurements of cell density were done by diluting yeast cultures 10-fold in water and determining the optical density at 600 nm (OD600) using a Beckman DU-640 spectrophotometer.
Table 2 .
Composition of synthetic dextrose (SD) minimal and synthetic complete (SC) media.
| Component* | SD | SC1 | SC2 |
|---|---|---|---|
| D-glucose | 20 g/L | 20 g/L | 20 g/L |
| Ammonium sulfate | 5 | 5 | 5 |
| Yeast nitrogen base (-AS/-AA) | 1.7 | 1.7 | 1.7 |
| Essential | |||
| L-Histidine | 20 mg/L | 20 mg/L | 73 mg/L |
| L-Leucine | 30 | 30 | 367 |
| L-Lysine | 30 | 30 | 73 |
| Uracil | 20 | 20 | 73 |
| Non-essential | |||
| Adenine | 20 | 18 | |
| L-Alanine | 73 | ||
| L-Arginine | 20 | 73 | |
| L-Asparagine | 73 | ||
| L-Aspartic acid | 100 | 73 | |
| L-Cysteine | 73 | ||
| L-Glutamine | 73 | ||
| L-Glutamic acid | 100 | 73 | |
| L-Glycine | 73 | ||
| Inositol | 73 | ||
| L-isoleucine | 30 | 73 | |
| L-Methionine | 20 | 73 | |
| p-Amino benzoic acid | 73 | ||
| L-Phenylalanine | 50 | 73 | |
| L-Proline | 73 | ||
| L-Serine | 400 | 73 | |
| L-Threonine | 200 | 73 | |
| L-Tryptophan | 20 | 73 | |
| L-Tyrosine | 30 | 73 | |
| L-Valine | 150 | 73 |
Concentrations of glucose, ammonium sulfate, and yeast nitrogen base are listed in g/L. Concentrations of other supplements are listed in mg/L. Yeast nitrogen base does not contain ammonium sulfate (AS) or amino acids (AA). SD and SC1 are prepared according to Sherman Sherman, 2002). SC2 is prepared according to Fink Amberg et al., 2005;Styles, 2002). In some experiments, as indicated in the text, different combinations and/or amounts of supplements were present in synthetic minimal media at concentrations equal to, or three-fold higher than, those shown in the SC1 column.
Chronological life span measurements
Four days prior to the start of a CLS experiment (on day -4), yeast strains from frozen stocks at −80°C were patched onto YPD agar plates. After 2 days of growth at 30°C (on day -2) cells from patches were inoculated into 5 ml of SD medium in 15 ml Falcon 2059 polypropylene tubes (Fisher Scientific) and grown overnight at 30°C in a drum rotator at ~15 rpm. After ~24 hr of growth (on day -1) overnight cultures were diluted 1/100 into 5 ml of the medium to be used in the experiment (e.g., SD, SC1, SC2) and grown overnight at 30°C in a drum rotator. After ~24 hr of growth (on day 0) overnight cultures were diluted 1/100 into 5 ml of the medium to be used in the experiment. Following ~24 hr of growth (on day 1), the number of colony forming units (CFU) per ml was determined by serial dilution. For this, 10 µl of culture were diluted into 190 µl of YPD in a sterile 96-well plate. Five five-fold serial dilutions were done in a 96-well plate, and a 48-pin stamp was used to transfer ~4 µl each dilution to a YPD agar plate. After 2–3 days of growth at 30°C, colonies were counted and CFU values from the two highest dilutions were averaged. Plasmid-transformed strains were freshly prepared and streaked immediately prior to each experiment, and were pin-stamped onto selective SD agar medium in order to collect CFU data for plasmid-bearing cells. The process of diluting and pin-stamping cells was repeated every 2–3 days. A running tally of the culture volume removed each day was maintained and sterile water was added back as needed to compensate for evaporation. As the number of CFU/ml declined over time, a larger volume of culture was diluted in the 96-well plate. Near the end of the experiment, 100 µl of culture were directly spread on agar medium to determine viability. Zero viability is defined as <10 CFU/ml (i.e., 0 CFU in 0.1 ml of culture volume). Viability is expressed in terms of CFU per ml of culture and is plotted as the log of the percent of viability on day 1. OD600 measurements were routinely done on days 1, 3, and 5. Most strains cultured in SD medium reached saturation (typically ~1–3 × 107 CFU/ml) by day 1. However, certain strains (e.g., leu3Δ) and/or growth conditions (e.g., SD+ITV) did not yield a saturated culture on day 1, in which case data were normalized to days 2 or 3, at which point saturation was achieved. FUN-1 staining was done as described by the supplier (Invitrogen), using at least 200 individual cells per strain per time point to calculate per cent viability.
Western blotting
Western blotting was used to assay activation of autophagy and measure Gcn4p levels during chronological aging. To assay autophagy, yeast strains were transformed as described above with plasmid pCuGFPAUT7(416) (Kim et al., 2001). pCuGFPAUT7(416) is based on plasmid pRS416 (URA3) (Sikorski and Hieter, 1989) and expresses a GFP-Atg8p fusion protein under the control of the CUP1 promoter. Freshly prepared and streaked transformant colonies were used to inoculate 50 ml of SD medium in Erlenmeyer flasks on day -1 and grown in an incubator shaker at 30°C and ~175 rpm. The essential supplements histidine, lysine, and leucine were present at standard or three-fold increased concentrations (Sherman, 2002). We found that the amount of copper sulfate (40 µg/L) present in yeast nitrogen base was sufficient to drive expression of the GFP-Atg8p fusion protein (data not shown). Therefore, no supplemental copper was added to growth media to induce expression from pCuGFPAUT7(416). To measure Gcn4p levels, the same approach was used with transformants carrying plasmids p164 and p238 grown in SD medium with or without supplemental isoleucine, threonine, and valine. To prepare protein extracts, equivalent amounts of yeast (e.g., 1.25 OD units; 1 ml of culture at OD600 = 1.25) were collected by centrifugation, washed with TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8), and stored as frozen pellets at −80°C. Subsequently, total protein extracts were prepared as described (Yaffe and Schatz, 1984), and equivalent amounts of samples, based on OD units, were loaded and separated on 10.5–14% Criterion pre-cast SDS gels (Bio-Rad), followed by semi-dry transfer to nitrocellulose membrane, air drying, and staining with India ink. Blots were probed with anti-GFP antibody ab290 (Abcam Inc.), anti-Nop1p mAb B15 (Aris and Blobel, 1988), or affinity purified anti-Gcn4p antibody (Zhang et al., 2008, from A. G. Hinnebusch) and visualized using chemiluminescence. A lysate from a gcn4Δ strain (BY4742 gcn4Δ::KanMX4) was used to identify GCN4-dependent protein bands. Quantitative analysis of western blotting results was done using ImageJ software (Rasband, 2008).
Results
Autophagy is required for chronological longevity in synthetic media
For the reasons discussed in the Introduction, we investigated the role of autophagy in chronological longevity using synthetic media. We compared the CLS of autophagy-competent and autophagy-deficient strains in four media: synthetic dextrose (SD) minimal medium containing only essential amino acids (e.g., histidine, leucine, and lysine); synthetic complete (SC1) medium prepared according to Sherman (Sherman, 2002); synthetic complete (SC2) medium prepared according to Fink and colleagues (Amberg et al., 2005; Styles, 2002); and YPD medium (Sherman, 2002) (see Table 2). Four deletion strains in the BY4742 background (Brachmann et al., 1998) were used: hoΔ, atg1Δ, atg7Δ, and atg11Δ (see Table 1). The hoΔ strain possesses a “wild type” capacity for autophagy, and will be referred to as “WT” hereafter. atg1Δ and atg7Δ mutants were selected because the functions of Atg1p and Atg7p are well-characterized. Atg1p is a protein serine/threonine kinase required for autophagy and may be a convergence point for signaling pathways that regulate its induction (Stephan and Herman, 2006). Atg7p is a member of the E1 family of ubiquitin-activating enzymes, and carries out two steps in autophagosome formation: conjugation of Atg12p (a ubiquitin-like modifier) to Atg5p; and covalent attachment of phosphatidylethanolamine to Atg8p (Ohsumi, 2001). Atg1p and Atg7p are essential for all known forms of non-selective and selective autophagy. Therefore, we also used an atg11Δ mutant to specifically evaluate the role of selective autophagy in chronological longevity. Atg11p is not required for starvation-induced autophagy, but is essential for the constitutive CVT pathway and nutrient-adaptive pexophagy (He et al., 2006).
