Abstract
Purpose
To screen ten genes for mutations in 32 Chinese patients with microphthalmia and/or coloboma.
Methods
Genomic DNA was prepared from 32 unrelated patients with microphthalmia (nine probands) and uveal coloboma (23 probands). Cycle sequencing was used to detect sequence variations in ten genes, including BMP4, VSX2, CRYBA4, GDF6, OTX2, RAX, SIX3, SIX6, SOX2, and LRP6. Variations were further evaluated in 96 unrelated controls by using restriction fragment length polymorphism (RFLP) or heteroduplex-single strand conformation polymorphism (HA-SSCP) analysis.
Results
In the ten genes, a novel c.751C>T (p.H251Y) in BMP4 was detected in a patient with bilateral microphthalmia and unilateral cataract. The c.751C>T variation is also present in his healthy brother (and possibly one of the normal parents). In addition, a novel c.608G>A (p.R203Q) in SIX6 was identified in an internal control for optimizing experimental conditions. The internal control was from a girl with typical aniridia and an identified c.718C>T (p.R240X) mutation in PAX6, suggesting the c.608G>A variation in SIX6 was unlikely to play a role in her ocular phenotype. The c.751C>T in BMP4 and the c.608G>A in SIX6 were not present in the 96 normal controls. In addition, 16 nucleotide substitutions, including eight known SNPs and eight new synonymous changes, were detected.
Conclusions
Although the genetic etiology for microphthalmia and/or coloboma is still elusive, rare variations in the related genes, such as c.608 G>A in SIX6 and c.751C>T in BMP4, may not be causative. These results further emphasize the importance of careful clinical and genetic analysis in making mutation-disease associations.
Introduction
Microphthalmia and coloboma are important cause of congenital blindness, with a prevalence at 1.9–3.5/10,000 live births [1,2]. Microphthalmia and coloboma may be isolated or syndromic with the extraocular phenotype in one or both eyes. The disease exhibits diverse patterns of genetic inheritance, and the severity is variable, due to the genetic heterogeneity of the ocular malformation [3-8]. Mutations in several genes have been reported in patients with microphthalmia and/or coloboma [3,9-14], including BMP4 (OMIM 112262), VSX2 (CHX10; OMIM 142993), CRYBA4 (OMIM 123631), OTX2 (OMIM 600037), RAX (OMIM 601881), SIX6 (OMIM 606326), and SOX2 (OMIM 184429). Of these, mutations in SOX2 account for about 10% of microphthalmia, anophthalmia, and coloboma [3,15,16]. However, mutations in VSX2, CRYBA4, OTX2, and RAX have been detected in about 2%–3% patients with microphthalmia, anophthalmia, and coloboma [10,11,15,17,18]. Mutations in SIX3 and GDF6 have been identified only in a few cases [19,20]. Recently, mutations in BMP4 mutations have been detected in patients with anophthalmia-microphthalmia [9]. In addition, knockout of LRP6 in mice resulted in microphthalmia and coloboma, but has not yet been reported in humans [21].
Because most of these genes were usually studied individually, and mutation analysis of Chinese patients is rare so far, we screened 32 unrelated patients with microphthalmia and/or coloboma for mutations in ten related genes, including BMP4, VSX2, CRYBA4, GDF6, OTX2, RAX, SIX3, SIX6, SOX2, and LRP6, through sequencing analysis of the coding and adjacent intronic regions of the ten genes.
Methods
Patients and controls
Thirty-two unrelated patients were recruited from our Pediatric and Genetic Eye Clinic, Zhongshan Ophthalmic Center. Clinical diagnoses of the 32 patients were microphthalmia (nine cases) and uveal coloboma (23 cases). Diagnosis of microphthalmia was based on criteria previously described [15], that is, a corneal diameter less than 10 mm and an axial length less than 20 mm. Of the nine cases with microphthalmia, eight met the criteria but one did not, although a small cornea and short axial length were recorded (details in results). On the other hand, the inclusion criteria for uveal coloboma were 1) congenital cleft in the inferior part of the iris and/or choroid (Figure 1) and 2) exclusion of aniridia or macular uveal coloboma. Besides this, an internal control sample for optimizing PCR and sequencing conditions was from a girl with aniridia and an identified PAX6 mutation. Furthermore, 96 unrelated controls were collected from normal volunteers. A previously established procedure was used for collecting subjects and obtaining informed consent [22]. This study was approved by the Institutional Review Board of Zhongshan Ophthalmic Center, China. Genomic DNA was prepared from venous blood from each participating individual [23].
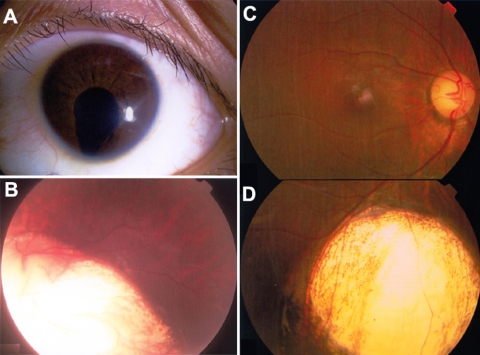
Figure 1.
