Abstract
Recent emerging evidence suggests that ING family proteins play roles in carcinogenesis both as oncogenes and tumor suppressor genes depending on the family members and on cell status. Previous results from non-physiologic overexpression experiments showed that all five family members induce apoptosis or cell cycle arrest, thus it had been thought until very recently that all of the family members function as tumor suppressor genes. Therefore restoration of ING family proteins in cancer cells has been proposed as a treatment for cancers. However, ING2 knockdown experiments showed unexpected results: ING2 knockdown led to senescence in normal human fibroblast cells and suppressed cancer cell growth. ING2 is also overexpressed in colorectal cancer, and promotes cancer cell invasion through an MMP13 dependent pathway. Additionally, it was reported that ING2 has two isoforms, ING2a and ING2b. Although expression of ING2a predominates compared with ING2b, both isoforms confer resistance against cell cycle arrest or apoptosis to cancer cells, thus knockdown of both isoforms is critical to remove this resistance. Taken together, these results suggest that ING2 can function as an oncogene in some specific types of cancer cells, indicating restoration of this gene in cancer cells could cause cancer progression. Because knockdown of ING2 suppresses cancer cell invasion and induces apoptosis or cell cycle arrest, ING2 may be an anticancer drug target. In this brief review, we discuss possible clinical applications of ING2 with the latest knowledge of molecular targeted therapies.
Keywords: ING2, oncogene, molecular targeted therapy, siRNA, small molecular compounds, cancer vaccine, cancer/testis antigen, oncoantigen
INTRODUCTION
Molecular Targeted Therapies
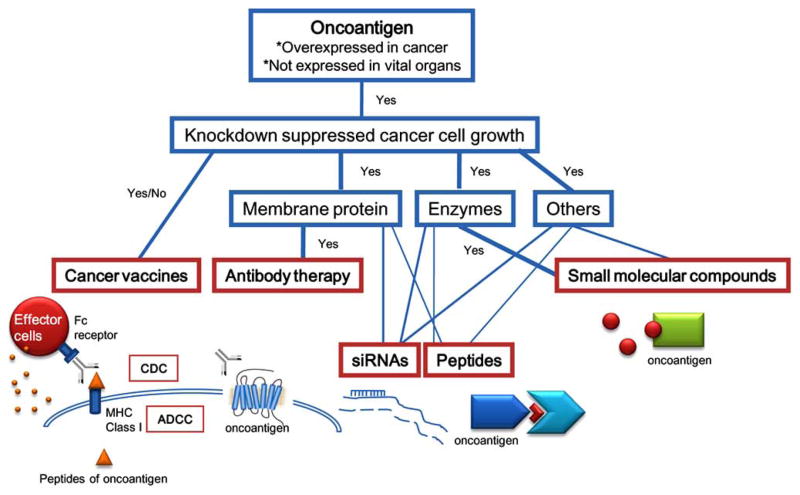
Evidence-based molecular targeted therapies, which interfere with specific targeted molecules necessary for cancer progression, have been developed intensively during the past decade. These compounds may have fewer side effects compared with current standard cytotoxic anticancer drugs, such as interference in rapidly dividing stem cells that are producing daughter cells for self-renewal in normal tissues. The target proteins for these therapies are required to be specifically expressed only in cancer cells. Additionally, inhibition of the target proteins ideally causes apoptosis, senescence, or cell cycle arrest to induce involution of the cancer, although this feature may not be necessary for oncoantigens for cancer vaccines. Molecular targeted therapies, which include small molecular compounds, monoclonal antibodies, small interference RNAs (siRNAs), and permeable dominant negative peptides, can be developed depending on subcellular localization, and functional/structural features of the target proteins (Fig. 1). For example, if the target protein has an enzymatic activity which associates with cancer malignancy, small molecular compounds can be developed to inhibit that activity. If the target protein is a membrane protein and transmits cell proliferation signals, an antibody against the protein might inhibit the growth signals from the protein, or cause antibody-dependent cellular cytotoxicity (ADCC) or complement-dependent cytotoxicity (CDC) [1]. Even if antibodies against these cancer-specific membrane proteins do not possess a cell growth suppressive effect or do not induce ADCC or CDC, these antibodies are still useful as a cancer-specific delivery tool for cytotoxic reagents such as radioisotopes or chemotherapy reagents [2]. siRNAs can be applied for all types of targets, but more sophisticated drug delivery systems (DDSs) are required and are still under development [3]. Permeable dominant negative peptides that inhibit a protein-protein interaction can also be effective [4], if the interference effectively inhibits cancer cell proliferation or leads to apoptosis. Imatinib mesylate (Gleevec) [5], Gefitinib (Iressa) [6], Erlotinib (Tarceva) [7], and Bortezomib (Velcade) [8] are the representative examples of small molecular compounds in clinical practice. Rituximab [9], Trastuzumab (Herceptin) [10], Cetuximab (Erbitux) [11], and Bevacizumab (Avastin) [12] are the representative examples of monoclonal antibodies in clinical use.
Fig. (1).
General strategy for targeted therapies.
Cancer Vaccines
Besides these anticancer drugs, a new generation of anti-cancer therapy utilizing the intrinsic immune response of cancer patients, are the cancer vaccines [13]. Oncoantigens, which are only expressed in cancer tissue, or cancer/testis antigens, which are highly expressed in cancer cells and reproductive tissues but not in normal cells, can be good immunogens that stimulate a patient’s T-cells which then recognize cancer cells presenting the oncoantigens on MHC class I molecules [14,15]. Because reproductive tissues do not express MHC class I molecules, these tissues are theoretically immune from T-cell recognition, thus no side effects are predicted when cancer/testis antigens are used to stimulate the innate immune system of cancer patients [16]. Cancer vaccines have had limited success so far. One of the reasons is that the therapy has been applied to cancer patients in the late stage. In these patients, the tumor burden is too large for the innate immune cells of the patients, causing difficulty in attacking all cancer cells. Now, cancer vaccine protocols are being revised. When a cancer vaccine is used for cancer patients at an early stage or patients in the setting of minimal residual disease states, more effect is expected. There is a potential concern regarding the generation of auto-immunity when the immune system does not shut down appropriately after vaccine stimulation. Many clinical trials are ongoing worldwide to evaluate the effect of this therapy.
ING Family Proteins
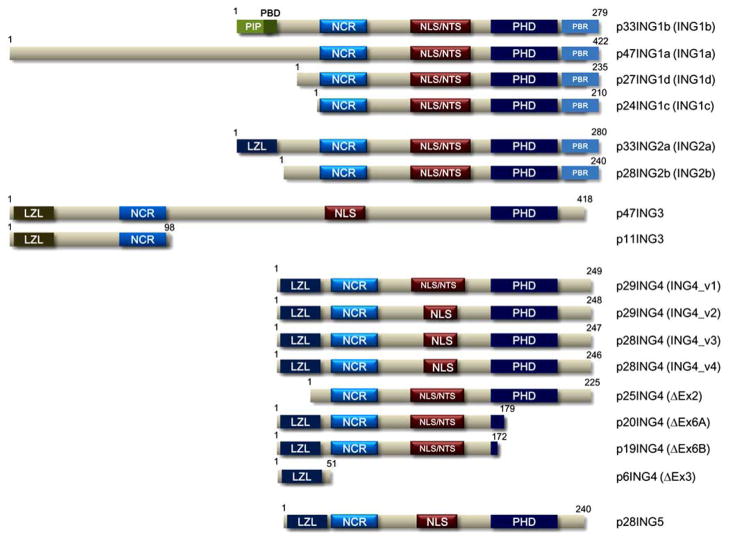
The inhibitor of growth (ING) family proteins are a well conserved family that is composed of five members, ING1 to ING5, with many splice variants or isoforms (Fig. 2) [17]. Non-physiological overexpression of these family members causes apoptosis, cell cycle arrest, and/or senescence, and thus, ING family proteins have been thought of as tumor suppressors for more than a decade [17]. However, recent results using siRNA and knockout mice are changing the previous implicit understanding of the ING family members, especially ING2. Results of analyses using ING1 knockout mice revealed that ING1 has a protective effect on apoptosis [18,19]. Our recent results showed that ING2 was overexpressed in colorectal cancer (Fig. 3A), and induced colon cancer cell invasion through an MMP13-dependent pathway (Fig. 4A) [20]. Knockdown of ING2 by siRNA induced premature senescence in normal fibroblast cells [21], and apoptosis or cell cycle arrest in various adherent cancer cells [22]. Taken together, these results suggest that ING2 may have roles in cancer progression and/or malignant transformation. Additionally, knockdown of ING4 and ING5 by siRNA showed an inhibitory effect on the transition of the cell cycle from the G2/M phase to the G1 phase, and DNA replication, respectively [23], suggesting that these proteins may play a role during cell proliferation. ING family proteins may play dual roles similarly to transforming growth factor beta (TGF-β) or p27, which can be either tumor suppressive or promoting depending on subcellular or tissue locations of these proteins and cellular status [24,25]. In this review, we mainly focus on possible clinical application of ING2-targeted therapy, and also explore the possibilities of other ING proteins in clinical application.
