Abstract
In this paper, we study a new physical mechanism to generate an activator field which signals the extreme margin of the contact area between an adherent cell and the substrate. This mechanism is based on the coupling between the adhesive bridges connecting the substrate to the cytoskeleton and a cytosolic activator. Once activated by adhesion on the adhesive bridges, this activator is free to diffuse on the membrane. We propose that this activator is part of the mecano-transduction pathway which links adhesion to actin polymerization and, thus, to cellular motility. Consequences of our model are as follows: (a) The activator is localised at the rim of the contact area; (b) The adhesion is reinforced at the margin of the contact area between the cell and the substrate; (c) Excitable waves of activator can propagate along the adhesion rim.
Keywords: Actins; metabolism; Biophysical Phenomena; Cell Adhesion; Cells; cytology; metabolism; Cytoskeleton; metabolism; Diffusion; Models, Biological
Keywords: actin, adhesion, model
1. Introduction
Cells must sense not only biochemical but also physical aspects of their environment [35, 16, 29, 50, 39] to perform complex functions such as migration, differentiation or apoptosis[18, 42]. The biochemical reactions involved in the continuous remodelling of the actin cytoskeleton are triggered by cell-substrate or cell-cell contacts [28]. The signals are usually mediated by transmembrane protein receptors at the cell surface that activate in a conformation dependent manner phosphorylation cascades [12] with or without activation of regulatory GTPases[49]. In all these functions, the dynamic rearrangement of the actin cytoskeleton is induced by local recruitment of activator proteins [38] where rapid signaling by diffusible factors are mechanical forces dependent[51, 48, 40]. In this paper, we introduce a physical model to describe the initial stages of cell spreading by recruiting a cytosolic molecule which reinforces adhesion.
Recent works have concentrated on receptor regulation and their link with actin dynamics during early and late events triggered by cell substratum contacts[15, 19, 20, 14, 21, 34, 37, 48, 11]. Biological and biophysical studies showed that cell adhesion, spreading and migration crucially depends on receptors of the integrin family[20] which performs bi-directional inside-out and outside-in signalization through the plasma membrane[25]. Upon binding to an extra cellular or intracellular ligand, integrins can undergo a conformational change and participate to the actin network reorganization that underlies cell spreading and migration. The transduction centers of these signaling pathways are associated with the integrin adhesion receptor family whose affinity towards their external ligand is regulated. A key observation is that change in their affinity is often preceded by changes of intracellular proteins which modulates their affinity [21]. All these findings have motivated theoretical physical works which have mainly focused on the dynamic reinforcement of the adhesion scaffold in connection with the acto-myosin driven protrusion machinery[4, 34, 33, 36]. Recent studies have however shown that the initial adhesion does not depend on myosines II which mediate adhesion maturation and it has been proposed that the initial cell spreading is separable from the focal contact maturation[9, 54].
More specifically, dynamic actin-based profusions such as ruffles, lamellopodia or filopodia [8] are observed at the the border of the cell-substrate contact area. Since actin polymerization is triggered by nucleating factors such as the Arp2/3 complex which are constitutively inactive, there is a need for activation factors such as the proteins from the WASP/wave family to promote[24, 8] or impair[17] actin polymerization with a possible regulation by membrane curvature or tension[45, 26, 7, 6]. Indeed experiments performed as earlier as in 1999 have shown that local recruitment of activated WASP’s induces the formation of actin-based membrane profusions [5]. Motivated by this findings, we introduce a physical model that relies on the generation of a diffusible regulatory molecule that is activated by the integrin substratum interaction. As described in the discussion section, the introduction of this activator field fits with experimental data where integrin occupency allows the activation of actin polymerizing factors.
This paper is organized as follows. The first section is concerned with the physical model for the activator field and gives emphasis on the role of the stretching energy which builds up at the margin of the contact area. This section is divided into two parts. The first deals with a characteristic biochemical cycle for the activator, which upon chemisorption followed by activation, modulates the affinity constant of adhesive bridges for the substrate. The second is concerned with the reaction-diffusion equation for the activator field which can diffuse on the membrane. The next section reports the main properties of the model in the case where the diffusion length of the activator field is small with respect to the width of the adhesive belt. Finally, in the discussion, we put our work in perspective with the biological context. Three appendices follows the conclusion. The first details the chemical reaction pathway experienced by the activator in the adhesive belt. The second and the third give additional numerical results concerning this non-linear problem.
2. The model
2.1. The affinity of adhesive bridges depends on a diffusing field
We consider a biochemical cycle where an unactivated regulatory protein with concentration ϕc gets activated by contact with the adhesion receptor where they are chemisorbed (concentration ϕi). Once activated with concentration , they are desorbed on the membrane with concentration ϕm where they diffuse with a finite life time 1/b after which they desorbed back to the cytosol (see Appendix A). The discussion at the end of the paper gives examples of such activator fields. Here, we will only summarized this scheme as follows:
| (1) |
Note that the initial and the final states of this biochemical cycle are the same. Thus, for an isolated system, the cycle would run clock wise and anti-clock wise with the same probability. In this work, however, we assume that the system is open to an energy flux, and that the reaction ϕ→ϕ★ is non-specifically driven by, e.g., phosphate hydrolysis. The system being open to an energy flux, the cycle runs only one way.
