Abstract
Rationale
Cardiac hypertrophy occurs in response to a variety of extrinsic and intrinsic stimuli that impose increased biomechanical stress. The PI3K/Akt pathway has previously been strongly associated with hypertrophic signaling in the heart, and with the control of cell size in multiple contexts. This pathway is tightly regulated by many factors, including a host of kinases and phosphatases that function at multiple steps in the signaling cascade. For example, the PTEN tumor suppressor protein is a phosphoinositide 3-phosphatase that, by metabolizing PtdIns(3,4,5)P3, acts in direct antagonism to growth factor stimulated PI3K. Inhibition of PTEN leads to cardiomyocyte hypertrophy. Another polyphoinositide phosphatase, inositol polyphosphate-5-phosphatase F (Inpp5f) has recently been implicated in regulation of cardiac hypertrophy. Like PTEN, this phosphatase can degrade PtdIns(3,4,5)P3 and thus modulates the PI3K/Akt pathway.
Objective
To characterize the role of Inpp5f in regulating cardiac hypertrophy.
Methods and Results
We generated homozygous Inpp5f knockout mice and cardiac specific Inpp5f overexpression transgenic mice. We evaluated their hearts for biochemical, structural and functional changes. Inpp5f knockout mice have augmented hypertrophy and reactivation of the fetal gene program in response to stress when compared to wild type littermates. Furthermore, cardiac over-expression of Inpp5f in transgenic mice reduces hypertrophic responsiveness.
Conclusions
Our results suggest that Inpp5f is a functionally important endogenous modulator of cardiac myocyte size and of the cardiac response to stress.
Keywords: Inpp5f, Hypertrophy, Akt, GSK3β, Hdac2
Introduction
Cardiovascular disease remains the number one cause of mortality in the Western world, with heart failure representing the fastest growing subclass over the past decade1. In many cases, cardiac hypertrophy precedes cardiac dilation and heart failure. Cardiac hypertrophy can occur in response to normal physiological stimuli or may be maladaptive leading to cardiac dilatation and congestive heart failure. However, the cellular mechanisms that regulate the hypertrophic response to agonists or to stretch remain poorly understood, and the transition from hypertrophy to failure is also ill-defined. On the other hand, evidence accumulating over the last few decades confirms that intercellular signaling pathways and gene expression are fundamentally altered in states of hypertrophy and failure and a thorough understanding of these changes will provide therapeutic targets2.
Recently, we showed that Hdac2 deficient mice are resistant to cardiac hypertrophy when exposed to hypertrophic stimuli3. Resistance to hypertrophy in Hdac2−/− mice is associated with increased expression of the gene encoding inositol polyphosphate-5-phosphatase f (Inpp5f) and constitutive activation of Gsk3β via inactivation of Akt and Pdk1. Conversely, transgenic over-expression of Hdac2 in the heart reactivates fetal genes and induces cardiac hypertrophy. In these transgenic hearts, Inpp5f is significantly down-regulated, Akt is activated, and Gsk3β is inactive3. Further studies suggest that Hdac2 is a direct regulator of Inpp5f3. Thus, we hypothesized that Inpp5f might functionally contribute to cardiac regulation of the Akt/Gsk3β pathway in response to stress.
Inpp5f is one of several polyphosphoinositide phosphatases that have been identified and partially characterized4. Prior work demonstrates that Inpp5f can degrade both phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2, PIP2) and phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3, PIP3) by removing the 5’ phosphate from the inositol ring5. Although the PIP3 3’ phosphatase PTEN has previously been shown to regulate cardiac myocyte hypertrophy and Akt signaling in the heart6, 7, a functional role for a PIP3 5’ phosphatase in the heart has been less clear. However, in various non-cardiac model systems, both 3’ and 5’ inositol phosphatases are important regulators of PIP3 activity and downstream signaling. For example, the SH2 domain-containing inositol 5'-phosphatase (SHIP) and PTEN both play important roles to regulate PIP3 and Akt in immune cells8.
In order to determine the role of Inpp5f in the adult heart, we have created both gain- and loss-of-function models. Our results indicate that Inpp5f transgenic mice are unable to reactivate fetal genes or to exhibit normal hypertrophic responses to adrenergic agonists, while Inpp5f knockout mice exhibit augmented hypertrophy and exaggerated reactivation of the fetal gene program under stress.
Materials and methods
Inpp5f−/− knockout mice
An Inpp5f gene-trap ES clone was obtained from BayGenomics (now the International Gene Trap Consortium, ES clone no. XL0571). The Inpp5f genomic locus is interrupted by insertion of the pGT01xf vector, which integrated into intron 6. Chimeric mice were produced by blastocyst injection according to standard protocols. Mice were genotyped by Southern blotting analysis of ScaI digested genomic DNA. Inpp5f+/+ and Inpp5f−/− mice were subsequently genotyped by PCR (See Online materials and methods). The loss of Inpp5f was confirmed by qRT-PCR (See Online materials and methods) and by Western blot analysis using a rabbit polyclonal antibody targeted to the C terminus of Inpp5f (generated by our lab, see below).
Inpp5f-transgenic mice
A cDNA encoding human Flag-tagged Inpp5f was cloned into an expression plasmid containing the Myh6 (encoding α-Mhc) promoter9, and transgenic mice were generated by standard techniques. Genotyping was performed by PCR analysis of genomic DNA. Cardiac specific expression of Inpp5f was revealed by qRT-PCR (See Online materials and methods) and Western blot analysis using antibodies to Flag (Sigma, F3165) or Inpp5f (generated by our lab, see below).