Fig. (2). Structure of the ING family proteins.
PHD, plant homeo domain; LZL, leucine zipper like domain; NLS, nuclear localization signal; NTS, nucleolar translocation sequence; NCR, novel conserved region; PIP, PCNA-interacting protein motif; PBD, partial bromo domain; PBR, poly basic region.
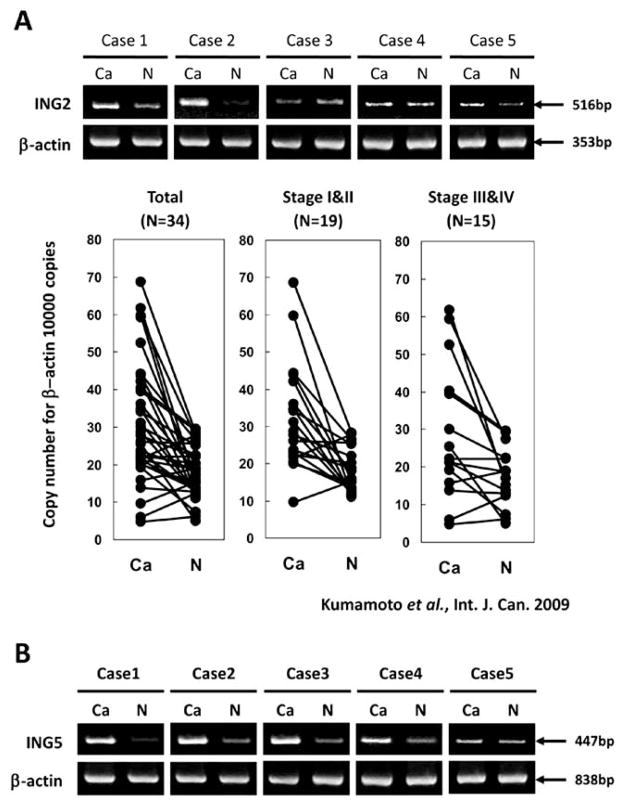
Fig. (3). Expression of ING2 and ING5 in colorectal cancers.
A. Upper panel, Semi-quantitative RT-PCR analysis of ING2 mRNA in five colon cancer samples, which were purchased from BD Bioscience. β-actin was used as the internal control. Lower panel, Results of real-time RT-PCR analyses of mRNA levels of ING2 in 34 human colon cancer tissues and nonmalignant mucosa. Cancerous tissues (Ca) and nonma-lignant mucosa (N) were prepared from the same patient. A paired t test was performed to ascertain statistical significance between the amount in cancer tissue and in nonmalignant mucosa. The ING2 and _-actin expression vector were used for calculating the copy number. Y-axis shows the copy number of ING2 per 10000 copies of β-actin. B. Semi-quantitative RT-PCR analysis of ING5 mRNA in five colon cancer samples, which were purchased from BD Bioscience. β-actin was used as the internal control.
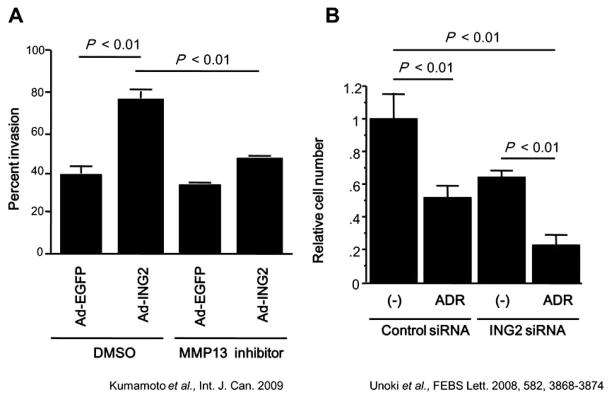
Fig. (4). ING2 in carcinogenesis and treatment.
A. MMP13 expression was associated with tumor invasion. Using the HCT116 cells infected with adenoviral constructs expression either GFP or ING2, a cell invasion assay was performed using a 24-well BD BioCoat Tumor Invasion System as described in Material and Methods. Statistical analysis was performed by Student’s t test. Columns, average of three independent experiments; Bars, SD. B. ING2 common siRNA enhanced growth suppression by adriamycin treatment. A549 cells were trans-fected with control siRNA or ING2 common siRNA (ING2 siRNA) twice every 3 days. After the second transfection, cells were treated with adriamycin (ADR) for 2 days. The combination of adriamycin and ING2 siRNA suppressed cell growth more effectively compared with adriamycin treatment alone or ING2 siRNA transfection alone.
2. ING2 MAY BE A GOOD ANTICANCER DRUG TARGET
Oncogenic Roles of ING2 in Cancer Cells
ING2 is one of the ING family proteins that was identified as a tumor suppressor candidate by computational homology search with ING1, which has also been thought to be a tumor suppressor [26,27]. ING2 has been reported to associate with numbers of proteins including p300, p53, the mSin3A-HDAC complex, the Brg-based SWI-SNF complex, BRMS1, trimethylated histone H3 lysine 4 (H3K4me3), and also with phosphatidylinositol 5-phosphate (Table 1) [23,28–32]. H3K4me3 modification is known to activate transcription [33]. There is a report that ING2 suppresses gene expression by binding to the H3K4me3 as a member of the mSin3A-HDAC1/2 complex [31]. ING2 also recruits histone methyltransferase (HMT) activity [34]. This report showed that ING2 recognizes H3K4me2 or me3 and facilitates methylation in a region around residues 1–20 of histone H3. The report did not specify a residue that was methylated in the region, but showed that it was not the well-studied histone H3K4 or H3K9, indicating that HMT methylates other amino acid residues in this region. Further analysis is required to determine the specific residue(s) methylated by the HMT activity and the effect of the methylation. Although it is unclear whether or not ING2 can also activate target genes through methylation, ING2 seems to play a significant role in transcriptional regulation through epigenetic histone modifications.
Table 1.
