SUMMARY
Allelic loss of the essential autophagy gene beclin1 occurs in human cancers and renders mice tumor-prone suggesting that autophagy is a tumor-suppression mechanism. While tumor cells utilize autophagy to survive metabolic stress, autophagy also mitigates the resulting cellular damage that may limit tumorigenesis. In response to stress, autophagy-defective tumor cells preferentially accumulate p62/SQSTM1 (p62), endoplasmic reticulum (ER) chaperones, damaged mitochondria, reactive oxygen species (ROS), and genome damage. Moreover, suppressing ROS or p62 accumulation prevented damage resulting from autophagy defects indicating that failure to regulate p62 caused oxidative stress. Importantly, sustained p62 expression resulting from autophagy defects was sufficient to alter NF-κB regulation and gene expression and to promote tumorigenesis. Thus defective autophagy is a mechanism for p62 upregulation commonly observed in human tumors that contributes directly to tumorigenesis likely by perturbing the signal transduction adaptor function of p62 controlling pathways critical for oncogenesis.
Keywords: autophagy, beclin1, atg5, genomic instability, p62, NF- κB, DNA damage, cancer
INTRODUCTION
Macroautophagy (autophagy) targets cellular proteins, protein aggregates and organelles for degradation in lysosomes (Levine and Kroemer, 2008). Autophagy is induced by starvation or stress where double membrane vesicles (autophagosomes) capture intracellular cargo, and then fuse with lysosomes and are degraded. This lysosome-mediated cellular self-digestion sustains cell metabolism during starvation and eliminates damaged proteins and organelles that accumulate during stress. In mice, autophagy enables survival to neonatal starvation by preventing energy depletion (Kuma et al., 2004). Brain-targeted autophagy-deficiency (atg5−/− or atg7−/−) causes damaged mitochondria and polyubiquitin-containing protein aggregate accumulation, and neuronal degeneration (Hara et al., 2006; Komatsu et al., 2006). Liver-targeted autophagy deficiency (atg7−/−) similarly results in protein aggregate accumulation, hepatocyte cell death and liver injury (Komatsu et al., 2005). These findings support a prosurvival role for autophagy in sustaining energy homeostasis and maintaining protein and organelle quality control by eliminating damaged proteins and organelles during stress and aging (Levine and Kroemer, 2008).
Autophagy is induced by and localizes to hypoxic tumor regions where it supports cell survival (Degenhardt et al., 2006; Karantza-Wadsworth et al., 2007; Mathew et al., 2007b). Paradoxically, autophagy defects due to allelic loss beclin1 or constitutive activation of the autophagy-suppressing PI-3 kinase/mTOR pathway are common in human tumors (Levine and Kroemer, 2008). How loss of autophagy survival promotes tumor growth is being gradually reconciled (Jin and White, 2007, 2008; Mathew et al., 2007a).
Analogous to a wound-healing response, chronic cell death in response to stress and induction of inflammation and cytokine production may provide a non-cell-autonomous mechanism by which autophagy defects promote tumorigenesis (Degenhardt et al., 2006). Autophagy-defective tumor cells also display elevated genome damage with stress, suggesting that damage mitigation by autophagy is a cell-autonomous mechanism of tumor suppression (Karantza-Wadsworth et al., 2007; Mathew et al., 2007a; Mathew et al., 2007b). Possible non-mutually exclusive mechanisms by which autophagy limits cellular damage include maintenance of energy homeostasis, reduction of oxidative stress, and elimination of damaged proteins and organelles.
The importance of autophagy in cellular garbage disposal is clear, as autophagy is the only known means for turnover of large cellular structures such as organelles and protein aggregates (Levine and Kroemer, 2008). How organelles are recognized and directed to autophagosomes for degradation is not known, but may involve organelle-specific processes such as mitophagy and ER-phagy (Bernales et al., 2006; Kim et al., 2007). Damaged proteins that accumulate during stress can be refolded, ubiquitinated and degraded by the proteasome pathway, or aggregated and degraded by autophagy. To direct damaged or unfolded proteins to the autophagy pathway, p62 binds to polyubiquitinated proteins and aggregates by oligomerization, and binds to Atg8/LC3 on the autophagosome membrane to target aggregates to autophagosomes for degradation (Figure 1A) (Pankiv et al., 2007; Wooten et al., 2008). Protein aggregation may be a protective mechanism to limit cellular exposure to toxic proteins through sequestration, and an efficient packaging and delivery mechanism that collects and directs damaged proteins to autophagosomes. Liver-specific autophagy defects in mice cause accumulation of p62 aggregates, oxidative stress and p62-dependent hepatocyte cell death (Komatsu et al., 2007). Thus, the inability to eliminate p62 through autophagy can be toxic to normal tissues, but whether this is related to the tumor suppression by autophagy was not known.
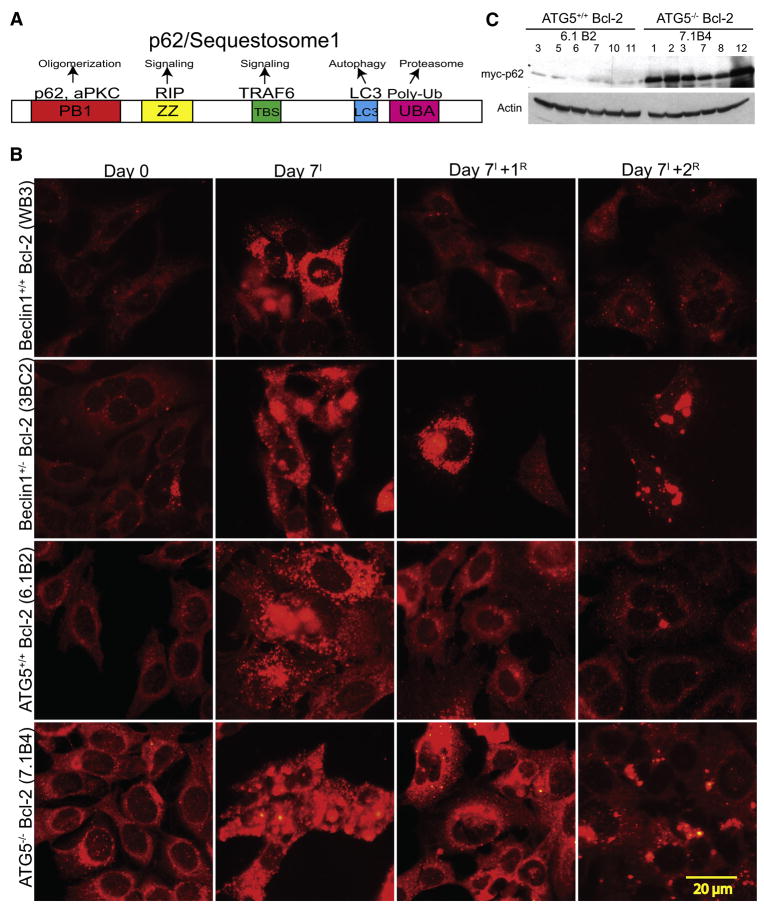
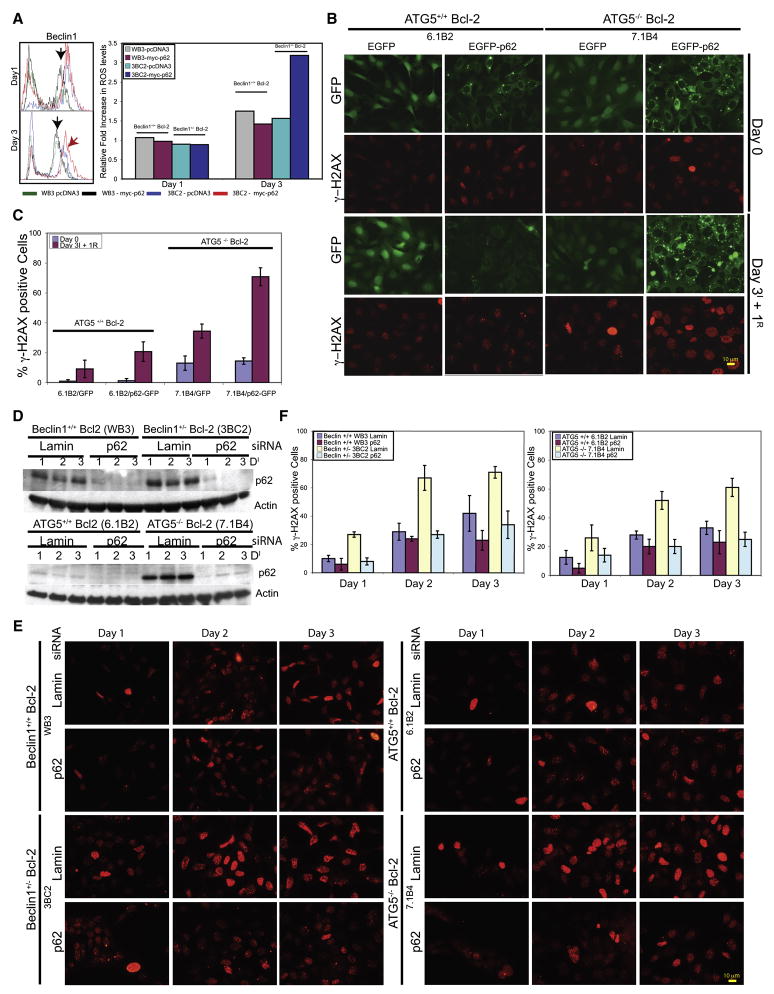
Figure 1. Elevated and persistent p62 in autophagy-defective tumor cells under metabolic stress.
