Abstract
BACKGROUND
Stromal-epithelial signaling plays an important role in prostate development and cancer progression. Study of these interactions will be facilitated by the use of suitable prostate cell lines in appropriate model systems.
METHODS
We have isolated an immortalized prostate mesenchymal cell line from the mouse E16 urogenital sinus (UGS). We characterized its expression of stromal differentiation markers, response to androgen stimulation, ability to induce and participate in prostate morphogenesis, response to Shh stimulation, and interaction with prostate epithelial cells.
RESULTS
UGSM-2 cells express vimentin and smooth muscle actin, but not the mature smooth muscle markers myosin and desmin. This expression profile is consistent with a myofibroblast phenotype. Unlike other fibroblasts such as 3T3, UGSM-2 cells express androgen receptor mRNA and androgen stimulation increases proliferation. UGSM-2 cells are viable when grafted with embryonic UGS under the renal capsule and participate in glandular morphogenesis, but are not capable of inducing prostate morphogenesis of isolated UGS epithelium. Co-culture of UGSM-2 cells with human BPH-1 cells or co-grafting in vivo results in organized clusters of BPH-1 cells surrounded by a mantle of UGSM-2 cells. UGSM-2 cells are responsive to Sonic hedgehog (Shh), an important signaling factor in prostate development, and mimic the transcriptional response of the intact UGS mesenchyme. In co-cultures with BPH-1, UGSM-2 cells exhibit a robust transcriptional response to Shh secreted by BPH-1.
CONCLUSIONS
UGSM-2 is a urogenital sinus mesenchyme cell line that can be used to study stromal-epithelial interactions that are important in prostate biology.
Keywords: stromal-epithelial interactions, androgen, Sonic Hedgehog, prostate development, mesenchyme
INTRODUCTION
The prostate develops from a specific region of the endodermal urogenital sinus (UGS) termed the prostatic anlagen. Formation of the prostatic ducts begins at embryonic day 17 (E17) in the mouse when epithelial buds evaginate into the surrounding mesenchymal sheath. Discrete groups of buds define the origins of the anterior, dorsal and ventral lobes of the prostate. At the time of ductal budding, the UGS mesenchyme is composed of undifferentiated fibroblasts and myofibroblasts. As the buds elongate, they lumenalize to form true secretory ducts connected to the urethral lumen and branch to form a highly complex ductal tree. As the ducts grow, they are surrounded by a sheath of mesenchyme which differentiates to a periductal stroma comprised of smooth muscle cells and fibroblasts (1).
The embryonic mesenchyme and its adult descendent stroma have emerged as key regulators of prostatic growth and differentiation. In the UGS, mesenchymal cells express androgen receptors and act under the influence of androgens to induce prostatic differentiation of the endodermal epithelium (2,3). Tissue recombination experiments have shown that the mesenchyme is the primary determinant of epithelial growth and differentiation (4). In the adult prostate, there is regional heterogeneity within the ducts: the distal tips are encased in a delicate fibroblastic sheath, while the more proximal segments are surrounded by thicker sheaths rich in smooth muscle (5). Androgen receptor expression is localized to the dense smooth muscle sheath surrounding epithelial ducts, whereas fibroblasts rarely express androgen receptors (6). Smooth muscle is required for maintenance of epithelial secretory function (7) and loss of smooth muscle in the adult prostate is associated with cancer lesions and de-differentiation of epithelium (8).
Primary stromal cells from human prostate tissue have been used to discover factors that regulate smooth muscle differentiation and proliferation of prostate stroma, and to identify stromal-derived factors that regulate epithelial functions. Several prostate stromal cell lines have been generated, including rat NbF-I, mouse PSMC1, rat PS-1, human WPMY-1, human DuK50 and human PS30 cells (9–14). Two rat UGS mesenchymal cell lines have been generated: rUGM and U4F1 (15,16). To our knowledge, none of these cell lines is able to induce or participate in prostate morphogenesis.
The signaling interactions that regulate prostate ductal budding and branching morphogenesis have received considerable attention as the paradigm for understanding normal prostate growth regulation. These studies have demonstrated that the UGS mesenchyme is the target of several key signals, including testosterone, estrogen and sonic hedgehog (17–19). UGS mesenchyme is also the origin of several key morphogens including BMP-4, FGF-10, TGFβ (20–23) and Shh target genes such as IGFBP-6 (Lipinski et al., submitted), which may regulate both epithelial and mesenchymal proliferation and differentiation. The complexity of these interactions is daunting. For the Shh pathway alone, there are three different Gli genes expressed in the UGS mesencyhme and each of these plays a unique role in the transcriptional response to Hh signaling (18). Similar complexities exist in the multiplicity of receptor subtypes for BMP, TGFβ and FGF signaling. To elucidate the complex regulation and crosstalk between these pathways in mesenchymal cells, we have developed an immortalized UGS mesenchymal cell line and demonstrated that it phenocopies the UGS mesenchyme response to Shh stimulation.
Several unique characteristics distinguish the mesenchyme of the urogenital sinus mesenchyme. These include responsiveness to androgen, the ability to induce prostate differentiation of isolated urogenital sinus epithelium, and responsiveness to morphogens such as Sonic hedgehog. UGSM-2 cells were found to be androgen responsive and to mimic the canonical response of urogenital sinus mesenchyme to Sonic Hedgehog. UGSM-2 cells did not induce morphogenesis of isolated UGS epithelium sheets, but when grafted together with the E16 UGS they did proliferate and become incorporated into the periductal stroma during glandular morphogenesis.