Inpp5f antibody generation
GST-Inpp5f fusion protein was expressed in bacteria, purified by standard techniques and used to immunize rabbits (Cocalico Biologicals Inc). A region of mouse Inpp5f cDNA encoding 61 C-terminal amino acids was amplified by PCR and cloned into the pGEX-2T vector (See Online materials and methods). The GST fusion protein was expressed in BL21 bacteria, extracted and purified using Glutathione Sepharose 4B (GE Healthcare) according to the manufacturer’s instructions. Purified protein was used to immunize rabbits (Cocalico Biologicals Inc).
Western blotting
Tissue lysates were prepared in lysis buffer consisting of 20 mM Tris HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton X-100, 1 µg/ml leupeptin, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4, 1 mM β-glycerophosphate. 1mM PMSF was added before use. Samples were separated by SDS-PAGE and transferred to PVDF membranes. We used antibodies to Gapdh (1:5,000 dilution, Chemicon, MAB374), phospho-Gsk3β (Ser 9), total Gsk3β, phospho-Akt (Ser 473), total Akt (1:1000 dilution, Cell Signaling, #9336, #9315, #4058, #9272). Primary antibody binding was visualized by using the Western Breeze Kit (Invitrogen) according to the manufacturer’s instructions. Inpp5f antibody was purified by Melon Gel IgG Spin Purification kit (Thermo Scientific) and then diluted 1:100 in 5% milk.
Treatment with isoproterenol
Isoproterenol (Sigma, I5627) was delivered by implanting a micro-osmotic pump (Alzet, Durect; model 1002) subcutaneously under pentobarbital anesthesia. ISO (30 mg/kg/d) or vehicle (Dulbecco’s phosphate buffered saline, Gibco) was infused subcutaneously for 14 days as described previously10.
Histology
Adult mouse hearts were collected in ice-cold PBS, fixed overnight in 4% paraformaldehyde at 4°C, washed with PBS, and dehydrated through an ethanol series prior to paraffin embedding. Masson’s Trichrome (to reveal fibrosis) and H&E stains were performed according to standard protocols.
Apoptosis analysis
Apoptosis was measured by TUNEL assay (Roche, 1684795). The total number of cells was quantified using ImageJ software.
Quantitative real time PCR
Total RNA was isolated from dissected mouse hearts using Trizol (Invitrogen). RNA was reverse-transcribed using random hexamers and the Superscript First Strand Synthesis Kit (Invitrogen). Gene expression was then evaluated by qRT-PCR (ABI PRISM 7900) using the SYBR Green (Applied Biosystems). Signals were normalized to their corresponding Gapdh controls and the ratios expressed as fold changes compared to wild-type. PCR conditions and primer set sequences are available upon request.
IGF-1 treatment
Hearts from one-month old mice were minced and incubated with 100nM Insulin like growth factor 1 (IGF-1) (Sigma, I8779) for 30 min at 37°C followed by lipid extraction.
PIP3 ELISA
Mouse heart tissue was pulverized under liquid nitrogen using a mortar and pestle, followed by lipid extraction and ELISA assays as per manufacture’s instruction (Echelon Biosciences Inc, K-2500). ELISA measurements were performed in triplicate after combining tissue from 4 hearts.
Statistic analysis
All data are expressed as the mean ± SD. Student’s t test was used to compare heart to body weight ratios and heart weight to tibia length ratios. Probability values <0.05 were considered statistically significant.
Results
Inactivation of Inpp5f
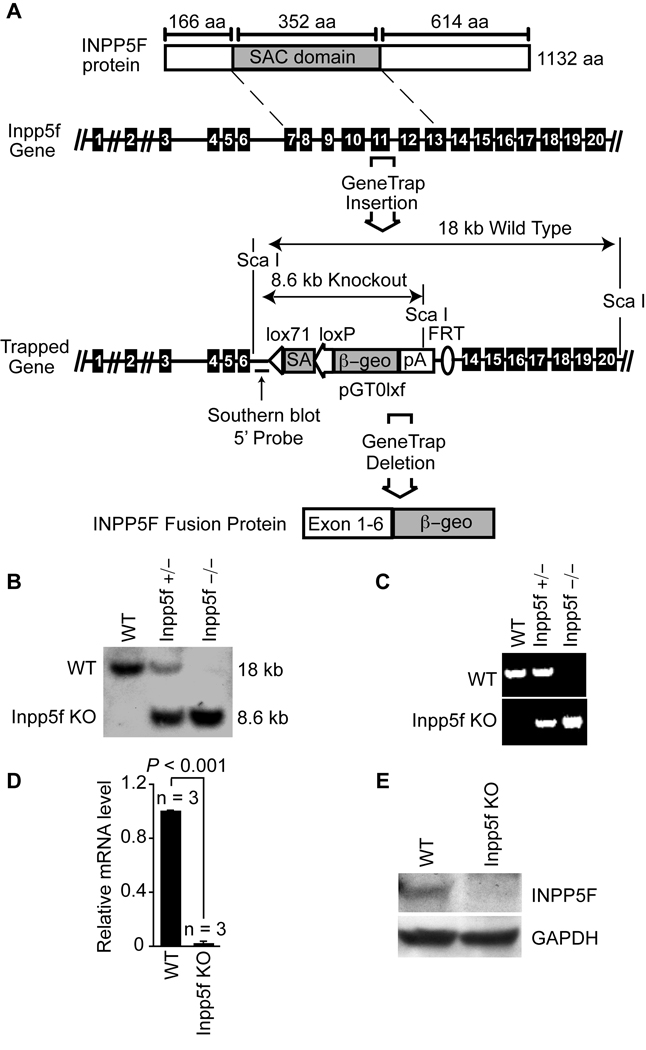
In order to produce mice lacking Inpp5f, we identified a gene-trap embryonic stem (ES) cell line in which the endogenous Inpp5f locus has been disrupted within intron 6, upstream of the exons encoding the catalytic phosphatase domain. This gene-trap insertion is predicted to result in the production of a fusion protein composed of the amino-terminal region of Inpp5f fused to β-geo (Fig. 1A and Online Figure I). Heterozygous gene-trap ES cells were injected into blastocysts to create chimeric mice, which were bred to produce germ-line offspring. Resulting heterozygous mice were inter-crossed to produce homozygous Inpp5f−/− offspring, which were born in expected Mendelian ratios (Table 1), survived into adulthood and appeared healthy. Genotype was confirmed by Southern blot (Fig. 1B) and by PCR (Fig. 1C).