Binding Partners of ING Family Proteins
| Name | Interacting Proteins | Description | Refs. |
|---|---|---|---|
| ING1 | p53 | a tumor suppressor | [49] |
| PCNA | an essential factor for DNA polymerases, functioning in both DNA replication and nucleotide excision repair. | [52, 57] | |
| GADD45 | binds to PCNA, stimulated DNA excision repair and inhibited entry of cells into S phase. | [57] | |
| hSir2 | links chromatin silencing, metabolism, and cell aging, belongs to the class III family of HDACs | [87] | |
| p42, p35 | novel proteins that are involved in the ING1 complex | [57] | |
| RBP1 | allow s recruitment of mSin3-HDAC complex by retinoblastoma tumor suppressor family pocket proteins to induce cell cycle arrest by reppressing E2F-dependent transcription and DNA replication origins. | [57] | |
| mSin3, HDAC1/2, SAP30, RbAp46/48 | members of mSin3-HDAC complex | [57] | |
| Brg1, BAF47/53/60/155/170/250 | members of Brg1-based Swi-Snf complex | [57] | |
| p300 | a member of acetyltransferase complex | [57] | |
| TRRAP, PCAF, CBP | members of NuA4/Tip60 complex that acetylates histone H4 | [59] | |
| DMAP1 | associates with DNMT1 and is involved in maintaining histone modification and HP1 binding at the heterochromatin | [59] | |
| p15PAF | competes with p21 for PCNA binding | [87] | |
| 14-3-3 | central regulators of the cell cycle | [87] | |
| ARF | a tumour suppressor protein that plays a critical role in the activation of p53 in response to oncogenic stress | [87] | |
| H3K4me2/3 | di- and trimethyl Histone H3 lysine 4 | [31, 32] | |
| HMT activity | undefined histone methyltransferase | [34] | |
| LaminA | fibrous proteins providing structural function and transcriptional regulation in the cell nucleus | [60] | |
| ING2 | PtdIns(5)P | may be involved in cell-cycle progression | [32] |
| p53 | a tumor suppressor | [28] | |
| p300 | a member of acetyltransferase complex | [28] | |
| mSin3A, HDAC1/2, RbAp46/48, SAP30, SAP130, SDS3 | members of mSin3-HDAC complex | [23] | |
| BRMS1/BRMS1-like | suppresses metastasis of multiple cancer cells and a member of mSin3-HDAC complex. | [23] | |
| RBP1/RBP1-like | allows recruitment of mSin3-HDAC complex by retinoblastoma tumor suppressor family pocket proteins to induce cell cycle arrest by reppressing E2F-dependent transcription and DNA replication origins. | [23] | |
| BAF47/53a/155/170 | members of Brg1-based Swi-Snf complex | [23] | |
| H3K4me2/3 | di- and trimethyl Histone H3 lysine 4 | [30, 31] | |
| HMT activity | undefined histone methyltransferase | [34] | |
| ING3 | TIP60, p400, TRRAP, Brd8, EPC1/2, DMAP1, RUVBL1/2, MRG15, hEaf6 | members of NuA4/Tip60 complex that acetylates histone H4 | [23] |
| BAF53a | a member of Brg1-based Swi-Snf complex, and also involved inTip60 complex | [23] | |
| GAS41 | a common subunit of the TIP60 and SRCAP complexes and is essential for cell growth and viability | [23] | |
| AcK5-H4, AcK8-H4, AcK12-H4, AcK16-H4 | acethylated histone H4 lysine 5, 8, 12, 16 | [23] | |
| H3K4me1/2/3 | mono-, di-, and trimethyl Histone H3 lysine 4 | [30, 31] | |
| ING4 | p53 | a tumor suppressor | [42] |
| p300 | a member of acetyltransferase complex | [42] | |
| NF-κB | is involved in regulating the adaptive immune responses, cell survival, angiogenesis, tumorigene-sis and metastasis | [80] | |
| HPH-2 | mediates HIF stabilityas a function of oxygen availability | [79] | |
| HBO1 | responsible for histone H4 acetylation, and required for cell cycle progression | [23] | |
| JADE1/2/3 | cofactors for HBO1-mediated histone H4 acetylation | [23] | |
| hEaf6 | a member of NuA4/Tip60 complex | [23] | |
| AcK5-H4, AcK8-H4, AcK12-H4 | acethylated histone H4 lysine 5, 8, 12 | [23] | |
| liprin α1 | required for the trafficking of synaptoc vesicles and involved in the development and maintenace of excitatory synapses, and enhances migration in cancer cells | [81, 62] | |
| G3BP2a | implicated in Ras signaling, NF-κB signaling and the ubiquitin proteasome pathway | [81, 62] | |
| H3K4me1/2/3 | mono-, di- and trimethyl Histone H3 lysine 4 | [30, 31] | |
| ING5 | p300 | a tumor suppressor | [42] |
| p53 | a member of acetyltransferase complex | [42] | |
| MOZ/MORF | responsible for histone H3 acetylation | [23] | |
| HBO1 | responsible for histone H4 acetylation, and required for cell cycle progression | [23] | |
| JADE1/2/3 | cofactors for HBO1-mediated histone H4 acetylation | [23] | |
| BRPF1/2/3 | bind histones and acts by multiple mechanisms to mediate Moz-dependent histone acetylation | [23] | |
| MCM2/4/6 | Helicases | [23] | |
| AcK5-H4, AcK8-H4, AcK12-H4, AcK14-H4 | acethylated histone H4 lysine 5, 8, 12, 14 | [23] | |
| H3K4me3 | trimethyl Histone H3 lysine 4 | [30, 31] |
ING2 has been thought to be a tumor suppressor for more than a decade because of its homology with ING1. However, recent data suggest that ING2 can also function as an oncogene. Two groups found that expression of ING2 is upregulated in colorectal cancer (Fig. 3A) [20,26]. The oncomine database (http://www.oncomine.org/main/login.jsp) shows that ING2 is also overexpressed in Burkitt lymphoma and cervical carcinoma. However, the same database shows that expression of ING2 is decreased in other types of cancer including cutaneous melanoma and head and neck squamous cell carcinoma, concordant with recent two publications [35,36]. It suggests that the functions of ING2 differ depending on cancer type. There are two more reports about ING2 expression in cancers. One reported that expression of ING2 decreased in lung cancer [37]. However, in this report, the expression level of ING2 in only eight lung cancer cell-lines was examined compared with a relatively normal human bronchial epithelium cell line, BET2A. It is too early to conclude that ING2 expression is decreased in all lung cancers based on this result. Another report was about the expression of ING2 in hepatocellular carcinoma (HCC) [38]. Their result is somewhat confusing. They found that the expression of ING2 decreased in HCC at the mRNA level, but not significant at the protein level. Moreover, positive ING2 expression correlated with poor survival by their Kaplan-Meier analysis. As they mentioned in their article, they found that ING2 was mostly expressed in cytoplasm even in non-cancerous tissues, although previous results have shown that ING2 is a nuclear protein, indicating the possibility that their antibody did not recognize endogenous ING2 and might have recognized non-specific proteins in cytoplasm. Although expression of ING2 in additional types of cancer should be examined, the expression levels of ING2 vary among different types of cancers, suggesting various roles for ING2 in different contexts. Recently, it was reported that ING2 is a novel mediator of TGF-β-dependent responses in epithelial cells [39]. Since TGF-β is a protein which has tumor suppressor-like functions in normal epithelium and also has oncogenic functions in invasive metastatic cancers, ING2 probably mediates different signals from TGF-β in normal cells and in cancers [24].
It was also reported that the tumor suppressor p53 down-regulates expression of ING2 by direct binding to the promoter region of ING2 [20]. Because approximately 50% of sporadic human tumors harbor somatic mutations in the p53 gene locus [40], up-regulation of ING2 in colorectal cancer may be partially caused by aberrant p53 expression. It was also reported that NF-κB, whose expression is activated in several cancers including colorectal cancer and is anti-apoptotic, up-regulates expression of ING2 by direct binding to the ING2 promoter. Additionally, the ING2-HDAC1-mSin3A complex increases the invasion ability of cancer cells by enhanced expression of matrix metalloproteinase 13 (MMP13), which plays a crucial role in tumor cell invasion through digestion of basement membrane and extracellular matrix components (Fig. 4A) [20,41]. Knockdown of ING2 using siRNAs suppressed the expression of MMP13 [20]. Because surgical techniques have improved remarkably, prevention of metastasis from minimal residual disease is the most important subject after surgery of cancers currently. Knockdown of ING2 in the residual cancer cells may prevent invasion, metastasis, and relapse of cancers.
Recently, a novel ING2 isoform, ING2b, was identified [22]. The original ING2 is also called ING2a when required, to distinguish it from ING2b. ING2b is transcribed from the middle of ING2a’s intron 1. The ING2b promoter does not possess an apparent p53 binding site in contrast to the promoter of ING2a. Consistently, activation of p53 only led to suppression of ING2a, and not ING2b. ING2a knockdown suppressed cell growth only when p53 was present, but not when it was absent, indicating that p53 is activated by knockdown of ING2a. Although further analysis is required, it seems that there may be a negative feedback loop between ING2 and p53; p53 suppresses transcription of ING2a, and ING2a may suppress p53 through protein-protein interaction of the N-terminal region that ING2b lacks [29]. In contrast to knockdown of ING2a, knockdown of ING2b did not show any effects on cell growth. However, knockdown of both isoforms suppressed cell growth and induced cell cycle arrest or apoptosis even in p53 aberrant cells, in which ING2a specific siRNA did not induce cell cycle arrest or apoptosis, indicating that ING2a and ING2b may have compensatory roles that protect cells from cell cycle arrest or apoptosis. Because these results suggested that a combination of the knockdown of both ING2 isoforms can improve current chemotherapies, a combination therapy was tried using cancer cells in vitro. The ING2 siRNA, which suppresses both ING2a and ING2b, and adriamycin, which is one of the most widely used anticancer drugs for chemotherapy, were used for the test. The combination of adriamycin and ING2 siRNA was more effective than either single agent alone, suggesting that targeting ING2 in combination with DNA damaging agents might be a viable therapeutic strategy (Fig. 4B).