We assume that the membrane and the cytoskeleton form a complex with the adhesive bridges which can be described using the continuum elasticity valid for elastic shells [27]. The connectors comprise both the integrin receptors and their adaptor proteins [33]. In this framework, the complex formed by the membrane and the cytoskeleton is described by a height variable h(x) which depends on the position x. As for elastic shells [27], the elasticity entails a stretching and a curvature term. In our model, the streching energy term accounts for the connections between the cytoskeleton and the substrate. Let the number of connected links per unit line with length h(x) and be the activator concentation field bound to the adhesive bridges. We assume local equilibrium between the molecules engaged to their ligands and the unbound ones with
| (2) |
so that is given by the usual Bell’s law (β is a short hand notation for 1/kBT where T is the temperature and kB the Bolzmann constant):
| (3) |
where n0 is the total number of adhesive bridges. First, the B > 0 term describes the increase with in the number of connected adhesive bridges, so that the activator field favors adhesion. Second, the A > 0 factor mimics the penalty due to the stretching elasticity when the bridges have length h(x). This length is equivalent to a displacement in elasticity theory and correspond to the distance between the cell and the substrate. When h(x) increases as at the border of the adhesion zone, since the cell leaves the substrate, tends rapidly to zero. Thus, the affinity constant is both modulated by the action of the field ϕ(x,t) which favors adhesion and a penalty term due to stretching.
Eq. (3) gives a streching energy per unit line for the one-dimensionnal problem we study here. The total elastic free energy ℰ is the sum of this stretching component with the one which originates from curvature:
| (4) |
In the case where is a step like function, Eq. (4) gives that the height profile is exponential like and we will use henceforth h(x) ~ h0 exp[x/λ] where λ ≈ 0.1μm is the typical width of the adhesive belt. Finally, the order of magnitude for h0 can be estimated from the maximal elongation length of an integrin like protein (≈ 25 nm[53]). In what follows, we will take this numerical value for the maximal height field which comprises both the deformations of the integrin like proteins and of the elastic medium which connects the integrins to the cytoskeleton.
2.2. Equation of motion for the field ϕ(x, t)
To get an equation of motion for , we remark that the derivative of with respect to gives the elastic part of chemical potential of the protein as:
| (5) |
This chemical potential influences the reaction between the two species and ϕm. We show in Appendix A that adding diffusion for ϕm and a lifetime 1/b leads to a reaction-diffusion equation:
| (6) |
where Γ is a kinetics coefficient (∝ b).
To make progress, we assume that the processes which govern the transformation ϕm to in the cytosol are sufficiently fast compared with the typical time scale set by the equilibration time of a diffusing field on the membrane. This assumption is consistent with the larger diffusion coefficient in the cytosol (Dc ≈ 10−5cm2s−1) compared with the diffusion coefficient of a small protein on membrane (D ≈ 10−8,−10cm2s−1[52]). Thus, we will assume henceforth that the kinetics between ϕm and is at equilibrium and we will take (see Appendix A). For convenience, we will write:
| (7) |
Typical order of magnitude of the coefficients appearing in our model are as follows: b ≈ a few tenth s−1 and which has the dimension of a speed is of the order of 10 nm.s−1. Thus, the diffusion length is typically smaller than the width of the adhesive belt λ and will assume for mathematical convenience that . When times are expressed in units of b−1, and lengths in unit of λ, we thus introduce the dimensionless parameter:
| (8) |
with ε < 1. This leads to the equation of motion:
| (9) |
where g(ϕ, h) is a source term which depends on the height h(x) at position x:
| (10) |
In this equation, Γ has been renormalized as ΓR and 1/β is an effective temperature.
In order to describe actin polymerization at the border of the cell, Eq. (6) can be complemented by the kinetics of growth of actin cortex. In this work, we assume that actin polymerization takes place on unactivated bonds by the ϕ field, e.g. talin free bonds, of density n0 − nb(ϕ, h).
| (11) |
where kon[ϕ] is a kinetic coefficient which increases with ϕ, and cg the concentration of globular actin supposed to be constant. C is a constant and γ is an exponent which lumps all non-linearities together. Eq. (11) gives that the rate of actin polymerization depends on the concentration of activator field and that it takes place at the interface between the precursor region and the adhesive belt.
Eqs.(9-10-11) give a physical model we analyse in the next section.
3. Results
For the sake of clarity, we will distinguish between the two geometries represented in Fig. 1. The first is the flat geometry when the distance between the membrane and the cytokeleton can be taken as constant and it corresponds, for example, to the basal surface of the cell. The second is the curved one and it corresponds to the adhesive belt where the cell leaves the substrate.
Figure 1.
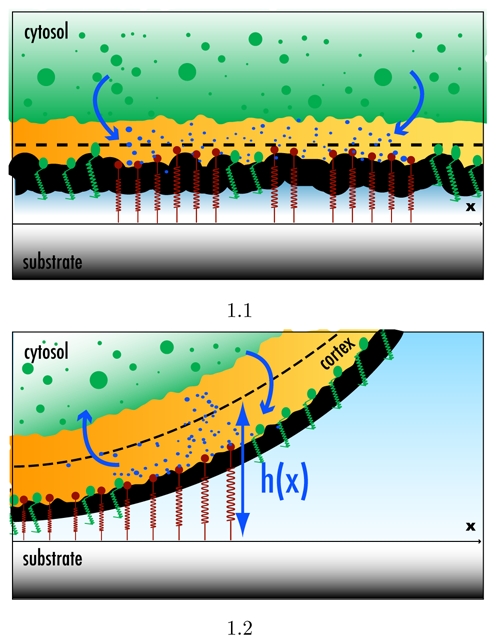
Artist’s representation of the two geometries studied in the text. Bound integrins with their actin cortex deformations are represented as red springs. Unbound integrins are green. Activator proteins are represented as blue points. Case (a) is the flat geometry where a wave of activation can propagate in the x-direction. Case (b) is the curved geometry corresponding to the adhesive belt where the activation wave is pinned in the adhesive belt. The dashed line corresponds to the height field h(x).
3.1. Flat geometry
To understand the properties of the solutions of Eq. (9), it is useful to consider the source term g(ϕ, h) defined in (10) which depends on the normal height h(x). Fig. 2 gives a family of plots for g(ϕ, h) as a function of ϕ for h(x) = Cste. For sufficiently large values of the height h(x), each curve possesses three zeros ϕi(h), i = 1, 2, 3 where ϕ1(h) ≈ 0.
Figure 2.