Figure 1. Inactivation of Inpp5f.
(A) Schematic representation of Inpp5f protein (top) and gene structure. The wild type and gene trap alleles are shown; exons are represented by black boxes. The sizes of the expected restriction fragments recognized by the Southern probe (indicated) are shown. (B) Southern blot of adult mice tail DNA resulting from a cross between Inpp5f+/− heterozygotes. (C) PCR genotyping of offspring resulting from a cross between Inpp5f+/− heterozygotes. (D) Real-time quantitative PCR of mRNA from adult wild-type and Inpp5f−/− hearts shows the absence of Inpp5f mRNA in the knockout hearts. (E) Western blot of wild-type and Inpp5f−/− adult heart tissue shows loss of Inpp5f protein in mutant hearts.
Table 1.
Loss of Inpp5f does not lead to embryonic or perinatal lethality.
| Genotype (offspring of heterozygous crosses) |
P0~P21 | |
|---|---|---|
| Expected | Observed | |
| Inpp5f+/+ | 105 | 123 |
| Inpp5f+/− | 210 | 192 |
| Inpp5f−/− | 105 | 105 |
| Total | 420 | 420 |
χ2=4.629, 2 DOF, p=0.10
Further in depth analysis of the Inpp5f genomic locus affected by the gene-trap revealed that the insertion event was accompanied by a loss of endogenous genomic sequence spanning exons 7–13 (Fig. 1A and Online Figure II). Thus, the affected allele cannot produce a full-length protein or one with functional phosphatase activity. We confirmed the lack of full length Inpp5f transcripts by real-time PCR using mRNA from adult Inpp5f−/− and wild-type hearts (Fig. 1D). Western blot using antibody recognizing the C-terminus of Inpp5f demonstrated a lack of protein expression in the knockout (Fig. 1E). Thus, we conclude that we have generated a null allele for Inpp5f.
Inpp5f null hearts appear normal
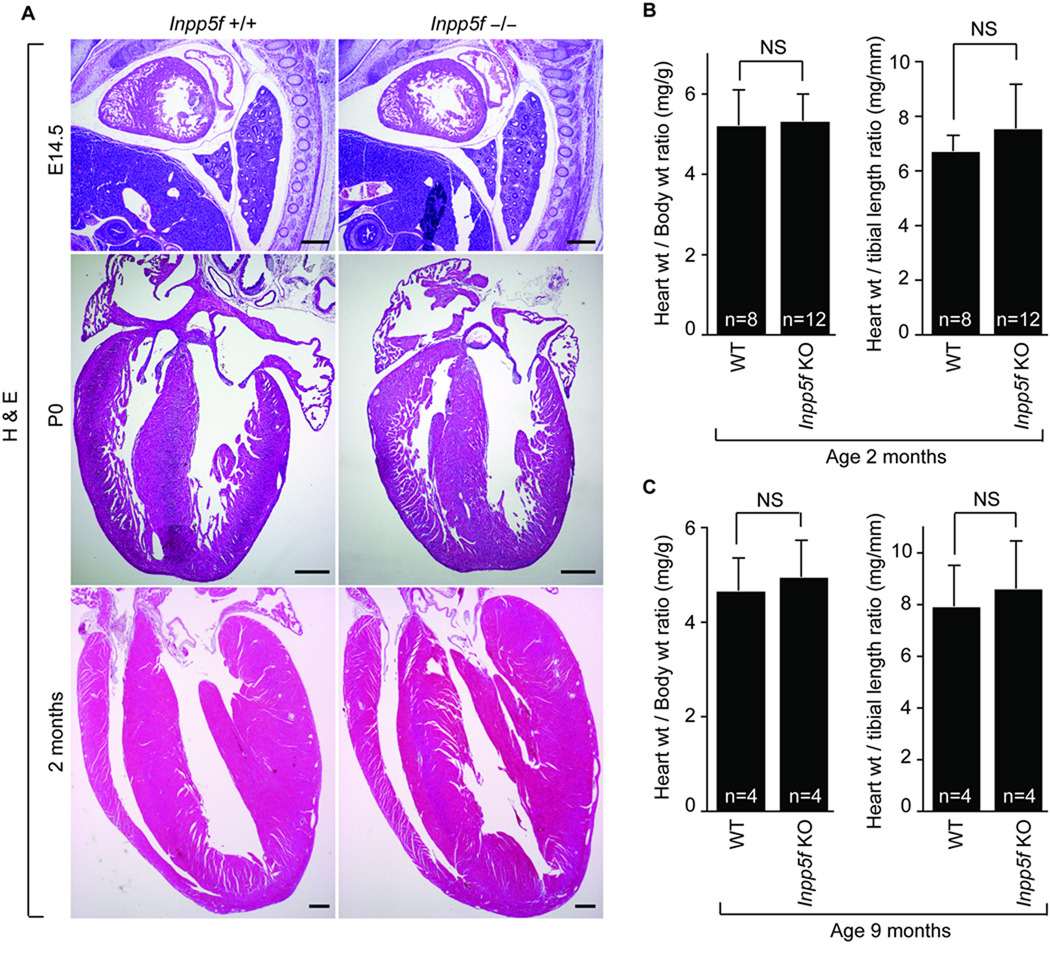
We examined cardiac histology, size and function in Inpp5f−/− and litter mate control mice. H&E staining of hearts was performed at E14.5, P0 and 2 months of age. At these time points, we could not identify any differences and Inpp5f null mice appear normal by gross and histological assessment (Fig. 2A). We measured cardiac weight in relation to total body weight to determine relative cardiac size. At both 2 and 9 months of age, there was no difference in heart to body weight ratio between wild type and Inpp5f−/− littermates (Fig 2B,C).