MATERIALS AND METHODS
Animals and Cell Lines
Balb/c 3T3 fibroblasts were obtained from ATCC and cultured according to ATCC guidelines. BPH-1 cells were obtained from Simon Hayward (Vanderbilt University, Nashville, TN) and maintained in RPMI + 25mM HEPES + 10% FBS. UGSM-2 cells were maintained in DMEM/F12 + ITS + 10% FBS + 10−8M DHT. Wild-type CD-1 and CD-1 nude mice were obtained from Charles River (Wilmington, MA). INK4a−/−, β-actin-tva transgenic mice were obtained from Bart Williams (Van Andel Research Inst, Grand Rapids, MI). All animals were housed according to institutional animal use and care guidelines.
Isolation of UGSM-2 Cells
Immortalized UGSM-2 cells were derived from the urogenital sinus of an E16 male INK4a−/− tva transgenic mouse embryo. INK4a−/−, β-actin-tva transgenic mice were provided by Bart Williams (Van Andel Research Institute, Grand Rapids, MI). UGS epithelium was separated from mesenchyme following trypsin digestion as described previously (24). Mesenchyme was further dissociated into single cells by digestion in 0.5% collagenase. Dissociated mesenchymal cells were grown in DMEM + 15% FBS + 1% pen/strep until they reached confluence in a 6-well plate. Thereafter cells were grown in DMEM/F12 + 10% FBS + 1% pen/strep + 1% ITS + 10−8M DHT (INK4 culture medium). The UGSM-2 clone was isolated from the mixed UGSM population by dilution cloning followed by ring cloning.
Growth Curve Analysis
UGSM-2 and 3T3 cells were plated at a density of 4×104 cells per well in 6-well plates in normal culture media containing 10% charcoal-stripped, dextran-treated fetal bovine serum, csFBS (Hyclone, Logan, UT). After 48 hours, cells were treated with 10−8M R1881 or 0.1% ethanol in normal culture media containing 10% csFBS. Each day, cells were trypsinized, diluted 1:100 in Isoton II solution (Beckman, Fullerton, CA) and counted in triplicate using a ViCell XR viable cell counter (Beckman Coulter). No significant difference in cell viabilities between treatments was noted. Doubling time was calculated by determining the time required to double the number of cells in linear mid-log phase.
Ploidy Analysis
UGSM-2 cells were determined to be tetraploid by comparison to ploidy number of known diploid cells: freshly isolated splenocytes from the spleen of a CD-1 mouse (Charles River, Wilmington, MA). Splenocytes and UGSM-2 cells were combined in the following 3 ways: (1) 2×106 splenocytes, (2) 0.5×106 splenocytes + 2×106 UGSM-2 cells, and (3) 2×106 UGSM-2 cells. Cells were pelleted and fixed in ice cold 70% ethanol for 30 minutes. Cells were then pelleted and resuspended in 33 ug/ml propidium iodide + 1 mg/ml RNase A + 0.2% Nonidet P-40 in PBS. DNA content of cells was determined using a FACScan cytometer and analyzed using ModFitLT V3.0 software.
Immunocytochemistry
UGSM-2 cells were grown on Lab-Tek II chamber slides (Fisher, Pittsburgh, PA) and immunostained for vimentin, smooth muscle actin (SMA) or pan-cytokeratin (pan-CK). Slides were fixed in 4% paraformaldehyde and blocked in 5% normal goat serum in PBS. Anti-vimentin clone LN-6 (Sigma, St. Louis, MO), anti-smooth muscle actin monoclonal antibody clone 1A4 (Sigma) or anti-pan-cytokeratin monoclonal antibody (Zymed, South San Francisco, CA) was applied at a dilution of 1:200. Staining was visualized by incubating with goat anti-mouse Alexa 546 conjugated antibody (Molecular Probes, Eugene, OR) at a dilution of 1:200. Slides were mounted with Vectashield Hardset + DAPI mounting media (Vector, Burlingame, CA) and imaged using an Olympus model BX51 fluorescent microscope and Spot Advanced software v. 3.5.2.
RT-PCR
RNA was isolated from confluent cells using RNeasy mini kit (Qiagen, Valencia, CA) with optional on-column DNase digestion to eliminate contaminating DNA. 1 ug of total RNA was reverse transcribed to generate cDNA using M-MLV reverse transcriptase (Invitrogen). Relative mRNA quantity was determined by real-time RT-PCR using iCycler instrumentation and software (BioRad, Hercules, CA). Primer sequences are listed in Table 1. Primer sets whose name starts with ‘m’ are mouse-specific, while primer sets whose name starts with ‘h’ are human-specific. All sequences are listed in 5′ to 3′ orientation.
Table 1.
Sequences of RT-PCR primers.