(A) Domain organization of p62 illustrating the Phox and Bem1p (PB1) oligomerization domain (p62 and atypical Protein Kinase C [aPKC]), the Zinc finger (ZZ) Rip 1 binding domain, the TRAF6 binding site (TBS), the microtubule associated protein light chain 3 (LC3) domain (LC3/ATG8 binding), and the ubiquitin-associated (UBA) (poly-ubiquitin binding).
(B) IF of endogenous p62 showing accumulation and persistence of p62 in autophagy-defective cells under normal growth conditions (Day 0), 7 days of metabolic stress (Day 7I), and 1 (Day 7+1R) and 2 (Day 7+1R) days of recovery.
(C) Autophagy-defective cells express constitutively higher levels of exogenous Myc-p62. Six independent cell lines of Bcl-2-expressing atg5+/+ and atg5−/− iBMK cells stably expressing Myc-tagged p62 were evaluated for p62 expression levels by Western blotting with an antibody to Myc tag.
We report here that metabolic stress caused autophagy-defective tumor cells to preferentially accumulate p62, ER chaperones and protein disulphide isomerases (PDIs), indicative of protein quality control failure. Moreover, autophagy defects caused accumulation of damaged mitochondria, elevated oxidative stress and DNA damage response activation, which were attributed directly to persistence of p62. Cytotoxic effects due to defective autophagy were suppressed by ROS scavengers or by p62 elimination, indicating that persistence of p62 and oxidative stress caused cellular damage. This failure of autophagy-defective cells to eliminate p62 was sufficient to alter NF-κB regulation and gene expression and to promote tumorigenesis, indicating that autophagy suppresses tumorigenesis by limiting p62 accumulation. As p62 is commonly upregulated in human tumors (Zatloukal et al., 2007), this is due to defective autophagy and plays a causal role in cancer.
RESULTS
Autophagy-defective tumor cells accumulate p62 in response to stress
To address the role of autophagy-dependant protein quality control in tumor suppression, we assessed p62 modulation during metabolic stress and recovery in autophagy-competent (beclin1+/+ and atg5+/+) and -defective (beclin1+/− and atg5−/−) immortalized baby mouse kidney (iBMK) cells. Cells were engineered to express Bcl-2, as assessment of autophagy is facilitated in an apoptosis-defective background and expression of Bcl-2 is functionally equivalent to loss of Bax and Bak in the context of autophagy modulation and tumorigenesis (Degenhardt et al., 2006; Lum et al., 2005; Nelson et al., 2004). Under normal growth conditions, endogenous p62 levels were low by indirect immunofluorescence (IF) in beclin1+/+ and atg5+/+ cells and slightly elevated in autophagy-defective iBMK cells (Figure 1B). Following 7 days of metabolic stress p62 was induced in beclin1+/+ and atg5 +/+ cells and further elevated in autophagy-defective cells in a punctate pattern indicative of aggregation. In beclin1+/+ and atg5+/+ cells p62 was eliminated within 24 hr of recovery, whereas p62 persisted in autophagy-defective cells often in aggregates (Figure 1B). Higher p62 levels were observed in autophagy-deficient atg5−/− as compared to atg5+/+ iBMK cells stably expressing myc-tagged p62 (myc-p62) (Figure 1C). Thus, metabolic stress-induced p62 accumulation required autophagy for elimination, consistent with the absence of p62 gene induction in beclin1+/− and atg5−/− tumors (see below).
Autophagy-defective tumor cells accumulate damaged mitochondria, ER chaperones and PDIs
Apoptosis-deficient atg5+/+ iBMK cells respond to prolonged stress by undergoing progressive autophagy, yielding cells less than one-third their original size (Degenhardt et al., 2006) with well-preserved mitochondria (M) and ER (E) (Figures 2A and 2C). In contrast, Bcl-2-expressing atg5−/− (Figure 2B) and beclin1+/− (data not shown) iBMK cells contained mitochondria with structural abnormalities (M) and abnormal cytoplasmic structures (A) resembling protein aggregates (Figures 2B and 2D), consistent with p62 accumulation (Figure 1B). Thus, autophagy functions to prevent accumulation of damaged organelles and protein aggregates during metabolic stress.
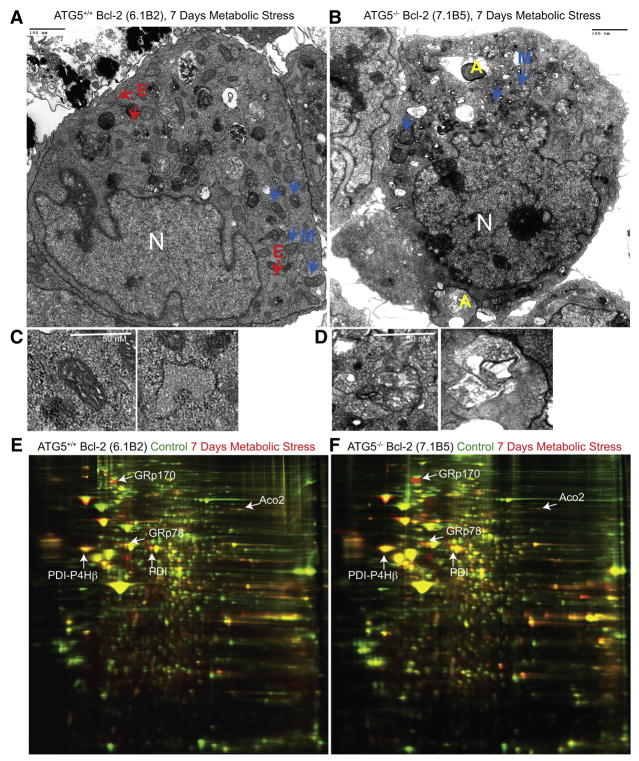
Figure 2. Metabolic stress promotes organelle damage and ER chaperones and PDI upregulation in autophagy-deficient cells.
(A–D) Representative electron micrographs of Bcl-2-expressing atg5+/+ (A) or atg5−/− (B) cells following 7 days of metabolic stress. Bcl-2 expressing atg5+/+ iBMK cell (A) showing mitochondria (M, blue arrows and C, left panel), ER (E, red arrows and C, right panel), mutant cell in (B) showing mitochondria (M, blue arrows and D, left panel), and protein aggregates (A, yellow and D, right panel).