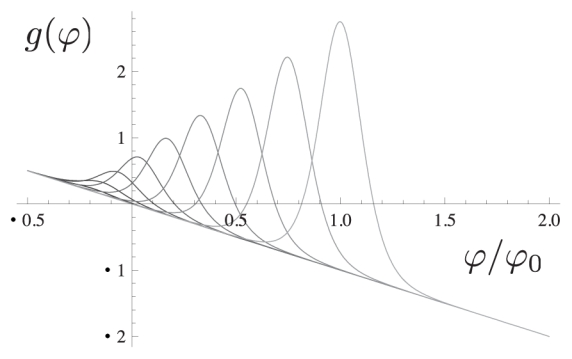
Plot of the source function g(ϕ, h) (see Eq. (10)) for different values of the height variable h(x) = Cst.. The maximal excitation occurs at the most stressed adhesive bridges (large value of h(x) on the right). The field ϕ has been renormalized by ϕ0 so that the maximum of the last curve occurs at ϕ/ϕ0 = 1.
Thus, for h(x) = Cste, we recover the classical picture of a non-linear reaction-diffusion equation where the source term does not depend explicitly on x. In this case, Eq. (9) possesses unique wavefront solutions ϕ(x, t) = f(ξ = x − ct) which interpolate between the two stable fixed points ϕi(h), i = 1, 3. These wavefronts solutions propagate at a velocity c determined by the condition ‡:
| (12) |
where f(ξ) → ϕ3,1 (h) as ξ → ±∞.
We conclude that the self enhancement mechanism for the activator field due to adhesion leads to propagating waves for the activator field. This conclusion applies to any dimension. In particular:
For flat adherent parts of the cell where the receptors are not fully activated. The model predicts basal propagation of receptors activation which, in turn, strengthens adhesion. This phenomenon has been analysed in experiments using fluorescent techniques [51, 48].
For adherent cells. Lateral excitable waves of activator can propagate along the rim of the adhesion zone. This a one-dimensional situation. The speed of these waves depends on the curvature of the contact line between the cell and the substrate [1].
For local mechanical excitation of the membrane-cytoskeleton adhesion. The model predicts the existence of a radial waves which propagate away from the stimulation point. Such an activation wave has been for instance observed for Src[54].
3.2. Curved geometry at the cell border
Stationary solutions of Eq. (9) can be numerically obtained by perturbing the ϕ = 0 state and by looking to the t → +∞ limit. Fig. 3 gives one example of these in the case where the dimensionless diffusion length ε in (9) is small. Additional numerical results concerning these solutions are provided in the appendices. We will assume in this section that the position of the cell border is fixed by the adhesive properties of the substrate.
Figure 3.
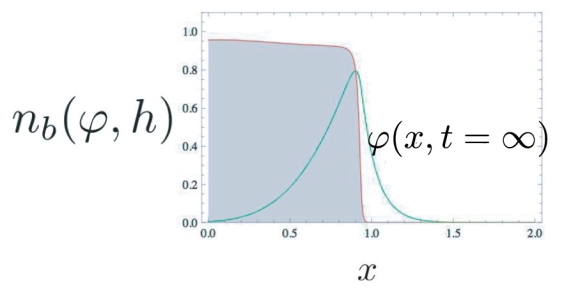
Stationary solution of the activator field starting from perturbing the ϕ(x) = 0 state. This plot is obtained in the large affinity limit and it shows that the maximum of ϕ(x) colocalizes with the border of the adhesive belt.
As foreseen, Fig. 3 demonstrates that the solution is strongly peaked at the border of the cell and that the variations of the activator field can be divided into two distinct domains.
A dorsal part where diffusion does not play any role and where the solution of the differential equation is very well approximated by the largest solution of the algebraic equation g(ϕ(x), h(x)) = 0. In the large β limit (small effective temperature), this root can be reasonably approximated by h(x)2 + C/β where C is a constant which depends on the affinity constant and on the kinetic coefficient (see Appendix).
A precursor part where the diffusion length ε sets the relevant length scale. In this domain, the solution is stiff and decreases abruptly from its maximum value ϕmax at xmax to zero. The decay is exponential like ~ exp [−(x − xmax/(ελ)]. We call this domain the precursor domain, since the activator field ϕ (x) begins to rise before the density of connected bounds nb(ϕ, h) changes abruptly from zero to a finite value set by the affinity constant . Since this rise depends on the value of the activator field in the precursor region where the actin polymerization takes place, we can say that the precursor is a guide for the activation of the adhesive bridges.
3.3. Consequences
First, it is useful to consider the variations of nb(ϕ(x), h(x)) of bound adhesive bridges for different values of the affinity constant . In the limit of small , almost no adhesive bridges are bound to the cytoskeleton far from the adhesive belt where ϕ(x) is almost zero. The self-enhancement mechanism of the activator reinforces markedly the adhesion at the extreme border of the contact area. This is examplified in Fig. 4 where we note that all curves have almost the same value at the border of the contact zone independently of the affinity constant.
Figure 4.
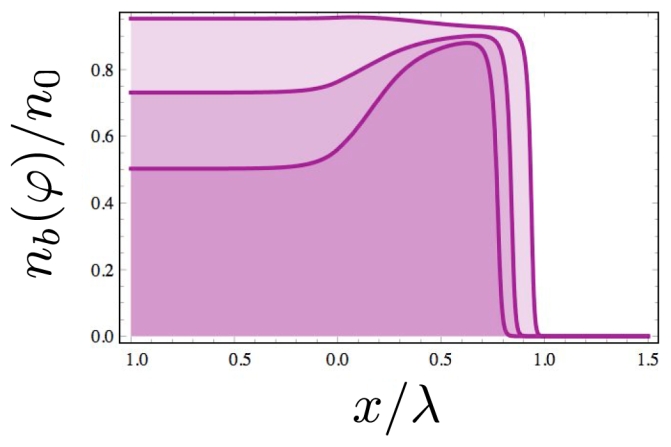
Plot of nb(ϕ) according to Eq. (3) for different values of the affinity constant in the low limit ( from the top to the bottom curve). These curves contrast with the case (c) of Fig. 3 and they show a marked reinforcement of the adhesion at the rim of the contact area.