Expression and Roles of ING2 in Normal Tissues
Molecular targeted therapies are proposed to have fewer side effects compared with current cytotoxic chemotherapy, because expression of the target proteins may be limited to cancer tissue. Expression of ING2 is ubiquitous, but relatively low in most normal tissues according to GNF-SymAtlas (http://symatlas.gnf.org/SymAtlas/) and “Expression Profile” provided by Unigene (http://www.ncbi.nlm.nih.gov/UniGene), except in testis. In testis, high expression of ING2 was observed [22,26]. Moderate expression was also observed in ovary, skeletal muscle, and pancreas [22,26]. In normal human fibroblast, endogenous p53, which is activated by an MDM2 inhibitor Nutlin3a, directly down-regulates endogenous ING2 expression via binding to two p53 binding sites on the ING2 promoter, causing senescence [21]. Decreased expression of ING2 by siRNA also induced premature senescence. The percentage of senescent cells that was induced by ING2 knockdown, Nutlin3a-induced p53 activation, and a combination of ING2 knockdown and Nutlin3a-induced p53 activation was around 25%, 40%, and almost 100%, respectively. Taken together, possible side effects, when ING2 functions are prohibited, may be minor atrophy of testis, ovary, and skeletal muscles, which are not vital organs. Because ING2 is also expressed in pancreas, patients may become glucose intolerant due to altered expression of insulin and/or experience malabsorption due to alterations in digestive enzyme secretion from the pancreas. The exact cell types in the pancreas which express ING2 should be examined to investigate possible side effects. Patients whose pancreas has been surgically removed can be maintained in good health, if treated properly. Therefore, we consider ING2-targeted therapy as a viable anticancer drug strategy with minimal potential side effects.
Possible Therapies that Target ING2
Antibody Therapy
As was described in the introduction, target proteins for antibody therapy should be membrane proteins that are recognized by specific antibodies. Because subcellular localization of ING2 is mainly to the nucleus [22], antibody therapy cannot be applied.
Cancer Vaccine
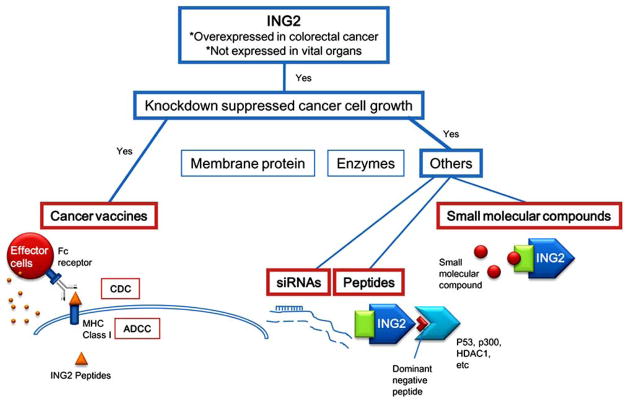
Although ING2 is moderately expressed in pancreas, ING2 is basically a cancer/testis antigen. Many cancer/testis antigens are digested into peptides in cells and presented on cytoplasmic membrane with HLA class I molecules, even if the antigen localizes to nucleus. Thus, it is possible that ING2 peptides may be presented on the cytoplasmic membrane of cancer cells which are overexpressing ING2, such as colon cancer cells, and possibly Burkitt lymphoma and cervical cancer as we mentioned above. If so, an ING2 peptide can be used as a cancer vaccine to stimulate the immune system of cancer patients (Fig. 5). Because the immune response towards peptides is different between mice and humans, it is impossible to validate the potential of the peptides as antigens in mice. Of course, because patients should not be exposed to any danger without merit, more detailed expression data of ING2 in cancers and normal tissues should be collected. Heterogeneity of cancer cells in a cancer mass can be a serious problem. Recently, the existence of cancer stem cells has been proposed. A cancer mass may be derived from the small population of cancer stem cells in the mass. Pathologists have been seeing heterogenous cancer cell masses for decades. Individual cancer cells in a mass may acquire additional, secondary changes during progression of the cancer. These secondary changes can include overexpression of some proteins, which are not necessary for carcinogenesis. When we develop a cancer vaccine, we need to select an oncoantigen, which expresses in all cancer cells in a mass to avoid recurrence from any remaining cells which do not express the oncoantigen. ING2 is expressed in almost all cancer cells in a mass of colorectal cancer, which we examined by immunohistochemistry [20]. Although further analysis using more samples is required, an ING2 peptide has potential as a cancer vaccine.
Fig. (5). ING2 and possible targeted therapies.
ING2 can be a target for cancer vaccines, siRNAs, peptides, and small molecular compounds.
Small Molecular Compounds
Another possibility is a small molecule that targets the ING2 PHD domain, which plays an important role in binding to H3K4me3 [30,31], and also functions as a nuclear phosphoinositide receptor (Fig. 5) [32]. The H3K4me3 binding region of the domain forms relatively deep grooves [30]. The grooves can be targeted by small molecular compounds, which inhibit ING2 binding to H3K4me3 and/or phosphoinositide. It is not yet known if inhibition of the functions of the PHD domain is enough to interfere with all the oncogenic functions of ING2 such as promotion of cancer cell invasion, and chemoresistance against anticancer drugs. It should be studied before these molecules are invented. The specificity of the small molecular compounds to the ING2 PHD domain may also be important, because the domain is well conserved among the family proteins [42], which may have different roles in carcinogenesis. If all ING family proteins work as oncogenes in cancer cells, specificity may not be so critical. As discussed below, further analyses are required to make a judgment. Small molecular compounds are usually used for inhibition of enzymatic activity, but the design of effective compounds that inhibit a protein-protein interaction is not impossible. There are several examples of small molecular compounds which inhibit protein-protein interactions. For example, small-molecule inhibitors of all the major Bcl-2 antiapoptotic family proteins such as GX15-070, which inhibits binding of Mcl-a and Bcl-xL to Bak, have been developed as anticancer and antiangiogenic agents and are in clinical trials [43]. If an effective small molecule that can inhibit the oncogenic functions of ING2 is developed, the molecule can be applied in combination therapy with other anticancer drugs such as adriamycin, for which we showed the combination to be effective, and can also be used for preventing micro metastasis after surgery. Many DDSs are being developed for small molecular compounds. Among these, a system utilizing the Enhanced Permeability and Retention (EPR) effect is now one of the most-watched systems [44]. The EPR effect is the property by which certain sizes of molecules, typically liposomes or macromolecular drugs, tend to accumulate in tumor tissue much more than they do in normal tissues. The general explanation that is given for this phenomenon is that, in order for cancer cells to grow rapidly, they need to stimulate the production of blood vessels. These newly formed tumor vessels are usually abnormal in form and architecture. They are poorly-aligned, defective endothelial cells with wide fenestrations, lacking a smooth muscle layer, or innervations with a wider lumen. Furthermore, tumor tissues usually lack effective lymphatic drainage. All these factors will lead to abnormal molecular and fluid transport dynamics especially for macromolecular drugs. Thus, once an effective small molecular compound is developed, the compound may be delivered using the EPR system in the future.
siRNAs
Because siRNA against ING2 induced apoptosis or cell cycle arrest in cancer cells, siRNA therapy can also be applied to the treatment of cancer patients (Fig. 5). siRNA is very unstable in blood because of rapid degradation by serum nucleases, and is excreted from blood into urine from glomera quite rapidly, probably because of its small molecular weight, linear structure, negative electric charge, and solubility. The negative electric charge prevents siRNAs from getting into target cells, as well. To deliver siRNA to a targeted location is also an area of active research. Thus, establishment of good DDSs is the biggest hurdle to overcome prior to using siRNA as a therapeutic strategy. Although there are many hurdles as we described above, significant progress has been made in recent years in the delivery of siRNA to tumors, and several promising siRNA delivery platforms have begun to emerge utilizing the EPR effect that is described above. These platforms include liposomes, in which siRNA is encapsulated in a lipid vesicle; polyplexes, in which a cationic carrier is used to bind siRNA to form siRNA-containing nanoparticles; liposome-polycation-DNA (LPD) complexes, in which an siRNA-containing polyplex is encapsulated in a lipid vesicle; and siRNA conjugates, in which siRNA is coupled to a targeting moiety that carries the siRNA into target cells via receptor-mediated endocytosis [45]. A number of groups have been investigating chemical modifications and alternative backbones, which improve the stability of siRNAs [46]. As a result of the continuous efforts of scientists, the stability of siRNAs has been increasing, and we may hope it will not be a problem soon. It seems that good DDSs and stable siRNAs will probably be established within the next decade, and siRNA therapy will bring tremendous benefits to cancer patients. The ING2 siRNA therapy can be applied alone, but is probably more effective in a combination with other current anticancer drugs to enhance their effects and overcome the resistance of cancer cells against the drugs. Because combinations of adriamycin and the ING2 siRNA further suppressed cancer cell growth (Fig. 4B) [22], the possibility of combination therapy is the most promising.