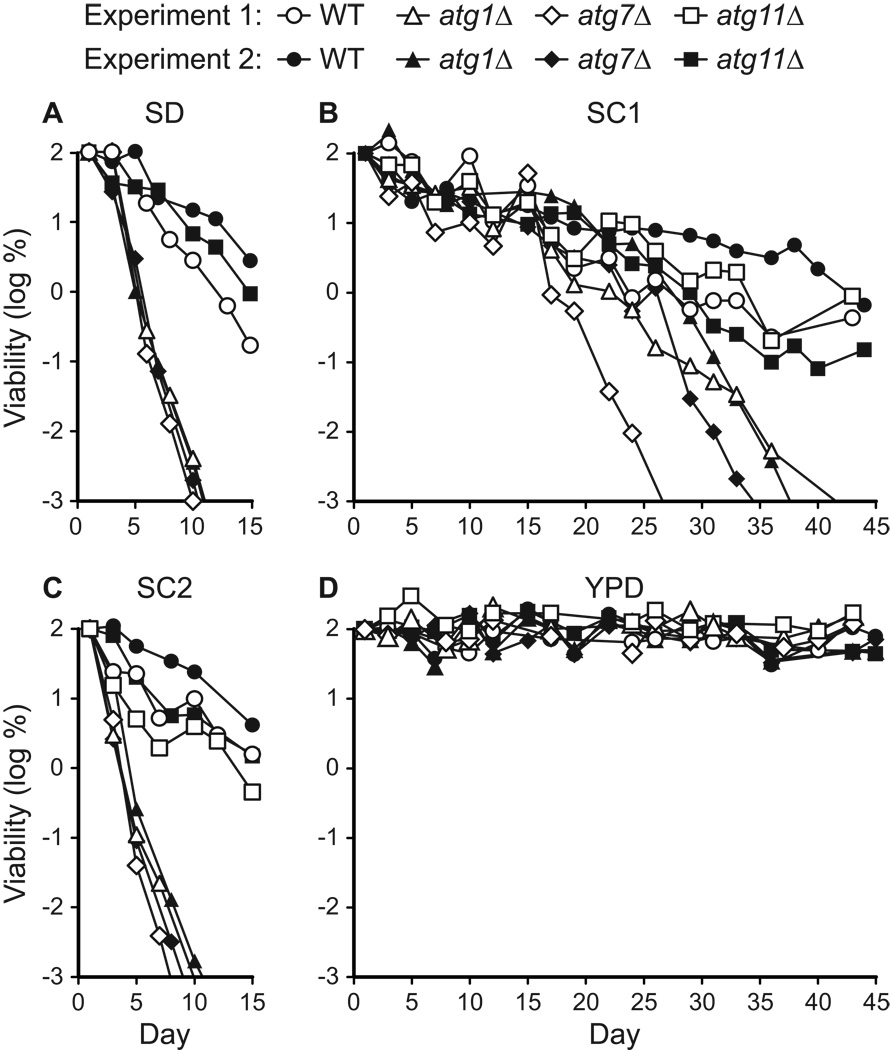
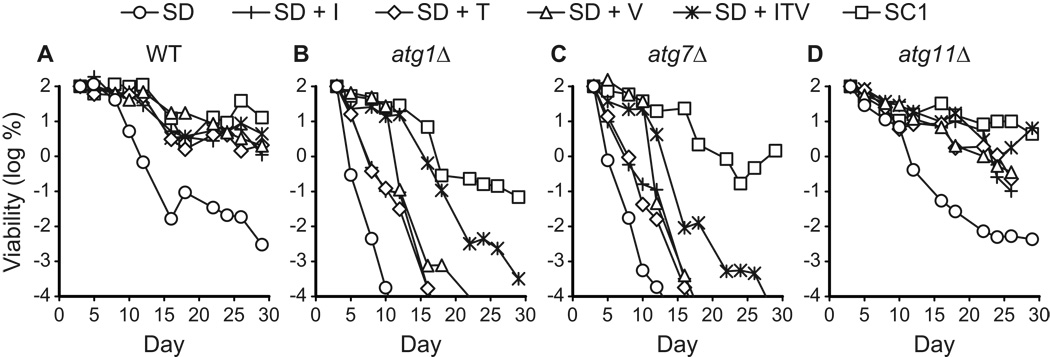
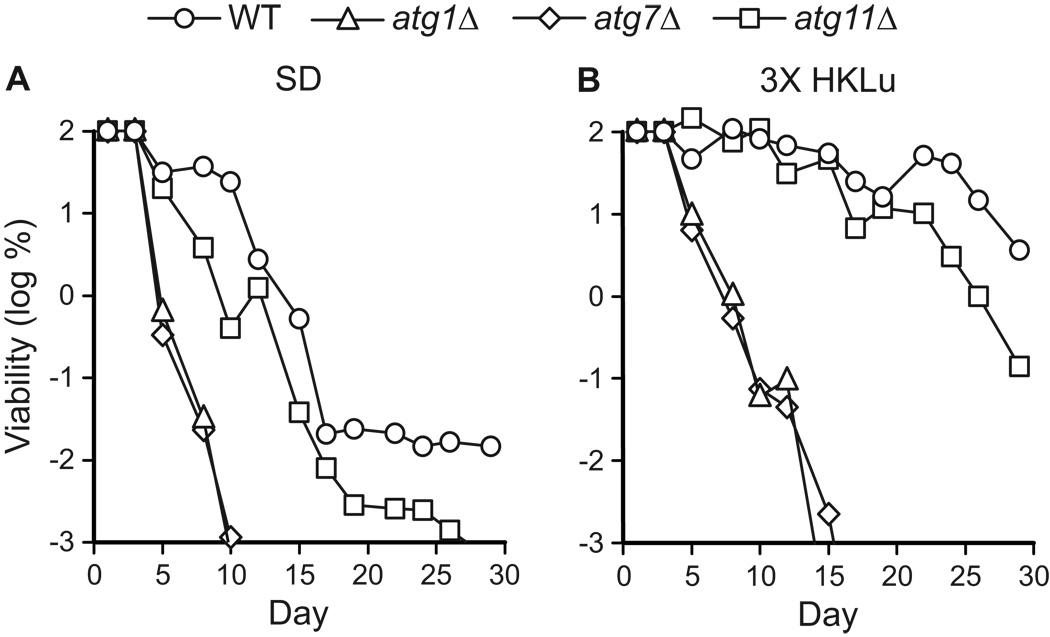
We observed that the chronological longevity of atg1Δ and atg7Δ autophagy-deficient strains in synthetic media was reduced compared to WT and atg11Δ controls (Fig. 1A–C). After ~10 days in either SD or SC2 medium, atg1Δ and atg7Δ strains reached “0” viability, which is defined as <10 CFU/ml (see Materials and Methods). Control strain cultures remained viable beyond day 15 in SD and SC media, and in most cases retained >103 CFU/ml on day 30 (Fig. 1B and data not shown). In SC1 medium, all four strains exhibited longer CLS, but the atg1Δ and atg7Δ strains lost viability prior to the WT and atg11Δ controls (Fig. 1B). By day 40 in SC1 medium, the autophagy-deficient strains had lost viability, whereas autophagy-competent control strains remained viable (Fig. 1B). In contrast to the synthetic media, we observed no difference between autophagy-deficient and control strains in YPD rich medium, which supported the longest CLS, with little loss in viability over 45 days (Fig. 1D). After 60 days in YPD medium, the viability of all four strains was reduced to ~1%, but there was no substantial difference between autophagy-competent and autophagy-deficient strains (data not shown).
Figure 1.
Autophagy is required for chronological longevity in synthetic media. CLS was measured by determining cell viability over time in synthetic dextrose minimal medium (SD, panel A), two different synthetic complete media (SC1 and SC2, panels B and C), and non-defined rich medium (YPD, panel D). SC1 refers to Sherman’s formulation (Sherman, 2002) and SC2 refers to Fink’s formulation (Amberg et al., 2005; Styles, 2002) (see Table 2). atg1Δ and atg7Δ strains are deficient in autophagy. The atg11Δ strain carries out autophagy, but is deficient in pexophagy and the CVT pathway. The WT strain is “wild type” for autophagy. Viability is expressed in terms of colony forming units (CFU) per ml of culture and is plotted as the log of the percent of viability on day 1. Two independent experiments are shown.
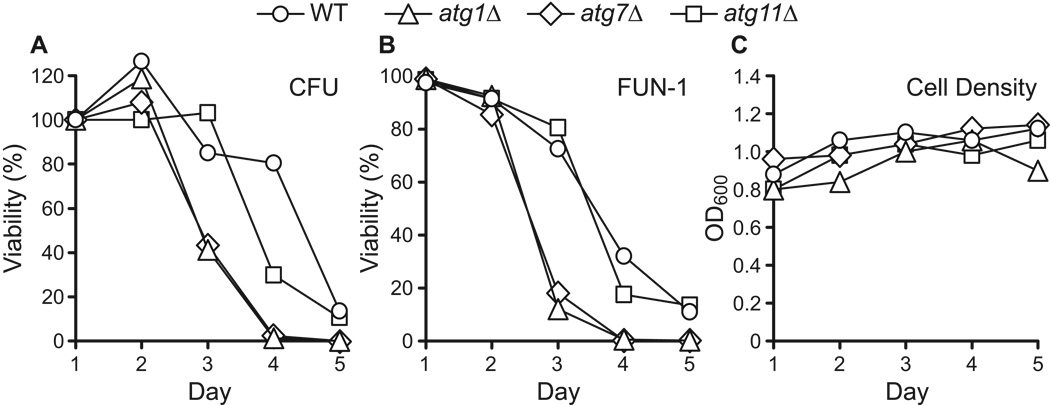
The data shown in Figure 1 were obtained by measuring cell viability in terms of colony forming units (CFU) per unit volume of culture (see Materials and Methods). This is a standard approach for measuring cell viability, but it does not address the possibility that yeast cells in minimal medium enter a senescent state in which they are viable but unable to divide. In addition, it is conceivable that atg mutations might interfere with exit from stationary phase, which could confound our measurements of viability based on colony formation. To address this concern, cell viability was assayed with the vital dye FUN-1 (Millard et al., 1997). We find that cell viability measurements using FUN-1 show good agreement with viability based on colony formation (Fig. 2), indicating that yeast do not enter a post-mitotic senescent state in minimal media. Similar results have been obtained using the vital dye erythrocin B (data not shown).
Figure 2.
Chronologically aging yeast do not exhibit senescence prior to cell death. CLS in synthetic dextrose minimal medium was determined using two measures of cell viability: colony forming units (CFU) (panel A) and staining with the vital dye FUN-1 (panel B). Yeast strains competent (WT, atg11Δ) or deficient (atg1Δ, atg7Δ) in autophagy were analyzed. Viability is expressed in terms of the percent of viability on day 1 and is plotted on a linear scale over a 5-day time period. FUN-1 staining was done as described in Materials and Methods. Cell density (OD600) measurements are shown (panel C).
We routinely monitor cell density during chronological aging experiments by measuring OD600 (see Materials and Methods). Cell densities typically reach 80–90% of their maximum value after ~24 hours of growth in SD medium (on day 1). An additional increase in cell density of 10–20% is usually observed over the course of the next few days (i.e., through days 2 to 4). By day 5, little change in cell density is usually observed. The results shown in Fig. 2C are typical in this regard. In some experiments, cell density was measured through day 10 or 12, without detection of a substantial change in OD600 (data not shown). Consistent with this, formaldehyde-fixed yeast cells from different samples over a 2-week time period showed no marked differences in cell size or shape (budding index), when viewed using phase contrast microscopy (data not shown). We conclude that loss of viability during chronological aging is not accompanied by detectable cell lysis or significant changes in cell morphology.