It is also interesting to plot the actin polymerization rate defined by Eq. (11), see Fig. 5. The actin polymerization rate being the product of an increasing by a decreasing function of ϕ(x), the self-enhancement of the activator field is shifted outside the adhesive belt where the density of bound connector molecules is almost zero. The height of the maximum of the actin polymerization rate is a non-monotonous function of the diffusion constant for the activator field. The presence of an optimum is understood by noting that the diffusion coefficient D is necessary to have a precursor region, but increasing to much D enlarges the domain of variations of the activator at total constant concentration. Thus, the activator field escapes from the adhesive belt but its concentration is everywhere low in the precursor domain. Theses two antithetic scenarios give an optimal value of D for the maximum of the actin polymerization rate. Thus, taking the product as in Eq. (11) with the help of unbound integrins as co-activators gives emphasis on the role of the precursor region.
Figure 5.
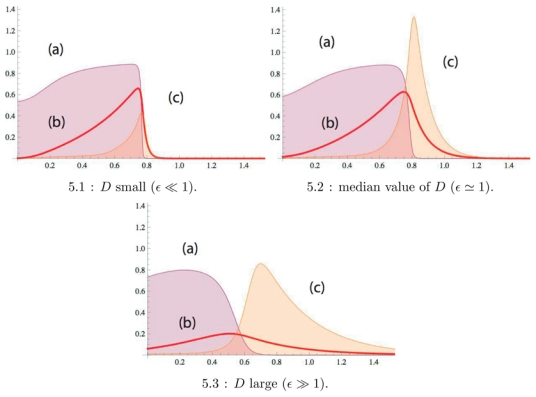
For all curves: (a) Plot of the density of bound connector molecules nb(ϕ(x)) as a function of x; (b) Plot of ϕm(x) as a function of x. In (c), plot of actin polymerisation rate as in Eq. (11) where ∂tf = ϕ (1 − nb(ϕ)) − 0.1. From cases (1) to (3) the diffusion coefficient is increased by a factor 10. Note that by increasing D, the maximum of the actin polymerization rate (curve (c)) reaches a maximum and then decreases. For all curves, the affinity is set to 0.9.
To conclude this section, we show that our model provides a simple actin based feedback mechanism. A model-free way to do it is to assume that the cell edge advances at a constant speed V because of actin polymerization. We will not discuss what is the relationship between the activator field and the speed V, but we will simply assume to lower order in complexity that the solution of Eq. (9) is a stationary solution in frame which moves at a velocity V with respect to the substrate. Introducing the coordinate ξ = x − Vt, we look for a solution ϕ(x, t) = ϕ (ξ = x − Vt) with:
| (13) |
so that that the actin polymerization rate which sets V influences the profile of the activator ϕ through the convective term V∂ϕ/∂x. Since V is set by the the activator field itself, our model is self-consistent.
To demonstrate that actin dynamics controls in turn the variations of ϕ(x, t) through the speed V, let consider Fig. 6 where the maximum of the profile of ϕ(ξ) is plotted against V (V > 0 in the direction set by the outwards pointing normal). This maximum ϕmax is determined from the stationary solution of Eq. (9) for different values of V as shown in the inset. This demonstrates that ϕmax decreases when V increases. Thus if the speed is too high, the maximum of ϕ(ξ) will get down, entailing a less efficient enhancement of polymerization and thus a decrease in speed. Conversely, if the speed is to small, ϕmax will increase and this will enhance more the actin polymerization and thus the speed.
Figure 6.
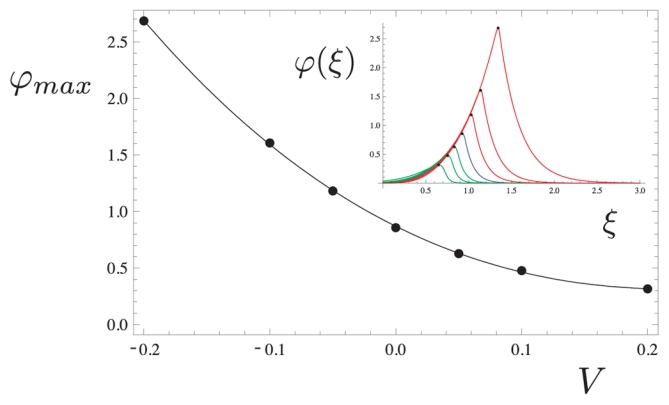
Plot of the maximum ϕmax of ϕ(ξ) as a function of the speed V of the stationary solution of Eq. (13) shown in the inset (the solutions for V > 0 are shown in green and the ones for V < 0 shown in red). As V increases, the maximum ϕmax decreases and so the actin polymerization rate.
4. Discussion
Our model describes the self-enhancement of an activator field on stressed adhesive bridges which is reinforced by recruiting a cytosolic molecule. It can describe the initial stages of cell spreading to extra cellular matrix associated with pathways that stimulate protusions whereas mature adhesion involve focal adhesions and actin stress fibers [13]. This early stage was recently found to be independent on talin 1 and 2 [54]. Talin interaction with β integrin cytosolic tail allows the integrin conformational switch between low to high affinity[44] and it represents the first stage of focal adhesion assembly that sustains long term adhesion. Thus, cell spreading and focal adhesion assembly are separable processes and our study applies only to the early times signaling pathways where actin polymerization is activated by ligand-bound integrins.
Indeed, integrin occupancy at the initial stage allows the activation of Src family kinases[54] and thereby monomeric downstream GTPases activation of Racl and Rap1[13, 3] and likely a PIP2 burst [30]. Both PIP2 and Rho family GTPases are required to recruit WASp/WAVE members. These proteins are inactive in their cytosolic state but they are in their activated state on the membrane where they stimulate actin nucleation[47, 32, 54]. These signaling pathways are the experimental clues which allow to introduce an the activator field. Note, however, that this field may not account for the activation of a single molecule but for a complete signaling pathway which includes Src or WASp/WAVE family proteins.