Uveal coloboma. A and B demonstrate iris coloboma (A) and choroid coloboma involving the optic disc (B). C and D show inferior choroid coloboma. (D did not align well with C).
Mutation detection
PCR was used to amplify the coding exons and adjacent intronic sequences of the ten genes. Reference sequences for the ten genes are listed in Table 1. The primer sequences used to amplify the coding exons of the ten genes are listed in Table 2. The PCR products from individual exons from each individual were sequenced with the ABI BigDye Terminator cycle sequencing kit v3.1 (Applied Biosystems, Foster City, CA) and an ABI 3100 Genetic Analyzer (Applied Biosystems). Sequencing results and consensus sequences from the NCBI human genome database were compared by using the SeqManII program of the Lasergene package (DNAstar Inc., Madison, WI). Each mutation was confirmed by bidirectional sequencing. Mutation description followed the nomenclature recommended by the Human Genomic Variation Society.
Table 1. Genomic information of the 10 genes referred in this study.
| Gene | Genomic DNA | mRNA | Protein |
|---|---|---|---|
|
BMP4 |
NC_000014.8 |
NM_001202.3 |
NP_001193.2 |
|
VSX2 |
NC_000014.8 |
NM_182894.2 |
NP_878314.1 |
|
CRYBA4 |
NC_000022.10 |
NM_001886.2 |
NP_001877.1 |
|
GDF6 |
NC_000008.10 |
NM_001001557.1 |
NP_001001557.1 |
|
OTX2 |
NC_000014.8 |
NM_021728.2 |
NP_068374.1 |
|
RAX |
NC_000018.9 |
NM_013435.2 |
NP_038463.2 |
|
SIX3 |
NC_000002.11 |
NM_005413.2 |
NP_005404.1 |
|
SIX6 |
NC_000014.8 |
NM_007374.2 |
NP_031400.2 |
|
SOX2 |
NC_000003.11 |
NM_003106.2 |
NP_003097.1 |
| LRP6 | NC_000012.11 | NM_002336.2 | NP_002327.2 |
The genomic DNA information was based on NCBI human genome build 36.3.
Table 2. Primers used for polymerase chain reaction amplification and sequencing of BMP4, VSX2, CRYBA4, GDF6, OTX2, RAX, SIX3, SIX6, SOX2, and LRP6.
| Gene | Exon | Primer sequence (5'-3') | Amplicon size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
|
BMP4 |
3-F |
CCATCTTGCCCCTCCATTTCTA |
570 |
65 |
| |
3-R |
CTTCTTCCCCAGGGCTTTCACT |
|
|
| |
4a-F |
TGCTTATTTTCCCCCAGTAGGT |
704 |
62 |
| |
4a-R |
GGCGCCGGCAGTTCTTATTCTT |
|
|
| |
4b-F |
GGGCCAGCATGTCAGGATTAGC |
575 |
62 |
| |
4b-R |
TGTGGGTGAGTGGATGGGAACG |
|
|
| |
RFLP-F |
CGGGAGAAGCAGCCAAACTATG |
237 |
65 |
| |
RFLP-R |
CTTCTTCCTGGCCCGCTGTGAG |
|
|
|
VSX2 |
1-F |
AGGGGGCACCTGGGACCAAC |
586 |
72 |
| |
1-R |
CCCGGCCTGGCAGGAACTTT |
|
|
| |
2-F |
CGGCGCGGGAGAGGTCAG |
427 |
66 |
| |
2-R |
GCAGATTCCGCCAAACAAC |
|
|
| |
3-F |
TGCCCAGGAGACACAGAGG |
347 |
64 |
| |
3-R |
ACATGATCATAGGCAAGCACA |
|
|
| |
4-F |
AGGACGCCCTGCTGGAGAAA |
380 |
68 |
| |
4-R |
CCAAGTGCCCCTGCCTCAA |
|
|
| |
5-F |
CTGCGGTGTGGGGAGTAAG |
574 |
65 |
| |
5-R |
GTGGGGAACAGGGAGGATG |
|
|
|
CRYBA4 |
2-F |
ACTCCTGGACTCCCTATGTG |
319 |
60 |
| |
2-R |
ATTCAACCTCCCTGTATGTG |