(E and F) 2-DIGE gels showing differential regulation of ER chaperones (GRp170, GRp78), PDI, PDI-P4Hβ and ACO2 in Bcl-2-expressing atg5−/− iBMK cells in response to metabolic stress (7 days). Total protein from unstressed or metabolically stressed (7 days) Bcl-2-expressing atg5+/+ and atg5−/− cell lines were labeled with Cy3 (Green-unstressed) or Cy5 (Red-stressed) and analyzed by 2-DIGE. Images show 2-DIGE gels with proteins that are induced (Red), repressed (Green) or unchanged (Yellow) under metabolic stress. Protein spots (n=106) that were differentially expressed were selected and identified by mass spectrometry.
Since tumor cells with defective autophagy displayed failure of protein quality control, we performed two-dimensional difference in gel electrophoresis (2-DIGE) coupled with mass spectrometry to characterize the cellular proteome. Autophagy-competent, apoptosis-defective (bax−/−/bak−/−) D3 iBMK cells manage long-term metabolic stress by activating autophagy (Degenhardt et al., 2006) and induced ER chaperones (glucose-regulated proteins 170 [GRp170 and GRp78] and calreticulin), PDIs, metabolism and mitochondrial proteins (Table S1). Some of these proteins (TPI-1, PGK-1, PK-3 and GPDH) are targets of hypoxia inducible factor-1 α (HIF-1α) indicative of metabolic adaptation (Table S1), and HIF-1α is induced in iBMK cells by metabolic stress in vitro and in tumors in vivo (Nelson et al., 2004). Similarly, Bcl-2-expressing autophagy-competent beclin1+/+ and atg5+/+ iBMK cells induced ER chaperones GRp170, GRp78, calreticulin and calnexin indicating that metabolic stress response was independent of the means of apoptosis inactivation (Figures 2C and 2D, Table S1). To determine if autophagy status altered this stress response, Bcl-2-expressing, apoptosis and autophagy-defective (beclin1+/− and atg5−/−) iBMK cells were similarly examined. Autophagy-defective cells displayed preferential upregulation of ER chaperones compared to beclin1+/+ and atg5+/+ cells (Figures 2E and 2F, Table S1). Allelic loss of beclin1 was associated with a marked, differential increase in GRp170, GRp78, calreticulin and calnexin while atg5−/− cells showed differential increase in GRp170, GRp78 and calnexin compared to autophagy-competent controls (Table S1). GRp170 levels were induced by 3- to 9-fold in autophagy-competent (D3, beclin1+/+ and atg5+/+) cells whereas induction was 20-fold in autophagy-deficient (beclin1+/− and atg5−/−) cells under metabolic stress (Figure 2F, Table S1). A similar but less striking differential increase in GRp78 was also observed in beclin1+/− and atg5−/− cells (Table S1). PDI family members (PDI and PDI-P4Hβ) instrumental in folding proteins in the ER were induced under metabolic stress and were further elevated by autophagy-deficiency (Figures 2E and 2F, Table S1). The lack of induction of calreticulin and PDI-P4Hβ by metabolic stress in atg5−/− iBMK cells (Table S1) may be due to their increased susceptibility to metabolic stress (Mathew et al., 2007b). Interestingly, levels of cytoskeletal- and protein synthesis-related proteins were repressed with stress in all cell lines (Table S1). This enhanced induction of the protein folding machinery in the autophagy-deficient cells under stress suggests a role for autophagy in mitigating ER stress by eliminating unfolded proteins through lysosomal degradation.
As individual proteins can be represented by multiple spots in 2-DIGE, complicating estimation of protein levels by spot volume ratios, quantitation was validated by Western blotting. p62 and ER chaperones GRp170, GRp78, calnexin and PDI, showed higher induction in beclin1+/− and atg5−/− compared to beclin1+/+ and atg5+/+ cells under stress (Figure 3A) consistent with p62 IF and proteomic analysis. As with p62, elevated or persistent levels of these proteins were more evident in atg5−/− and beclin1+/− cells (Figure 3A), supporting that autophagy defects elevated demand for protein folding under metabolic stress that persists during recovery.
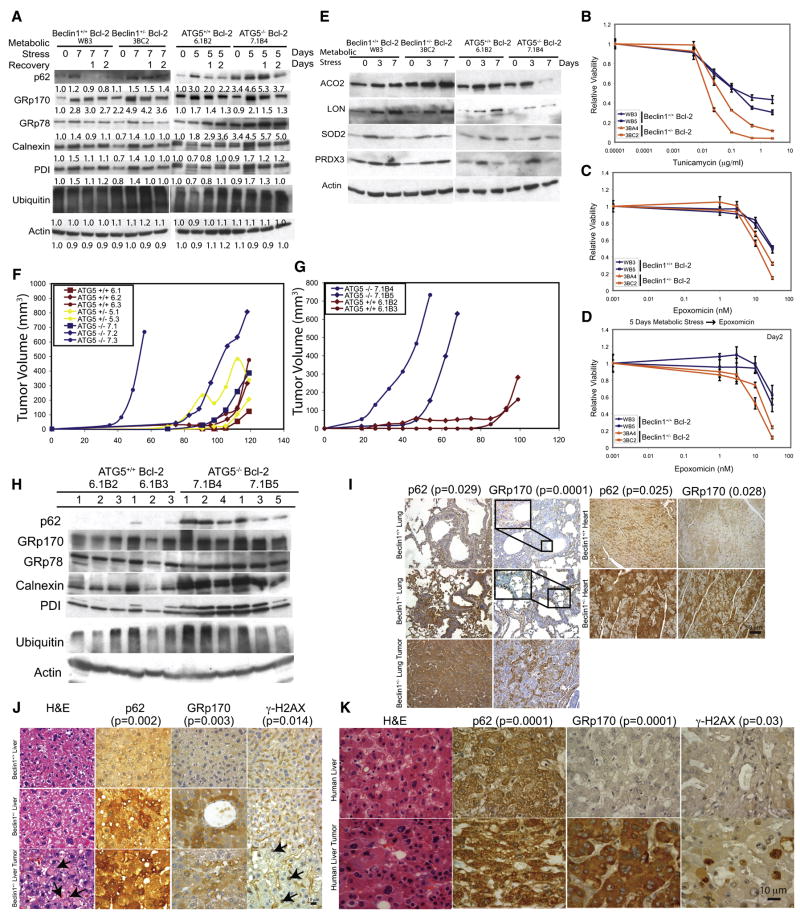
Figure 3. Autophagy defects promote ER stress and elevated DNA damage response in tumors in vivo.
(A) Western blots showing levels of p62, ER chaperones, PDI and ubiquitin in Bcl-2-expressing beclin1+/+, beclin1+/−, atg5+/+ and atg5−/− iBMK cells following 5 or 7 days of metabolic stress, and 1 and 2 days of recovery. Values below the bands represent relative signal intensities compared to the basal protein level in the first lane in each group (untreated Bcl-2-expressing beclin1+/+ and atg5+/+ control).
(B–D) Allelic loss of beclin1 sensitizes cells selectively to ER stress and proteasome inhibition. MTT assays showing sensitivity of Bcl-2-expressing beclin1+/+(Blue) and beclin1+/− (Red) iBMK cells in response to increasing concentrations of (B) tunicamycin (3 days), or (C) epoxomycin (2 days), and (D) following 5 days of metabolic stress. Data are presented as mean ± SD.
(E) Autophagy deficiency causes mitochondrial damage. Western blots showing levels of mitochondrial proteins (ACO2, LON, SOD2 and PRDX3) in Bcl-2-expressing beclin1+/+, beclin1+/−, atg5+/+ and atg5−/− iBMK cells, following metabolic stress.
(F) Deficiency in atg5 in iBMK cells promotes tumorigenesis. Tumor allograft growth of atg5+/+ (red), atg5+/− (yellow) and atg5−/− (blue) iBMK cell lines in nude mice.
(G) Deficiency in atg5 cooperates with defective apoptosis enhances tumor growth. Tumor allograft growth of Bcl-2-expressing atg5+/+ (red), and atg5−/− (blue) iBMK cell lines in nude mice.