In view of the large number of integrin partner proteins which link adhesion to actin polymerization, it is unlikely that cells use only one activation factor as the ϕm(x, t) of our model. We thus ask what matters if instead of a unique scalar field we use a multicomponent field (ϕ0 = ϕm, ϕ1, …, ϕn) to describe a full reaction pathway. Since integrins can be either in an unactivated or in an activated state, our model suggests to write the chemical potential for the activated state as:
| (14) |
where μ0 is a reference and B > 0 gives that the chemical potential of the activated state decreases upon binding with the species ϕm but that it increases with stretching (compare with (3)). This change in affinity with elasticity is consistent with the change in size of the extracellular domain of the integrins which extend from 5 to 25 nm when fully activated [21].
Eq. (14) describes a mechanistic trade-off between the potency of being activated by binding to ϕm and the cost of streching. The sum of this to terms alone is able to provide stress induced enhancement of activation, since the excitation goes larger with the extension h(x) (see Fig. 2 where the largest functioning point, i.e. zero, corresponds to the largest value for h(x)). Thus taking definition (14) translates the ability for the integrins to be activated into a positive feedback loop for the self-excitation of the ϕm(x, t) field. It seems to us that this condition is necessary but not sufficient for the network (ϕ0 = ϕm, ϕ1, …, ϕn) to have two or more functioning points, one of which corresponding to dentitric polymerization§.
Our model also offers a theoretical basis for the effect of topographic characteristic on cell migration and spreading. Although the effects of topography have been extensively investigated, the mechanism determining the cell-surface reaction are largely unknown [10, 2]. It should be also useful in situations where adhesive substrate is micro-patterned on a flat area and force the cells to be at the border of an adhesive and non-adhesive zone[46, 22]. In this case, one is interested in quantifying the activity which takes place along the adhesion rim. The predicted result reported in this paper is the co-localization of an activator field with the border of the adhesive area. Moreover, this diffusion mechanism describes a long range receptor-receptor interaction mediated by the activator field. Fluorecent biosensors for localization of the activator field should match the pattern of microtextured cell substrates.
5. Conclusion
In this paper, we have introduced a simple reaction-diffusion model for an activator field of actin polymerisation. The dynamic of this field entails a positive loop which depends on the steching elasticity which builds up at the margin of the contact area between the cell and the substrate. This approach should useful to describe the short times following the contact between a cell and a substrate when focal adhesions and stress fibers did not have time to complete their development. This is the case where one studies, for example, cell spreading. It should be also useful in situations where micro-structuration of patterned substrates forces the cell to be at the borderline between an adhesive and a non-adhesive zone. In this case, one is interested in quantifying the activity which takes place along the adhesion rim[15].
The main results reported in this paper as follows. First, there is a co-localization of an activator field with the border of the adhesive area. Second, there exists progressive waves for the activator field with a speed scaling as where D is a typical diffusion constant and b−1 the life time of an activated state. This co-localization follows the rules of a reaction-diffusion mechanism where the propagating diffusing wave is pinned at the border of the adhesive zone. On the experimental side, our mechanism suggests to probe using biofluorescence assays on textured micro-patterns both the activation of the receptors involved in cell adhesion with the proteins involved in actin polymerization.
Acknowledgments
We thank F. Bruckert, O. Destaing, E. Faurobert and E. Planus for very useful comments and discussions. The group belongs to the CNRS consortium CellTis.
Appendix A. Modeling the cell edge as a chemical reactive surface
Introduction
We consider a simple model system where a cytosolic protein ϕc can adsorb on the adhesive bridges I with reaction rate m+,−. The concentration of adsorbed molecules will be noted as ϕi. When adsorbed, we assume that it is transformed to a high energy state, i.e. activated, with concentration by an enzymatic coupling to an other chemical reaction which is used as an energy source. The back reaction is supposed to be negligible if the concentration of the corresponding exchange factor is sufficiently low. Finally, undergoes desorption on the membrane where it goes back to the cytosol with a life time 1/b. Let I denotes an integrin or a receptor associated with an adhesive bridge, the total reaction pathway is:
| (A.1) |
where ϕi is the concentration of activators bound to adhesive bridges. We note this quantity ϕi. Assuming quasi-stationary solutions, we find:
| (A.2) |
which implies that the chemical potential of is a function of the concentration ϕm. Eq. (A.2) is valid when the kinetic coefficients are independent of the coordinate x along the membrane and it assumes that the diffusion of the species ϕm can be neglected.
Including elasticity
To find the stationary and local distribution of membranous ϕm(x, t), we generalize this reaction pathway to include elasticity. From now on, the two kinetic coefficients are denoted by K+,−. First, if the diffusion in the cytosol is fast enough, we can assume that the concentration ϕc in the reaction layer just above the membrane is independent of the coordinate x along the membrane. Second, Van’t Hoff’s law implies that the ratio of the kinetic coefficients is a function of difference of chemical potential between the bound phases, ϕi (x, t), and the membranous one, ϕm (x, t):
| (A.3) |
where βΔμ is a small parameter.
To find Δμ, we note that for bound activators , the chemical potential is the derivative of the free energy including the stretching energy. Thus,
| (A.4) |
which gives Δμ since the chemical potential of the cytosolic phase is constant.
In what follows, we generalize the kinetic equation for the field ϕm(x, t). We will assume that the activation from ϕc to is fast so that . This gives a relationship equivalent to (A.2):
| (A.5) |
which is valid if the diffusion for ϕm is neglected. Note that k−/k+ is given by Van’t Hoff’s law (A.3) and that (A.5) is non-linear.
Including diffusion on the membrane
Now, we postulate that ϕm(x, t) solves the diffusion reaction equation:
| (A.6) |
Eq. (A.6) implies that when the energy of bound molecules will be larger than the one of the membranous proteins ϕm(x), the molecules will desorb from the adhesive bridges. We solve (A.6) to leading order in .