| TARGET GENE | FORWARD PRIMER | REVERSE PRIMER |
|---|---|---|
| mGAPDH | AGCCTCGTCCCGTAGACAAAAT | CCGTGAGTGGAGTCATACTGGA |
| mSMA | ATCATGCGTCTGGACTTGG | AATAGCCACGCTCAGTCAGG |
| mVim | CCCCCTTCCTCACTTCTTTC | AAGAGTGGCAGAGGACTGGA |
| mDesmin | GTGAAGATGGCCTTGGATGT | TTGAGAGCAGAGAAGGTCTGG |
| mHCM | GCAGCTTCTACAGGCAAACC | CAAAGCGAGAGGAGTTGTCG |
| mAR | GTGAAGCAGGTAGCTCTGGG | GAGCCAGCGGAAAGTTGTAG |
| mCD31 | CTGAGCCTAGTGTGGAAGCC | TACATCCATGTTCTGGGGGT |
| mGli1 | GGAAGTCCTATTCACGCCTTGA | CAACCTTCTTGCTCACACATGTAAG |
| mPtc1 | CTCTGGAGCAGATTTCCAAGG | TGCCGCAGTTCTTTTGAATG |
| mIGFBP6 | AGCTCCAGACTGAGGTCTTCC | GAACGACACTGCTGCTTGC |
| mHIP | CCTGTCGAGGCTACTTTTCG | TCCATTGTGAGTCTGGGTCA |
| hGAPDH | CCACATCGCTCAGACACCAT | GCAACAATATCCACTTTACCAGAGTTAA |
| hGli1 | AATGCTGCCATGGATGCTAGA | GAGTATCAGTAGGTGGGAAGTCCATAT |
| hPtc1 | CGCTGGGACTGCTCCAAGT | GAGTTGTTGCAGCGTTAAAGGAA |
Co-cultures
UGSM-2 and BPH-1 cells were plated at equal densities (1×106 cells each) in 25cm2 flasks coated with neutralized rat tail collagen (25). Morphology of cells was observed and photographed over a 1-week period using a Nikon Eclipse TS100 inverted light microscope with a Spot Insight QE digital camera. RNA was prepared from 48 hour co-cultures as described above. Expression of Shh signaling targets Gli1 and Ptc1 was examined by RT-PCR.
Renal Capsule Grafts
For UGE + UGSM-2 grafts, E16 UGSs were separated into epithelium (UGE) and mesenchyme (UGM) using the method described previously(24). UGE + UGSM-2 were combined and allowed to adhere together overnight on 0.6% agar plates containing INK4 culture medium. For UGS+UGSM-2 grafts, UGSs were dissected from E16 male CD-1 mouse embryos and chopped into 5–6 pieces, combined with UGSM-2 cells, and incubated overnight on agar plates prepared with INK4 culture media. For BPH-1 + UGSM-2 grafts, 500,000 UGSM-2 and 100,000 BPH-1 cells were resuspended in cold Matrigel (BD Biosciences, Becton, MA) and allowed to gel in sterile culture dishes. After 30 minutes, Matrigel beads containing cells were covered with INK4 culture medium and placed in a CO2 incubator overnight. Recombinants were placed under the renal capsule of CD-1 adult male nude mice using the method outlined by Cunha, et al (http://mammary.nih.gov/tools/mousework/Cunha001/Pages/Navigation.html). After 1–4 weeks, grafts were harvested, fixed and paraffin-embedded sections were prepared.
BrdU Pulse and Immunolabeling
BrdU labeling was used to trace UGSM-2 cells in renal grafts. Subconfluent UGSM-2 cells were incubated with 10uM bromodeoxyuridine (BrdU) in normal culture media overnight. Overnight incubation with BrdU resulted in approximately 50% of cells with BrdU incorporated. Immunolabeling of cells in formalin fixed paraffin-embedded sections was accomplished using the BrdU Labeling and Detection Kit II (Roche, Indianapolis, IN). We used goat anti-mouse-Alexa 546 conjugated antibody (Molecular Probes) to visualize BrdU stained cells. Sections were co-stained for pan-cytokeratin (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:50 dilution. Pan-CK was visualized by incubating with goat anti-rabbit Alexa 488 conjugated antibody (Molecular Probes, Eugene, OR) at a dilution of 1:200. Sections were mounted with Vectashield Hardset mounting media + DAPI counterstain (Vector).
Shh Treatment
UGSM-2 cells were plated in 6-well plates at a density of 4×105 cells/well in complete media and allowed to attach overnight. The next day, cells were treated with 1nm octylated N-Shh peptide (Curis Inc., Cambridge, MA). After 48 hours, cells were lysed and RNA was collected. RNA was purified and prepared for RT-PCR as described above.
Shh Overexpression
A mammalian expression vector expressing human Shh driven by CMV promoter (pIRES2-hShh-EGFP) was constructed as described previously(26). BPH-1 cells were transfected with pIRES2-hShh-EGFP vector or pIRES2-EGFP vector control (Clontech, Palo Alto, CA) using Lipofectamine 2000 (Invitrogen). BPH-1 cells stably overexpressing Shh/GFP were derived by fluorescence-activated cell sorting for GFP for 2 months after transfection. BPH-Shh cells stably express 50,000-fold more Shh mRNA than BPH-GFP or parent BPH-1 cells.
Statistical Analysis
An unpaired t-test was used to determine if significant differences exist between cell growth rates for untreated, testosterone or dihydrotestosterone treated cells. The Wilcoxon Rank Sum test was used to determine if there were significant differences in the gene expression responses to Shh treatment.
RESULTS
Isolation and Characterization of UGSM-2 Cells
Immortalized UGS mesenchymal (UGSM) cells were derived from a subline of the INK4a mouse, a transgenic knockout that lacks p16INK4a and p19ARF. Both p16INK4a and p19ARF are specific inhibitors of cyclin-dependent kinases Cdk4 and Cdk6 that regulate cell cycle progression (27). Loss of p16INK4a and p19ARF allows mouse embryonic fibroblasts (MEFs) to escape cellular senescence. INK4a−/− MEFs spontaneously immortalize in culture (28). UGSM cells were isolated by dissecting UGS mesenchyme from an E16 INK4a−/− mouse embryo (Figure 1A). UGSM cells obtained in this fashion were propagated continuously without evidence of crisis. Immortalized mouse cells are typically tetraploid and these cells remained stably tetraploid for over 100 passages (data not shown). Several ring clones were derived and characterized. All exhibited a similar growth rate and morphology in culture and all responded to treatment with Sonic Hedgehog by upregulating transcription of the conserved Hh target genes Ptc and Gli1. One representative clonal cell line, UGSM-2, was selected for use in subsequent experiments. Like the parent mixed cell population, UGSM-2 cells were found to be stably tetraploid (Figure 1B). Recent studies revealed that INK4a−/− MEFs can acquire chromosomal rearrangements at high passage (29). To assess tumorigenicity, both the parent UGSM cell line and UGSM-2 cells were co-injected with Matrigel into the flanks of nude mice. No tumor formation was observed in any of 12 injections for each group of cells over 6 months observation, whereas co-injection of LNCaP cells with Matrigel at the same time yielded tumor formation at over 80% of sites injected within 6 weeks (data not shown). Sarcoma formation was observed when a mixed population of UGSM cells at high passage (>30) were injected into nude mice, however, we have never observed sarcoma formation with the UGSM-2 clone.