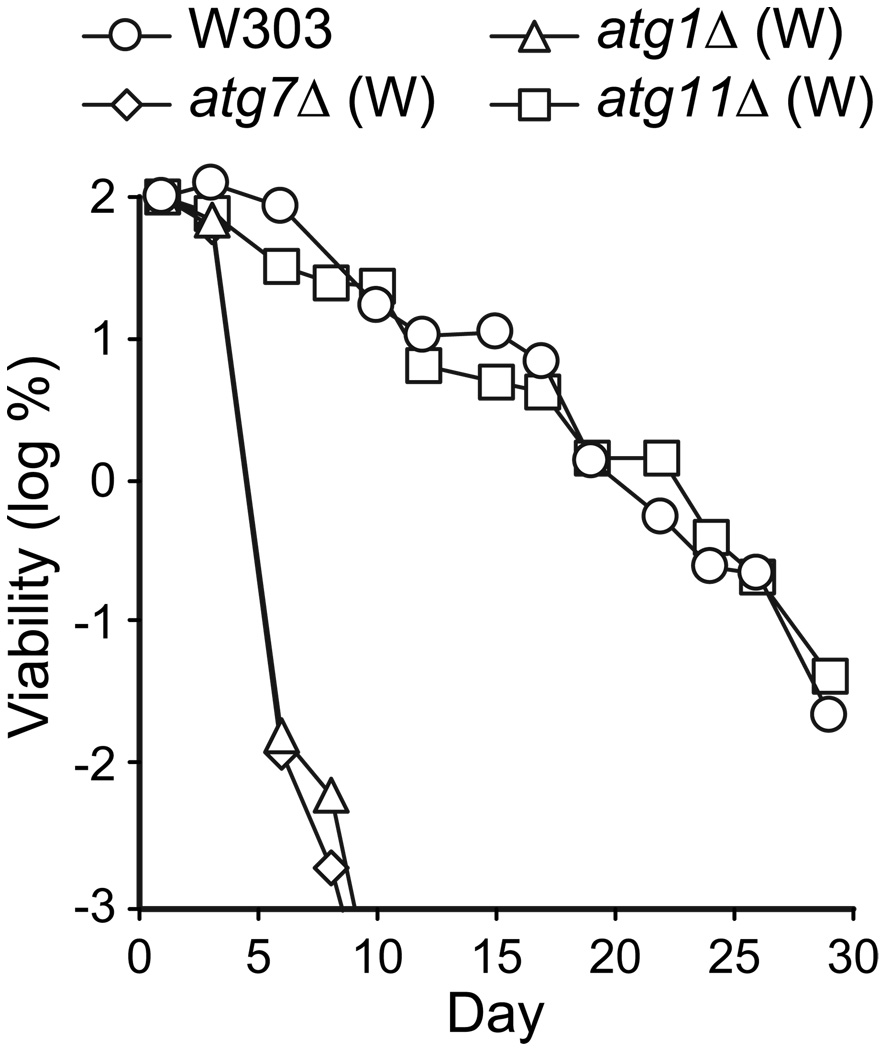
The results above were obtained with yeast strains in the BY4742 genetic background. To extend these findings to a different genetic background, we measured the CLS of strains containing the same atg alleles in the W303 genetic background (Table 1). In SD medium, the autophagy-deficient atg1Δ and atg7Δ W303 strains had a significantly reduced CLS compared to the W303 and atg11Δ control strains (Fig. 3). The W303 atg1Δ and atg7Δ mutants appeared somewhat more short-lived than their BY4742 counterparts, while the control strains in both strain backgrounds exhibited similar chronological longevities (compare Fig. 3 to Fig. 1A). Because the effects of autophagy-deficient alleles are not specific for the BY4742 strain background, we conclude that yeast exhibit a general requirement for autophagy for chronological longevity.
Figure 3.
Autophagy is required for chronological longevity in the W303 strain background. CLS was measured by determining cell viability over time in synthetic dextrose minimal medium. The W303 parental strain is competent for autophagy. atg1Δ (W) and atg7Δ (W) are autophagy deficient strains constructed in the W303 parent. The atg11Δ (W) strain carries out autophagy, but is deficient in the pexophagy and the CVT pathway. Viability in colony forming units (CFU) per ml culture is plotted as the log of percent viability on day 1.
Chronological longevity is modulated by the composition of synthetic media
Figure 1 reveals clear differences in chronological longevity of autophagy-competent and autophagy-deficient strains in SC1 and SC2 media, but chronological longevity in SC2 was similar to that in SD medium. These results were unexpected given the similarities between the two SC media and the differences between SC and SD media (Table 2). Importantly, SC1 does not yield a longer CLS simply because it contains more amino acids than SC2 medium. SC2 contains all of the amino acids found in SC1 plus 6 amino acids not present in SC1 (i.e., SC2 contains 20 amino acids, whereas SC1 contains 14). Both SC media contain adenine and uracil, but SC2 alone contains inositol and p-amino benzoic acid. Of the 14 amino acids in SC1, only 5 (D, E, S, T, and V) are present at higher concentrations in SC1 compared to SC2. SC1 and SD contain a significantly lower concentration of leucine (0.23 mM in SC1 and SD vs. 2.8 mM in SC2). Furthermore, SC2 and SD yield similar chronological life spans despite the fact that SD contains only essential amino acids. Clearly, the life spans of autophagy-deficient and control strains do not directly correlate with the presence, number, and amount of non-essential amino acids in synthetic media. In addition, although there are differences in chronological longevity in different synthetic media, all three synthetic media demonstrate a requirement for autophagy for maximum chronological longevity (Fig. 1 and Fig.3). Given that one of the primary functions of autophagy is to maintain intracellular amino acid levels, these results indicated that specific non-essential amino acids, or combinations thereof, contributed to chronological longevity.
Specific non-essential amino acids extend chronological longevity of autophagy-deficient yeast
We were intrigued by the observation that SC1 medium, but not SC2 medium, extended chronological longevity compared to SD medium. SC1 medium is prepared by adding 12 non-essential supplements to SD medium (Table 2), which suggested that one or more of these supplements extended CLS. To investigate this possibility, we surveyed the non-essential supplements in SC1 for their effects on CLS of autophagy deficient strains. For this experiment, atg1Δ and atg7Δ strains were cultured in SD media to which the 11 non-essential amino acids present in SC1 or adenine were added one-at-a-time.
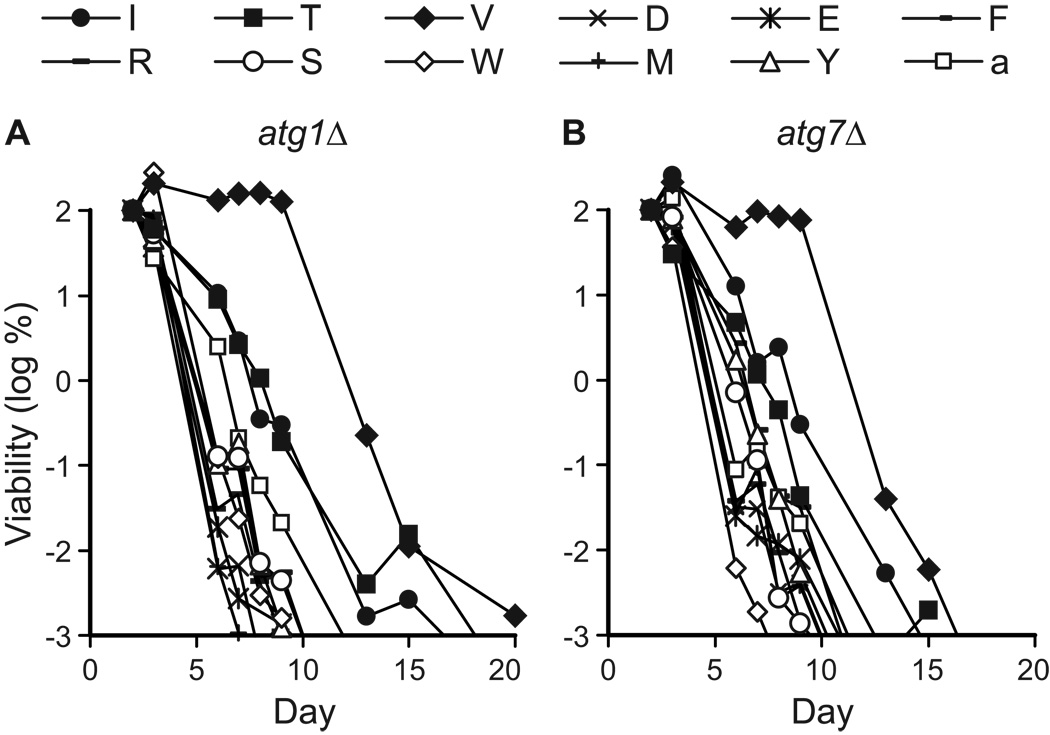
We observed that isoleucine (I), threonine (T), or valine (V) had a more positive effect on the life spans of autophagy-deficient strains than other amino acids or adenine in synthetic minimal medium (Fig. 4). The effects of isoleucine, threonine, or valine on life span are not due to the fact that they are present in higher amounts than other amino acids in SC1. For example, serine is present in the highest amount in SC1 (see Table 2), but had no discernable effect on CLS when added to SD medium (Fig. 4). Supplemental isoleucine, threonine, or valine increased cell densities in SD medium (OD600 = 1.5–2), compared to most other supplements in SD (OD600 = ~1–1.5). However, there was no strict correlation between cell density and longevity. Phenylalanine, serine, or tyrosine did not extend CLS, but yielded cell densities in the same range as isoleucine, threonine, or valine (data not shown). Thus, the effects of isoleucine, threonine, or valine do not simply correlate with concentration of the non-essential amino acid or the final cell density of the saturated culture. This indicates that the effects of isoleucine, threonine, or valine are not simply due to nitrogen supplementation, in agreement with our conclusions regarding differences in CLS observed in SC1 and SC2 media (see above).
Figure 4.
Specific non-essential amino acids promote chronological longevity. The CLS of autophagy-deficient (atg1Δ, atg7Δ) yeast was measured in synthetic dextrose minimal medium containing individual non-essential amino acids (I, T, V, D, E, F, R, S, W, M, or Y) or adenine (a) added to final concentrations as described in Table 2. Viability in colony forming units (CFU) per ml of culture is plotted as the log of the percent of viability on day 2.
Next, we measured life spans of autophagy-deficient and control strains grown in SD minimal medium plus supplemental isoleucine, threonine, or valine as well as in SC1 synthetic complete medium (Fig. 5). The purpose of this experiment was to examine the combined effects of these three amino acids and make a side-by-side comparison to SC1 medium for both autophagy-deficient and autophagy-competent cells. For the WT control strain, growth in SD containing supplemental isoleucine, threonine, or valine yielded chronological life spans comparable to life spans in SC1 medium (Fig. 5A). Similar results were obtained with the atg11Δ control strain, although individual amino acid additions were not quite as effective as the combination of all three amino acids or SC1 medium (Fig. 5D). The atg1Δ and atg7Δ autophagy-deficient strains showed extended CLS in media containing isoleucine, threonine, or valine as expected, but the extension was not as great as that observed in SC1 medium (Fig. 5B, C). To some extent in autophagy deficient strains, the effects of isoleucine, threonine, or valine were additive, insofar as a combination of these three amino acids (i.e., SD+ITV) extended CLS to a greater degree than observed with each amino acid individually. The fact that CLS is extended by supplemental isoleucine, threonine, or valine demonstrates that autophagy-deficient strains do not have an inherently short life span (i.e., < 2 weeks) in minimal medium.
Figure 5.
Non-essential amino acids isoleucine, threonine, and valine promote chronological longevity. The CLS of yeast strains competent (WT, atg11Δ) or deficient (atg1Δ, atg7Δ) in autophagy was measured in synthetic dextrose (SD) minimal or synthetic complete (SC1) media prepared as described in Table 2. Isoleucine (I), threonine (T), and valine (V) were added individually or in combination (ITV) to SD medium to achieve final concentrations present in SC1 medium (Table 2). Viability is expressed in colony forming units (CFU) per ml of culture and is plotted as the log of the percent of viability on day 3.