(H) Western blot showing elevated p62 and ER chaperones and PDI in Bcl-2-expressing atg5+/+ and atg5−/− iBMK tumors in
(I) Elevated p62 and GRp170 levels in lung and heart tissues and spontaneous lung tumors from beclin1+/− mice. Lung, heart and spontaneous lung tumor sections from two 1.5-year-old beclin1+/+ and four 1.5-year-old beclin1+/− mice were stained for p62 and GRp170 by IHC. Sections were independently scored and analyzed by Students t-test (lung) or Mann-Whitney test (heart) and a p < 0.05 was considered statistically significant.
(J) Elevated p62 and GRp170 in liver tissue and p62, Mallory-Denk bodies (H&E, arrows) and γ-H2AX (arrows) in spontaneous liver tumors from beclin1+/− mice (1.5 years). Sections were independently scored and analyzed by Students t-test for significance (p < 0.05).
(K) Elevated p62, GRp170, and γ-H2AX positive nuclei in human HCC. Representative images from a human liver TMA (46 samples), showing H&E, p62, GRp170 and γ-H2AX accumulation in HCCs. Representative images from a normal human liver TMA (14 samples) are shown for comparison. Sections were independently scored and analyzed by Students t-test for significance for significance (p < 0.05).
Autophagy defects cause sensitivity to ER stress
Since autophagy defects upregulated protein folding regulators, we compared the sensitivity of beclin1 cells to pharmacological induction of ER stress. Tunicamycin induces ER stress by inhibiting protein glycosylation and allelic loss of beclin1 increased sensitivity to this drug (Figure 3B). To investigate if autophagy deficiency also elevated the burden on the ubiquitin-proteasome system, sensitivity to the proteasome inhibitor epoxomycin was assessed. Autophagy-defects sensitized to proteasome inhibition, and metabolic stress increased this sensitivity (Figures 3C and 3D) suggesting an elevated dependency of autophagy-defective cells on proteasome pathway. Thus, autophagy maintains protein quality control cooperatively with the ubiquitin-proteasome pathway, consistent with suppression of proteasome function activating autophagy, the inhibition of which promotes cell death (Ding et al., 2007).
Electron Microscopy (EM) revealed preferential accumulation of morphologically abnormal mitochondria in autophagy-deficient cells (Figures 2A–2D) and stressed atg5−/− cells showed aberrant regulation of mitochondrial proteins such as aconitase (ACO2) (Figures 2E and 2F) consistent with mitochondrial deterioration. ACO2 is susceptible to mitochondrial oxidative stress and upon oxidation is either stabilized or degraded (Fariss et al., 2005). Autophagy-deficient cells showed increased (beclin1+/−) or reduced (atg5−/−) levels of ACO2 after 7 days of metabolic stress compared to autophagy-competent controls (Figure 3E). Furthermore, LON, which degrades the oxidized form of ACO2 (Fariss et al., 2005), increased under metabolic stress, as did other oxidative stress markers. The accelerated deterioration of mitochondria in atg5−/− compared to beclin1+/− cells may account for the reduction in ACO2, superoxide dismutase (SOD2) and peroxiredoxin (PRDX3) at 7 days of stress (Figure 3E). Thus autophagy defects are associated with accumulation of damaged mitochondria under stress.
Defects in autophagy upregulate p62 and ER chaperones in tumors
To assess whether the differential accumulation of p62, ER chaperones and PDI was also a feature of autophagy defects in tumors, Bcl-2-expressing atg5+/+ and atg5−/−, and beclin1+/+ and beclin1+/− iBMK tumor allografts, and spontaneous tumors from beclin1+/− mice were examined. As in iBMK cells with allelic loss of beclin1 (Degenhardt et al., 2006; Mathew et al., 2007b), deficiency in atg5 increased tumorigenesis and cooperated with defects in apoptosis to accelerate tumor growth (Figure 3F and G). Bcl-2-expressing atg5−/− tumors displayed elevated p62, GRp170, GRp78, calnexin, and PDI compared to atg5+/+ tumors by Western blotting (Figure 3H) as did Bcl-2-expressing beclin1+/− and atg5−/− tumors by immunohistochemistry (IHC) (data not shown). p62 was not transcriptionally upregulated in beclin1+/− and atg5−/− tumors as determined in the gene expression analysis (p62 mRNA expression varied between 0.9- and 1.3-fold among wild type and autophagy-defective tumors) (data not shown). Upregulation of ubiquitinated proteins, although common in tissues of autophagy-defective mice (Hara et al., 2006; Komatsu et al., 2006), was not striking in atg5−/− tumors (Figure 3H). Additionally, tissues (lung, heart, and liver) and spontaneous lung and liver tumors from 1.5 year-old beclin1+/− mice showed significant accumulation of p62 and GRp170 when compared with age-matched tissues from beclin1+/+ littermates (Figures 3I and 3J). Thus, allelic loss of beclin1 and impaired autophagy caused elevated ER chaperone levels as a compensation mechanism and this phenotype is manifested in tissues and spontaneous tumors.
While p62 and ER chaperone upregulation are common in human tumors and are associated with poor prognosis, the cause is unknown (Ni and Lee, 2007; Zatloukal et al., 2007). Human hepatocellular carcinoma (HCC) is associated with p62 accumulation in Mallory-Denk bodies, and beclin1+/− mice display p62 accumulation in liver in association with steatohepatitis and spontaneous liver tumors (Figure 3J) (Komatsu et al., 2007; Qu et al., 2003; Yue et al., 2003), suggesting that autophagy defects may play a prominent role in HCC etiology. Indeed, liver and spontaneous liver tumors from beclin1+/− mice also showed higher levels of p62, GRp170 and DNA damage response activation (γ-H2AX) compared to normal liver tissue from age-matched beclin1+/+ mice (Figure 3J). p62, GRp170 and γ-H2AX were examined in a panel of human liver and HCCs in a tissue microarray (TMA). p62, GRp170, and γ-H2AX were upregulated with high frequency in HCC compared to normal liver (Figure 3K). Thus, accumulation of p62 and GRp170 in human tumors may be biomarkers for defective autophagy manifesting as accumulation of unfolded proteins associated with activation of the DNA damage response. Moreover, failure of protein and organelle quality control caused by autophagy defects may cause oxidative stress that is genotoxic.
Autophagy mitigates oxidative stress and progression to aneuploidy
Activation of the DNA damage response is a hallmark of oxidative stress caused by ROS. Protein re-folding in the ER by PDIs can elevate oxidative stress through redox reactions involving free radicals (Tu and Weissman, 2004), and mitochondrial stress and damage can also be a source of ROS (Fariss et al., 2005) in autophagy-deficient cells. Since autophagy deficiency causes accumulation of damaged mitochondria and the oxidative protein folding machinery and activated DNA damage response, we examined ROS levels in beclin1+/+ and beclin1+/− cells. Under normal growth conditions ROS levels were slightly elevated in the beclin1+/− iBMK cells compared to the beclin1+/+ cells, however, 5 days of metabolic stress caused a marked ROS increase in beclin1+/− cells (Figures 4A and 4B). During recovery, beclin1+/− iBMK cells showed increased ROS that persisted for 24 hr compared to beclin1+/+ cells (Figures 4A and 4B). Thus allelic loss of beclin1 was associated with elevated ROS production with stress.