Figure 1.
Isolation of UGSM-2 cells. (A) Schematic of UGSM-2 cell isolation from the E16 UGS-mesenchyme of an INK4a−/−; tva transgenic mouse. (B) The clonal cell line UGSM-2 is tetraploid on flow cytometric analysis. Freshly isolated splenocytes were provided as a diploid comparison.
UGSM-2 cells display a myofibroblast phenotype in culture
The mesenchymal identity of UGSM-2 cells was established by characterizing expression of selected differentiation markers by RT-PCR and immunocytochemistry (Figure 2). UGSM-2 cells express the stromal differentiation markers smooth muscle actin (SMA) and vimentin, and do not express either cytokeratins or the endothelial marker CD31/PECAM. The prostatic stroma contains cells that are classified as fibroblasts or smooth muscle, as well as cells termed myofibroblasts, which exhibit an intermediate phenotype. The profile of four stromal markers has been used to characterize cells as fibroblast (SMA−, vimentin+, desmin−, HCM−), myofibroblast (SMA+, vimentin+, desmin−, HCM−), or smooth muscle (SMA+, vimentin−, desmin+, HCM+)(30). According to this classification UGSM-2 cells which express SMA and vimentin, but do not express either desmin or heavy chain myosin (HCM) would be considered to exhibit a myofibroblast phenotype.
Figure 2.
(A) UGSM-2 cells immunostained for smooth muscle actin (SMA), vimentin (VIM), and high molecular weight cytokeratin (panCK). (B) Real-time RT-PCR analysis for expression of stromal differentiation markers by UGSM-2 cells and the freshly isolated intact E16 UGS. Genes studied include the endothelial marker CD31, SMA, VIM, desmin, heavy chain myosin (HCM) and androgen receptor (AR). Each bar represents the mean +/− sem of at least 2 independent determinations.
Growth characteristics of the UGSM-2 cell line
Growth of many cell lines in culture is characterized by 3 phases: a lag phase while cells attach to the substrate; a log phase of exponential growth; and a plateau phase triggered by confluence and contact inhibition. UGSM-2 cell growth in culture exhibits all 3 phases of growth. The typical doubling time for UGSM-2 cells in normal culture media is 13 hours (Figure 3A). The presence of a plateau phase shows that UGSM-2 cells are contact inhibited and, indeed, the cells at confluence adopt a tight monolayer appearance (Figure 5A).
Figure 3.
Unique characteristics of UGSM cells. (A) Growth of UGSM-2 cells in culture. UGSM-2 cells were grown in normal groth medium (see methods). Cell growth was monitored by daily Coulter counts for 1 week. Growth in culture exhibits three phases of growth: lag, log and plateau. (B) Growth of UGSM-2 cells is stimulated by androgen. UGSM-2 cells were grown in charcoal-stripped serum media or with 10−8M synthetic testosterone (R1881). Cell growth was monitored by daily Coulter counts for 1 week. Data are shown as fraction of cells relative to untreated controls. No significant differences in cell viability were noted by Trypan blue exclusion (not shown). *Significant increase in proliferation rate, P<0.05. All data points represent the mean of 3 determinations.
Figure 5.
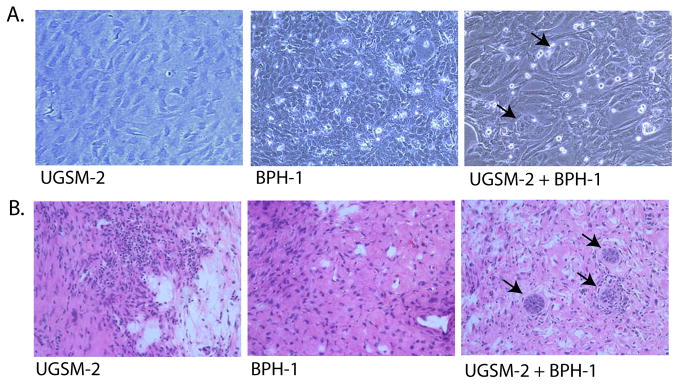
Interaction of UGSM-2 and BPH-1 epithelial cells. (A) Photomicrograph of co-culture of BPH-1 and UGSM-2 on collagen gels showing small clusters of BPH-1 cells (arrows) surrounded by UGSM-2 cells. (B) Grafting of UGSM-2 cells alone under the renal capsule yields only stromal tissue. BPH-1 cells grafted alone do not produce identifiable viable grafts. Grafting UGSM-2 and BPH-1 cells together results in small clusters of BPH-1 cells (arrows) surrounded by UGSM-2 cells.