Figure 2. Inpp5f−/− hearts appear normal.
(A) H&E-stained sections of wild-type and Inpp5f−/− hearts at E14.5, P0 and P65 (scale bar, 400 µm) reveals normal heart structure. (B) Heart to body weight and heart to tibia length ratios of Inpp5f+/+ (n = 8) and Inpp5f−/− (n = 12) mice at 2 month of age. (C) Heart to body weight and heart to tibia length ratios of Inpp5f+/+ (n = 4) and Inpp5f−/− (n = 4) mice at 9 month are shown. NS: not significant.
In order to evaluate the functional impact of the loss of Inpp5f, we performed echocardiography on Inpp5f−/− and wild type litter mates at 2 and 9 months (Table 2). We did not observe any significant difference in the inter-ventricular septum (IVS), left ventricular posterior wall (LVPW) and the left ventricular internal diameter (IVID) measurements at either end-diastole or end-systole. Left ventricular (LV) ejection fraction (EF) and fractional shortening (FS) also was not different between groups. Thus, under basal conditions, Inpp5f−/− mice appear to have normal cardiac form and function.
Table 2.
Inpp5f−/− mice have normal cardiac function by echocardiography.
| 2 months | 9 months | |||||
|---|---|---|---|---|---|---|
| wild type (n=3) |
Inpp5f KO (n=3) |
p Value | wild type (n=4) |
Inpp5f KO (n=4) |
p Value | |
| IVSd (mm) | 0.69±0.06 | 0.66±0.03 | ns | 0.66±0.06 | 0.67±0.07 | ns |
| IVSs (mm) | 1.13±0.12 | 1.00±0.04 | ns | 0.96±0.10 | 1.02±0.09 | ns |
| LVPWd (mm) | 0.69±0.06 | 0.64±0.07 | ns | 0.65±0.03 | 0.70±0.06 | ns |
| LVPWs (mm) | 1.02±0.13 | 0.90±0.07 | ns | 0.95±0.14 | 1.04±0.07 | ns |
| LVIDd (mm) | 3.92±0.16 | 3.76±0.13 | ns | 4.13±0.22 | 4.13±0.48 | ns |
| LVIDs (mm) | 2.57±0.10 | 2.54±0.17 | ns | 2.98±0.12 | 2.92±0. 40 | ns |
| LVEF (%) | 64±2 | 60±4 | ns | 56±4 | 57±4 | ns |
| LVFS (%) | 34±1 | 31±3 | ns | 29±3 | 29±2 | ns |
ns: not significant
Inpp5f regulates stress-induced hypertrophy
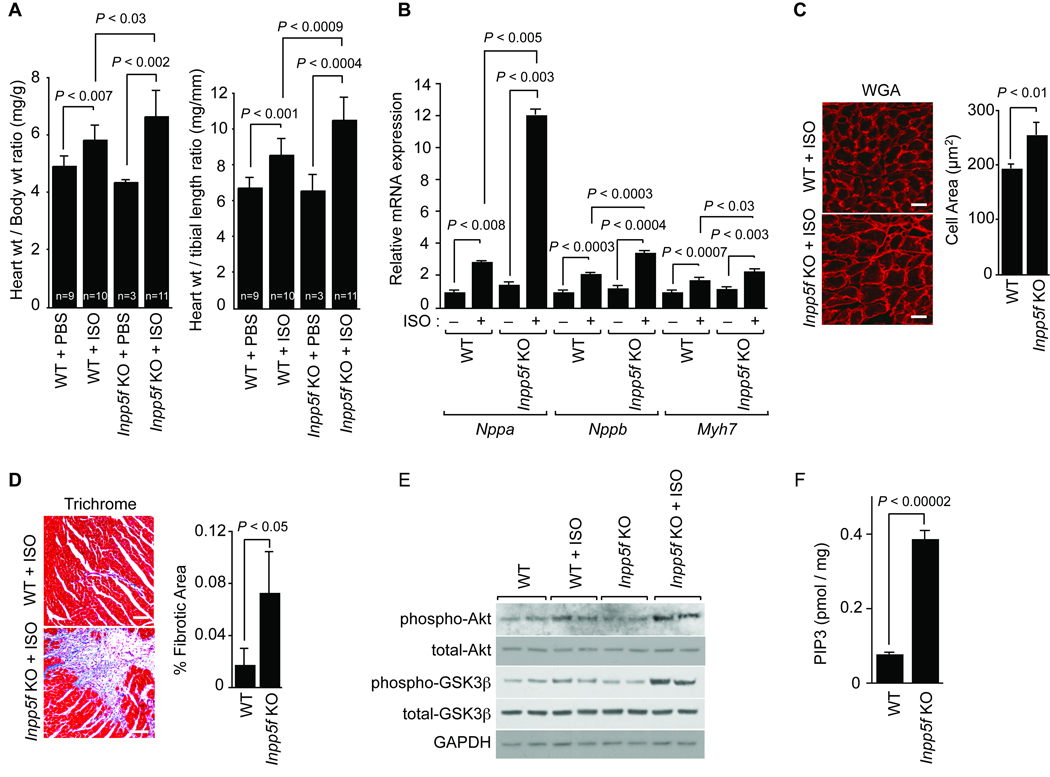
Next, we sought to determine if adult mice lacking Inpp5f would be more susceptible to agonist-induced cardiac hypertrophy. Wild-type and Inpp5f−/− litter mates at 2 months of age were treated with a constant infusion of saline or isoproterenol (ISO) delivered by osmotic mini-pump for 14 days. Animals lacking Inpp5f showed an increase in ISO-induced cardiac hypertrophy as measured by either the heart to body weight ratio or the heart weight to tibia length ratio (Fig. 3A). In response to ISO, Inpp5f−/− heart also showed more potent reactivation of the fetal program of gene expression than wild type litter mates; transcripts for Nppa (encoding atrial natriuretic factor, ANF), Myh7 (encoding β-MHC) and Nppb (encoding BNP) increased more dramatically in the knockout hearts (Fig. 3B). Cellular hypertrophy, as