Permeable Dominant Negative Peptides
A permeable dominant negative peptide, which is a partial region of ING2 or that of its binding proteins, can also be utilized for interfering with ING2 bindings to its partners (Fig. 5). As an example, a peptide derived from AMAP1 specifically blocked AMAP1/cortactin binding and effectively inhibited breast cancer invasion and metastasis [47]. Because ING2 interacts with various proteins including p300, p53, mSin3A-HDAC1 complex, Brg-based SWI-SNF complex, BRMS1, and trimethylated histone H3 lysine 4 (H3K4me3) [23,28–32], if inhibition of some of the binding can cause apoptosis or cell cycle arrest, peptide therapy can be applied. The biggest problem is stability at this time. For a cancer vaccine, a peptide can be applied with an adjuvant that increases the stability of the peptide. In this case, a DDS that increases the stability of the peptide and provides cancer specific delivery is required.
3. OTHER MEMBERS OF THE ING FAMILY AS ANTICANCER DRUG TARGETS
ING1, the Closest ING Family Member of ING2
Using subtractive hybridization between cDNAs from a normal mammary cell-line and several transformed breast cancer epithelial cell-lines and the subsequent selection, ING1 was identified as a tumor suppressor that associates with the tumor suppressor p53 [48,49]. Since the identification of ING1, experiments have usually been performed under non-physiological conditions in vitro. There is a possibility that non-physiological overexpression of ING1 might just disturb the homeostasis of cells, leading to cell cycle arrest, apoptosis, and senescence [50–53]. However, these overexpression results have led to ING1 being considered to be a tumor suppressor. On the other hand, ING1 was identified as a breast cancer antigen by serological analysis of recombinant tumor cDNA expression libraries (SEREX) [54]. High expression of ING1 was reported to associate with poor survival of bladder cancer patients [55]. There is another noteworthy report that overexpression of p37ing1, which is a counterpart of human major ING1 isoform p33ING1b, suppressed p53 expression and subsequent p53-induced senescence [56]. In the article, the authors warned that a simplified view of the potential role of ING1 in cancer should be revised [56]. Thus, there is a possibility that ING1 cannot be defined simply as a tumor suppressor gene, much like TGF-β or p27, which have completely different roles under different circumstances [24,25].
Recently, two groups generated Ing1 knockout mice [18,19]. Both groups were unable to detect any correlation between Ing1 and p53, although that association has been used as evidence that ING1 is a tumor suppressor. They found that loss of p37ing1 induces Bax expression and increases DNA damage-induced apoptosis in primary cells and mice irrespective of p53 status, suggesting that ING1 can work as an oncogene by suppressing apoptosis in some situations, although Ing1 cannot be defined categorically as an oncogene because loss of Ing1 was associated with earlier onset and higher incidence of lymphomas [18,19].
ING1 is involved with an mSin3-HDAC complex, a Brg-based Swi-Snf complex, and a NuA4/Tip60 complex (Table 1). ING1 also associates with various other proteins including PCNA, GADD45, p53, DMAP1, and trimethylated histone H3K4 (H3K4me3) [30,31,57–59]. H3K4me3, H3K4me2, and H3ac modifications are tightly associated with the transcriptional starting sites of genes [33]. Additionally, because the p33ING1-mSin3-HDAC and DMAP1-DNMT1 complexes are recruited to pericentric heterochromatin regions and required for deacetylation of histones and methylation of histone H3K9, which is a mark well known for repression, ING1 is probably involved in a dynamic system that connects histone deacetylation, histone methylation, and DNA methylation in maintaining pericentric heterochromatin structure throughout cell divisions [59]. Interestingly, it was also reported that ING1 interacts with lamin A [60]. In lamin A deficient cells, which have invaginations of the nuclear membrane, endogenous ING1 expression was decreased, suggesting that the interaction is important for stabilization of ING1 and also keeping structures of the nuclear membrane intact. Mutation of lamin A results in several laminopathies including Hutchinson-Gilford progeria syndrome (HGPS), which is a severe premature aging disorder. There is no doubt that ING1 is a fundamentally important gene, but thus, it is very difficult to define ING1 as a tumor suppressor or an oncogene based in what is known.
There are several reports that expression of ING1 is decreased, or that the ING1 gene was mutated or deleted in cancers including breast, ovarian, esophageal squamous cell and human head and neck cancers [17]. Interpretation of genetic or epigenetic changes should be done carefully, because there are mainly two types of changes: one is a change that is involved in cancer development directly, and the other is a change that occurs just collaterally following the primary change such as instability of the genome, which is often observed in cancer cells. If these changes are mutations that have occurred in a genome, they are called “driver mutation” and “passenger mutation”, respectively. Rare single nucleotide polymorphisms (SNPs) and minor splice events should be considered also, when a mutation search is conducted: there is an example that a reported mutation in the ING4 gene was just a splice variant of ING4 [61,62]. There is an opposing report using microarray data showing that high expression of ING1 was found to be significantly associated with poor survival of patients with bladder tumors [55]. The tumor suppressor p53 accumulates in cancer cells because of a mutation that avoids protein degradation; thus expressional change of a protein in cancer tissues sometimes occurs as a result of very different causes [63]. Further analyses are perhaps required to conclude that low expression of ING1 was actually related with carcinogenesis directly.
Expression levels of ING1 in normal tissues are very low according to the GNF SymAtlas (http://symatlas.gnf.org/SymAtlas/). This would be a desirable situation, if ING1 is an oncogene. However, if ING1 is a tumor suppressor, it does not help in the development of anticancer drugs. Generally, to utilize tumor suppressor activity to cure cancer, restoration of the tumor suppressor’s activity in cancer cells is required, and often virus vectors are used for this purpose, because of a high infection efficiency to cells. Cancer specific infection of the tumor suppressor expression virus vectors is the biggest subject in this case, and such virus vectors have been improved to make it possible [64]. At this time, identification of the exact functions of ING1 in different contexts is the most urgent subject. To think of clinical usages of ING1 is the next step.
ING3, Still a Mysterious Protein
ING3 was identified as the third member of the ING family by computational homology search [65]. The amino acid sequence of ING3 is the most distinctive among the five ING family members evolutionarily [66]. ING3 possesses the same domains as the other ING family proteins. However it has the largest molecular weight, and thus it has the longest unique regions, although no domains have been predicted in these regions. Therefore, ING3 may have distinctive roles compared with other family members. Down-regulation and aberrant subcellular localization of ING3 have been reported in head and neck cancers, and cutaneous melanoma [67–69]. ING3 may work as a tumor suppressor in these types of cancers. However, it was described above, interpretation of the down-regulation of a gene should be done carefully, because it can simply occur collaterally following a primary change in cancer. Therefore we need to understand the precise functions of ING3, which are almost unknown, using siRNA and/or knockout mice to interpret these expression data. Although non-physiological overexpression of ING3 induced apoptosis [65,70], these results may not reflect the physiological functions of ING3 in cells. However, it seems that knowledge of the binding partners of ING3 might help to determine its functions [23,31,58,71]. ING3 works mainly as a member of a NuA4/Tip60 HAT complex that acetylates the N-terminal tails of histone H4 and H2A, and binds to trimethylated H3K4 similarly to the other ING family members (Table 1) [23,31,58,71]. Because the NuA4/Tip60 complex acetylates histones H4 and H2A, ING3 may bind to trimethylated lysines of histone H4 and H2A as well. It would be very interesting to study these molecular interactions. ING2 is reported to recruit the mSin3A-HDAC complex to H3K9me3 and represses transcription [31], on the other hand, ING3 has an ability to recruit the HAT complex [23]. Therefore, even though both proteins bind to H3K4me3, the biological effects led to by these bindings can be very different. At this time, because only minimal information concerning ING3 is available, the possibility of clinical application of ING3 is still unclear.
ING4, which has Various Splice Variants
The original ING4, ING4_v1, was identified by computational homology search [35]. Recently, eight splice variants of ING4, ING4_v1, ING4_v2, ING4_v3, ING4_v4, ΔEx2, ΔEx3, ΔEx6A, and ΔEx6B, have been reported (Fig. 2) [62,72,73]. Alternative splicing is a main source of transcriptome and proteome diversity. It has been reported that splicing acceptors with the NAGNAG motif can cause NAG insertion-deletion (InDel) in transcripts and play complex roles in switching protein conformation and functions [74,75]. Besides the NAGNAG-based tandem repeat in splice acceptor sites, a splice donor site (GTNGT) can also be used to generate such InDel variants. This splicing mechanism is called “wobble alternative splicing” or “subtle alternative splicing”.