Figure 4 and Figure 5 demonstrate that individual non-essential amino acids can exert a positive, partially cooperative, effect on chronological longevity of both autophagy-competent and autophagy-deficient cell types. The effect of supplemental non-essential amino acids is somewhat puzzling, given that the yeast strains used in our studies are capable of synthesizing these amino acids. The amino acids isoleucine and valine (along with leucine) are synthesized by a superpathway for branched side chain amino acid biosynthesis, in which catalytically similar steps are carried out by common enzymes (i.e., Ilv2p, Ilv3p, Ilv5p, Ilv6p, Bat1p, and Bat2p). Threonine is the upstream biosynthetic precursor for isoleucine. Given this, one possibility is that isoleucine, valine, and threonine exert their effect via a regulatory mechanism rather than fulfilling a strictly nutritional role. To test this scenario, we took advantage of the central role of the transcription factor Gcn4p in regulating amino acid biosynthesis.
Extension of chronological longevity by isoleucine, valine, and threonine is suppressed by constitutive expression of Gcn4p
The results above suggested that the non-essential amino acids isoleucine, threonine, and valine influence CLS via a non-nutritional regulatory mechanism. A well-known pathway for global regulation of amino acid homeostasis in yeast is the general amino acid control (GAAC) pathway (Hinnebusch, 2005). A hallmark of GAAC is that reduction in the abundance of a single amino acid can exert a global effect on biosynthetic pathways for amino acids, as well as purines and other metabolites (Niederberger et al., 1981). GAAC is also activated by amino acid imbalances that result from differential regulation of enzymes shared by more than one biosynthetic pathway (Niederberger et al., 1981).
We hypothesized that supplemental isoleucine, threonine, and valine extend CLS by downregulating the GAAC pathway, which is activated by nutrient limitation during chronological aging. If so, then extension of CLS by isoleucine, threonine, and valine should be suppressed by constitutive upregulation of the GAAC pathway. GAAC is mediated by the transcription activator Gcn4p, levels of which are subject to translational regulation (Niederberger et al., 1981). To express Gcn4p at constitutively high levels, we transformed yeast with plasmid p238 which carries the GCN4C gain-of-function allele (Mueller and Hinnebusch, 1986). The GCN4C allele contains five site-directed changes in the untranslated 5’ leader that eliminate the four start codons in the four upstream ORFs that are required for translational repression (Mueller and Hinnebusch, 1986). In the absence of these upstream ORFs, GCN4 is expressed constitutively, but is not over-expressed. Plasmid p164 (Hinnebusch, 1985), carrying wild type GCN4, and the parent vector YCp50 (Rose et al., 1987) were used as controls.
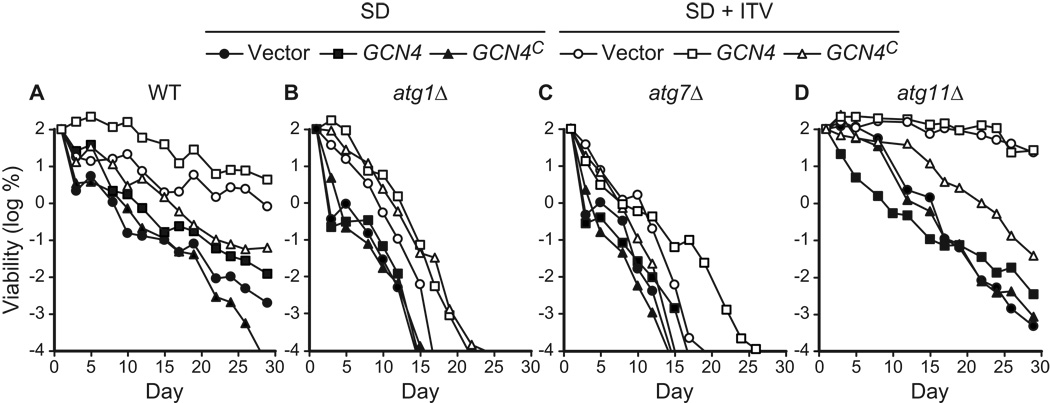
We observed that strains transformed with the control plasmids p164 (GCN4) and YCp50 showed the expected extension of CLS due to supplemental isoleucine, threonine, and valine (Fig. 6A, D). The WT and atg11Δ strains showed a more pronounced extension of CLS than the autophagy-deficient atg1Δ and atg7Δ strains (Fig. 6B, C), similar to what was observed previously (see Fig. 5). However, three strains bearing p238 (GCN4C) did not show the full extension of CLS due to isoleucine, threonine, and valine (Fig. 6). The life spans of the WT and atg11Δ control strains bearing p238 (GCN4C) were only partially extended by supplemental isoleucine, threonine, and valine (Fig. 6A, D). Similarly, the atg7Δ strain transformed with p238 (GCN4C) did not show the expected small extension of CLS in medium containing isoleucine, threonine, and valine (Fig. 6C). The atg1Δ strain bearing p238 (GCN4C) did show extension of CLS comparable to the controls (Fig. 6B), and was an exception in this regard. A complicating factor in these studies is that supplemental isoleucine, threonine, and valine increase life span in atg1Δ and atg7Δ strains to a smaller extent than in control strains, which makes it difficult to evaluate the significance of small changes in CLS in autophagy-deficient plasmid transformants (Fig. 6B, C). For example, the absence of an effect of the GCN4C allele on longevity may reflect a more severe effect of atg1Δ on life span in this strain, which is consistent with other results (see below, Fig. 10). The atg1Δ results notwithstanding, these findings support the interpretation that supplemental isoleucine, threonine, and valine extend longevity by repressing expression of GCN4 and downregulating the GAAC response.
Figure 6.
Constitutive expression of Gcn4p suppresses longevity conferred by non-essential amino acids isoleucine, threonine, and valine. Yeast strains competent (WT, atg11Δ) or deficient (atg1Δ, atg7Δ) in autophagy were transformed with three plasmids: YEp50 (vector control), p164 (GCN4), or p238 (GCN4C, which constitutively expresses Gcn4p). CLS was measured in synthetic dextrose minimal media without (SD) or with the non-essential amino acids isoleucine, threonine, and valine (SD + ITV) added to final concentrations present in SC1 (see Table 2). Viability is expressed in terms of colony forming units (CFU) per ml of culture and is plotted as the log of the percent of viability on day 1.
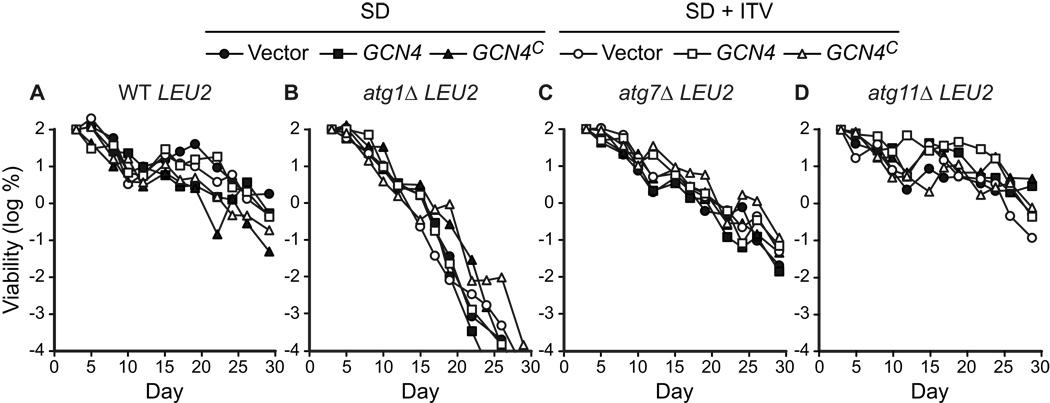
Figure 10.
LEU2 promotes chronological longevity and blocks the effects of constitutive expression of Gcn4p. LEU2 strains competent (WT, atg11Δ) or deficient (atg1Δ, atg7Δ) in autophagy were transformed with three plasmids: YEp50 (vector control), p164 (GCN4), or p238 (GCN4C, which constitutively expresses Gcn4p). The LEU2 strains YAA1, YAA3, YAA5, and YAA7 are described in Table 1. CLS was measured in synthetic dextrose minimal media without (SD) or with (SD + ITV) the non-essential amino acids isoleucine, threonine, and valine added to final concentrations present in SC1 (Table 2). Viability is expressed in terms of colony forming units (CFU) per ml of culture and is plotted as the log of the percent of viability on day 3.
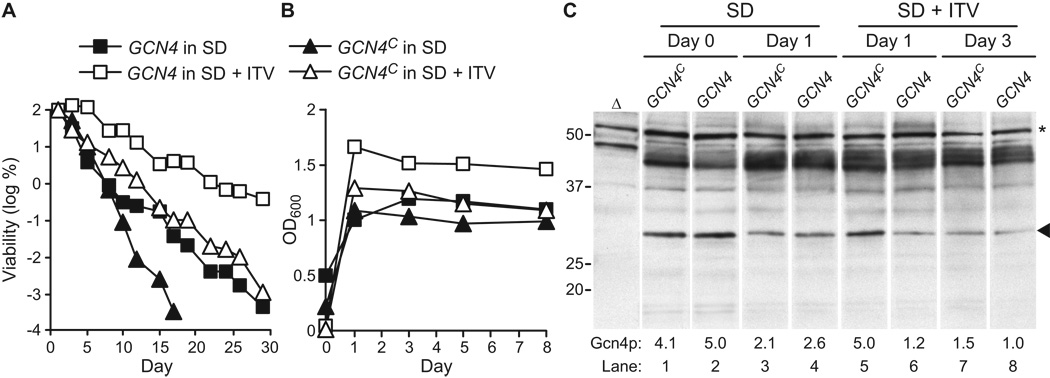
To confirm that GCN4 expression was downregulated is SD medium supplemented with isoleucine, threonine, and valine, Gcn4p protein levels were measured using western blotting. WT yeast were transformed with plasmids p238 (GCN4C) and p164 (GCN4) as described above and CLS data were collected. As expected, SD+ITV supported a longer CLS than unsupplemented SD, and constitutive expression of GCN4 from plasmid p238 suppressed this extension of CLS (Fig. 7A). In addition, constitutive expression of GCN4 reduced the CLS in SD medium (Fig. 7A), which is consistent with a smaller effect of the GCN4C allele observed in SD medium in Fig. 6A. Constitutive expression of GCN4 also suppressed the ~40% increase in cell density at saturation observed in SD+ITV medium (Fig. 7B).
Figure 7.