revealed by wheat germ agglutinin (WGA) staining followed by quantification, was more pronounced in the Inpp5f−/− hearts compared to wild-type hearts after ISO (Fig. 3C) (256.5 ± 21.8 µm2 for Inpp5f −/− mice treated with ISO, n = 446 cells from 3 hearts; 194.8 ± 6.8 µm2 for wild type mice treated with ISO, n = 514 from 3 hearts). Patchy areas of fibrosis were more evident in ISO-treated Inpp5f−/− hearts when compared to controls (Fig. 3D) (Quantification of fibrotic area was 7.3% for Inpp5f −/− mice, 25 sections, three hearts; 1.8% for wild type mice, 27 sections, three hearts). We also observed more apoptotic cells in the Inpp5f−/− hearts after ISO treatment compared to wild type (0.37% of 16,804 cells from 4 Inpp5f−/− hearts, 0.24% of 21,558 cells from 5 wild type hearts, p = 0.01). Functional assessment by echocardiography after 2 weeks of ISO treatment revealed relative preservation fractional shortening and ejection fraction, with thickening of the posterior wall in Inpp5f−/− animals at this time point (Online Table I).
Figure 3. Inpp5f-deficient mice are susceptible to agonist-induced cardiac hypertrophy.
(A) Wild-type and Inpp5f−/− littermates were subjected to chronic infusion of saline or ISO for 14 days. Heart weight to body weight and heart weight to tibia length ratios (± s.d.) were determined. (B) Transcripts for Nppa, Nppb and Myh7 were detected by real-time quantitative PCR in hearts from wild-type and Inpp5f−/− mice following chronic infusion of saline or ISO. The values are expressed as the fold change in transcript abundance (± s.d.) compared to wild-type mice. (C) Wheat germ agglutinin (WGA) staining shows increased myocyte size in Inpp5f−/− compared to wild-type hearts (scale bar, 25 µm). (D)Trichrome staining demonstrates more fibrosis (blue) in Inpp5f−/− hearts treated with ISO (scale bar, 150 µm,) compared to control. (E) Western blots from neonatal cardiomyocytes treated with or without 10 µM ISO for 10 min are shown; Gapdh is shown as a loading control. (F) PIP3 levels, normalized to heart weight, in the Inpp5f−/− and wildtype hearts after 30 min of 100 nM IGF-1 stimulation were assayed.
Neonatal myocytes isolated from Inpp5f−/− and wild type littermates revealed similar levels of Akt and Gsk3β phosphorylation under basal conditions (Fig. 3E). Treatment with ISO resulted in increased phosphorylation of Akt and Gsk3β in knockout cells when compared to controls (Fig. 3E), while no significant differences in Erk activation were noted (data not shown). IGF-1 treatment of myocytes revealed similar results (Online Figure III). Taken together, our data suggest that cardiac myocytes lacking Inpp5f are sensitive to stress-induced cardiac hypertrophy and activation of the Akt pathway.