|
|
| |
3-F |
TCTTGCCTTCCTGGCTCCTG |
430 |
62 |
| |
3-R |
TGCGCAACCTGCATAATCTT |
|
|
| |
4-F |
CCCCTGAATGGTTGTGACT |
396 |
62 |
| |
4-R |
AACCGAGGCTTGGAGAGGAA |
|
|
| |
5-F |
AAGGGCAAATGGCAAGGTT |
415 |
62 |
| |
5-R |
TGGGCATCAGAGCACAAAAG |
|
|
|
GDF6 |
1-F |
GGCGGGGCCGGGGTTTGT |
635 |
72 |
| |
1-R |
TAGCCTCCAGCGGGAACAGC |
|
|
| |
2a-F |
CGGCCGACCTGCCCCCACTC |
504 |
70 |
| |
2a-R |
CCGGCCGAAGCCCAGACTCC |
|
|
| |
2b-F |
GCCGGCCGGCTGGGAAGTCT |
557 |
70 |
| |
2b-R |
GCGAGCGCAGCGGGAAGTCG |
|
|
| |
2c-F |
GCGCACGGCCTTCGCCAGTC |
469 |
70 |
| |
2c-R |
CCAGCGCCAGCTTCCTCCTC |
|
|
|
OTX2 |
3-F |
TTTGCTTTGCCCTTAGTTCC |
424 |
62 |
| |
3-R |
CCCTGTTCTCTGCTTGGTCA |
|
|
| |
4-F |
ACGGTGGGGAGAGCATTGGT |
445 |
62 |
| |
4-R |
CCTGGCCCCTTAGTGAGTGA |
|
|
| |
5a-F |
CTGCCCATGTAGGATAGATT |
440 |
61 |
| |
5a-R |
ATGCCCCCAAAGTAGGAAGT |
|
|
| |
5b-F |
GCTTCCATCTCCCCACTGTC |
562 |
61 |
| |
5b-R |
GGCCCTTCGTTTTTCCTTCT |
|
|
|
RAX |
1-F |
TTCGCCCGCGGAGCTTGACCT |
527 |
66 |
| |
1-R |
CCCCAACCCCGCGCCCAGTT |
|
|
| |
2-F |
CCATCGCCGCCCTCACCA |
522 |
66 |
| |
2-R |
ACTCTGGGCATGCCAAGTCG |
|
|
| |
3-F |
TTGAGGGGGACGGAGTGGAG |
716 |
69 |
| |
3-R |
GCAGGCGACAGGGAAAGAGG |
|
|
|
SIX3 |
1a-F |
TCATCGCCCCTCTCCTCCTCTT |
398 |
70 |
| |
1a-R |
GCCGCTCGATGTCGCCCGTCTC |
|
|
| |
1b-F |
CGGCGGCGGCGGCTCCAG |
387 |
72 |
| |
1b-R |
CGCACGCGGTACTTGTCCAC |
|
|
| |
1c-F |
GCGTGCGAGGCCATCAACAA |
552 |
65 |
| |
1c-R |
CACGGCTTCCCTGGCTCTCA |
|
|
| |
2-F |
GCTCGGGTTCTGCCTCTC |
457 |
64 |
| |
2-R |
TCGGTTTGTTCTGGGGATGG |
|
|
|
SIX6 |
1a-F |
TGTGTCCCGCTGCCCCAATC |
514 |
65 |
| |
1a-R |
TTCTGTTCGCCGTCCCAAATG |
|
|
| |
1b-F |
CCTTTCACGGTGGCAACTAC |
514 |
65 |
| |
1b-R |
GACAGACCGCGCTCCCAACTC |
|
|
| |
2-F |
CGCCTTGCCGAGTAATCCT |
447 |
70 |
| |
2-R |
AGCCCGCGGGTCCCTGGTCAC |
|
|
| |
HA-SSCP-F |
TCGCCTTAACTGCTGGGGTCTT |
253 |
67 |
| |
HA-SSCP-R |
AGTGGCCGCCTTGCTGGATA |
|
|
|
SOX2 |
1-F |
CGCCTCCCCTCCTCCTCTC |
443 |
68 |
| |
1-R |
CGCCGGGGCCGGTATTTAT |
|
|
| |
2-F |
GGGCGCCGAGTGGAAACTT |
473 |
65 |
| |
2-R |
GGGTGCCCTGCTGCGAGTA |
|
|
| |
3-F |
CACGGCGCAGCGCAGATGC |
459 |
65 |
| |
3-R |
TTTGCACCCCTCCCATTTC |
|
|
|
LRP6 |
1-F |
CTCCTCGCCTCCCCCACTTCTG |
279 |
72 |
| |
1-R |
CTGCTCCCGGGCCCCTTTCTCT |
|
|
| |
2-F |
ATTTTCGACAGTCTTTGCTCAC |
589 |
65 |
| |
2-R |
TTCTTTTCTCATAGGGGTCAGG |
|
|
| |
3-F |
GCGCGGCCTGAGCTTTCTTTA |
410 |
64 |
| |
3-R |
CTTCTTCCCCTCTGGCACTTAG |
|
|
| |
4-F |
ATTTTAATGGGAGAGGTGACG |
395 |
60 |
| |
4-R |
TTTATTCCCGCCAACTATCTTT |
|
|
| |
5-F |
AATTTTGGCTTATCACAGTT |
330 |
58 |
| |
5-R |
GGTCTCCCAAAGCAGTAT |
|
|
| |
6-F |
TTTTATATTTATTTTTCAGTTC |
575 |
50 |
| |
6-R |
ATGTTATCTTAGTCAATGTTTT |
|
|
| |
7-F |
GGGATGGATCTCACCTTTAG |
478 |
58 |
| |
7-R |
GATCAGCAGCCATTTCTCA |
|
|
| |
8-F |
GGGGGAAAAGTGGTCAAA |
538 |
56 |
| |
8-R |
GGGGGCAGTAAAGAAGGT |
|
|
| |
9-F |
TGGGAGCAAGACATAATCATAG |
690 |
64 |
| |
9-R |
TGGCACGCACCTGTAGTCCT |