In the approximation where the activation reaction is supposed to be fast with respect to all other processes, we use (A.5) to write as a function of ϕm. Since (A.6) is already first order in , it is enough to use (A.2) instead of (A.5)
| (A.7) |
with:
| (A.8) |
Assuming again that the concentration ϕc in the cytosol is much larger than ϕm, we have to leading order in ϕm in (A.6)
| (A.9) |
Using this result for the equation of motion (A.6) gives the equation of motion for ϕm(x, t) as quoted in the text when ϕm(x, t) is replaced by its linear deviation δϕ (x, t) around
| (A.10) |
Appendix B. Staircase approximation for the height field
One key specificity of our model is the position dependance of the source term g = g(ϕ(x), h(x)). However, when looking at small scale features, one can assume the cell profile h(x) to be constant and thus map our problem to a simpler and well known spatial independant reaction-diffusion problem. Albeit giving a simple criterion for the existence of stationary concentration profile, the results of this appendix will be used in the next one.
Consider case (a) of Fig. B1 where h(x) ~ ex is plotted with its piece wise approximation. This approximation is designed to fit the original function using a series of plateaux with a width larger than the diffusion length (D/b)1/2. On each plateau, the height field h(x) is constant. Approximating the original function by its plateau approximation consists of assuming that the height field h(x) doe not vary on the scale (D/b)1/2 where the variations of the activator field are stiff. Using Eq.(12) to calculate the speed c of the equivalent wave solution for the diffusion reaction, we see that there is a critical height hmax at which the speed c passes from positive to negative values. In this limit, the maximum height h(xmax) at which the activator reaches its maximum ϕ(xmax) corresponds to the condition c(h) = 0 in (12) Thus, a criterion on the equivalent plateau problem gives a criterion for the maximum height, and thus the maximum value of the activator field. This criterion can be used to get the numerical values of ϕ(xmax) as a function of the parameters entering into the problem (see next Appendix).
Figure B1.
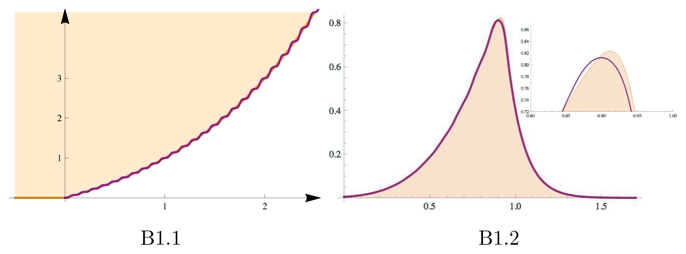
In (a), plot of the function h(x) = ex − 1 with its plateau approximation when the width of each plateau is given by (D/b)1/2. In (b), plot of the solutions for the activator field ϕ(x) when the height h(x) corresponds the two curves of (a). Note that the plateau approximation gives a reasonable estimate of the maximum value for the activator.
Appendix C. Additional results
To understand how the variations of the activator field depends on the parameters of the problem, it is useful to write the reaction-diffusion equation (9) as:
| (C.1) |
where:
| (C.2) |
with constants Ci defined in table C1 in terms of the physical constants defined in text. Henceforth, we will divide the variations of ϕ(x, t) into its slow varying component corresponding to the dorsal part of Fig. C1 and its fast component. The latter corresponds to the precursor region ahead of the adhesive belt.
Table C1.
Table of symbols used in the Appendix:τ, ϕ0are arbitrary normalization factors.
| Symbols | Meaning | |
|---|---|---|
| Cl | b·τ | |
| C2 | ||
| C3 | βAφ0 | |
| C4 | ||
| C5 |
Figure C1.
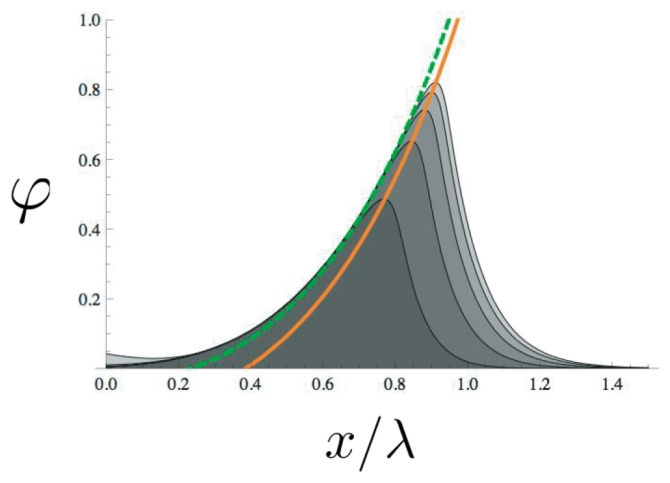
Plot of the activator concentration ϕ(x, t) starting from an initial perturbation of the ϕ = 0 state. The upper curve is the t → +∞ limit which has converged to the stationary solution of Eq. (C.1). The dashed curve is the slow varying component ϕc(x) (see C.5). The curve corresponds to the maximum of ϕ(x, t) according to the condition (C.9).
In the limit of small diffusion coefficient, and in a region sufficiently far away from the maximum of ϕ(x), the effect of a diffusion coefficient is small [1]. Thus, we set D = 0 in (C.1) and solve the algebraic equation for the slow variations of ϕ(x) (we call ϕs(x)):
| (C.3) |
whose solution can be approximated to leading order in 1/β as the solution of:
| (C.4) |
Because of Eq. (C.2), Eq. (C.4) is nothing but a second order polynomial equation for exp [ϕs(x)] which can be solved to get ϕs(x).
| (C.5) |
This equation corresponds to the dashed curve of Fig. C1 where it approximates very well the numerical solution.