PIP3 levels are altered in Inpp5f null mice
Although a prior report has indicated that Inpp5f exhibits 5-phosphatase activity when PtdIns(3,4,5)P3 and PtdIns(4,5)P2 are used as substrates in vitro5, we sought to determine directly if loss of Inpp5f alters endogenous cardiac PIP3 levels in vivo. Endogenous PtdIns(3,4,5)P3 levels are low, and we are unaware of prior studies that directly measure levels of this important signaling molecule in vivo in genetically engineered animals. Therefore, we adapted an ELISA-based sensitive assay that has been used for cultured cells and compared PIP3 levels in control and Inpp5f−/− hearts before and after ligand stimulation intended to augment PIP3 levels. Heart tissue from 4 mice age 3–5 weeks was combined for each condition, and some samples were treated ex vivo with IGF-1, a potent activator of PI3K and AKT in the heart. Our data shows that PIP3 levels in Inpp5f−/− mice are ~1.6 fold greater than wild type littermates. However, upon IGF-1 stimulation, PIP3 levels in the knockout hearts increased to 4.9 fold those of stimulated wild type littermates (Fig. 3F). Thus, loss of Inpp5f sensitizes the heart to hypertrophic stimulation.
Cardiac-specific Inpp5f transgenic mice
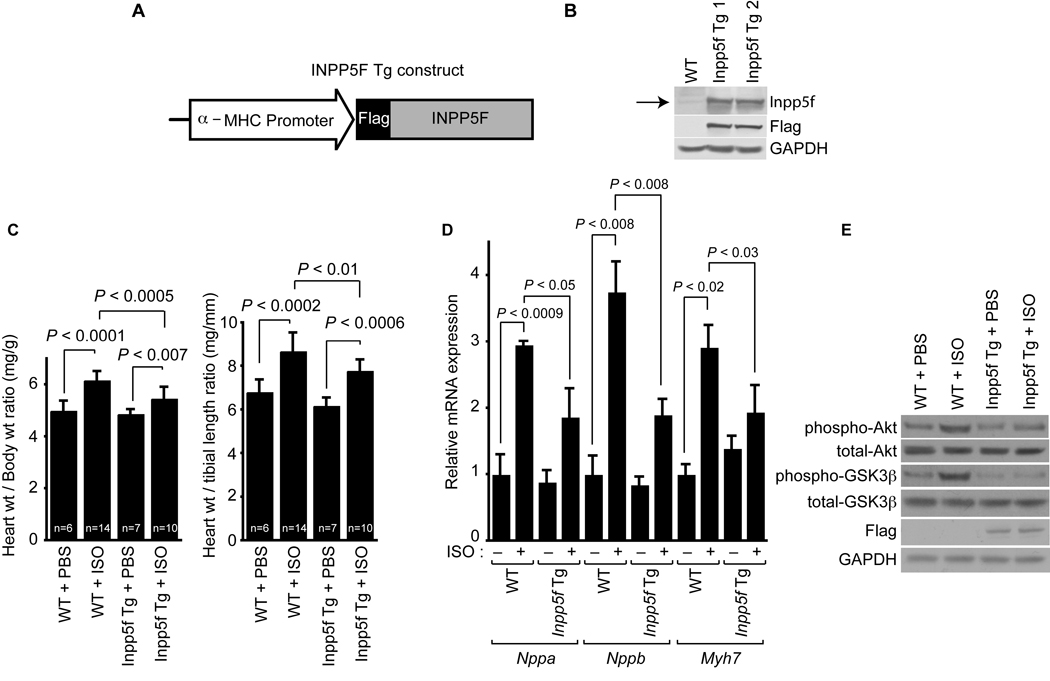
We used the well-characterized α-myosin heavy chain (α-MHC) promoter to direct cardiac-restricted expression of Inpp5f in transgenic mice (Fig. 4A). We evaluated two independent transgenic lines of mice to control for effects mediated by sites of insertion. Germline transmission of the transgene was verified by PCR (data not shown) and the transgene mRNA level was determined by real-time PCR (data not shown). Transgenic expression of Inpp5f was verified by Western blotting (Fig. 4B) which revealed similar levels of protein expression in the two independent lines (Fig. 4B).
Figure 4. Inpp5f-Tg mice are resistant to agonist-induced cardiac hypertrophy.
(A) Schematic representation of the Inpp5f transgenic construct. (B) Western blot analysis of myocardium from 2-month-old wild-type and Inpp5f-Tg mice. Transgenically expressed Inpp5f was epitope-tagged and was detected using an Inpp5f antibody and a Flag antibody. Arrow: Inpp5f protein. (C) Wild-type and Inpp5f-Tg littermates were subjected to chronic infusion of saline or ISO for 14 days. Heart weight to body weight and heart weight to tibia length ratios (± s.d.) were determined. (D) Transcripts for Nppa, Nppb and Myh7 were detected by real-time quantitative PCR from hearts of wild-type and Inpp5f-Tg mice following chronic infusion of saline or ISO. The values are expressed as the fold change in transcript abundance (± s.d.) compared to wild-type mice. (E) Western blots of myocardium from 2-month-old wild-type and Inpp5f-Tg mice subjected to infusion of saline or ISO for 14 days. Gapdh is shown as a loading control.
Inpp5f transgenic mice are resistant to stress-induced hypertrophy
Adult Inpp5f transgenic mice appeared healthy. Heart to body weight ratios of transgenic and wild type littermates were not significantly different at P60 (Fig. 4C). Control and Inpp5f transgenic littermates at 2 months of age were treated with a constant infusion of saline or ISO for 14 days. As predicted, wild-type mice exhibited cardiac hypertrophy, as revealed by an increase in both the heart to body weight ratio and the heart weight to tibia length ratio (Fig. 4C). Hypertrophy of Inpp5f transgenic mice in response to ISO was significantly blunted (Fig. 4C). Likewise, reactivation of the fetal gene program that accompanied hypertrophy in control animals was markedly attenuated in transgenic animals (Fig. 4D). After treatment with ISO, we noted activation of the Akt pathway with enhanced phosphorylation of Akt and Gsk3β in the control hearts (Fig. 4E). However, ISO-induced increases in phospho-Akt and phospho-Gsk3β were blunted in transgenic hearts.