|
|
| |
10-F |
GGATCCTCTTGCCCCTGACA |
560 |
62 |
| |
10-R |
TAACCCATTCCCCTCTTTCTTC |
|
|
| |
11-F |
ATTGTAGCCGTGATTTTGTTTA |
577 |
58 |
| |
11-R |
TCAGGAGTATCTAGGGAGTTAT |
|
|
| |
12-F |
AAGCATGGGGTCAGAAGATAGA |
777 |
62 |
| |
12-R |
AAAGTGCTGGGTTACGGACATA |
|
|
| |
13-F |
TGAGGGCATGCCAAAGAAT |
499 |
58 |
| |
13-R |
AATAAGCTACCAGGTCCAGAAT |
|
|
| |
14-F |
GTGTGCCCATGTAGGTGTAAGC |
511 |
64 |
| |
14-R |
TAGTGGCCCAGGAAAGAAAGTC |
|
|
| |
15-F |
CCGCCTCAGTCTCCCAAAGT |
491 |
68 |
| |
15-R |
TGCCAAGAAATGTGCCAAAAAC |
|
|
| |
16-F |
TATCTAGTTTATTGGCTGTT |
506 |
52 |
| |
16-R |
CTAAAAGTGCATGAAAGTCT |
|
|
| |
17-F |
AAGCTGATTATACATTTGATTT |
403 |
64 |
| |
17-R |
GGGCAGGGTGGCAGAGAA |
|
|
| |
18-F |
TAAAGGAAGTAATGTGAAAACC |
521 |
58 |
| |
18-R |
TGAAAAACCCCAACTGAC |
|
|
| |
19-F |
AGGCACCTTTTGATTCTTG |
495 |
61 |
| |
19-R |
CGCCCGGCTGATTTCTATGTAT |
|
|
| |
20-F |
TTCAGGGCGTGGTATGTATGT |
578 |
58 |
| |
20-R |
TATCTAAGGCCTTCTGTGTAAA |
|
|
| |
21-F |
AGCTATTCTTGGCCTTGTTCTA |
508 |
61 |
| |
21-R |
AGTCCTTTGAGCCTTTTATGC |
|
|
| |
22-F |
TTTTAGCCATGATGAGGTCTTA |
373 |
64 |
| |
22-R |
GGGGCTATATCAGGTCCACAAC |
|
|
| |
23-F |
GAAAATTGCCTCTTGGTCTGTG |
550 |
65 |
| 23-R | TGGTCTGCCTCATCCTTCTCTA |
Heteroduplex-single strand conformation polymorphism analysis
The c.608G>A variation detected in SIX6 was further evaluated in 96 normal controls by heteroduplex-single strand conformation polymorphism (HA-SSCP) analysis, as previously described [24], using an extra pair of primers (Table 2). Briefly, PCR products were mixed with an equal volume of formamide dye loading buffer. Then 1–4 μl of the mixture was loaded on 40 cm×30 cm×1 mm 8% polyacrylamide gels containing 10% glycerol. The DNA samples were separated by electrophoresis for 8–9 h at room temperature without temperature control. The DNA fragments were visualized by silver staining.
Restriction fragment-length polymorphism analysis
The variation detected in BMP4 c.751C>T was further evaluated in available family members, as well as in 96 normal controls, by restriction fragment-length polymorphism (RFLP) analysis using an extra pair of primers (Table 2). Since the c.751C>T variation in BMP4 erased an enzyme recognition site of CviAII, wild amplicons were digested into four fragments (78, 76, 68, and 15 bp) while the variant amplicons were cut into three pieces (154, 68, and 15 bp).
Results
Mutation analysis
Eighteen nucleotide substitutions (Table 3), including two novel missense variations, eight known SNPs, and eight new synonymous changes, were detected upon complete sequencing analysis of the coding exons and the adjacent intronic regions of BMP4, VSX2, CRYBA4, GDF6, OTX2, RAX, SIX3, SIX6, SOX2, and LRP6. Of the two novel heterozygous missense variations, one was c.608G>A (p.R203Q) in SIX6 and the other was c.751C>T (p.H251Y) in BMP4.