Non-essential amino acids isoleucine, threonine, and valine suppress GCN4 expression. The WT strain was transformed with plasmids p164 (GCN4) or p238 (GCN4C, which constitutively expresses Gcn4p) and CLS was measured in synthetic dextrose minimal medium without (SD) or with (SD + ITV) the non-essential amino acids isoleucine, threonine, and valine added to final concentrations listed for SC1 (Table 2). A. Viability is expressed in colony forming units (CFU) per ml of culture and is plotted as the log of the percent of viability on day 1. B. Cell density (OD600) was measured on days 0, 1, 3, 5, and 8. C. Measurement of Gcn4p levels was done during chronological aging. WT cells transformed with p164 (GCN4) or p238 (GCN4C) and grown in SD or SD + ITV were harvested on days 0, 1, or 3. Equal amounts of whole cell lysates (based on OD600 units) were analyzed by Western blotting with an affinity-purified antibody to Gcn4p (see Materials and Methods). The intensities of Gcn4p bands (arrowhead) were quantified using ImageJ software and normalized to the intensities of bands (asterisk) detected non-specifically in a gcn4Δ strain (Δ). Values for the normalized Gcn4p band intensities relative to lane 8 are shown below each lane (Gcn4p). Positions of molecular weight markers are shown on the left (in kDa).
Over the course of this CLS experiment, cells were harvested and total protein lysates were analyzed by western blotting with an anti-Gcn4p antibody. Because strains consistently grew more slowly in SD+ITV medium (OD600 = 0.01–0.06 on day 1) than in SD medium (OD600 = 0.2–0.5 on day 1), we compared samples from days 0 and 1 in SD medium to samples from days 1 and 3 in SD+ITV medium. On day 0 in SD medium, Gcn4p protein levels were similar in GCN4 and GCN4C transformants (Fig. 7C, lanes 1 and 2), indicating that GCN4 expression is fully induced in SD medium during mid-log to late-log growth phase. In freshly saturated cultures grown in SD+ITV, the GCN4C transformant contained a higher level of Gcn4p than the GCN4 transformant (Fig. 7C, lanes 5 and 6), indicating that GCN4 expression was repressed by the presence of isoleucine, threonine, and valine relative to constitutive expression. At later times, on day 1 in SD medium and on day 3 in SD+ITV medium, Gcn4p levels in the GCN4 and GCN4C transformants were similar (Fig. 7C, compare lane 3 to 4, and lane 7 to 8). However, Gcn4p protein levels for both transformants were higher in SD on day 1 than in SD+ITV on day 3 (Fig. 7C, compare lanes 3 and 4 to lanes 7 and 8), consistent with the notion that SD+ITV suppresses GCN4 expression and GAAC. Taken together, these data confirm that growth in SD induces expression of GCN4 and GAAC, and that supplemental non-essential isoleucine, threonine, and valine suppress GCN4 expression and GAAC.
Extra essential amino acids extend chronological longevity in autophagy-deficient yeast
The observation that non-essential amino acids modulated GAAC was unexpected. GAAC is best understood in terms of induction by limiting amounts of essential amino acids, which prompted us to ask: do essential amino acids affect chronological longevity? Most of the strains used above were in the BY4742 genetic background, which is auxotrophic for histidine, leucine, lysine, and uracil. Therefore, we assayed the chronological longevity of WT, atg1Δ, atg7Δ, and atg11Δ strains in synthetic minimal media with normal (Table 2) and three-fold elevated levels of essential amino acids and uracil. We observed that extra essential supplements extended the chronological longevity of these four strains (Fig. 8). However, only a relatively small extension of CLS was observed in autophagy-deficient strains. This extension was similar in extent to what was observed with addition of the individual non-essential amino acids isoleucine, threonine, or valine (compare Fig. 8 with Fig. 1, Fig.4, Fig.5). Thus, in autophagy-deficient strains, increased concentrations of all essential supplements resulted in an extension of CLS that was no greater than the extension observed with the non-essential amino acids isoleucine, threonine, and valine.
Figure 8.
Essential amino acids promote chronological longevity. The CLS of yeast competent (WT, atg11Δ) or deficient (atg1Δ, atg7Δ) in autophagy was measured in synthetic dextrose minimal medium containing histidine, lysine, leucine, and uracil supplements added to standard (SD) or three-fold elevated (3X HKLu) final concentrations as described in Table 2. Viability is expressed in terms of colony forming units (CFU) per ml of culture and is plotted as the log of the percent of viability on day 1.
To address the possibility that elevated levels of essential supplements promote chronological longevity by supporting additional cell growth prior to stationary phase, we compared the cell densities of cultures shown in Figure 8. We found that both longer-lived control (WT and atg11Δ) and shorter-lived autophagy-deficient (atg1Δ and atg7Δ) strains exhibited elevated cell densities in SD + 3X HKLu compared to SD (i.e., containing 1X HKLu). Specifically, all four strains attained OD600 = 2.1–2.3 at saturation on day 3 in SD + 3X HKLu whereas the same strains achieved OD600 = 0.9–1.0 at saturation on day 3 in SD. This indicates that the extended longevity in SD + 3X HKLu is not due to enhanced growth. Furthermore, the absence of a substantial effect on chronological longevity in the atg1Δ and atg7Δ strains despite the elevated cell density suggests that uptake and utilization of essential supplements does not compensate for the deficiency in autophagy in these strains.
Supplemental leucine and LEU2 prototrophy extend chronological longevity
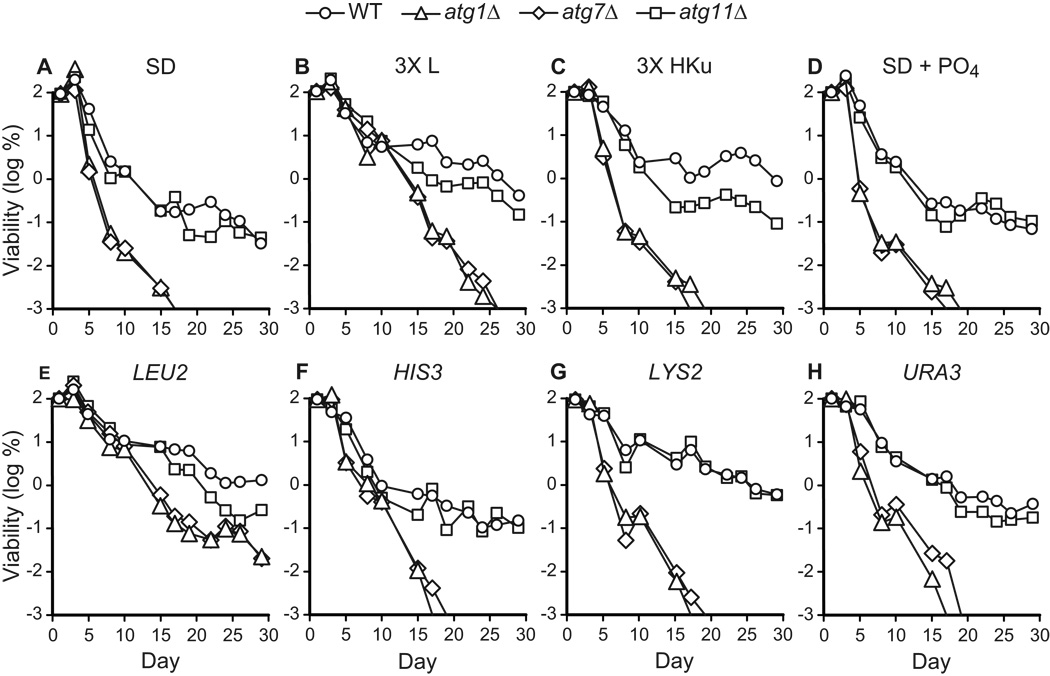
The fact that non-essential amino acids do not promote chronological longevity with equal efficacy raised the possibility that essential amino acids may also make different contributions to chronological longevity. This was tested in CLS studies comparing the effects of extra supplemental leucine, histidine, lysine, and uracil on chronological longevity (Fig. 9A–C). Extra supplemental leucine extended chronological longevity of both control and autophagy-deficient strains (Fig. 9B), whereas extra supplemental histidine, lysine, and uracil extended CLS in wild type, but not autophagy-deficient strains (Fig. 9C). The relative degree of extension of CLS in control and autophagy-deficient strains appears similar in media containing extra supplemental leucine (compare Fig. 9A and B), unlike elevated levels of all essential supplements, which appear to affect control strains more than autophagy-deficient strains (compare Fig. 8A and B). This may suggest that histidine, lysine, and uracil attenuate the effects of leucine. Nevertheless, our studies clearly show that leucine is the essential supplement that makes the greatest contribution to extension of CLS in control and autophagy-deficient strains.
Figure 9.
Leucine promotes chronological longevity. The CLS of autophagy-competent (WT, atg11Δ) or autophagy-deficient (atg1Δ, atg7Δ) yeast strains was measured in synthetic dextrose minimal media containing standard amounts of supplements (SD), a three-fold elevated level of leucine (3X L), a three-fold elevated levels of histidine, lysine, and uracil (3X HKu), or 10 mM dibasic potassium phosphate (SD + PO4) (Table 2). Transformants carrying low-copy number, centromeric plasmids pRS513 (HIS3), pRS315 (LEU2), pRS316 (URA3), or pRS317 (LYS2) were also analyzed. Viability is expressed in terms of colony forming units (CFU) per ml of culture and is plotted as the log of the percent of viability on day 1.
The concentrations of essential amino acids in these experiments reached 1–2 mM, which could potentially have affected CLS due to increased buffering capacity. To examine this possibility, 10 mM dibasic potassium phosphate (K2HPO4) was added to SD medium, raising the pH to ~7. However, supplemental buffering capacity had no effect on CLS (Fig. 9D).
Given these results, we wished to determine the relative importance of each of the nutritional auxotrophies in the BY4742 strain background. To examine nutritional auxotrophies, control and autophagy-deficient strains were transformed with stably-inherited centromeric plasmids carrying each of the corresponding auxotrophic markers (i.e., HIS3, LEU2, LYS2, or URA3). The LEU2 plasmid had the most pronounced effect on both wild type and autophagy deficient strains and extended CLS to the greatest extent (Fig. 9E), in agreement with the effects of additional supplemental leucine (Fig. 9B). Plasmids carrying HIS3 and URA3 were found to have a negligible effect on chronological longevity in both wild type and autophagy deficient strains (Fig. 9A, F, H). The plasmid bearing LYS2 promoted longevity in autophagy competent cells, but had no discernable effect on autophagy-deficient strains (Fig. 9A, G). We conclude that leucine availability, either through de novo biosynthesis or uptake from the environment, extends CLS in autophagy-competent and autophagy-deficient yeast. Lysine availability also appears to promote CLS, but in an autophagy-dependent manner.