Recently, two groups found that the exon 4-5 boundary of ING4 has GC(N)7GT and NAGNAG motifs that generate four of the eight splice variants, ING4_v1, ING4_v2, ING4_v3, and ING4_v4, which have 0, 3, 9, and 12 bp skips, respectively [62,73]. Among these four splice variants, ING4_v4 was reported as a common mutation in ING4 only observed in cancer cells, but it turned out that it is just one of the splice variants [61]. Now both groups have shown that all four splice variants are expressed ubiquitously in various tissues with keeping their composition ratios; ING4_v1 and _v2 are the major transcripts and _v3 and _v4 are the minor transcripts [62,76]. It was shown that a single nucleotide mutation in the splicing donor or the acceptor sites can avoid generation of a certain type of splice variant depending on the mutation locus in the splice donor or acceptor sites [62]. Because many single nucleotide polymorphisms (SNPs) have been discovered in the NAGNAG sequences that are spread widely throughout the human genome [77], and a change in these sequences can affect the composition of splice variants, SNP and/or mutation search at the ING4 exon4-5 boundary may be an interesting approach for examining involvement of the ING4 variants in congenital and acquired genetic diseases including carcinogenesis.
The roles of each of the endogenous variant are still unclear, because the NAGNAG-based tandem repeat prevents designation of a specific siRNA against each variant. ING4_v2, ING4_v3, and ING4_v4 have 1, 3, and 9 amino acids deleted compared with ING4_v1. The deletions cause a partial lack of the nuclear localization signal (NLS) that is important for p53 binding [78] as well as nuclear localization. Localization of these splice variants is somewhat controversial; Raho et al. showed that the ING4 splice variants including ING4_v4, ΔEx2, ΔEx3, ΔEx6A, and ΔEx6B localized in nucleus [72], Unoki et al. showed that ING4_v4 has an increased tendency to localize in cytoplasm compared with ING4_v1 whose localization is mostly in nucleus [62], and Tsai et al. also showed that ING4_v1 and _v2 localize in nucleolus, whereas ING4_v3 and _v4 localize in nucleoplasm [76]. Collectively, the splice variants of ING4 may distribute differentially in different cell types and/or different conditions. The ING4 splice variants may enrich the roles of ING4 as an ensemble through the different subcellular localization and different interacting proteins.
ING4 has been reported to interact with a variety of proteins including HPH-2 that regulates HIF-α stability, the p65 subunit of NF-κB, p53, p300, and liprin α1 (Table 1) [42,78–82]. These reports showed that ING4 works as a tumor suppressor by regulating various pathways through interactions with these proteins. ING4 also binds to mono-, di-, and tri-methyl H3K4 [31,58]. Histone H4 and H2A were immunoprecipitated with a NuA4/Tip60 complex including ING3, while histone H4 and H3 were immunoprecipitated with a complex including ING4, HBO1, hEaf6, and JADE1/2/3 paralogs [23]. The NuA4/Tip60/ING3 complex acetylates histone H4 lysine 5, 8, and 12, but not 16, on the other hand, the complex including ING4 and HBO1 acetylates all these 4 residues, indicating each ING family protein has different elaborate roles in transcriptional regulation through “histone code” modifications.
Although non-physiological overexpression of ING4 also induced cell cycle arrest and apoptosis in cells like other members of the ING family [42,62], there is a report using siRNA to knockdown endogenous ING4. The knockdown led the cells to specifically accumulate in G2/M [23]. Additionally, expression of endogenous ING4 was induced in cells at G2/M arrest [83]. Thus, ING4 has some important roles in the transition of the cell cycle from the G2/M phase to the G1 phase, suggesting that ING4 is involved in cell cycle progression. However, in contrast, non-physiological overexpression of ING4 also induced G2/M cell cycle arrest and inhibited cell proliferation [62,84]. In this case, the excess amount of ING4 might have prevented its physiological functions. Restoration of ING4 in cancer tissues using some virus vectors may inhibit cell proliferation, but it may also promote G2/M progression depending on its expression level in cells. Therefore, the detailed roles of ING4 in the G2/M phase and carcinogenesis should be further analyzed before clinical application of this protein. If all splice variants have different roles in carcinogenesis, ING4 may not be the best target for clinical application, because of the difficulty of specific knockdown of a particular variant as described above.
ING5 may be an Anticancer Drug Target?
ING5 was identified together with ING4 by computational homology search [42]. There have been just a few reports about this protein. ING5 binds to trimethylated histone H3K4 like other ING family proteins [31,58,85]. ING5 is involved in two different HAT complexes (Table 1) [23]. One is a complex that includes ING5, HBO1 and JADE, similar to ING4, and binds to histone H4. Another complex includes ING5, MOZ/MORF, and BRPF, and binds to histone H3. ING5 also associates with minichromosome maintenance (MCM) proteins, which play an essential role in eukaryotic DNA replication through formation of a prereplicative complex at the origins of replication. ING4 does not make complexes with MOZ/MORF-BRPF nor MCM proteins, although ING4 and ING5 share high homology of amino acid sequence. Although non-physiological overexpression of ING5 also induced cell cycle arrest and apoptosis in cancer cells [42], there is a report using siRNA to knockdown endogenous ING5. The knockdown of ING5 completely abolished DNA synthesis. Additionally knockdown of HBO1 increased cells in S-phase. Therefore, there has been speculation that the HBO1-JADE-ING5 HAT complex has an important role during DNA replication in cooperation with the MCM complex. We found that expression levels of ING5 are significantly high in colorectal cancer (Fig. 3B). Taking these results together, ING5 may be involved in carcinogenesis by regulating DNA replication. Although further analyses are required, if ING5 is involved in carcinogenesis as an oncogene, ING5 may be a good anticancer drug target in addition to ING2. In this case, all possible clinical applications for ING2 can be also applied to this protein.
4. CONCLUSION
Recent emerging results suggest that ING2 may be a cancer/testis antigen in some types of cancers, and thus may be a drug target for small molecular compounds, dominant negative peptides, and siRNAs. Additionally, ING2 peptides can be utilized as a cancer vaccine to stimulate the innate immune system of cancer patients. Further analyses are required to determine the roles of other ING family proteins in carcinogenesis. ING family proteins have been studied for more than a decade and were implicated as tumor suppressor genes based on a sequence of non-physiological overexpression experiments. Although there were few data in earlier times that addressed the question of whether the ING family proteins were actually tumor suppressors or not, reports of these findings [24, 47, 48] have not been well considered. We are afraid of biased interpretations based on the overexpression data that were obtained in an era before the development of siRNA technology. The recent data obtained from Ing1 knockout mice show that Ing1 is a negative regulator of apoptosis [18,19]. Knockdown of ING2 induced senescence in normal cells and apoptosis and/or cell cycle arrest in cancer cells [21,22]. Knockdown of ING4 and ING5 also indicates that these proteins cannot be simply classified as tumor suppressors [23]. These knockdown data show completely opposite results compared with the results of the previous overexpression experiments. ING proteins may work similarly to TGF-β or p27, which have completely opposite roles in different contexts. There are reviews that imply using ING family proteins for cancer therapy as tumor suppressors [17,86]. But if the hypothesis that they are tumor suppressors is wrong, cancer patients would not get any benefits. More data using siRNAs against ING family mRNA should be collected to help understand the physiological functions of the various family proteins. We propose to hold off any clinical trials that target ING family proteins until these data are properly evaluated.
Acknowledgments
We thank Dr. Yo Matsuo and Mr. Akira Togashi for helpful advice regarding the structure of the ING2 PHD domain and current molecular targeting therapies, and Dr. Tom Holroyd for editorial help.