Table 3. Sequence variations found in BMP4, CRYBA4, GDF6, LRP6, RAX, SIX3, SIX6, SOX2, and VSX2.
| Gene | Exon | Sequence variation | Amino acid change | Patient number | Result |
|---|---|---|---|---|---|
| BMP4 |
4 |
c.455T>C |
S155S |
TT:TC:CC = 19:10:3 |
rs17563 |
| |
4 |
c.751C>T |
H251Y |
1 |
Novel variation |
| CRYBA4 |
Intron 2 |
c.40-71C>T |
No splice site change |
2 |
rs2071860 |
| |
Intron 3 |
c.158+58C>T |
No splice site change |
3 |
rs58707060 |
| |
Intron 3 |
c.159-20A>G |
No splice site change |
AA:A/G:GG = 8:19:13 |
rs59023621 |
| |
Intron 5 |
c.444-18g>a |
No splice site change |
4 |
|
| GDF6 |
1 |
c.255G>T |
P85P |
1 |
|
| LRP6 |
5 |
c.867C>T |
D289D |
1 |
|
| |
11 |
c.2450C>G |
S817C |
1 |
rs2302686 |
| |
14 |
c.3184G>A |
V1062I |
5 |
rs2302685 |
| RAX |
1 |
c.132C>A |
D44E |
6 |
rs2271733 |
| |
3 |
c.882A>G |
Q294Q |
4 |
|
| SIX3 |
1 |
c.90G>T |
A30A |
4 |
|
| SIX6 |
1 |
c.421C>A |
N141K |
7 |
rs33912345 |
| |
2 |
c.637C>T |
P213S |
1 |
|
| |
2 |
c.608G>A |
R203Q |
1 |
Novel variation |
| SOX2 |
1 |
c.573A>G |
A191A |
1 |
|
| VSX2 | 4 | c.750G>A | P250P | 1 |
No variation was identified in OTX2.
For internal quality control, the c.608G>A variation in SIX6 was detected in an individual when her sample was used to optimize the experimental condition, but was not present in 96 unrelated normal controls. She was a three-month-old girl who had typical congenital aniridia with normal cornea size (a bilateral cornea diameter of 10 mm at the age of 3 months, within the normal range at this age) and a previously determined novel PAX6 mutation (c.718C>T, p.R240X). This suggested that the c.608G>A variation in SIX6 did not play additive effect and, therefore, might not be causative.
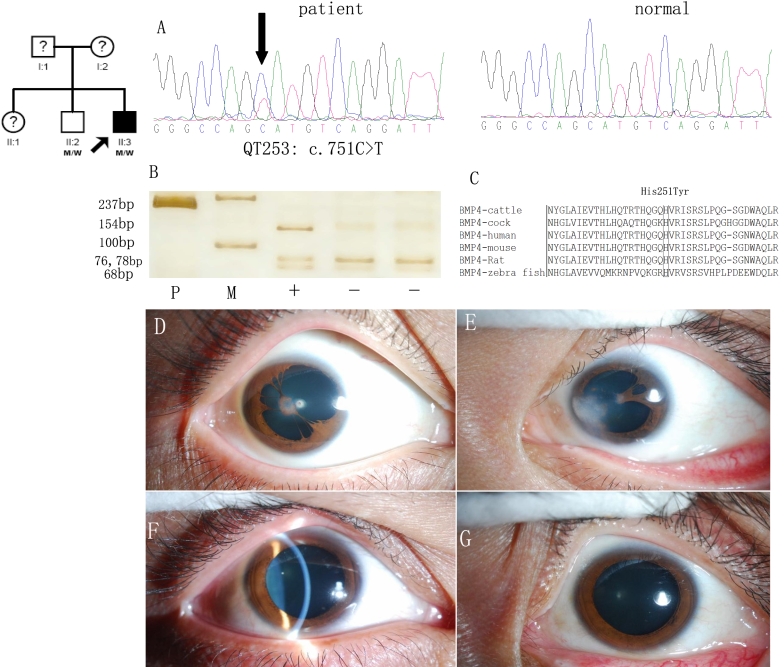
The c.751C>T variation in BMP4 was detected in a proband suspected for microphthalmia (Figure 2), but was not detected in 96 unrelated normal controls. BMP4 alignment among six different species showed that the residue at 251 of BMP4 protein is highly conserved (Figure 2C). This ocular biometry measurement did not fully meet the criteria for micropthalmia, but did demonstrate an obviously small cornea and short axial length (Table 4). Besides this, he had bilateral corneal opacities, multiple pupils, an persistent iris membrane, and anterior pole cataract (Figure 2D-E). Unexpectedly, the c.751C>T variation was also present in his healthy brother with a normal ocular phenotype, including a normal anterior segment and normal axial length (Table 4 and Figure 2F-G). His sister (II:1 in Figure 2) and parents (I:1 and I:2 in Figure 2) were reported to be normal, but were unavailable to have ocular biometry. The variation was present in both the proband and in his healthy brother, and at least one of the parents (in whom only the proband had an abnormal ocular phenotype).