Longevity conferred by LEU2 prototrophy is not suppressed by constitutive expression of Gcn4p
The extension of CLS observed in LEU2 strains or in media containing extra supplemental leucine suggested that leucine levels were limiting for BY4742-derived strains grown in SD medium, and that limiting leucine levels may activate the GAAC pathway. To test this, the effect of GCN4 expression on CLS was measured in a series of LEU2 strains. We carried out an experiment analogous to that shown in Figure 6, but used strains in which the chromosomal leu2Δ0 allele was restored to LEU2 (see Materials and Methods). This experiment revealed that both autophagy-competent and autophagy-deficient LEU2 stains showed no significant effect of supplemental isoleucine, threonine, and valine on CLS (Fig. 10). The WT and atg11Δ LEU2 transformants were substantially as long-lived in the absence of supplemental isoleucine, threonine, and valine as the corresponding leu2 transformants in the presence of the supplements (compare Fig. 10A, D to Fig. 6A, D). The LEU2 atg1Δ and atg7Δ autophagy-deficient strains exhibited chronological longevities greater than that observed with supplemental isoleucine, threonine, and valine (compare Fig. 10B, C with Fig. 6B, C). Leucine prototrophy extended CLS to a greater extent in the context of the atg7Δ mutation than in the context of the atg1Δ mutation in this experiment (Fig. 10B, C). However, such a difference was not observed in the previous experiment (Fig. 9A, E), which was done with different strains and media (i.e., with plasmid-transformed strains in medium containing uracil).
In addition, constitutive expression from the GCN4C allele did not have a pronounced effect on longevity in any of the LEU2 strains (Fig. 10). Transformation with each of the three plasmids (YEp50, p164, and p238) yielded comparable chronological longevities for each of the four strains (i.e., WT, atg1Δ, atg7Δ, and atg11Δ), unlike what is observed in the BY4742 (leu2) strain background (see Fig. 6). The only exception was observed in the WT LEU2 strain, in which a small decrease in CLS was observed (Fig. 10 A). These results agree with the western blotting results (Fig. 7C) and suggest that normal levels of supplemental leucine in synthetic media (Table 2) trigger a GAAC response in leu2 strains that reduces CLS.
Deletion of LEU3 restores chronological longevity
The observation that supplemental isoleucine, threonine, and valine extend CLS in a leu2 strain, but not in a LEU2 strain, suggested that these non-essential amino acids compensate in some manner for reduced availability of the essential amino acid leucine. One mechanism by which isoleucine, threonine, and valine may do this is by altering cross-pathway regulation within the superpathway for branched side chain amino acid biosynthesis. Transcription of multiple genes in this superpathway is regulated by LEU3, which encodes a zinc finger transcription factor (Kohlhaw, 2003; MacPherson et al., 2006). Transcription of LEU3 is subject to regulation by GAAC and Gcn4p. This raised the possibility that LEU3 is a relevant downstream target of GCN4, and that downregulation of LEU3 may mediate extension of CLS by isoleucine, threonine, and valine, which we have already shown involves GCN4.
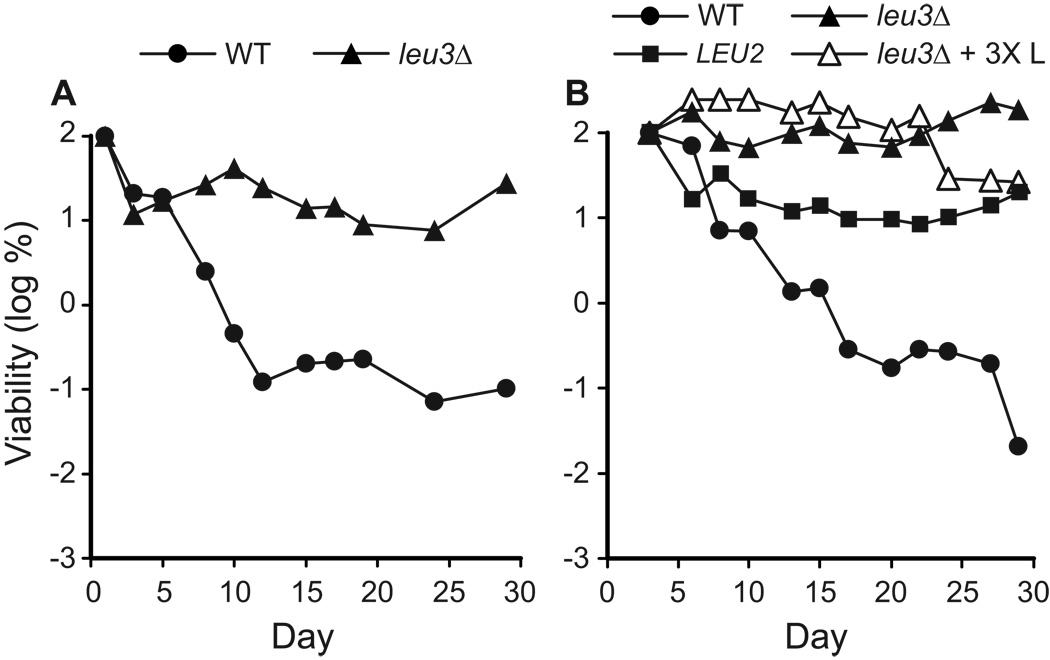
To test this possibility, we measured the CLS of a leu3Δ deletion strain. Although Leu3p is a potent transcription activator, it is not required for transcription of target genes, and low level transcription takes place in a leu3Δ strain (Brisco and Kohlhaw, 1990). Interestingly, deletion of LEU3 results in a profound extension of CLS (Fig. 11A). The longevity of the leu3Δ (leu2Δ) strain is greater than the longevity of LEU2 (LEU3) control strains (compare Fig. 11 to Fig. 9 and Fig.10), and supplemental leucine does not further extend CLS in the leu3Δ strain (Fig. 11B). leu3Δ strains grow more slowly than control strains, but reach cell densities comparable to LEU3 strains at saturation (OD600 values of ~1.5 and ~1.6, respectively, on day 3).
Figure 11.
Deletion of the regulatory gene LEU3 extends CLS. Control (WT), leu3Δ, and LEU2 (YAA1) strains (Table 1) were grown in synthetic dextrose minimal medium (SD) with standard or three-fold elevated levels of leucine (3X L) (Sherman, 2002, Table 2). CLS was measured and viability is expressed in terms of colony forming units (CFU) per ml of culture and is plotted as the log of the percent of viability on day 1 or day 3.
The observation that the leu3Δ0 mutation suppresses the life span shortening effects of the leu2Δ0 mutation is interesting because it indicates that the standard amount of leucine available in SD medium is sufficient for extended chronological longevity as long as Leu3p is absent. This implies that the underlying mechanism by which leucine availability affects CLS is regulatory in nature rather that strictly tied to leucine uptake, utilization, and metabolism. The extended chronological life span of the leu3Δ strain points to the possibility that branched side chain amino acid synthesis is misregulated in the BY4742 strain, likely as a result of the leu2Δ0 mutation, and that this imbalance adversely affects chronological life span (see Discussion).
Dynamics of induction of autophagy during early stationary phase
The findings presented above indicate that autophagy and leucine availability play key roles in chronological longevity in synthetic media. If so, then autophagy should be activated during chronological aging, and this activation should be influenced by amino acid availability. Also, autophagy should be induced even in the absence of amino acid limitation (i.e., because autophagy promotes longevity in LEU2 strains and in the presence of extra supplements). To test these predictions, we measured induction of autophagy during chronological aging. Autophagy was assayed using a GFP-Atg8p chimeric reporter protein that is proteolytically processed in an autophagy-dependent manner (Kim et al., 2001; Klionsky et al., 2007). Following induction of autophagy, the GFP-Atg8p reporter, like Atg8p, is conjugated at its C-terminus to phosphatidylethanolamine and is tethered to the autophagosomal membrane. GFPAtg8p bound to the internal membrane of the autophagosome is delivered to the vacuole interior where Atg8p is degraded. GFP is relatively resistant to degradation and persists in the vacuole. Thus, the appearance of a GFP band on a western blot is an indirect, yet reliable marker for autophagic activation (Klionsky et al., 2007).
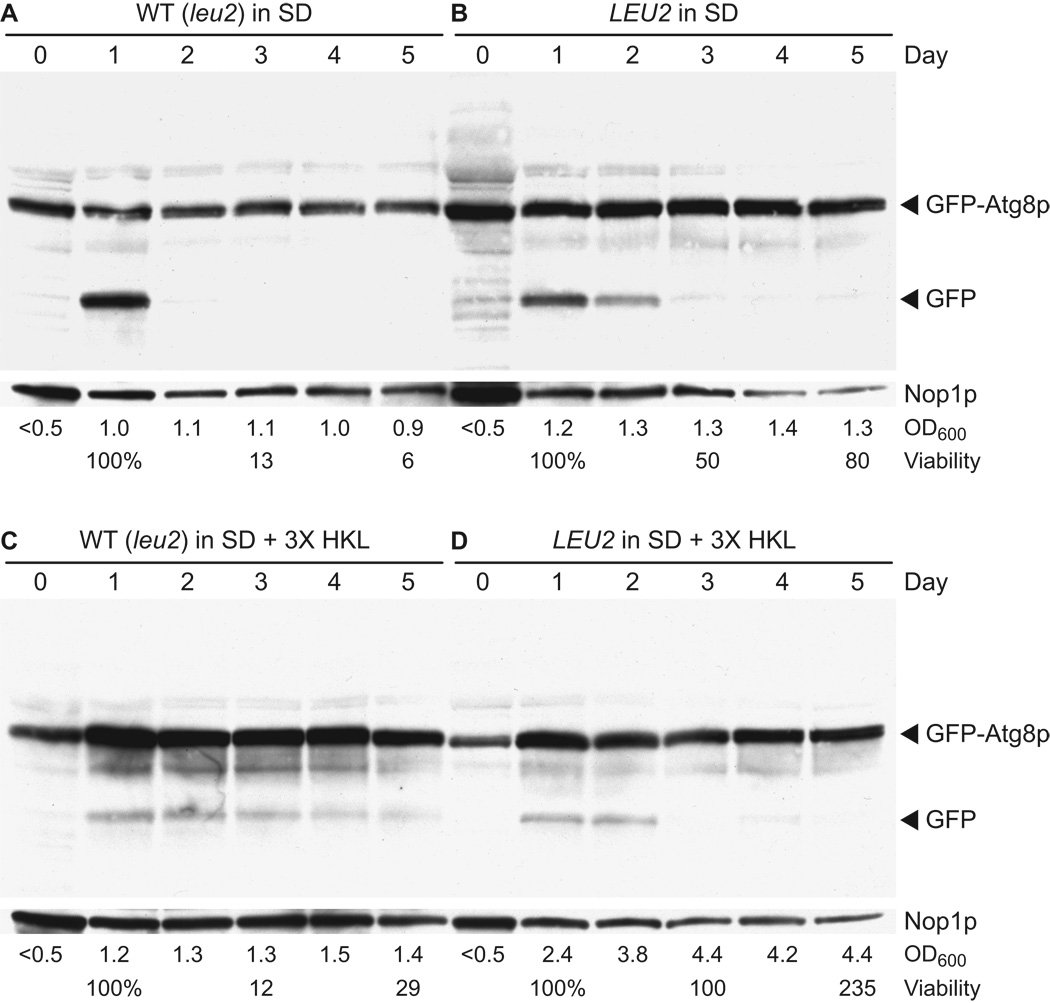
Autophagy was assayed in the WT (leu2Δ) strain and the WT LEU2 strain following transformation with pCuGFPAUT7(416) (Kim et al., 2001) and growth in minimal media with standard amounts of supplements (SD) or with three-fold increased amounts of histidine, lysine, and leucine (SD + 3X HKL). A standard CLS experiment was done as described above. Cultures reached saturation on day 1 (OD600 = 1.0–1.2), with the exception of YAA1 in SD + 3X HKL, which reached saturation on day 3 (OD600 = 4.4). CFU viability was measured on days 1, 3, and 5 to confirm the expected patterns of chronological aging for these strains and growth conditions. Cells were collected on days 0, 1, 2, 3, 4, and 5, and whole cell extracts were prepared and analyzed by western blot detection of GFP.