References
- 1.Weiner GJ. Immunol Res. 2007;39(1–3):271–278. doi: 10.1007/s12026-007-0073-4. [DOI] [PubMed] [Google Scholar]
- 2.Chapuy B, Hohloch K, Trumper L. Biotechnol J. 2007;2(11):1435–1443. doi: 10.1002/biot.200700043. [DOI] [PubMed] [Google Scholar]
- 3.Durcan N, Murphy C, Cryan SA. Mol Pharm. 2008;5(4):559–566. doi: 10.1021/mp070048k. [DOI] [PubMed] [Google Scholar]
- 4.Harbour JW, Worley L, Ma D, Cohen M. Arch Ophthalmol. 2002;120(10):1341–1346. doi: 10.1001/archopht.120.10.1341. [DOI] [PubMed] [Google Scholar]
- 5.Carpiuc KT, Stephens JM, Botteman MF, Feng W, Hay JW. Expert Opin Pharmacother. 2007;8(16):2775–2787. doi: 10.1517/14656566.8.16.2775. [DOI] [PubMed] [Google Scholar]
- 6.Cappuzzo F, Finocchiaro G, Metro G, Bartolini S, Magrini E, Cancellieri A, Trisolini R, Castaldini L, Tallini G, Crino L. Crit Rev Oncol Hematol. 2006;58(1):31–45. doi: 10.1016/j.critrevonc.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Gridelli C, Rossi A, Maione P, Colantuoni G, Del Gaizo F, Ferrara C, Nicolella D, Guerriero C. Expert Opin Pharmacother. 2007;8(15):2579–2592. doi: 10.1517/14656566.8.15.2579. [DOI] [PubMed] [Google Scholar]
- 8.Utecht KN, Kolesar J. Am J Health Syst Pharm. 2008;65(13):1221–1231. doi: 10.2146/ajhp070272. [DOI] [PubMed] [Google Scholar]
- 9.Schuster SJ, Venugopal P, Kern JC, McLaughlin P. Leuk Lymphoma. 2008;49(9):1681–1692. doi: 10.1080/10428190802216731. [DOI] [PubMed] [Google Scholar]
- 10.Ismael G, Rosa DD, de Azambuja E, Braga S, Piccart-Gebhart M. Hematol Oncol Clin North Am. 2007;21(2):239–256. doi: 10.1016/j.hoc.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Rossi A, Bria E, Maione P, Palazzolo G, Falanga M, Gridelli C. Rev Recent Clin Trials. 2008;3(3):217–227. doi: 10.2174/157488708785700276. [DOI] [PubMed] [Google Scholar]
- 12.McCormack PL, Keam SJ. Drugs. 2008;68(4):487–506. doi: 10.2165/00003495-200868040-00009. [DOI] [PubMed] [Google Scholar]
- 13.Ho C, Ochsenbein AF, Gautschi O, Davies AM. Clin Lung Cancer. 2008;9(Suppl 1):S20–27. doi: 10.3816/clc.2008.s.004. [DOI] [PubMed] [Google Scholar]
- 14.Chiarle R, Martinengo C, Mastini C, Ambrogio C, D’Escamard V, Forni G, Inghirami G. Nat Med. 2008;14(6):676–680. doi: 10.1038/nm1769. [DOI] [PubMed] [Google Scholar]
- 15.Romero P. Clin Lung Cancer. 2008;9(Suppl 1):S28–36. doi: 10.3816/clc.2008.s.005. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa N, Takano A, Yasui W, Inai K, Nishimura H, Ito H, Miyagi Y, Nakayama H, Fujita M, Hosokawa M, Tsuchiya E, Kohno N, Nakamura Y, Daigo Y. Cancer Res. 2007;67(24):11601–11611. doi: 10.1158/0008-5472.CAN-07-3243. [DOI] [PubMed] [Google Scholar]
- 17.Ythier D, Larrieu D, Brambilla C, Brambilla E, Pedeux R. Int J Cancer. 2008;123(7):1483–1490. doi: 10.1002/ijc.23790. [DOI] [PubMed] [Google Scholar]
- 18.Kichina JV, Zeremski M, Aris L, Gurova KV, Walker E, Franks R, Nikitin AY, Kiyokawa H, Gudkov AV. Oncogene. 2006;25(6):857–866. doi: 10.1038/sj.onc.1209118. [DOI] [PubMed] [Google Scholar]
- 19.Coles AH, Liang H, Zhu Z, Marfella CG, Kang J, Imbalzano AN, Jones SN. Cancer Res. 2007;67(5):2054–2061. doi: 10.1158/0008-5472.CAN-06-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumamoto K, Fujita K, Kurotani R, Saito A, Unoki M, Hagiwara N, Shiga H, Bowman E, Yanaihara N, Okamura S, Nagashima M, Takenoshita S, Yokota J, Harris CC. Int J Cancer. 2009 doi: 10.1002/ijc.24437. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S, Yokota J, Harris CC. Cancer Res. 2008;68(9):3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unoki M, Kumamoto K, Robles AI, Shen JC, Zheng ZM, Harris CC. FEBS Lett. 2008;582(28):3868–3874. doi: 10.1016/j.febslet.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. Mol Cell. 2006;21(1):51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Roberts AB, Wakefield LM. Proc Natl Acad Sci USA. 2003;100(15):8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moller MB. Leuk Lymphoma. 2000;39(1–2):19–27. doi: 10.3109/10428190009053535. [DOI] [PubMed] [Google Scholar]
- 26.Shimada Y, Saito A, Suzuki M, Takahashi E, Horie M. Cytogenet Cell Genet. 1998;83(3–4):232–235. doi: 10.1159/000015188. [DOI] [PubMed] [Google Scholar]
- 27.Nagashima M, Shiseki M, Miura K, Hagiwara K, Linke SP, Pedeux R, Wang XW, Yokota J, Riabowol K, Harris CC. Proc Natl Acad Sci USA. 2001;98(17):9671–9676. doi: 10.1073/pnas.161151798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedeux R, Sengupta S, Shen JC, Demidov ON, Saito S, Onogi H, Kumamoto K, Wincovitch S, Garfield SH, McMenamin M, Nagashima M, Grossman SR, Appella E, Harris CC. Mol Cell Biol. 2005;25(15):6639–6648. doi: 10.1128/MCB.25.15.6639-6648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Wang J, Li G. FEBS Lett. 2006;580(16):3787–3793. doi: 10.1016/j.febslet.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 30.Pena PV, Hom RA, Hung T, Lin H, Kuo AJ, Wong RP, Subach OM, Champagne KS, Zhao R, Verkhusha VV, Li G, Gozani O, Kutateladze TG. J Mol Biol. 2008;380(2):303–312. doi: 10.1016/j.jmb.2008.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. Nature. 2006;442(7098):96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. Cell. 2003;114(1):99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 33.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. Genome Res. 2007;17(6):691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goeman F, Otto K, Kyrylenko S, Schmidt O, Baniahmad A. Biochim Biophys Acta. 2008;1783(10):1673–1680. doi: 10.1016/j.bbamcr.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 35.Lu F, Dai DL, Martinka M, Ho V, Li G. Br J Cancer. 2006;95(1):80–86. doi: 10.1038/sj.bjc.6603205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borkosky SS, Gunduz M, Nagatsuka H, Beder LB, Gunduz E, Al Sheikh Ali M, Rodriguez AP, Cilek MZ, Tominaga S, Yamanaka N, Shimizu K, Nagai N. J Cancer Res Clin Oncol. 2008 doi: 10.1007/s00432-008-0507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okano T, Gemma A, Hosoya Y, Hosomi Y, Nara M, Kokubo Y, Yoshimura A, Shibuya M, Nagashima M, Harris CC, Kudoh S. Oncol Rep. 2006;15(3):545–549. [PubMed] [Google Scholar]
- 38.Zhang HK, Pan K, Wang H, Weng DS, Song HF, Zhou J, Huang W, Li JJ, Chen MS, Xia JC. Cancer Lett. 2008;261(2):183–192. doi: 10.1016/j.canlet.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 39.Sarker KP, Kataoka H, Chan A, Netherton SJ, Pot I, Huynh MA, Feng X, Bonni A, Riabowol K, Bonni S. J Biol Chem. 2008;283(19):13269–13279. doi: 10.1074/jbc.M708834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meulmeester E, Jochemsen AG. Curr Cancer Drug Targets. 2008;8(2):87–97. doi: 10.2174/156800908783769337. [DOI] [PubMed] [Google Scholar]
- 41.Leeman MF, Curran S, Murray GI. Crit Rev Biochem Mol Biol. 2002;37(3):149–166. doi: 10.1080/10409230290771483. [DOI] [PubMed] [Google Scholar]
- 42.Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S, Onogi H, Higashimoto Y, Appella E, Yokota J, Harris CC. Cancer Res. 2003;63(10):2373–2378. [PubMed] [Google Scholar]
- 43.Zeitlin BD, Zeitlin IJ, Nor JE. J Clin Oncol. 2008;26(25):4180–4188. doi: 10.1200/JCO.2007.15.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iyer AK, Khaled G, Fang J, Maeda H. Drug Discov Today. 2006;11(17–18):812–818. doi: 10.1016/j.drudis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Shen Y. IDrugs. 2008;11(8):572–578. [PubMed] [Google Scholar]