Androgen response of UGSM-2 cells
The fetal urogenital sinus mesenchyme expresses androgen receptor and the androgen response of UGS mesenchyme is an important aspect of prostate biology. We examined androgen receptor expression by RT-PCR and found that UGSM-2 cells express the androgen receptor at levels comparable to the E16 UGS. Another fibroblast cell line that is not derived from the embryonic urogenital sinus, 3T3 fibroblasts, do not express androgen receptor (Figure 2B). UGSM-2 cells are not dependent on androgen for survival or proliferation (data not shown), however, their proliferation in culture is androgen sensitive. When we compared UGSM-2 growth in charcoal stripped serum supplemented medium without exogenous steroid hormone or with 10−8M synthetic androgen R1881, we found that UGSM-2 cells cultured in the presence of androgen grow at a significantly faster rate. 3T3 fibroblasts do not increase their proliferation rate in response to androgen (Figure 3B). The same effects were seen with either 10−8M testosterone or dihydrotestosterone (data not shown).
Participation of UGSM-2 in prostate morphogenesis
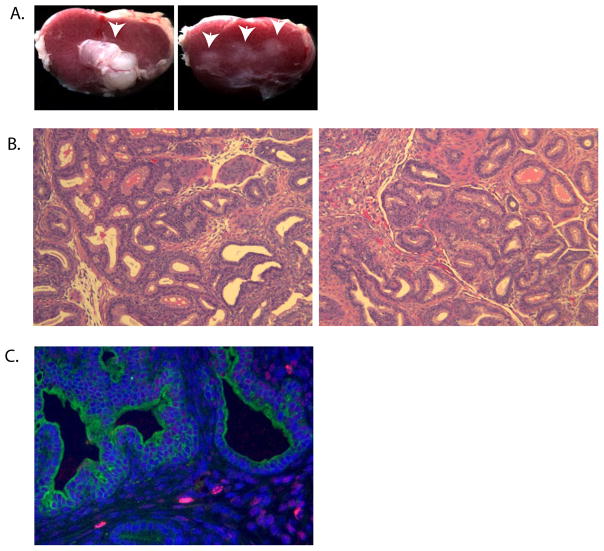
Our goal in developing the UGSM-2 cell line was to create a genetically modifiable cell line that could be used to study specific stromal-epithelial signaling interactions in prostate development. We therefore examined the ability of UGSM-2 cells to mimic three attributes of E16 UGS mesenchyme (UGM): the capacity to induce prostatic differentiation in the UGS epithelium (UGE), the potential to form the stromal component of prostatic glands, and the ability to mimic the signaling interactions of urogenital sinus mesenchyme. To examine the ability of UGSM-2 cells to induce prostate morphogenesis we grafted UGSM-2 cells together with isolated E16 UGE sheets under the renal capsule of adult male nude mice. When retrieved 1 month later, the resulting grafts were much smaller than grafts composed of E16 UGE and E16 UGM (Figure 4A) and histologic examination did not reveal any evidence of glandular morphogenesis (not shown). Therefore, UGSM-2 cells are unable to induce prostate development in this model system. To determine whether UGSM-2 cells can participate in glandular morphogenesis during prostate development, UGSM-2 cells were grafted with minced E16 UGS under the renal capsule of adult male nude mice. UGSM-2 cells were pre-labeled with BrdU to trace their fate in matured UGS/prostate. The fate of UGSM-2 cells was examined after 1, 2, and 3 weeks of growth in vivo. UGS + UGSM-2 grafts had a similar size, gross morphology (data not shown) and histology (Figure 4B) to minced E16 UGS implanted alone. Immunohistochemical staining for BrdU showed that BrdU-labeled UGSM-2 cells were present within the periductal stroma of the mature prostate tissue (Figure 4C). The BrdU staining in nuclei of UGSM-2 cells exhibited varying degrees of speckling that increased from 1–3 weeks (data not shown). This was interpreted as indicating active UGSM-2 proliferation during growth of the grafted tissue.
Figure 4.
Developmental fate of UGSM-2 cells in renal subcapsular grafts. (A) Gross morphology of 1 month renal grafts of E16 UGE combined with UGM (UGE+UGM, left) or UGSM-2 (UGE+UGSM-2, right). Arrows indicate position of grafts. (B) H&E stained sections of UGS+UGM (left) or UGS+UGSM-2 (right) recombinants 2 weeks after grafting. (C) BrdU staining (red) of 2 week UGS+UGSM-2 renal grafts identifies UGSM-2 cells in the graft. DAPI counterstained nuclei are blue. Pan-cytokeratin identifies ductal epithelium in green.
To assess the interaction of UGSM-2 cells with adult prostate epithelial cells, UGSM-2 cells were co-cultured with human prostate epithelial BPH-1 cells. After 24 hours in coculture, BPH-1 cells became organized into tight clusters surrounded by elongated UGSM-2 cells (Figure 5A). When UGSM-2 cells were grafted together with BPH-1 cells under the renal capsule of adult male nude mice and the grafts examined one month later, the BPH-1 cells were organized into clusters surrounded by stromal cells very similar to those observed in co-culture (Figure 5B). Mitotic figures were common in clusters, indicating active cell proliferation. Since BPH-1 cells injected alone do not form viable grafts, these observations suggest that UGSM-2 cells and BPH-1 can participate in a rudimentary process of cellular organization and that allows BPH-1 cells to survive and proliferate.