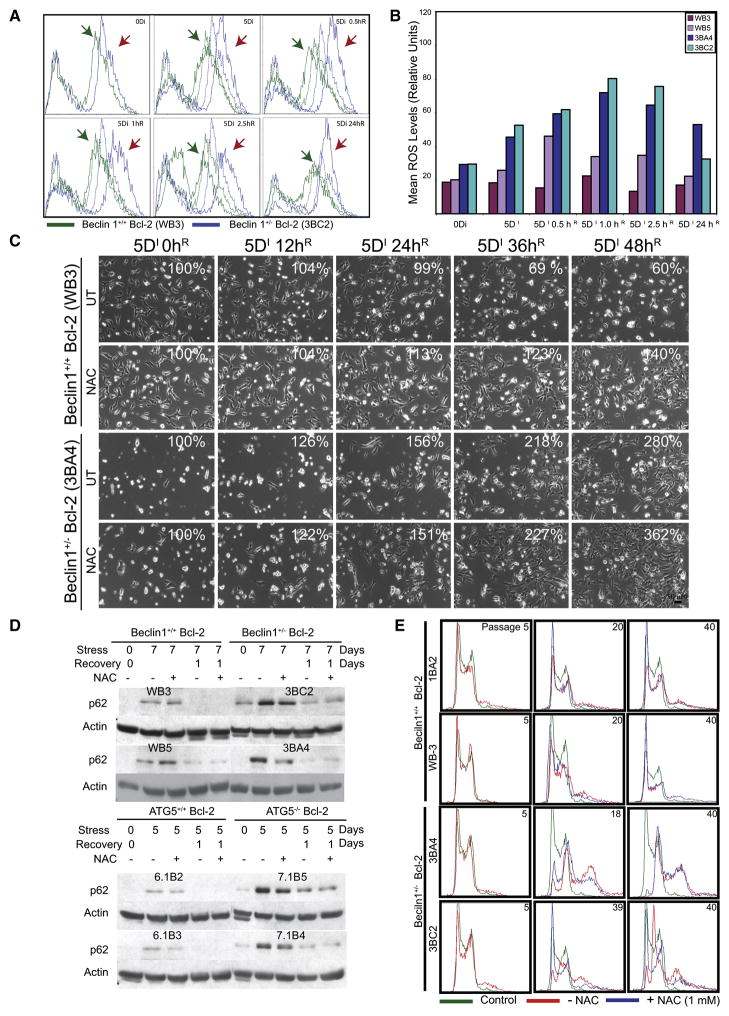
Figure 4. Metabolic stress promotes elevated ROS production and chromosomal instability in autophagy-deficient cells.
(A) Autophagy-deficiency leads to elevated ROS production. Overlays show ROS levels in Bcl-2-expressing beclin1+/+ (Green) and beclin1+/− iBMK cells (Blue) (x-axis, log scale) under normal growth (0Di) and at 0.5, 1, 2.5 and 24 hr (beclin1+/+, green arrows and beclin1+/−, red arrows) during recovery (0.5–24hR) from 5 days of metabolic stress (5Di) by flow-cytometry using the ROS sensor DCF-DA. The ROS levels in untreated cells are shown in dotted lines for comparison.
(B) Representative histogram from three independent experiments measuring the mean ROS levels in Bcl-2-expressing beclin1+/+ and beclin1+/− iBMK cells shown in (A).
(C) ROS scavenging partially rescues the susceptibility to metabolic stress and recovery due to allelic loss of beclin1. Representative time-lapse images of Bcl-2-expressing beclin1+/+ and beclin1+/− iBMK cells during recovery following 5 days of metabolic stress in presence (NAC) and absence (UT) of the ROS scavenger NAC (1mM) (relative percentage of adherent cells compared to time 0 is shown).
(D) ROS scavenging suppresses p62 accumulation in autophagy-deficient (beclin1+/− and atg5−/−) cells. Western blot analysis of p62 levels in Bcl-2-expressing beclin1+/+, beclin1+/−, atg5+/+ and atg5−/− iBMK cell lines following 0 or 7 days of metabolic stress followed by 1 day recovery without and with NAC.
(E) ROS scavenging limits progression to aneuploidy associated with allelic loss of beclin1. Flow-cytometry analysis of DNA content in diploid, Bcl-2-expressing beclin1+/+ and beclin1+/− iBMK cells grown in presence (blue) or absence (red) of the ROS scavenger NAC (1mM). Numbers represent passage numbers at which ploidy was determined.
To determine if the elevated ROS in autophagy-deficient cells contributes to cellular damage, the stress response without and with the ROS scavenger, N-acetyl cysteine (NAC) was examined. Following 5 days of metabolic stress, beclin1+/− and atg5−/− iBMK cells showed increased susceptibility to stress compared to control cells (Figure 4C and S2) (Degenhardt et al., 2006; Karantza-Wadsworth et al., 2007; Mathew et al., 2007b). The presence of NAC during metabolic stress improved survival, and this protective effect was more profound in beclin1+/− and atg5−/− cells, suggesting that elevated ROS contribute to increased susceptibility of autophagy-defective cells to stress (Figure 4C and S2). This enhanced survival provided by NAC was associated with decreased p62 accumulation during metabolic stress in the beclin1+/− and atg5−/− iBMK cells (Figure 4D and S2), suggesting that ROS-mediated oxidative stress leads to protein damage and accumulation of p62. A feature of genomic instability associated with autophagy defects is the accelerated progression to aneuploidy (Mathew et al., 2007b). To examine the role of increased ROS levels on genomic instability, we monitored the DNA content of early passage diploid beclin1+/+ and beclin1+/− iBMK cells without and with NAC. beclin1+/+ cells maintained diploidy after 40 passages and the presence of NAC had no effect. In contrast, beclin1+/− cells showed accelerated progression to aneuploidy by passages 18 and 39 (Figure 4E), and NAC delayed this progression (Figure 4E), indicating a causative role for basal ROS-mediated oxidative stress in progression to aneuploidy associated with autophagy defects. Thus, metabolic stress causes p62 accumulation mediated in part by elevated ROS production and the failure to suppress ROS in autophagy-deficient cells is associated with genomic instability.
p62 accumulation activates the DNA damage response
Since autophagy deficiency leads to accumulation of p62, oxidative stress and accelerated progression to aneuploidy, we examined if p62 accumulation was sufficient to induce ROS and the DNA damage response. Transiently expressed p62-EGFP formed aggregates (Figure S1B) and elevated ROS in the autophagy-deficient (beclin1+/− and atg5−/−) but not in beclin1+/+ and atg5+/+ iBMK cells, whereas EGFP expression alone did not (Figures 5A and S1A). Transient p62-EGFP expression was also associated with DNA damage response activation (γ-H2AX positive nuclear foci) in autophagy-deficient (beclin1+/− and atg5−/−) cells compared to beclin1+/+ and atg5+/+ cells (Figures S1B and S1C). To monitor accumulation and clearance of p62, apoptosis-deficient atg5+/+ and atg5−/− iBMK cells engineered to stably express either EGFP or p62-EGFP were subjected to metabolic stress followed by recovery. As with endogenous p62 (Figure 1B) and myc-p62 (Figure 1C), p62-EGFP-expressing atg5−/− iBMK cells displayed p62 aggregates that were induced by metabolic stress and persisted in recovery which atg5+/+ cells were able to eliminate (Figures 5B and 5C). Consistent with transient expression (Figure S1B), stable p62-EGFP expression activated the DNA damage response during stress and recovery as indicated by γ-H2AX positive nuclear foci. While the focal pattern of γ-H2AX staining characteristic of DNA double strand breaks (Balajee and Geard, 2004) was clear in majority of the γ-H2AX positive nuclei, there were also nuclei showing uniform γ-H2AX staining indicating variations in the levels of DNA damage. atg5−/− iBMK cells expressing EGFP showed markedly elevated levels of supernumerary centrosomes compared to atg5+/+ cells under normal conditions, but more strikingly so following stress and recovery. Expression of p62-EGFP increased centrosome abnormalities and multi-polar divisions, which were more dramatic under stress and recovery in atg5−/− compared to atg5+/+ cells (Figure S1E and S1F). Thus p62 accumulation was sufficient for ROS and DNA damage response induction under metabolic stress leading to cell division abnormalities and genomic instability in autophagy-defective cells. Indeed, RNAi-mediated knockdown of p62 during metabolic stress reduced DNA damage induction in autophagy-deficient beclin1+/− and atg5−/− iBMK cells, further suggesting that impaired p62 elimination was the cause of DNA damage response activation (Figure 5D–5F).
Figure 5. Failure to eliminate p62 by autophagy activates the DNA damage response.
(A) p62 expression leads to elevated ROS production in the autophagy-defective beclin1+/− cells. Bcl-2-expressing beclin1+/+ and beclin1+/− iBMK cells were transfected with myc-tagged p62 or control vector and ROS levels (DCF-DA) were measured by flow-cytometry at day 3 post-transfection. Histogram on the right is representative of three independent experiments measuring mean ROS level in each cell line on days 1 and 3 post-transfection.