To find the fast component of the activator, ϕf(x), valid in the precursor region, we note that the height h(x) does not vary on the scale where ϕf(x) varies. This means that h′(xmax) ≪ (D/b)1/2 and h(x) can be taken as a constant. This is the plateau approximation of the previous appendix (see Fig. B1).
For h = Cst., we assume a travelling wave solution at velocity c in the coordinate system ξ = x − ct. Thus, we look for a solution ϕ(x, t) = ϕ̃(ξ) which satisfies:
| (C.6) |
Since has three zeros as a function of ϕ̃ at h = Cst., we see after multiplying Eq. (C.6) by ∂ϕ̃/∂ξ and integrating between the two end zeros (0, ϕmax) that:
| (C.7) |
Since the maximum height hmax corresponds to the condition where c passes from positive to negative values, we have the pinning condition c = 0. Eq. (C.7) is a single equation for to the unknowns (ϕmax, h(xmax)). From the numerical point of view, a very good approximation consists in solving (C.7) together with the condition for ϕ(xmax) to be the inflexion point of the source source term:
| (C.8) |
This gives:
| (C.9) |
which corresponds to the maximum of the activator field in Fig. C1. Eqs. (C.7) and (C.9) gives a system of two equations with two unknown which can be numerically solved.
Footnotes
This property can be easily demonstrated by going to the reference frame ξ = x − ct in Eq. (9) with dϕ/dt = −cdϕ/dξ. After multiplying by dϕ/dξ and integrating, Eq. (12) follows[41].
The converse statement that a general network with more than two functioning point possesses a positive circuit has been proven by Soulé[43].
References
- 1.Ali O, Fourcade B. In preparation. [Google Scholar]
- 2.Berry CC, Campbell G, Spadiccino A, Robertson M, Curtis ASG. The influence of microscale topography on fibroblast attachment and motility. Biomaterials. 2004 Nov;25(26):5781–5788. doi: 10.1016/j.biomaterials.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 3.Bos JL. Linking rap to cell adhesion. Curr Opin Cell Biol. 2005 Apr;17(2):123–128. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Bruinsma R. Theory of force regulation by nascent adhesion sites. Biophys J. 2005;89(1):87–94. doi: 10.1529/biophysj.104.048280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castellano F, Montcourrier P, Guillemot JC, Gouin E, Machesky L, Cossart P, Chavrier P. Inducible recruitment of cdc42 or wasp to a cell-surface receptor triggers actin polymerization and filopodium formation. Curr Biol. 1999 Apr 8;9(7):351–360. doi: 10.1016/s0960-9822(99)80161-4. [DOI] [PubMed] [Google Scholar]
- 6.Chamaraux F, Ali O, Keller S, Bruckert F, Fourcade B. Physical model for membrane protrusions during spreading. Phys Biol. 2008;5(3):36009. doi: 10.1088/1478-3975/5/3/036009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamaraux F, Fache S, Bruckert F, Fourcade B. Kinetics of cell spreading. Phys Rev Lett. 2005 Apr;94(15):158102–158102. doi: 10.1103/PhysRevLett.94.158102. [DOI] [PubMed] [Google Scholar]
- 8.Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007 Oct;9(10):1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 9.Choi C, Vicente-Manzanares M, Zareno J, Whitmore L, Mogilner A, Horwitz A. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin ii motor-independent manner. Nat Cell Biol. 2008 Aug 17; doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis A, Wilkinson C. Topographical control of cells. Biomaterials. 1997 Dec;18(24):1573–1583. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 11.Cuvelier D, Thery M, Chu YS, Dufour S, Thiery JP, Bornens M, Nassoy P, Mahadevan L. The universal dynamics of cell spreading. Curr Biol. 2007;17(8):694–699. doi: 10.1016/j.cub.2007.02.058. [DOI] [PubMed] [Google Scholar]
- 12.Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008 Aug 1;121(Pt 15):2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeMali KA, Wennerberg K, Burridge K. Integrin signaling to the actin cytoskeleton. Curr Opin Cell Biol. 2003 Oct;15(5):572–582. doi: 10.1016/s0955-0674(03)00109-1. [DOI] [PubMed] [Google Scholar]
- 14.Dobereiner HG, Dubin-Thaler BJ, Giannone G, Sheetz MP. Force sensing and generation in cell phases: analyses of complex functions. J Appl Physiol. 2005;98(4):1542–6. doi: 10.1152/japplphysiol.01181.2004. [DOI] [PubMed] [Google Scholar]
- 15.Dubin-Thaler BJ, Hofman JM, Cai Y, Xenias H, Spielman I, Shneidman AV, David LA, Dobereiner HG, Wiggins CH, Sheetz MP. Quantification of cell edge velocities and traction forces reveals distinct motility modules during cell spreading. PLoS ONE. 2008;3(11):e3735. doi: 10.1371/journal.pone.0003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006 Aug 25;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 17.Filippi BM, Mariggio S, Pulvirenti T, Corda D. Src-dependent signalling regulates actin ruffle formation induced by glycerophosphoinositol 4-phosphate. Biochim Biophys Acta. 2008 Dec;1783(12):2311–2322. doi: 10.1016/j.bbamcr.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Friedl P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr Opin Cell Biol. 2004 Feb;16(l):14–23. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Giannone G, Dubin-Thaler BJ, Rossier O, Cai Y, Chaga O, Jiang G, Beaver W, Dobereiner HG, Freund Y, Borisy G, Sheetz MP. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128(3):561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giannone G, Sheetz MP. Substrate rigidity and force define form through tyrosine phosphatase and kinase pathways. Trends Cell Biol. 2006 Apr;16(4):213–223. doi: 10.1016/j.tcb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Ginsberg MH, Partridge A, Shattil SJ. Integrin regulation. Curr Opin Cell Biol. 2005 Oct;17(5):509–516. doi: 10.1016/j.ceb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Guillou H, Depraz-Depland A, Planus E, Vianay B, Chaussy J, Grichine A, Albiges-Rizo C, Block MR. Lamellipodia nucleation by filopodia depends on integrin occupancy and downstream rac1 signaling. Exp Cell Res. 2008 Feb 1;314(3):478–488. doi: 10.1016/j.yexcr.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Lim CJ, Watanabe N, Soriani A, Ratnikov B, Calderwood DA, Puzon-McLaughlin W, Lafuente EM, Boussiotis VA, Shattil SJ, Ginsberg MH. Reconstructing and deconstructing agonist-induced activation of integrin alphaiibbeta3. Curr Biol. 2006 Sep 19;16(18):1796–1806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 24.Higgs H, Pollard T. Activation by Cdc42 and PIP2 of Wiskoot-aldrich syndrome (WASp) stimulates actin nucleation by Arp2/3 complex. J Cell Bio. 2000;150(6):1311–1320. doi: 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 26.Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat Cell Biol. 2008 Dec;10(12):1393–1400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landau L, Lifshitz E. Theory of elasticity. Pergamon: 1970. [Google Scholar]