Shh response of UGSM-2
To determine if the UGSM-2 cell line could accurately model the mesenchymal response to Shh signaling, we assayed gene expression in UGSM-2 cells treated with Shh peptide. When treated in cell culture with purified Shh peptide, UGSM-2 cells show robust activation of the conserved Hh target genes Gli1, Ptc1 and Hip. In addition, the Shh target gene IGFBP6, recently found to be upregulated in the UGS mesenchyme in response to Shh, was also induced (Figure 6A). The 3-fold increase in IGFBP6 expression after treatment with Shh is comparable to the response of the isolated E16 UGS mesenchyme to Shh (31). To determine whether UGSM-2 cells would respond to Shh secreted by prostate epithelial cells in co-culture, we transfected BPH-1 cells with a Shh overexpression construct or GFP control vector (described in Fan et al., 2004). We cocultured UGSM-2 cells with the BPH-1 overexpressing or GFP control cells and analyzed Shh target gene expression using species-specific primers. This showed that overexpression of Shh by BPH-1 cells increased Gli1 and Ptc1 expression specifically in the UGSM-2 cells. There was no induction of Ptc and Gli1 in the BPH-1 cells (Figure 6B). These experiments show that UGSM-2 cells in co-culture respond to a signaling ligand expressed by epithelial cells and can therefore mimic a stromal-epithelial interaction that plays an important role in prostate development.
Figure 6.
Response of UGSM-2 to Shh. (A) real-time RT-PCR of UGSM-2 cells treated with purified Shh peptide shows activation of Shh target genes Gli1, Ptc1, HIP, and IGFBP-6. (B) UGSM-2 cells were co-cultured with BPH-1 benign human prostate epithelial cells stably transfected with a human Shh overexpression vector or GFP control vector. RT-PCR analysis using species-specific primers shows significant activation of Shh signaling targets Gli1 and Ptc1 in mouse UGSM-2 cells (top), but not in human BPH-1 cells (bottom). *Significantly increased from untreated or UGSM-2 + BPH-GFP, P=0.06. Error bar represents mean +/− sem of at least 2 independent determinations.
CONCLUSIONS
Mechanistic studies of cell-cell interactions are facilitated by the use of genetically modified cell lines. Our long-term goal in developing the UGSM-2 cell line is to provide a tool for mechanistic studies of prostate development. We will use it to probe the mesenchymal signaling pathways that are important for prostate growth and differentiation. Urogenital sinus mesenchyme serves a critical role during prostate development as a medium for communication with developing epithelial glandular structures. Two of the signaling molecules involved in mesenchymal-epithelial communication during prostate development are androgen and Sonic hedgehog. The ability of UGSM-2 cells to respond to both of these molecules makes it an appropriate tool for mechanistic studies of androgen and Sonic hedgehog activities in prostate development.
We found that these cells could not induce prostate differentiation when co-transplanted with the isolated sheets of E16 UGS epithelium tissue. However, we cannot exclude the potential of these cells to exhibit inductive potential in other assays such as one that uses dissociated UGS epithelial cells grafted under the renal capsule (32). When UGSM-2 cells were mixed with and co-transplanted with the whole UGS, they clearly did populate the mesenchyme/stroma of the subcapsular graft. In these grafts, UGSM-2 cells took up various positions within the stroma of mature prostate. Some UGSM-2 cells were situated beside ductal epithelium, whereas others were embedded among other stromal cells in interductal stromal sheets. Although we have not analyzed stromal differentiation in these grafts, the ability of UGSM-2 cells to localize to different regions of the mature graft could indicate that they may take up both fibroblast and smooth muscle positions or functions in mature prostate tissue. Since UGSM-2 cells are able to occupy a stromal niche in developing UGS renal grafts, they may be used in in vivo gain and loss-of-function studies to examine the role of various gene products in early prostate development.
In addition to their ability to participate in prostate development, UGSM-2 cells form primitive acinar structures when either co-cultured or co-grafted with human BPH-1 prostate epithelial cells. Clustering of BPH-1 cells has been observed previously when co-cultured with primary fibroblasts derived from normal human prostate, but not with primary fibroblasts derived from human prostate tumors (Simon Hayward, personal communication). Cunha has shown that the inductive relationships between epithelium and mesenchyme are preserved between human and rodents(21). Since the interactions between human epithelial cells and rodent mesenchymal cells are preserved, recombinants composed of human epithelium and UGSM-2 cells provide a useful model system for studying the role of these interactions in prostate development. An additional strength of this model is that we can distinguish signaling in mesenchyme and epithelium using species-specific RT-PCR. This dual species cell-based model therefore allows manipulation and analysis of gene expression in both epithelial and mesenchymal components to examine mesenchymal-epithelial interactions in vitro and in vivo.
In addition to their use in co-culture and xenograft models, UGSM-2 cells can be used as a cellular model to study mesenchymal signaling pathways that are important in prostate development. The first and most obvious use is to probe the molecular mechanisms of specific pathways. For example, we have used UGSM-2 cells to examine the concentration dependence and kinetics of Gli gene activation by Shh signaling (unpublished observations). The second is to use UGSM-2 cells in microarray studies to identify specific target genes of selected inductive signals. Finally, the immortalized UGSM cells can be used for genetic gain- and loss-of-function studies. Overexpression of selected genes in UGSM-2 cells may be engineered to examine the gain-of-function effect. It should be noted that the INK4a mutant was created by insertion of a neomycin resistance gene and cell lines derived from this mouse are neomycin resistant. Therefore, an alternative method of selection must be used when these cells are transfected. We have successfully used adenovirus, retrovirus and plasmid vectors with hygromycin or zeocin resistance selection to express genes of interest in UGSM-2. UGSM cells are particularly useful in studying genetic changes that are lethal, since harvest of UGSM cells at E16 allows for isolation of cells even from non-viable mutants. Indeed, we have developed UGSM cell lines from INK4a−/− mice bred to transgenic lines with mutations in various Shh signaling pathway components.