- 46.Leung RK, Whittaker PA. Pharmacol Ther. 2005;107(2):222–239. doi: 10.1016/j.pharmthera.2005.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hashimoto S, Hirose M, Hashimoto A, Morishige M, Yamada A, Hosaka H, Akagi K, Ogawa E, Oneyama C, Agatsuma T, Okada M, Kobayashi H, Wada H, Nakano H, Ikegami T, Nakagawa A, Sabe H. Proc Natl Acad Sci USA. 2006;103(18):7036–7041. doi: 10.1073/pnas.0509166103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Nat Genet. 1996;14(4):415–420. doi: 10.1038/ng1296-415. [DOI] [PubMed] [Google Scholar]
- 49.Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV. Nature. 1998;391 (6664):295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 50.Helbing CC, Veillette C, Riabowol K, Johnston RN, Garkavtsev I. Cancer Res. 1997;57(7):1255–1258. [PubMed] [Google Scholar]
- 51.Shinoura N, Muramatsu Y, Nishimura M, Yoshida Y, Saito A, Yokoyama T, Furukawa T, Horii A, Hashimoto M, Asai A, Kirino T, Hamada H. Cancer Res. 1999;59(21):5521–5528. [PubMed] [Google Scholar]
- 52.Scott M, Bonnefin P, Vieyra D, Boisvert FM, Young D, Bazett-Jones DP, Riabowol K. J Cell Sci. 2001;114(Pt 19):3455–3462. doi: 10.1242/jcs.114.19.3455. [DOI] [PubMed] [Google Scholar]
- 53.Soliman MA, Berardi P, Pastyryeva S, Bonnefin P, Feng X, Colina A, Young D, Riabowol K. Aging Cell. 2008;7(6):783–794. doi: 10.1111/j.1474-9726.2008.00427.x. [DOI] [PubMed] [Google Scholar]
- 54.Jager D, Stockert E, Scanlan MJ, Gure AO, Jager E, Knuth A, Old LJ, Chen YT. Cancer Res. 1999;59(24):6197–6204. [PubMed] [Google Scholar]
- 55.Sanchez-Carbayo M, Socci ND, Lozano JJ, Li W, Charytonowicz E, Belbin TJ, Prystowsky MB, Ortiz AR, Childs G, Cordon-Cardo C. Am J Pathol. 2003;163(2):505–516. doi: 10.1016/S0002-9440(10)63679-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeremski M, Hill JE, Kwek SS, Grigorian IA, Gurova KV, Garkavtsev IV, Diatchenko L, Koonin EV, Gudkov AV. J Biol Chem. 1999;274(45):32172–32181. doi: 10.1074/jbc.274.45.32172. [DOI] [PubMed] [Google Scholar]
- 57.Feng X, Hara Y, Riabowol K. Trends Cell Biol. 2002;12(11):532–538. doi: 10.1016/s0962-8924(02)02391-7. [DOI] [PubMed] [Google Scholar]
- 58.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Nature. 2006;442(7098):100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xin H, Yoon HG, Singh PB, Wong J, Qin J. J Biol Chem. 2004;279(10):9539–9546. doi: 10.1074/jbc.M311587200. [DOI] [PubMed] [Google Scholar]
- 60.Han X, Feng X, Rattner JB, Smith H, Bose P, Suzuki K, Soliman MA, Scott MS, Burke BE, Riabowol K. Nat Cell Biol. 2008;10(11):1333–1340. doi: 10.1038/ncb1792. [DOI] [PubMed] [Google Scholar]
- 61.Kim S, Chin K, Gray JW, Bishop JM. Proc Natl Acad Sci USA. 2004;101(46):16251–16256. doi: 10.1073/pnas.0407158101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Unoki M, Shen JC, Zheng ZM, Harris CC. J Biol Chem. 2006;281(45):34677–34686. doi: 10.1074/jbc.M606296200. [DOI] [PubMed] [Google Scholar]
- 63.Soussi T. Ann N Y Acad Sci. 2000;910:121–137. doi: 10.1111/j.1749-6632.2000.tb06705.x. discussion 137–129. [DOI] [PubMed] [Google Scholar]
- 64.Jounaidi Y, Doloff JC, Waxman DJ. Curr Cancer Drug Targets. 2007;7(3):285–301. doi: 10.2174/156800907780618301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagashima M, Shiseki M, Pedeux RM, Okamura S, Kitahama-Shiseki M, Miura K, Yokota J, Harris CC. Oncogene. 2003;22(3):343–350. doi: 10.1038/sj.onc.1206115. [DOI] [PubMed] [Google Scholar]
- 66.He GH, Helbing CC, Wagner MJ, Sensen CW, Riabowol K. Mol Biol Evol. 2005;22(1):104–116. doi: 10.1093/molbev/msh256. [DOI] [PubMed] [Google Scholar]
- 67.Gunduz M, Ouchida M, Fukushima K, Ito S, Jitsumori Y, Nakashima T, Nagai N, Nishizaki K, Shimizu K. Oncogene. 2002;21(28):4462–4470. doi: 10.1038/sj.onc.1205540. [DOI] [PubMed] [Google Scholar]
- 68.Gunduz M, Beder LB, Gunduz E, Nagatsuka H, Fukushima K, Pehlivan D, Cetin E, Yamanaka N, Nishizaki K, Shimizu K, Nagai N. Cancer Sci. 2008;99(3):531–538. doi: 10.1111/j.1349-7006.2007.00708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Dai DL, Martinka M, Li G. Clin Cancer Res. 2007;13(14):4111–4116. doi: 10.1158/1078-0432.CCR-07-0408. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, Li G. J Biol Chem. 2006;281(17):11887–11893. doi: 10.1074/jbc.M511309200. [DOI] [PubMed] [Google Scholar]
- 71.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Mol Cell Biol. 2004;24(5):1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raho G, Miranda C, Tamborini E, Pierotti MA, Greco A. Oncogene. 2007;26(36):5247–5257. doi: 10.1038/sj.onc.1210335. [DOI] [PubMed] [Google Scholar]
- 73.Tsai KW, Lin WC. Genomics. 2006;88(6):855–864. doi: 10.1016/j.ygeno.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 74.Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. Nat Genet. 2004;36(12):1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
- 75.Tadokoro K, Yamazaki-Inoue M, Tachibana M, Fujishiro M, Nagao K, Toyoda M, Ozaki M, Ono M, Miki N, Miyashita T, Yamada M. J Hum Genet. 2005;50(8):382–394. doi: 10.1007/s10038-005-0261-9. [DOI] [PubMed] [Google Scholar]
- 76.Tsai KW, Tseng HC, Lin WC. Exp Cell Res. 2008;314(17):3130–3141. doi: 10.1016/j.yexcr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 77.Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. Am J Hum Genet. 2006;78(2):291–302. doi: 10.1086/500151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X, Wang KS, Wang ZQ, Xu LS, Wang QW, Chen F, Wei DZ, Han ZG. Biochem Biophys Res Commun. 2005;331(4):1032–1038. doi: 10.1016/j.bbrc.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 79.Ozer A, Wu LC, Bruick RK. Proc Natl Acad Sci USA. 2005;102(21):7481–7486. doi: 10.1073/pnas.0502716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, Barnett GH, Jain RK. Nature. 2004;428(6980):328–332. doi: 10.1038/nature02329. [DOI] [PubMed] [Google Scholar]
- 81.Shen JC, Unoki M, Ythier D, Duperray A, Varticovski L, Kumamoto K, Pedeux R, Harris CC. Cancer Res. 2007;67(6):2552–2558. doi: 10.1158/0008-5472.CAN-06-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nozell S, Laver T, Moseley D, Nowoslawski L, De Vos M, Atkinson GP, Harrison K, Nabors LB, Benveniste EN. Mol Cell Biol. 2008;28(21):6632–6645. doi: 10.1128/MCB.00697-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu E, Wu J, Cao W, Zhang J, Liu W, Jiang X, Zhang X. J Neurooncol. 2007;85(3):263–270. doi: 10.1007/s11060-007-9421-4. [DOI] [PubMed] [Google Scholar]
- 84.Zhang X, Xu LS, Wang ZQ, Wang KS, Li N, Cheng ZH, Huang SZ, Wei DZ, Han ZG. FEBS Lett. 2004;570(1–3):7–12. doi: 10.1016/j.febslet.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 85.Champagne KS, Saksouk N, Pena PV, Johnson K, Ullah M, Yang XJ, Cote J, Kutateladze TG. Proteins. 2008;72(4):1371–1376. doi: 10.1002/prot.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gunduz M, Gunduz E, Rivera RS, Nagatsuka H. Curr Cancer Drug Targets. 2008;8(4):275–284. doi: 10.2174/156800908784533454. [DOI] [PubMed] [Google Scholar]
- 87.Soliman MA, Riabowol K. Trends Biochem Sci. 2007;32(11):509–519. doi: 10.1016/j.tibs.2007.08.006. [DOI] [PubMed] [Google Scholar]