No evidence of autophagy (i.e., no GFP band) was visible in any sample on day 0 during logarithmic growth (Fig. 12). On day 1, all samples showed evidence of induction of autophagy, although the extent varied. The most prominent GFP band appeared on day 1 in the WT strain in SD medium containing standard amounts of supplements (Fig. 12A). GFP was barely or not detectable on days 2–5 in the WT strain in SD medium (Fig. 12A). The level of induction of autophagy on day 1 in panel A is similar to what is observed following transfer of logarithmic phase yeast to starvation media lacking ammonium sulfate and amino acids (data not shown). The LEU2 strain showed upregulation of autophagy on day 1 in SD medium, persistence of the GFP band on day 2, after which the GFP band remained visible at a low level through day 5 (Fig. 12 B). In medium containing extra supplemental amino acids (SD + 3X HKL), both the WT and LEU2 strains showed a lower level of activation of autophagy on day 1 (Fig. 12C, D) compared to standard SD medium (Fig. 12A, B). The WT strain grown in SD + 3X HKL showed the most uniformly persistent activation of autophagy though day 5 (Fig. 12C). Autophagy in the LEU2 strain grown in SD + 3X HKL was activated following log phase growth on days 1 and 2 prior to saturation on day 3, after which a low level of autophagy was observed (Fig. 12D).
Figure 12.
Induction of autophagy during chronological aging. Control (WT) and LEU2 (YAA1) strains were transformed with plasmid pCuGFPAUT7(416) (Kim et al., 2001), which expresses a GFP-Atg8p fusion protein that is proteolytically processed in an autophagy-dependent manner to yield GFP. Transformants were grown in synthetic dextrose minimal medium containing standard amounts of the essential amino acids histidine, lysine, and leucine (SD) or containing three-fold increased amounts of the same supplements (SD + 3X HKL) as described in Table 2. Protein lysates from cells collected on days 0–5 were analyzed by western blotting using a GFP-specific polyclonal antibody (see Materials and Methods). Blots were reprobed with monoclonal antibody directed against the nucleolar protein Nop1p, which served as a loading control. Cell density (OD600) and percent viability values are shown below the lane corresponding to the day on which data were collected.
We conclude that autophagy is induced following logarithmic growth as yeast make the transition into stationary phase, and that the degree of autophagic induction correlates inversely with the availability of essential amino acids. Activation was strongest in the leu2Δ control strain in standard SD medium (Fig. 12A) and weakest in the LEU2 strain in SD medium with elevated supplemental amino acids (Fig. 12D). However, it is important to note that autophagy is observed even under conditions in which essential amino acids are plentiful (Fig. 12D), suggesting that its activation is tied to metabolic changes associated with stationary phase survival rather than nitrogen starvation per se. Finally, the duration of autophagic activation correlates with CLS. The shortest chronological life span follows a transient burst of autophagy (Fig. 12A); the longest life span is associated with a long duration of low level autophagy activity (Fig. 12D).
Discussion
We have investigated the role of autophagy and amino acid homeostasis during chronological aging in yeast. We find that autophagy is required for the chronological longevity of yeast grown in synthetic dextrose (SD) minimal and synthetic complete (SC) media, but not in non-defined rich (YPD) medium. The requirement for autophagy in synthetic media likely reflects the importance of this recycling pathway for amino acid homeostasis. Autophagy is well-known as a mechanism by which amino acid homeostasis is maintained during cell growth and aging (Droge, 2004). However, autophagy may also be required for remodeling of the yeast proteome during the transition from a growth-based metabolism to a survival-based metabolism, and for organellar remodeling, which involves removal of organelles that become damaged or functionally superfluous (Dunn et al., 2005). Thus, although our studies demonstrate the importance of autophagy and amino acid homeostasis for chronological longevity, we recognize that “non-nutritional” functions of autophagy may also be important.
The absence of a requirement for autophagy in YPD has been reported by other laboratories, and likely reflects the abundance of alternative nitrogen and carbon sources in YPD medium. Powers et al conducted a genome-wide survey of viable single gene deletion mutations and found no significant effect of atg1Δ and atg7Δ mutations on CLS in YPD (Powers et al., 2006). This agrees with the fact that autophagy is known to be down-regulated under conditions in which nitrogen and amino acids are amply available, and the fact that YPD is known to suppress autophagy in yeast because of a reduced demand for amino acid recycling (Klionsky et al., 2007).
Although YPD is commonly used for CLS experiments, SD medium is a viable alternative that provides an opportunity to test the importance of nutritional factors during chronological aging. Our studies show that SD supplemented with sufficient essential amino acids or specific non-essential amino acids support CLS to a degree similar to YPD. Moreover, certain mutant strains (e.g., a leu3Δ strain) exhibit a long CLS in SD medium. In addition, SD medium containing 1.8% glucose and 0.2% lactate supports a CLS comparable to that observed in YPD (data not shown). These results indicate that a rich growth environment containing many non-essential components is not required for chronological longevity. This is not altogether surprising. In the natural environment, where a wide range of complex and alternative carbon and nitrogen sources are not necessarily available, wild yeast synthesize metabolites from simple carbon and nitrogen sources. As a result, yeast have evolved mechanisms to survive in the non-dividing state under conditions that are seldom as salubrious as YPD broth. Because the composition of minimal media resembles conditions in the natural environment, it should prove useful for investigating such survival mechanisms that promote chronological longevity.
It is clear that low levels of essential amino acids reduce CLS. We have shown that CLS is most sensitive to levels of the essential amino acid leucine; CLS is extended by both supplemental leucine and leucine prototrophy (i.e., conversion of leu2 to LEU2). Gomes et al reached similar conclusions by demonstrating that medium with five-fold higher than normal levels of essential amino acids extended life span (Gomes et al., 2007). leu2 auxotrophs grown in batch culture under conditions that are limiting for leucine exhibit a range of stress responses (Saldanha et al., 2004) and have reduced survival during chronological aging (Boer et al., 2008). Furthermore, leucine uptake is influenced by the composition of growth media, which may explain differences in CLS in the two synthetic complete (SC) media we have used. In particular, BY4741-based yeast strains have been found to exhibit a growth defect on SC agar medium prepared according to Fink (Amberg et al., 2005; Styles, 2002), but not on SC agar medium prepared according to Sherman (Sherman, 2002), which has also been referred to as SMM (Amberg et al., 2005), and this defect was attributed to a specific impairment in leucine uptake (Cohen and Engelberg, 2007). In the Results section, we refer to the Fink and Sherman media as SC2 and SC1, respectively (see Table 2). We observed rapid aging in SC2, but not SC1, which correlates with the leucine uptake defect in BY4741 strains, which are congenic with the BY4742 strains used in our experiments. Thus, the effects of amino acids on CLS likely reflect both the concentrations of amino acids in, and the efficiency of uptake of amino acids from, the external environment.
One of the most intriguing results that emerged in our studies was the extension of CLS by non-essential amino acids, notably isoleucine, its precursor threonine, and valine. We found that these amino acids influence CLS via the general amino acid control (GAAC) pathway, which regulates cellular amino acid homeostasis at a global level (reviewed in Hinnebusch, 2005). Specifically, increased availability of isoleucine, threonine, and valine extends CLS and suppresses expression of GCN4, which encodes a key transcriptional regulator of GAAC. Conversely, constitutively high level expression of GCN4 counteracts extension of CLS. This reveals a general relationship between GAAC and CLS. That is, induction of GAAC shortens CLS, while suppression of GAAC lengthens CLS. However, this relationship does not appear to extend to inactivation of GAAC, because deletion of GCN4 does not extend CLS (data not shown). This suggests that GCN4 function, perhaps at low levels, is required for chronological longevity during suppression of GAAC. Similarly, deletion of GCN4 does not affect replicative life span, but GCN4 is required for extension of replicative life span in specific 60S ribosomal protein mutants and during dietary restriction (Steffen et al., 2008).
The fact that leucine, isoleucine, and valine, were most important for CLS points to a special status for the branched side chain amino acids during chronological aging in minimal media. Consistent with this, we find that deletion of LEU3 extends CLS. LEU3 encodes the primary transcriptional regulator of the superpathway for branched side chain amino acid biosynthesis (Boer et al., 2005; Harbison et al., 2004; Kohlhaw, 2003; Tang et al., 2006). LEU3 is also subject to regulation by Gcn4p and GAAC. This suggests the existence of a more elaborate relationship between branched side chain amino acids, GAAC, and CLS. Our interpretation is that reduced levels of branched side chain amino acids induces GAAC and shortens CLS, while increased levels of branched side chain amino acids suppresses GAAC and lengthens CLS. Furthermore, balanced synthesis of branched side chain amino acids appears to be an important factor. Balanced synthesis of isoleucine, leucine, and valine depends on the extent to which the level of alpha-isopropylmalate (αIPM), an intermediate the leucine pathway, accurately reflects the levels of intermediates in the isoleucine and valine pathways. αIPM is a transcriptional coactivator for Leu3p and is the only intermediate in branched side chain amino acid synthesis that functions as such (Kohlhaw, 2003). In wild type cells, αIPM levels generally reflect metabolic flow through all three pathways (Kohlhaw, 2003). However, in a leu2 strain, αIPM levels may be altered. Leu2p functions immediately downstream of Leu1p, which consumes αIPM and is subject to feedback inhibition. This may cause a regulatory imbalance in the synthesis of branched side chain amino acids, which may be exacerbated by upregulation of GAAC and LEU3 during chronological aging in minimal media. Deletion of LEU3 may relieve such an imbalance. In the absence of Leu3p, transcription of target genes takes place at a basal level (Brisco and Kohlhaw, 1990), resulting in reduced, but balanced levels of branched side chain amino acids, which may underlie suppression of GAAC and extension of CLS.