Discussion
In this study, we investigated the effects of gain- and loss-of-function of Inpp5f. Our results suggest that Inpp5f modulates stress-induced hypertrophic responsiveness in the heart. Under basal conditions, Inpp5f knockout and transgenic mice appear normal, with preserved cardiac structure and function. However, in the setting of adrenergic stimulation produced by infusion of isoproterenol, Inpp5f−/− animals had elevated level of PIP3 and showed accentuated hypertrophy as measured by heart size, myocyte size and gene expression. Isolated myocytes lacking Inpp5f were hypersensitive to ISO and IGF-1 as reflected by accentuated activation of Akt compared to control myocytes. Conversely, Inpp5f transgenic mice were relatively resistant to hypertrophic stimulation.
Inpp5f encodes a 5’ PIP3 phosphatase that is predicted to reduce PIP3 levels and subsequent activation of Akt and downstream signals. Our biochemical findings in Inpp5f transgenic and knockout hearts are consistent with this mode of action. The Akt signaling network in the adult heart has been extensively examined and has been shown to contribute to both adaptive (physiologic) and maladaptive (pathologic) hypertrophy7. Akt1−/− mice have a 20% reduction in body size, with a concomitant reduction in heart size11 and they are defective in exercise-induced cardiac hypertrophy12. Cardiac specific over-expression of constitutively active or dominant negative forms of Akt lead to larger or smaller hearts respectively13–15. Enhanced Akt activity is associated with increased p70S6 kinase activity and increased phospho-Gsk3β13, 14. Cardiac-specific over-expression of constitutively active Gsk3β is associated with reduced agonist and pressure-overload hypertrophy confirming that Gsk3β functions as a negative regulator of hypertrophy in vivo16. Thus, the alterations in Akt and Gsk3β phosphorylation that we observed in Inpp5f knockout and transgenic hearts are consistent with a model in which Inpp5f regulates PIP3 levels, Akt and Gsk3β activity and subsequent hypertrophic responsiveness.
The inositol polyphosphate 5-phosphatases are a large family of enzymes comprising at least 10 mammalian and 4 yeast members17. Inpp5f has a Sac phosphatase domain which exhibits phosphatidylinositol polyphosphate phosphatase activity5. The Sac domain is approximately 400 amino acid residue in length and defined by seven conserved motifs which appear to define the catalytic and regulatory regions of the phosphatase18. The highly conserved sequence RXNCXDCLDRTN in the sixth motif is proposed to be the catalytic core of the SAC domain phosphatases. The CX5R(T/S) motif within this sequence is also found in a number of metal-independent protein phosphatases and inositol polyphosphate phosphatases and is known to be a phosphatase catalytic site18, 19. Interestingly, the SAC domain of the yeast synaptojanin-like protein Inp51p does not exhibit phosphatase activity, and the cysteine, arginine and threonine/serine residue are absent from CX5R(T/S) motif of this protein, being replaced by alanine, lysine and proline respectively. Furthermore, mutations of the first conserved aspartate residue in the RXNCXDCLDRTN sequence as seen in the yeast sac1–8 and sac1–22 mutant alleles were demonstrated to inactivate the Sac1p functions18, 20, 21.
Some 5-phosphatases, such as the Src homology 2 (SH2) domain-containing inositol polyphosphophate 5-phosphatases 1 and 2 (Ship and Ship2) have been extensively characterized22–26. Ship is expressed predominantly in hematopoietic cells where it is an important negative regulator of cytokine signaling. Ship−/− mice have a short life span associated with massive myeloid cell infiltration of the lungs and numerous hematopoietic abnormalities27, 28. Ship2 is more widely expressed, with high expression in brain, skeletal muscle and heart17. It plays an important role in insulin signaling and obesity regulation29, 30. Ship2 has been reported to be a negative regulator of Akt activation. Although loss of Ship2 is not sufficient to activate Akt, the absence of Ship2 allows for greater activity upon Akt stimulation31, 32. These findings are reminiscent of our results with regard to Inpp5f-mediated regulation of basal and agonist-induced activation of Akt in the heart.
Although PTEN and the 5-phosphatases can all degrade PIP3, the degradation products are distinct. Whereas PTEN converts PIP3 to PI(4,5)P2, the 5-phosphatases convert PIP3 to PI(3,4)P2, which can function as a second messenger33. Therefore, the production of PI(3,4)P2 from PIP3 by Inpp5f and Ship phosphatases may function in part via active signaling, whereas PTEN action appears to be mediated through loss of active PIP3 signaling34. For example, PI(3,4)P2 has been shown to activate reactive oxygen species (ROS)35 and the generation of ROS is a process that is increasingly recognized as an important contributor to depressed cardiac function and maladaptive remodelling36. Thus, this process could also contribute to the phenotype we observed. Moreover, PI(3,4)P2 activity has been shown to correlate with Akt phosphorylation and activity37. Inpp5f can also dephosphorylate PI(4,5)P25, and depletion of PI(4,5)P2 may contribute to cardiomyocyte apoptosis and subsequent heart failure38. Thus, regulation of PI(4,5)P2 levels may contribute to the mechanism by which Inpp5f regulates hypertrophy.