Figure 2.
BMP4 variation and associated phenotype. A: Sequence chromatogram demonstrated the c.751C>T variation in BMP4 from the patient (left) and normal sequence from a control (right). B: The c.751C>T variation in BMP4 detected by PCR-RFLP analysis (P: PCR products [237 bp] and M: marker, showing 100 bp and 250 bp, respectively; the plus sign [+] indicates CviAII-digested-products with heterozygous c.751C>T variation; the minus sign [-] indicates CviAII-digested products without the c.751C>T variation). C: Protein alignment of human BMP4 (residues 231–271) with other BMP4 orthologs from cattle, cock, mouse, rat, and zebra fish. D and E: Ocular phenotype of the proband showing bilateral microcornea, corneal opacities, multiple pupils, persistent iris membrane adhering to cornea (right eye) or lens capsule (left eye), and anterior pole cataract (right eye). F and G: Normal ocular phenotype of the proband’s healthy brother, which also carried the heterozygous c.751C>T variation in BMP4.
Table 4. Ocular biometry of the individuals with the BMP4 mutation.
| Individual | Eye | Best visual acuity | Cornea diameter (mm) | Anterior chamber depth (mm) | Ocular length (mm) |
|---|---|---|---|---|---|
| Proband |
OD |
0.5 |
10.7 |
2.25 |
20.28 |
| |
OS |
0.3 |
10.8 |
2.23 |
20.13 |
| Healthy brother |
OD |
1.5 |
11.4 |
3.39 |
22.78 |
| OS | 1.5 | 11.6 | 3.32 | 22.74 |
Ocular biometry was measured by using IOL Master V5 (Carl Zeiss Meditec AG, Jena, Germany). The proband had a small cornea and short axial length. His brother had normal ocular biometry (normal range in Chinese adults: 12.12±0.40 mm for cornea diameter and 23.60±0.79 mm for axial length).
Discussion
Normal development of the eye involves a complex process. Both genetic and environmental factors may play roles in the malformation of the eye. Although mutations in several genes have been detected in patients with microphthalmia or coloboma, such mutations are only detected in a small percentage of patients. In addition, these genes have not been analyzed simultaneously in any cohort of microphthalmia and/or coloboma cases.
In the present study, ten genes previously reported to be responsible for microphthalmia and/or uveal coloboma were analyzed simultaneously in 32 Chinese patients with microphthalmia and/or uveal coloboma. Upon complete screening of the coding exons and adjacent intronic regions of BMP4, VSX2, CRYBA4, GDF6, OTX2, RAX, SIX3, SIX6, SOX2, and LRP6, no causative mutation was detected. This is the first systemic analysis of all ten genes in a series of microphthalmia and coloboma patients, and is the first analysis of most such genes in Chinese patients. The results suggest that the genetic cause of microphthalmia and uveal coloboma in Chinese is largely unknown.
SIX6 encodes a nuclear homeoprotein and is expressed in the developing retina, optic stalk, and the hypothalamic and pituitary regions. Interstitial deletions at 14q22.3-q23, where SIX6 is located, were found in three patients with bilateral anophthalmia, absence of the optic nerve, and chiasm and pituitary abnormalities [25]. Gallardo et al. [14] identified a heterozygous c.493A>G (p.T165A) variation in SIX6 in a patient with bilateral microphthalmia, cataract, and nystagmus from among a series of 73 patients with syndromic or nonsyndromic sporadic clinical anophthalmia and microphthalmia. However, this variation was also present in the healthy father, although it was not detected in more than 160 chromosomes from normal individuals. Aijaz et al. [26] did not find SIX6 mutation 173 patients with microphthalmia, anophthalmia, and coloboma. In that study, the c.608G>A variation in SIX6 was detected in an individual with typical congenital aniridia and a previously determined PAX6 mutation (c.718C>T, p.R240X). Overall, there is no firm evidence that mutation in SIX6 alone can cause microphthalmia, anophthalmia, or coloboma.
The BMP4 gene product is a regulatory molecule functioning in mesoderm induction, tooth development, limb formation, bone induction, and fracture repair. BMP4 is located in 14q22-q23, where recurrent interstitial deletions have been associated with anophthalmia-microphthalmia [9]. Bakrania et al. [9] identified a c.226del2 (p.S76fs104X) mutation at BMP4 in a family whose carrier members had various phenotypes, including anophthalmia-microphthalmia, retinal dystrophy, myopia, brain anomalies, and polydactyly. In another family, a c.278A>G (p.E93G) mutation was found. However, these two mutations were also present in one of the phenotypically normal parents from each family [9]. On the other hand, three missense mutations in BMP4 were reported in patients with orofacial cleft 11 (OMIM 600625) [27]. In that study, the c.751C>T variation in BMP4 was present in a patient with microphthalmia, as well as in his healthy brother, and possibly in one of his normal parents. Therefore, further study is needed to reveal the role of BMP4 in micropthalmia.