The potential for immortalized stromal cell lines to become tumorigenic is well recognized. The INK4a−/− mutation produces impairment of G1 checkpoint control and the INK4a−/− mouse is prone to develop tumors in several mesenchymal tissues (28). A recent report shows that INK4a−/− mouse embryonic fibroblasts display chromosomal rearrangements at high passage and develop the potential for sarcoma formation (29) We have found that after 30 passages in culture, a mixed population of UGSM cells can form sarcomas when co-injected with Matrigel into nude mice. This can occur even while the cells remain contact inhibited and monolayer in culture (unpublished observations). However, we have never observed sarcoma formation with the UGSM-2 clonal cell line that was derived from the mixed UGSM population. Even so, we utilize the cells at low passage and perform sentinel grafts to monitor for sarcoma formation in all in vivo studies.
The UGSM-2 cell line and comparable cell lines derived from specific transgenic mutant mice will provide powerful tools to study signaling between prostate mesenchymal and epithelial cells. Using genetically modified UGSM cells in complementary cell-based assays, in vitro co-culture models and xenografts will allow detailed mechanistic studies of specific pathways and their influence on prostate development.
Acknowledgments
Grant sponsor: Department of Defense; grant number W81XWH-04-1-0263
Grant sponsor: National Institutes of Health; grant number DK56238
Footnotes
Disclosure Statement
Aubie Shaw: no disclosures
Wade Bushman: no disclosures
References
- 1.Cunha GR, Donjacour AA, Cooke PS, Mee S, Bigsby RM, Higgins SJ, Sugimura Y. The endocrinology and developmental biology of the prostate. Endocr Rev. 1987;8(3):338–362. doi: 10.1210/edrv-8-3-338. [DOI] [PubMed] [Google Scholar]
- 2.Boutin EL, Battle E, Cunha GR. The response of female urogenital tract epithelia to mesenchymal inductors is restricted by the germ layer origin of the epithelium: prostatic inductions. Differentiation. 1991;48(2):99–105. doi: 10.1111/j.1432-0436.1991.tb00248.x. [DOI] [PubMed] [Google Scholar]
- 3.Donjacour AA, Cunha GR. Assessment of prostatic protein secretion in tissue recombinants made of urogenital sinus mesenchyme and urothelium from normal or androgen-insensitive mice. Endocrinology. 1993;132(6):2342–2350. doi: 10.1210/endo.132.6.7684975. [DOI] [PubMed] [Google Scholar]
- 4.Cunha GR, Donjacour AA, Sugimura Y. Stromal-epithelial interactions and heterogeneity of proliferative activity within the prostate. Biochem Cell Biol. 1986;64(6):608–614. doi: 10.1139/o86-084. [DOI] [PubMed] [Google Scholar]
- 5.Donjacour AA, Cunha GR, Sugimura Y. Heterogeneity of structure and function in the mouse prostate. Prog Clin Biol Res. 1987;239:583–600. [PubMed] [Google Scholar]
- 6.Hayward SW, Baskin LS, Haughney PC, Foster BA, Cunha AR, Dahiya R, Prins GS, Cunha GR. Stromal development in the ventral prostate, anterior prostate and seminal vesicle of the rat. Acta Anat (Basel) 1996;155(2):94–103. doi: 10.1159/000147794. [DOI] [PubMed] [Google Scholar]
- 7.Hayward SW, Haughney PC, Lopes ES, Danielpour D, Cunha GR. The rat prostatic epithelial cell line NRP-152 can differentiate in vivo in response to its stromal environment. Prostate. 1999;39(3):205–212. doi: 10.1002/(sici)1097-0045(19990515)39:3<205::aid-pros9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 8.Wong YC, Tam NN. Dedifferentiation of stromal smooth muscle as a factor in prostate carcinogenesis. Differentiation. 2002;70(9–10):633–645. doi: 10.1046/j.1432-0436.2002.700916.x. [DOI] [PubMed] [Google Scholar]
- 9.Chung LW, Chang SM, Bell C, Zhau HE, Ro JY, von Eschenbach AC. Co-inoculation of tumorigenic rat prostate mesenchymal cells with non-tumorigenic epithelial cells results in the development of carcinosarcoma in syngeneic and athymic animals. Int J Cancer. 1989;43(6):1179–1187. doi: 10.1002/ijc.2910430636. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes MJ, Dang TD, Lu B, Larsen M, McBride L, Rowley DR. Androgen-regulated proliferation and gene transcription in a prostate smooth muscle cell line (PS-1) Endocrinology. 1996;137(3):864–872. doi: 10.1210/endo.137.3.8603596. [DOI] [PubMed] [Google Scholar]
- 11.Price DT, Rudner X, Michelotti GA, Schwinn DA. Immortalization of a human prostate stromal cell line using a recombinant retroviral approach. J Urol. 2000;164(6):2145–2150. [PubMed] [Google Scholar]
- 12.Roberson KM, Edwards DW, Chang GC, Robertson CN. Isolation and characterization of a novel human prostatic stromal cell culture: DuK50. In Vitro Cell Dev Biol Anim. 1995;31(11):840–845. doi: 10.1007/BF02634567. [DOI] [PubMed] [Google Scholar]