(B) Failure to eliminate p62 by autophagy under metabolic stress leads to DNA damage response induction. Bcl-2-expressing atg5+/+ or atg5−/− iBMK cells stably expressing EGFP or p62-EGFP were subjected to 3 days of metabolic stress, allowed to recover (1 day) and stained for γ-H2AX.
(C) Quantitation of the percentage cells with γ-H2AX positive foci in cells shown in (B). Data from 200 cells are presented as mean ± SD.
(D) Western blots of RNAi-knockdown of p62. Bcl-2-expressing wild-type (beclin1+/+ and atg5+/+) and autophagy-defective (beclin1+/− and atg5−/−) iBMK cells were transfected with either Lamin- or p62-siRNA and subjected to metabolic stress for 0, 24, 48 or 72 hr (24, 48, 72 and 96 hr post-transfection, respectively) and analyzed for p62 levels.
(E) p62 accumulation in autophagy-defective cells is responsible for activation of the DNA damage response. Cells in (D) were evaluated for γ-H2AX positive nuclear foci.
(F) Quantitation of percentage cells with γ-H2AX positive nuclei from the data shown in (E). Data from two hundred cells are presented as mean ± SD.
p62 promotes tumorigenesis of autophagy-defective cells
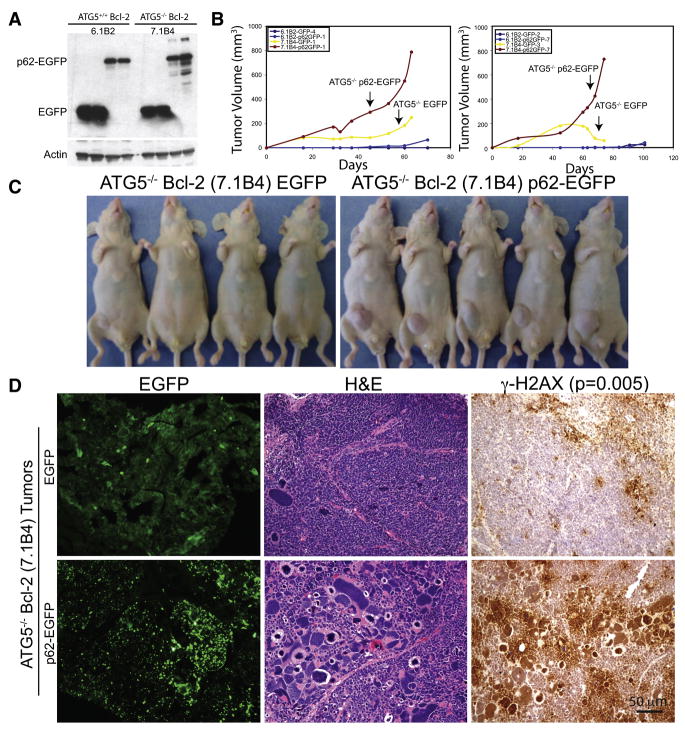
Since p62 accumulates in tissues and tumors from autophagy-deficient mice and in human cancers, we tested if p62 directly contributed to tumorigenesis. Apoptosis-deficient atg5+/+ and atg5−/− iBMK cell lines expressing EGFP or p62-EGFP (Figure 6A) were assessed for their tumorigenic potential. p62-EGFP expression in atg5+/+ cell lines did not substantially increase tumor growth (Figure 6B). In contrast, p62-EGFP expression in atg5−/− cells, dramatically increased tumor growth compared to EGFP-expressing atg5−/− controls (Figure 6B). In contrast to EGFP-expressing atg5−/− tumors displaying diffuse cytoplasmic EGFP localization and uniformly sized nuclei with occasional appearance of tumor giant cells, p62-EGFP-expressing atg5−/− tumors displayed dramatic p62 aggregate accumulation and numerous pleomorphic tumor cells with heterochromatic, giant nuclei by H&E staining indicative of polyploidy and aneuploidy (Figure 6D). Persistent p62 accumulation in p62-EGFP-expressing atg5−/− tumors was also associated with elevated DNA damage response induction (γ-H2AX) compared to EGFP-expressing atg5−/− tumors (Figure 6D), suggesting that inability to clear p62 through autophagy promoted tumor growth and elevated DNA damage and genomic instability.
Figure 6. p62 expression cooperates with autophagy-deficiency to promote tumor growth.
(A) Western blot for EGFP, in Bcl-2-expressing atg5+/+ and atg5−/− iBMK cells stably expressing EGFP or p62-EGFP.
(B) Tumor growth of cell lines in (A) showing enhanced tumor growth in p62-EGFP expressing atg5−/− tumors (red) compared to that of the control vector (yellow).
(C) Panel showing tumor-bearing mice injected with p62-EGFP- (right panel), and EGFP- expressing (left panel) atg5−/− iBMK cells from (B), at day 74 post-injection.
(D) Tumors from p62-EGFP expressing atg5−/− cells are associated with p62 aggregates and polymorphic and γ-H2AX positive nuclei. Representative photomicrographs of frozen tumor sections (left), and paraffin embedded sections stained by H&E (middle) or IHC for γ-H2AX (right) [Students t-test for significance for significance (p < 0.05)] in tumors from atg5−/− cells shown in (C).
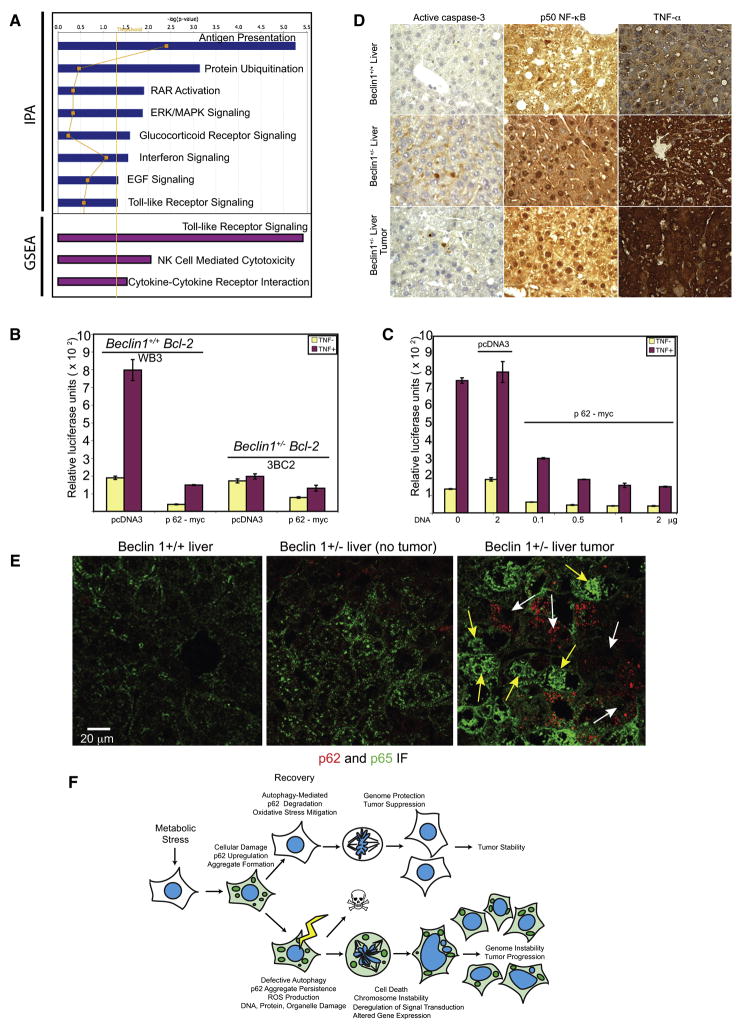
In addition to its role in binding and targeting polyubiquitinated proteins to autophagosomes for lysosomal degradation, p62 also has a role as an adaptor protein regulating signal transduction in the RANKL, TrkA, and aPKC pathways through interactions with TRAF6 and RIP1 that can regulate NF-κB (Moscat et al., 2007; Wooten et al., 2008). To address the possibility that deregulation of p62 in autophagy defective cells altered cancer signaling pathways, global patterns of gene expression where analyzed in atg5−/− EGFP- and EGFP-p62-expressing iBMK tumors (Figure 6). Of the 14,000 genes assessed, 893 genes (p=0.05) displayed differential expression in EGFP- compared with EGFP-p62-expressing tumors and were further subjected to Ingenuity Pathway Analysis (IPA) and Gene Set Enrichment Analysis (GSEA) (Figure 7A, supplemental Tables S2 and S3). Both analyses indicated down-regulation of host defense pathways including antigen presentation, Toll-like receptor and Natural Killer (NK) cell mediated cytotoxicity pathways in p62-EGFP- compared to EGFP-expressing tumors (Figure 7A and supplemental Table S2). Interaction map analysis indicated that a commonality in the pathways suppressed by p62 gain-of-function was the NF-κB pathway, and indeed NF-κB target genes were down regulated in p62-EGFP compared to EGFP-expressing tumors (Table S3).