- 28.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008 Apr;88(2):489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 29.Li S, Butler P, Wang Y, Hu Y, Han DC, Usami S, Guan JL, Chien S. The role of the dynamics of focal adhesion kinase in the mechanotaxis of endothelial cells. Proc Natl Acad Sci USA. 2002 Mar 19;99(6):3546–3551. doi: 10.1073/pnas.052018099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling K, Schill NJ, Wagoner MP, Sun Y, Anderson RA. Movin’ on up: the role of ptdins(4,5)p(2) in cell migration. Trends Cell Biol. 2006;16(6):276–84. doi: 10.1016/j.tcb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Martel V, Racaud-Sultan C, Dupe S, Marie C, Paulhe F, Galmiche A, Block MR, Albiges-Rizo C. Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J Biol Chem. 2001;276(24):21217–21227. doi: 10.1074/jbc.M102373200. [DOI] [PubMed] [Google Scholar]
- 32.Misra A, Lim RPZ, Wu Z, Thanabalu T. N-wasp plays a critical role in fibroblast adhesion and spreading. Biochem Biophys Res Commun. 2007 Dec 28;364(4):908–912. doi: 10.1016/j.bbrc.2007.10.086. [DOI] [PubMed] [Google Scholar]
- 33.Nicolas A, Besser A, Safran SA. Dynamics of cellular focal adhesions on deformable substrates: consequences for cell force microscopy. Biophys J. 2008 Jul;95(2):527–539. doi: 10.1529/biophysj.107.127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicolas A, Geiger B, Safran SA. Cell mechanosensitivity controls the anisotropy of focal adhesions. Proc Natl Acad Sci USA. 2004;101(34):12520–12525. doi: 10.1073/pnas.0403539101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005 Sep;8(3):241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Pathak A, Deshpande VS, McMeeking RM, Evans AG. The simulation of stress fibre and focal adhesion development in cells on patterned substrates. J R Soc Interface. 2008 May 6;5(22):507–524. doi: 10.1098/rsif.2007.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierres A, Benoliel A, Touchard D, Bongrand P. Biophys J. 2008. How cells tiptoe on adhesive surfaces before sticking. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollard TD, Borisy G. Cellular motility driven by assembly and diassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 39.Puklin-Faucher E, Sheetz MP. The mechanical integrin cycle. J Cell Sci. 2009 Jan 15;122(Pt 2):179–186. doi: 10.1242/jcs.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynolds AR, Tischer C, Verveer PJ, Rocks O, Bastiaens PIH. Egfr activation coupled to inhibition of tyrosine phosphatases causes lateral signal propagation. Nat Cell Biol. 2003;5(5):447–453. doi: 10.1038/ncb981. [DOI] [PubMed] [Google Scholar]
- 41.Scott A. Nonlinear Science, Emergence and Dynamics of Coherence Structure. Oxford University Press; 1999. [Google Scholar]
- 42.Shattil SJ. Integrins and src: dynamic duo of adhesion signaling. Trends Cell Biol. 2005 Aug;15(8):399–403. doi: 10.1016/j.tcb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Soulé C. Mathematical approaches to differentiation and gene regulation. CR Biologies. 206;329:13–20. doi: 10.1016/j.crvi.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Tadokoro S, Shattil SJ, Eto K, Tai V, Liddington RC, de Pereda JM, Ginsberg MH, Calderwood DA. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 2003 Oct 3;302(5642):103–106. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 45.Takenawa T, Suetsugu S. The wasp-wave protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007 Jan;8(l):37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 46.Thery M, Racine V, Pepin A, Piel M, Chen Y, Sibarita JB, Bornens M. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7(1465–7392 Print):947–53. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 47.Tsuboi S, Nonoyama S, Ochs HD. Wiskott-aldrich syndrome protein is involved in alphaiib beta3-mediated cell adhesion. EMBO Rep. 2006 May;7(5):506–511. doi: 10.1038/sj.embor.7400665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verveer PJ, Wouters FS, Reynolds AR, Bastiaens PI. Quantitative imaging of lateral erbb1 receptor signal propagation in the plasma membrane. Science. 2000;290(5496):1567–1570. doi: 10.1126/science.290.5496.1567. [DOI] [PubMed] [Google Scholar]
- 49.Vicente-Manzanares M, Choi CK, Horwitz AR. Integrins in cell migration - the actin connection. J Cell Sci. 2009 Jan;122(Pt 2):199–206. doi: 10.1242/jcs.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7(4):265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Visualizing the mechanical activation of src. Nature. 2005 Apr 21;434(7036):1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 52.Webb WW, Barak LS, Tank DW, Wu ES. Molecular mobility on the cell surface. Btochem Soc Symp. 1981;(46):191–205. [PubMed] [Google Scholar]
- 53.Xiong JP, Stehle T, Diefenbach B, Zhang R, Bunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alpha vbetaS. Science. 2001 Oct 12;294(5541):339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Jiang G, Cai Y, Monkley S, Critchley D, Sheetz M. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat Cell Biol. 2008 Aug 17; doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]