- 13.Salm SN, Koikawa Y, Ogilvie V, Tsujimura A, Coetzee S, Moscatelli D, Moore E, Lepor H, Shapiro E, Sun TT, Wilson EL. Transforming growth factor-beta is an autocrine mitogen for a novel androgen-responsive murine prostatic smooth muscle cell line, PSMC1. J Cell Physiol. 2000;185(3):416–424. doi: 10.1002/1097-4652(200012)185:3<416::AID-JCP12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 14.Webber MM, Trakul N, Thraves PS, Bello-DeOcampo D, Chu WW, Storto PD, Huard TK, Rhim JS, Williams DE. A human prostatic stromal myofibroblast cell line WPMY-1: a model for stromal-epithelial interactions in prostatic neoplasia. Carcinogenesis. 1999;20(7):1185–1192. doi: 10.1093/carcin/20.7.1185. [DOI] [PubMed] [Google Scholar]
- 15.Rowley DR. Characterization of a fetal urogenital sinus mesenchymal cell line U4F: secretion of a negative growth regulatory activity. In Vitro Cell Dev Biol. 1992;28A(1):29–38. doi: 10.1007/BF02631077. [DOI] [PubMed] [Google Scholar]
- 16.Zhau HE, Hong SJ, Chung LW. A fetal rat urogenital sinus mesenchymal cell line (rUGM): accelerated growth and conferral of androgen-induced growth responsiveness upon a human bladder cancer epithelial cell line in vivo. Int J Cancer. 1994;56(5):706–714. doi: 10.1002/ijc.2910560516. [DOI] [PubMed] [Google Scholar]
- 17.Cunha GR, Chung LW. Stromal-epithelial interactions--I. Induction of prostatic phenotype in urothelium of testicular feminized (Tfm/y) mice. J Steroid Biochem. 1981;14(12):1317–1324. doi: 10.1016/0022-4731(81)90338-1. [DOI] [PubMed] [Google Scholar]
- 18.Lamm ML, Catbagan WS, Laciak RJ, Barnett DH, Hebner CM, Gaffield W, Walterhouse D, Iannaccone P, Bushman W. Sonic hedgehog activates mesenchymal Gli1 expression during prostate ductal bud formation. Dev Biol. 2002;249(2):349–366. doi: 10.1006/dbio.2002.0774. [DOI] [PubMed] [Google Scholar]
- 19.Risbridger G, Wang H, Young P, Kurita T, Wang YZ, Lubahn D, Gustafsson JA, Cunha G. Evidence that epithelial and mesenchymal estrogen receptor-alpha mediates effects of estrogen on prostatic epithelium. Dev Biol. 2001;229(2):432–442. doi: 10.1006/dbio.2000.9994. [DOI] [PubMed] [Google Scholar]
- 20.Donjacour AA, Thomson AA, Cunha GR. FGF-10 plays an essential role in the growth of the fetal prostate. Dev Biol. 2003;261(1):39–54. doi: 10.1016/s0012-1606(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 21.Hayward SW, Haughney PC, Rosen MA, Greulich KM, Weier HU, Dahiya R, Cunha GR. Interactions between adult human prostatic epithelium and rat urogenital sinus mesenchyme in a tissue recombination model. Differentiation. 1998;63(3):131–140. doi: 10.1046/j.1432-0436.1998.6330131.x. [DOI] [PubMed] [Google Scholar]
- 22.Lamm ML, Podlasek CA, Barnett DH, Lee J, Clemens JQ, Hebner CM, Bushman W. Mesenchymal factor bone morphogenetic protein 4 restricts ductal budding and branching morphogenesis in the developing prostate. Dev Biol. 2001;232(2):301–314. doi: 10.1006/dbio.2001.0187. [DOI] [PubMed] [Google Scholar]
- 23.Raghow S, Shapiro E, Steiner MS. Immunohistochemical localization of transforming growth factor-alpha and transforming growth factor-beta during early human fetal prostate development. J Urol. 1999;162(2):509–513. [PubMed] [Google Scholar]
- 24.Cunha G, Donjacour A. Mesenchymal-Epithelial Interactions: Technical Considerations. In: Coffey D, Bruchovsky N, Gardner W, Resnick M, Karr J, editors. Current Concepts and Approaches to the Study of Prostate Cancer. New York: AR Liss, Inc; 1987. pp. 273–282. [PubMed] [Google Scholar]
- 25.Hallowes R, Bone E, Jones W. A New Dimension in the Culture of Human Breast. In: Richards R, Rajan K, editors. Tissue Culture in Medical Research (II) Oxford: Pergamon Press; 1980. pp. 213–220. [Google Scholar]
- 26.Fan L, Pepicelli CV, Dibble CC, Catbagan W, Zarycki JL, Laciak R, Gipp J, Shaw A, Lamm ML, Munoz A, Lipinski R, Thrasher JB, Bushman W. Hedgehog signaling promotes prostate xenograft tumor growth. Endocrinology. 2004;145(8):3961–3970. doi: 10.1210/en.2004-0079. [DOI] [PubMed] [Google Scholar]
- 27.Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9(10):1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 28.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85(1):27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 29.Robertson S, Schoumans J, Looyenga B, Yuhas J, Zylstra C, Koeman J, Swiatek P, Teh B, Williams B. Spectral karyotyping of sarcomas and fibroblasts derived from Ink4a/Arf-deficient mice reveals chromosomal instability in vitro. Int J Oncol. 2005 in press. [PubMed] [Google Scholar]
- 30.Tuxhorn JA, Ayala GE, Rowley DR. Reactive stroma in prostate cancer progression. J Urol. 2001;166(6):2472–2483. [PubMed] [Google Scholar]
- 31.Lipinski RJ, Cook CH, Barnett DH, Gipp JJ, Peterson RE, Bushman W. Sonic hedgehog signaling regulates the expression of insulin-like growth factor binding protein-6 during fetal prostate development. Dev Dyn. 2005;233(3):829–836. doi: 10.1002/dvdy.20414. [DOI] [PubMed] [Google Scholar]
- 32.Xin L, Ide H, Kim Y, Dubey P, Witte ON. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc Natl Acad Sci U S A. 2003;100 (Suppl 1):11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]