Figure 7. Accumulation of p62 in autophagy-defective cells alters signal transduction pathways involved in host and anti-oxidant defense.
(A) Pathways (p=0.05; yellow line) suppressed in tumors from atg5−/− iBMK cells expressing p62-EGFP compared to those expressing EGFP analyzed by IPA (blue bars) and by GSEA (purple bars).
(B) Luciferase-reporter assays in Bcl-2 expressing wild-type (beclin1+/+) and autophagy-defective (beclin1+/−) iBMK cells showing suppression of IL-6-Luciferase reporter (NF-κB activity) in autophagy-defective cells by p62.
(C) Luciferase-reporter assays in Bcl-2 expressing beclin1+/+ iBMK (WB3) cells showing that over-expression of p62 is sufficient to suppress NF-κB transcriptional activity in a concentration dependant manner.
(D) Impaired hepatocyte survival, activation of NF-κB and cytokine production in beclin1+/− liver and beclin1+/− liver tumors. IHC staining showing elevated levels apoptotic cell death (active caspase-3), NF-κB activation (nuclear p50), and cytokine production (TNF-α) in liver tissue and spontaneous liver tumors from beclin1−/− mice.
(E) Defective autophagy and accumulation of p62 suppress NF-κB activation and promotes HCC. Representative photomicrographs of frozen liver sections from beclin1+/+ and beclin1+/− and spontaneous HCC from beclin1+/− mice, immunostained for p62 (red) and p65 NF-κB (green) and analyzed by confocal microscopy. Note that those beclin1+/− hepatocytes that accumulate p62 do not display nuclear p65 (yellow arrows) whereas those without p62 display nuclear localization of p65 indicative of NF-κB activation.
(F) A model for the role of autophagy as a tumor suppressor mechanism by limiting p62 accumulation.
Allelic loss of beclin1 causes steatohepatitis, Mallory body formation (p62 aggregates), and HCC (Fig. 4E), which phencopies hepatocyte-targeted deficiency in NF-κB activators IKK-β or NEMO (Luedde et al., 2007; Maeda et al., 2005). To test the hypothesis that p62 gain-of-function was responsible for the suppression of NF-κB, IL-6-luciferase reporter assays were performed in autophagy-competent (beclin1+/+ and atg5+/+) and autophagy-deficient (beclin1+/− and atg5−/−) iBMK cells in response to TNF-α. Despite similar basal levels, autophagy-deficient cells showed reduced NF-κB activation in response to TNF-α, which was further inhibited by p62 expression (Figures 7B, 7C and S3) suggesting that p62 accumulation impaired NF-κB activation. In liver, deficient NF-κB canonical pathway activation leads to impaired hepatocyte survival, Kupffer cell activation and hepatomitogen (Il-6, TNF-α, hepatocyte growth factor) production leading to non-canonical NF-κB pathway activation and compensatory proliferation of healthy hepatocyes and to HCC. To test if defective autophagy and p62 accumulation caused the same phenotype, nuclear localization of p50 NF-κB indicative of activation was assessed in liver tissue from age-matched beclin1+/+ and beclin1+/− mice. Nuclear p50 and active caspase-3 were observed in many beclin1+/− liver and liver tumor hepatocytes but not in beclin1+/+ livers (Figure 7D). Similarly, p65 NF-κB was frequently nuclear in beclin1+/− liver tumor hepatocytes but was not nuclear in beclin1+/+ liver tissue (Figure 7E). Importantly, p62 accumulation in hepatocytes was heterogenous (Figure 3J). beclin1+/− hepatocytes that accumulated high p62 did not display nuclear p65 whereas those with less p62 showed nuclear localization of p65 (Figure 7E). This suggests that as in the IKKβ- and NEMO-deficient hepatocyes, defective autophagy and deregulation of p62 was associated with suppression of the canonical NF-κB pathway, impaired survival and hepatomitogen-driven oncogenesis through the non-canonical pathway. Indeed, beclin1+/− livers and liver tumors displayed increased TNF-α production (Figure 7D).
DISCUSSION
Accumulation of p62 in response to metabolic stress is a striking phenotype of autophagy-defective tumors cells, suggesting defective protein quality control may contribute to tumorigenesis and that autophagy is the main mechanism by which tumor cells turnover p62. Moreover, the failure of autophagy-defective tumor cells to eliminate p62 was sufficient for tumorigenesis. Unlike brain tissue (Hara et al., 2006; Komatsu et al., 2006), there was no accumulation of polyubiquitinated proteins in autophagy-defective tumor cells. There may be tissue-specific differences in autophagy-mediated protein elimination or dilution of polyubiquitinated proteins through cell proliferation in tumor cells may prevent their accumulation, which is not possible in post-mitotic neurons. Alternatively, proteasome-mediated turnover of polyubiquitinated proteins may be elevated in tumor cells compared to neuronal tissues. Indeed, autophagy defects sensitized cancer cells to proteasome inhibitors, suggesting a compensatory function of the two protein degradation pathways.
The persistence of p62 and accumulation of ER chaperones and the oxidative protein folding machinery in autophagy-deficient cells and tumors indicated a defect in the management of protein turnover. The inability to degrade damaged or misfolded proteins through autophagy may increase the burden on the ER protein folding machinery necessitating its upregulation. Both p62 and GRp170 were dramatically upregulated in beclin1+/− tissues as well as in spontaneous tumors, indicating that coping with unfolded proteins may be a biomarker for impaired autophagy that precedes tumor initiation. ER chaperone and PDI upregulation are common in human tumors (Goplen et al., 2006; Ni and Lee, 2007), and increased GRp170 expression is associated with poor prognosis in breast cancer (Tamatani et al., 2001). Although the cause of ER chaperone accumulation in tumors was not known, chaperones are stress-responsive and provide a protective function by suppressing the accumulation of unfolded proteins that may be an important compensatory mechanism for autophagy-defective cells. Protein folding is a source of oxidative stress (Tu and Weissman, 2004), particularly when cells are overburdened with damaged and unfolded proteins, in concordance with increased ROS in stressed beclin1+/− cells.