In summary, the c.608G>A variation in SIX6 and the c.751C>T variation in BMP4 might be reported as causative mutations if the SIX6 c.608G>A variation is detected in a patient without a confirmed genetic basis, and if cosegregation analysis is not performed for the BMP4 c.751C>T variation. Additional studies are expected to validate the association of microphthalmia and uveal coloboma with mutations in SIX6 and BMP4. Great care is needed in making mutation–disease associations based on marginal evidence, especially for those genes with only a few identified mutations. Future genetic analyses of additional patients, as well as of other candidate genes, may enrich our understanding of the molecular basis of microphthalmia and uveal coloboma, as well as of the genotype–phenotype correlation.
Acknowledgments
The authors thank all patients and controls for their participation. This study was supported by grant 30725044 from the National Science Fund for Distinguished Young Scholars (to Qingjiong Zhang).
References
- 1.Morrison D, FitzPatrick D, Hanson I, Williamson K, van Heyningen V, Fleck B, Jones I, Chalmers J, Campbell H. National study of microphthalmia, anophthalmia, and coloboma (MAC) in Scotland: investigation of genetic aetiology. J Med Genet. 2002;39:16–22. doi: 10.1136/jmg.39.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell H, Holmes E, MacDonald S, Morrison D, Jones I. A capture-recapture model to estimate prevalence of children born in Scotland with developmental eye defects. J Cancer Epidemiol Prev. 2002;7:21–8. doi: 10.1080/14766650252962649. [DOI] [PubMed] [Google Scholar]
- 3.Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–3. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 4.Sandler D, Mancuso A, Becker T, Zori R, Hellrung J, Silverstein J, Burton V, Hamosh A, Williams C. Association of anophthalmia and esophageal atresia. Am J Med Genet. 1995;59:484–91. doi: 10.1002/ajmg.1320590415. [DOI] [PubMed] [Google Scholar]
- 5.Seller MJ, Davis TB, Fear CN, Flinter FA, Ellis I, Gibson AG. Two sibs with anophthalmia and pulmonary hypoplasia (the Matthew-Wood syndrome). Am J Med Genet. 1996;62:227–9. doi: 10.1002/(SICI)1096-8628(19960329)62:3<227::AID-AJMG5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 6.Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–71. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 7.Guichet A, Triau S, Lepinard C, Esculapavit C, Biquard F, Descamps P, Encha-Razavi F, Bonneau D. Prenatal diagnosis of primary anophthalmia with a 3q27 interstitial deletion involving SOX2. Prenat Diagn. 2004;24:828–32. doi: 10.1002/pd.997. [DOI] [PubMed] [Google Scholar]
- 8.Forrester S, Kovach MJ, Reynolds NM, Urban R, Kimonis V. Manifestations in four males with and an obligate carrier of the Lenz microphthalmia syndrome. Am J Med Genet. 2001;98:92–100. [PubMed] [Google Scholar]
- 9.Bakrania P, Efthymiou M, Klein JC, Salt A, Bunyan DJ, Wyatt A, Ponting CP, Martin A, Williams S, Lindley V, Gilmore J, Restori M, Robson AG, Neveu MM, Holder GE, Collin JR, Robinson DO, Farndon P, Johansen-Berg H, Gerrelli D, Ragge NK. Mutations in BMP4 cause eye, brain, and digit developmental anomalies: overlap between the BMP4 and hedgehog signaling pathways. Am J Hum Genet. 2008;82:304–19. doi: 10.1016/j.ajhg.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferda Percin E, Ploder LA, Yu JJ, Arici K, Horsford DJ, Rutherford A, Bapat B, Cox DW, Duncan AM, Kalnins VI, Kocak-Altintas A, Sowden JC, Traboulsi E, Sarfarazi M, McInnes RR. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet. 2000;25:397–401. doi: 10.1038/78071. [DOI] [PubMed] [Google Scholar]
- 11.Billingsley G, Santhiya ST, Paterson AD, Ogata K, Wodak S, Hosseini SM, Manisastry SM, Vijayalakshmi P, Gopinath PM, Graw J, Heon E. CRYBA4, a novel human cataract gene, is also involved in microphthalmia. Am J Hum Genet. 2006;79:702–9. doi: 10.1086/507712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ragge NK, Brown AG, Poloschek CM, Lorenz B, Henderson RA, Clarke MP, Russell-Eggitt I, Fielder A, Gerrelli D, Martinez-Barbera JP, Ruddle P, Hurst J, Collin JR, Salt A, Cooper ST, Thompson PJ, Sisodiya SM, Williamson KA, Fitzpatrick DR, van Heyningen V, Hanson IM. Heterozygous mutations of OTX2 cause severe ocular malformations. Am J Hum Genet. 2005;76:1008–22. doi: 10.1086/430721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voronina VA, Kozhemyakina EA, O'Kernick CM, Kahn ND, Wenger SL, Linberg JV, Schneider AS, Mathers PH. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet. 2004;13:315–22. doi: 10.1093/hmg/ddh025. [DOI] [PubMed] [Google Scholar]