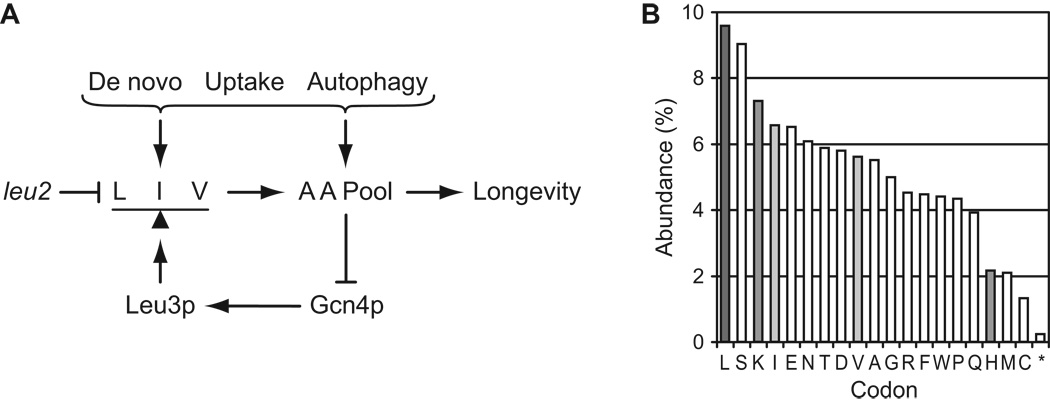
Our studies highlight relationships between autophagy, amino acid homeostasis, and chronological longevity that are summarized in Fig. 13A. Given the key role of branched side chain amino acids in determining CLS, we wondered: why are branched side chain amino acids more important than other amino acids? The simplest answer is that low levels of branched side chain amino acids have a disproportionate effect on protein synthesis. Consistent with this, leucine codons are the most numerous among the annotated open reading frames (ORFs) in the yeast genome (Fig. 13B). The relative abundance of leucine codons may cause yeast to be especially sensitive to reduced leucine availability. Of the other essential amino acids that we tested, histidine availability had relatively little effect on CLS, and histidine codons are relatively rare among yeast ORFs (Fig. 13B). Lysine availability exerted a greater effect than histidine on CLS, and lysine codons are more numerous than histidine codons (Fig. 13B). Isoleucine and valine are among the most abundant codons (Fig. 13B). Thus, low levels of branched side chain amino acids may directly affect CLS by inhibiting synthesis of proteins required for stationary phase survival, or may indirectly affect CLS indirectly via GAAC, as discussed above.
Figure 13.
Amino acid homeostasis and chronological longevity in yeast. A. Model highlighting relationships between amino acid homeostasis and chronological longevity in yeast. De novo synthesis, uptake, and autophagy contribute to the cellular amino acid (AA) pools, including the branched side chain amino acids leucine (L), isoleucine (I), and valine (V). The superpathway for branched side chain amino acids is regulated by the Leu3p transcription factor, which functions to balance synthesis of L, I, and V. Yeast strains containing a leu2 mutation are hypothesized to misregulate this superpathway in a manner involving Leu3p. Branched side chain amino acids from uptake or autophagic recycling compensate for superpathway misregulation in a manner dependent on Gcn4p, which mediates general amino acid control and regulates levels of Leu3p. Not depicted are pathways by which Gcn4p regulates de novo synthesis and uptake of amino acids and autophagy. Panel B. Bar graph of codon abundance in yeast (in percent of total codons). Data were derived from a codon usage table generated by CUSP (EMBOSS software suite). The FASTA dataset for ORFs (not including dubious ORFs and pseudogenes) was obtained from the Saccharomyces Genome Database. Similar results were obtained using the FASTA dataset for all ORFs, or publicly available codon usage tables prepared by J. M. Cherry that were generated using the GCG program CodonFrequency based on ORFs listed within SGD as of January 1999 (data not shown).
In summary, our findings reveal the importance of amino acid homeostasis in chronological aging and highlight the roles for autophagy and GAAC. These studies are the first to demonstrate that autophagy and supplemental non-essential amino acids can extend CLS. Furthermore, we suggest that autophagy promotes longevity by providing the cell with recycled amino acids. Thus, the aged cell is reliant on autophagy and cellular amino acid pools for survival during chronological aging.
Acknowledgements
The autophagy mutants in strain BY4742 and plasmid pCuGFPAUT7(416) were kindly provided by Daniel Klionsky (University of Michigan). Autophagy mutants in the W303 strain background were generously provided by Sebastian Schuck and Peter Walter (University of California, San Francisco). Plasmids p164 and p238 and the affinity purified antibody to Gcn4p were provided by Alan Hinnebusch (National Institute of Child Health and Human Development, NIH). Michael Cherry (Stanford University) provided assistance with codon abundance analysis. Access to the online EMBOSS molecular biology software suite was provided by the Interdisciplinary Center for Biotechnology Research at the University of Florida. This study was supported by NIH grant R21 AG023719 to JPA.
Abbreviations
- CLS
chronological life span
- GAAC
general amino acid control
References
- Allen C, Buttner S, Aragon AD, Thomas JA, Meirelles O, Jaetao JE, Benn D, Ruby SW, Veenhuis M, Madeo F, Werner-Washburne M. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J Cell Biol. 2006;174:89–100. doi: 10.1083/jcb.200604072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. p. 230. [Google Scholar]
- Aris JP, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- Boer VM, Amini S, Botstein D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci U S A. 2008;105:6930–6935. doi: 10.1073/pnas.0802601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer VM, Daran JM, Almering MJ, de Winde JH, Pronk JT. Contribution of the Saccharomyces cerevisiae transcriptional regulator Leu3p to physiology and gene expression in nitrogen- and carbon-limited chemostat cultures. FEMS Yeast Res. 2005;5:885–897. doi: 10.1016/j.femsyr.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brisco PR, Kohlhaw GB. Regulation of yeast LEU2. Total deletion of regulatory gene LEU3 unmasks GCN4-dependent basal level expression of LEU2. J Biol Chem. 1990;265:11667–11675. [PubMed] [Google Scholar]
- Chen Q, Thorpe J, Ding Q, El-Amouri IS, Keller JN. Proteasome synthesis and assembly are required for survival during stationary phase. Free Radic Biol Med. 2004;37:859–868. doi: 10.1016/j.freeradbiomed.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Chen Q, Thorpe J, Dohmen JR, Li F, Keller JN. Ump1 extends yeast lifespan and enhances viability during oxidative stress: central role for the proteasome? Free Radic Biol Med. 2006;40:120–126. doi: 10.1016/j.freeradbiomed.2005.08.048. [DOI] [PubMed] [Google Scholar]
- Cohen R, Engelberg D. Commonly used Saccharomyces cerevisiae strains (e.g. BY4741, W303) are growth sensitive on synthetic complete medium due to poor leucine uptake. FEMS Microbiol Lett. 2007;273:239–243. doi: 10.1111/j.1574-6968.2007.00798.x. [DOI] [PubMed] [Google Scholar]
- Droge W. Autophagy and aging--importance of amino acid levels. Mech Ageing Dev. 2004;125:161–168. doi: 10.1016/j.mad.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Dunn WA, Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gomes P, Sampaio-Marques B, Ludovico P, Rodrigues F, Leao C. Low auxotrophy-complementing amino acid concentrations reduce yeast chronological life span. Mech Ageing Dev. 2007;128:383–391. doi: 10.1016/j.mad.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. "Sleeping beauty": quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, Klionsky DJ. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman PK. Stationary phase in yeast. Curr Opin Microbiol. 2002;5:602–607. doi: 10.1016/s1369-5274(02)00377-6. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hung GC, Brown CR, Wolfe AB, Liu J, Chiang HL. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J Biol Chem. 2004;279:49138–49150. doi: 10.1074/jbc.M404544200. [DOI] [PubMed] [Google Scholar]
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Neufeld TP. Drosophila Atg7: required for stress resistance, longevity and neuronal homeostasis, but not for metamorphosis. Autophagy. 2008;4:357–358. doi: 10.4161/auto.5572. [DOI] [PubMed] [Google Scholar]
- Kim J, Huang WP, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I, Salin B, Schaeffer J, Bhatia S, Manon S, Camougrand N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007:3. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Kohlhaw GB. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol Mol Biol Rev. 2003;67:1–15. doi: 10.1128/MMBR.67.1.1-15.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Thompson CB. Macroautophagy versus mitochondrial autophagy: a question of fate? Cell Death Differ. 2005;12 Suppl 2:1484–1489. doi: 10.1038/sj.cdd.4401780. [DOI] [PubMed] [Google Scholar]
- Kvam E, Goldfarb DS. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 2007;3:85–92. doi: 10.4161/auto.3586. [DOI] [PubMed] [Google Scholar]
- MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard PJ, Roth BL, Thi HP, Yue ST, Haugland RP. Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl Environ Microbiol. 1997;63:2897–2905. doi: 10.1128/aem.63.7.2897-2905.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnson JR. Life spans of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Mueller PP, Hinnebusch AG. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986;45:201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280:41785–41788. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- Niederberger P, Miozzari G, Hutter R. Biological role of the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:584–593. doi: 10.1128/mcb.1.7.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. Bethesda, MD, USA: US National Institutes of Health; 2008. [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ, Brauer MJ, Botstein D. Nutritional homeostasis in batch and steady-state culture of yeast. Mol Biol Cell. 2004;15:4089–4104. doi: 10.1091/mbc.E04-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast life span extension by depletion of 60S ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan JS, Herman PK. The regulation of autophagy in eukaryotic cells: do all roads pass through Atg1? Autophagy. 2006;2:146–148. doi: 10.4161/auto.2.2.2485. [DOI] [PubMed] [Google Scholar]
- Styles C. How to set up a yeast laboratory. Methods Enzymol. 2002;350:42–71. doi: 10.1016/s0076-6879(02)50955-1. [DOI] [PubMed] [Google Scholar]
- Tang L, Liu X, Clarke ND. Inferring direct regulatory targets from expression and genome location analyses: a comparison of transcription factor deletion and overexpression. BMC Genomics. 2006;7:215. doi: 10.1186/1471-2164-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, Vellai T. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth SE, Julian D, Akin DE, Fried J, Toscano K, Leeuwenburgh C, Dunn WA., Jr Autophagy in the heart and liver during normal aging and calorie restriction. Rejuvenation Res. 2007;10:281–292. doi: 10.1089/rej.2006.0535. [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gaur NA, Hasek J, Kim SJ, Qiu H, Swanson MJ, Hinnebusch AG. Disrupting vesicular trafficking at the endosome attenuates transcriptional activation by Gcn4. Mol Cell Biol. 2008;28:6796–6818. doi: 10.1128/MCB.00800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]