Stressed autophagy-defective tumor cells accumulate damaged mitochondria as a potential additional source of oxidative stress. This accumulation of unfolded protein and protein aggregates and the persistence of damaged mitochondria may collectively lead to elevated ROS production in autophagy-defective cells. As ROS scavengers partially suppressed p62 accumulation and cell death in stressed beclin1+/− tumor cells, the elevated oxidative stress may contribute to p62 induction, cell damage and death. Although oxidative stress and DNA damage arise through multiple genotoxic events (Halazonetis et al., 2008), stress-mediated p62 accumulation in autophagy-defective cells was sufficient for ROS and DNA damage response induction that was prevented by knockdown of p62, establishing that that elevated oxidative stress was attributable directly to p62 accumulation. Enforced p62 expression induced ROS, suggesting a possible amplification loop where oxidative stress induces p62 accumulation, which in turn amplifies ROS generation. Thus, the inability of autophagy-defective tumor cells to eliminate p62 contributes to oxidative stress and likely to DNA damage. These observations are strikingly similar to the rescue of oxidative stress toxicity caused by atg7 deficiency with p62 deficiency in mouse liver (Komatsu et al., 2007). In normal tissues, toxicity due to p62 accumulation resulting from autophagy defect may trigger cell death, whereas in checkpoint-defective tumor cells this instead may also result in enhancement of mutations, genome instability and tumor progression.
atg5−/− tumors displayed pronounced p62 and p62 aggregate accumulation and this p62 expression was sufficient to activate the DNA damage response and to enhance tumor growth. p62 expression, p62 aggregates, and Mallory-Denk bodies containing p62 are common in steatosis and in HCC and other cancers (Zatloukal et al., 2007). Defects in autophagy may be a mechanism for sustained p62 accumulation and formation of Mallory-Denk bodies. As such, p62 accumulation is not merely a histologic marker for certain cancers, but rather, directly contributes to tumor growth. While the prevalence of autophagy defects in HCC is not yet known, mutations such as pten loss that constitutively activates the PI3-kinase pathway and mTOR that inhibits autophagy are common (Wong and Ng, 2008). Interestingly, pten (Watanabe et al., 2005), beclin1, or atg7 deficiency (Komatsu et al., 2007; Qu et al., 2003; Yue et al., 2003) produce liver steatosis in mice suggesting that suppression of autophagy and the resulting steatosis can lead to HCC.
How persistent p62 promotes oxidative stress and tumorigenesis appears to be related to its role as an adaptor protein regulating receptor signaling and the activation of NF-κB. p62 is also required for efficient oncogene activation in vitro and p62 deficiency suppresses spontaneous lung tumorigenesis by K-ras (Duran et al., 2008). Thus, p62 had been identified as an oncoprotein in both loss- (Duran et al., 2008) and gain-of-function situations (Figure 6). p62 gain-of-function caused by defective autophagy altered NF-κB signal transduction pathways that regulate host defense. Whether p62 is upregulated and either sequestered in non-functional aggregates inhibiting signal transduction as indicated here, or is retained in an active state enhancing signal transduction, may be cell type- or stress-specific. In liver, defects in NF-κB canonical pathway activation promote tumorigenesis by stimulation of inflammation and activation of the non-canonical NF-κB pathway. However, this function of NF-κB may be tissue specific, and as NF-κB signaling also regulates anti-oxidant defense, suppression of NF-κB by p62 may explain increased oxidative stress in other autophagy-defective tissues. In conclusion, defects in autophagy promote a failure of protein and organelle quality control in tumors this leads to p62 accumulation resulting in perturbation of gene expression, increased oxidative stress, genome damage and tumorigenesis (Figure 7F). As p62 upregulation is common in liver tissues of individuals at risk and hepatocellular carcinomas in patients, this suggests that facilitating the clearance of p62 by promoting autophagy may be a strategy for cancer chemoprevention.
EXPERIMENTAL PROCEDURES
Generation of stable cell lines and culture conditions
atg5+/+, atg5+/−, atg5−/−, beclin1+/+ and beclin1+/− iBMK cell lines were described previously (Degenhardt et al., 2006; Degenhardt et al., 2002; Mathew et al., 2008; Mathew et al., 2007b). Bcl-2-expressing atg5+/+ and atg5−/− iBMK cells, were engineered to stably express Myc-tagged p62 (pcDNA3-myc-p62), EGFP (pEGFPC1) or p62-EGFP (pEGFPC1-p62) (Rodriguez et al., 2006), or were co-transfected with pcDNA3-Zeo by electroporation as described previously (Nelson et al., 2004). Independent clones were selected in zeocin (500μg/mL) and expanded as stable cell lines in normal culture conditions (DMEM, 10% FBS, 1% Pen Strep (Invitrogen, Carlsband, CA) at 38.5°C and 8.5% CO2) (Mathew et al., 2008). To induce metabolic stress, cells were placed in glucose-free DMEM (Invitrogen) containing 10% FBS and incubated with a defined gas mixture containing 1% oxygen, 5% CO2 and 94% N2 (GTS-Welco, Allentown, PA) (Nelson et al., 2004). NAC (Sigma-Aldrich, St. Louis, MO) was used at a concentration of 1mM.
Supplementary Material
Acknowledgments
We thank Drs. Heintz, Yue and Jin for providing beclin1+/+ and beclin1+/− mice, Dr. Mizushima for providing atg5+/+ and atg5−/− mice, Dr. Zimmermann for GRp170 antibody and Dr. Moscat for myc-p62 and p62-EGFP plasmids. This work was supported by grants from the NIH (R37 CA53370 and RO1 CA130893) to E.W., (K99CA133181) to V.K.W., and DOD (W81XWH06-1-0514 and W81XWH05) to E.W. and R.S.D.
Footnotes
Supplemental data include supplemental experimental procedures, three figures, and three tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balajee AS, Geard CR. Replication protein A and gamma-H2AX foci assembly is triggered by cellular response to DNA double-strand breaks. Exp Cell Res. 2004;300:320–334. doi: 10.1016/j.yexcr.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Sundararajan R, Lindsten T, Thompson C, White E. Bax and Bak independently promote cytochrome C release from mitochondria. J Biol Chem. 2002;277:14127–14134. doi: 10.1074/jbc.M109939200. [DOI] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Fariss MW, Chan CB, Patel M, Van Houten B, Orrenius S. Role of mitochondria in toxic oxidative stress. Mol Interv. 2005;5:94–111. doi: 10.1124/mi.5.2.7. [DOI] [PubMed] [Google Scholar]
- Goplen D, Wang J, Enger PO, Tysnes BB, Terzis AJ, Laerum OD, Bjerkvig R. Protein disulfide isomerase expression is related to the invasive properties of malignant glioma. Cancer Res. 2006;66:9895–9902. doi: 10.1158/0008-5472.CAN-05-4589. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, White E. Tumor Suppression by Autophagy Through the Management of Metabolic Stress. Autophagy. 2008:4. [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata JI, Ezaki J, Murata S, et al. Homeostatic Levels of p62 Control Cytoplasmic Inclusion Body Formation in Autophagy-Deficient Mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the Pathogenesis of Disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedde T, Beraza N, Kotsikoris V, van Loo G, Nenci A, De Vos R, Roskams T, Trautwein C, Pasparakis M. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2007;11:119–132. doi: 10.1016/j.ccr.2006.12.016. [DOI] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Mathew R, Degenhardt K, Haramaty L, Karp CM, White E. Immortalized mouse epithelial cell models to study the role of apoptosis in cancer. Methods Enzymol. 2008;446:77–106. doi: 10.1016/S0076-6879(08)01605-4. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007a;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007b;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Nelson DA, Tan TT, Rabson AB, Anderson D, Degenhardt K, White E. Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes Dev. 2004;18:2095–2107. doi: 10.1101/gad.1204904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Duran A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, Serrano M, Auwerx J, Diaz-Meco MT, Moscat J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3:211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Tamatani M, Matsuyama T, Yamaguchi A, Mitsuda N, Tsukamoto Y, Taniguchi M, Che YH, Ozawa K, Hori O, Nishimura H, et al. ORP150 protects against hypoxia/ischemia-induced neuronal death. Nat Med. 2001;7:317–323. doi: 10.1038/85463. [DOI] [PubMed] [Google Scholar]
- Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Horie Y, Suzuki A. Hepatocyte-specific Pten-deficient mice as a novel model for nonalcoholic steatohepatitis and hepatocellular carcinoma. Hepatol Res. 2005;33:161–166. doi: 10.1016/j.hepres.2005.09.026. [DOI] [PubMed] [Google Scholar]
- Wong CM, Ng IO. Molecular pathogenesis of hepatocellular carcinoma. Liver Int. 2008;28:160–174. doi: 10.1111/j.1478-3231.2007.01637.x. [DOI] [PubMed] [Google Scholar]
- Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco MT, Moscat J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatloukal K, French SW, Stumptner C, Strnad P, Harada M, Toivola DM, Cadrin M, Omary MB. From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res. 2007;313:2033–2049. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.