- 14.Gallardo ME, Rodriguez De Cordoba S, Schneider AS, Dwyer MA, Ayuso C, Bovolenta P. Analysis of the developmental SIX6 homeobox gene in patients with anophthalmia/microphthalmia. Am J Med Genet A. 2004;129A:92–4. doi: 10.1002/ajmg.a.30126. [DOI] [PubMed] [Google Scholar]
- 15.Verma AS, Fitzpatrick DR. Anophthalmia and microphthalmia. Orphanet J Rare Dis. 2007;2:47. doi: 10.1186/1750-1172-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ragge NK, Lorenz B, Schneider A, Bushby K, de Sanctis L, de Sanctis U, Salt A, Collin JR, Vivian AJ, Free SL, Thompson P, Williamson KA, Sisodiya SM, van Heyningen V, Fitzpatrick DR. SOX2 anophthalmia syndrome. Am J Med Genet A. 2005;135:1–7. doi: 10.1002/ajmg.a.30642. [DOI] [PubMed] [Google Scholar]
- 17.Wyatt A, Bakrania P, Bunyan DJ, Osborne RJ, Crolla JA, Salt A, Ayuso C, Newbury-Ecob R, Abou-Rayyah Y, Collin JR, Robinson D, Ragge N. Novel heterozygous OTX2 mutations and whole gene deletions in anophthalmia, microphthalmia and coloboma. Hum Mutat. 2008;29:E278–83. doi: 10.1002/humu.20869. [DOI] [PubMed] [Google Scholar]
- 18.Lequeux L, Rio M, Vigouroux A, Titeux M, Etchevers H, Malecaze F, Chassaing N, Calvas P. Confirmation of RAX gene involvement in human anophthalmia. Clin Genet. 2008;74:392–5. doi: 10.1111/j.1399-0004.2008.01078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallis DE, Roessler E, Hehr U, Nanni L, Wiltshire T, Richieri-Costa A, Gillessen-Kaesbach G, Zackai EH, Rommens J, Muenke M. Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat Genet. 1999;22:196–8. doi: 10.1038/9718. [DOI] [PubMed] [Google Scholar]
- 20.Asai-Coakwell M, French CR, Berry KM, Ye M, Koss R, Somerville M, Mueller R, van Heyningen V, Waskiewicz AJ, Lehmann OJ. GDF6, a novel locus for a spectrum of ocular developmental anomalies. Am J Hum Genet. 2007;80:306–15. doi: 10.1086/511280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou CJ, Molotkov A, Song L, Li Y, Pleasure DE, Pleasure SJ, Wang YZ. Ocular coloboma and dorsoventral neuroretinal patterning defects in Lrp6 mutant eyes. Dev Dyn. 2008;237:3681–9. doi: 10.1002/dvdy.21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q, Li S, Xiao X, Jia X, Guo X. The 208delG mutation in FSCN2 does not associate with retinal degeneration in Chinese individuals. Invest Ophthalmol Vis Sci. 2007;48:530–3. doi: 10.1167/iovs.06-0669. [DOI] [PubMed] [Google Scholar]
- 23.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, Minoda K. Detection of congenital color vision defects using heteroduplex-SSCP analysis. Jpn J Ophthalmol. 1996;40:79–85. [PubMed] [Google Scholar]
- 25.Gallardo ME, Lopez-Rios J, Fernaud-Espinosa I, Granadino B, Sanz R, Ramos C, Ayuso C, Seller MJ, Brunner HG, Bovolenta P, Rodriguez de Cordoba S. Genomic cloning and characterization of the human homeobox gene SIX6 reveals a cluster of SIX genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics. 1999;61:82–91. doi: 10.1006/geno.1999.5916. [DOI] [PubMed] [Google Scholar]
- 26.Aijaz S, Clark BJ, Williamson K, van Heyningen V, Morrison D, Fitzpatrick D, Collin R, Ragge N, Christoforou A, Brown A, Hanson I. Absence of SIX6 mutations in microphthalmia, anophthalmia, and coloboma. Invest Ophthalmol Vis Sci. 2004;45:3871–6. doi: 10.1167/iovs.04-0641. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki S, Marazita ML, Cooper ME, Miwa N, Hing A, Jugessur A, Natsume N, Shimozato K, Ohbayashi N, Suzuki Y, Niimi T, Minami K, Yamamoto M, Altannamar TJ, Erkhembaatar T, Furukawa H, Daack-Hirsch S, L'Heureux J, Brandon CA, Weinberg SM, Neiswanger K, Deleyiannis FW, de Salamanca JE, Vieira AR, Lidral AC, Martin JF, Murray JC. Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip. Am J Hum Genet. 2009;84:406–11. doi: 10.1016/j.ajhg.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]