Under the sedentary conditions, we found that Inpp5f null mice have slightly higher level of cardiac PIP3, though phospho-Akt and phospho-Gsk3β are unaltered and the mice do not show abnormal hypertrophy. However, upon IGF-1 stimulation, PIP3 levels in the knockout mice increase dramatically and they are more sensitive to hypertrophic stimuli. Hence, unlike PTEN, which alters the basal level of PIP334, Inpp5f appears to modulate PIP3 levels primarily upon agonist-induced stimulation.
Our prior studies have suggested that Inpp5f is repressed at the transcriptional level by Hdac2. Inpp5f transcripts are elevated in Hdac2 knockout hearts and are diminished by over-expression of Hdac2 in the heart3. siRNA-mediated knockdown of Hdac2 or chemical Hdac inhibition in H9C2 myocytes results in increased levels of Inpp5f and decreased phosphorylation of Akt and Gsk3β. Although Hdac2 may modulate cardiac myocyte size and hypertrophic signaling via multiple pathways, our analysis of Inpp5f knockout and transgenic mice are consistent with a functional relationship between Hdac2 and Inpp5f in the setting of hypertrophic stimulation.
Taken together, the work presented here suggests that Inpp5f functions as a negative regulator of cardiac hypertrophy and Akt signaling. Loss of Inpp5f sensitizes the heart’s response to hypertrophic stimuli by modulating PI(3,4,5)P3 levels.
Supplementary Material
Acknowledgments
Sources of Funding:
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 HL071546 (to J. A. E.). W.Z. is supported by an American Heart Association Predoctoral Fellowship (0715297U). C.M.T was supported by an American Heart Association Postdoctoral Fellowship (0625427U).
Non-standard abbreviations and acronyms
- Gsk3beta
glycogen synthase kinase 3beta
- Hdac2
histone deacetylase-2
- Inpp5f
inositol polyphosphate-5-phosphatase f
- Akt
thymoma viral proto-oncogene
- Pten
phosphatase and tensin homolog
- PIP2
phosphatidylinositol-4,5-bisphosphate
- PIP3
phosphatidyl inositol-3,4,5-trisphosphate
- Nppa
Natriuretic peptide precursor type A
- Myh7
myosin heavy polypeptide 7
- Iso
Isoproterenol
- IGF
Insulin like growth factor
- MHC
Myosin Heavy chain
- SHIP
SH2-containing inositol phosphatase
Footnotes
Disclosures: None.
Subject codes: [15] Hypertrophy, [138] Cell signaling/signal transduction, [89] Genetics of cardiovascular disease, [130] Animal models of human disease
References
- 1.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 2.Dorn GW, 2nd, Force T. Protein kinase cascades in the regulation of cardiac hypertrophy. J Clin Invest. 2005;115:527–537. doi: 10.1172/JCI24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 4.Astle MV, Horan KA, Ooms LM, Mitchell CA. The inositol polyphosphate 5-phosphatases: traffic controllers, waistline watchers and tumour suppressors? Biochem Soc Symp. 2007:161–181. doi: 10.1042/BSS0740161. [DOI] [PubMed] [Google Scholar]
- 5.Minagawa T, Ijuin T, Mochizuki Y, Takenawa T. Identification and characterization of a sac domain-containing phosphoinositide 5-phosphatase. J Biol Chem. 2001;276:22011–22015. doi: 10.1074/jbc.M101579200. [DOI] [PubMed] [Google Scholar]
- 6.Crackower MA, Oudit GY, Kozieradzki I, Sarao R, Sun H, Sasaki T, Hirsch E, Suzuki A, Shioi T, Irie-Sasaki J, Sah R, Cheng HY, Rybin VO, Lembo G, Fratta L, Oliveira-dos-Santos AJ, Benovic JL, Kahn CR, Izumo S, Steinberg SF, Wymann MP, Backx PH, Penninger JM. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell. 2002;110:737–749. doi: 10.1016/s0092-8674(02)00969-8. [DOI] [PubMed] [Google Scholar]
- 7.Oudit GY, Sun H, Kerfant BG, Crackower MA, Penninger JM, Backx PH. The role of phosphoinositide-3 kinase and PTEN in cardiovascular physiology and disease. J Mol Cell Cardiol. 2004;37:449–471. doi: 10.1016/j.yjmcc.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Kashiwada M, Lu P, Rothman PB. PIP3 pathway in regulatory T cells and autoimmunity. Immunol Res. 2007;39:194–224. doi: 10.1007/s12026-007-0075-2. [DOI] [PubMed] [Google Scholar]
- 9.Heine HL, Leong HS, Rossi FM, McManus BM, Podor TJ. Strategies of conditional gene expression in myocardium: an overview. Methods Mol Med. 2005;112:109–154. doi: 10.1007/978-1-59259-879-3_8. [DOI] [PubMed] [Google Scholar]
- 10.Trivedi CM, Lu MM, Wang Q, Epstein JA. Transgenic overexpression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J Biol Chem. 2008;283:26484–26489. doi: 10.1074/jbc.M803686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276:38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 12.DeBosch B, Treskov I, Lupu TS, Weinheimer C, Kovacs A, Courtois M, Muslin AJ. Akt1 is required for physiological cardiac growth. Circulation. 2006;113:2097–2104. doi: 10.1161/CIRCULATIONAHA.105.595231. [DOI] [PubMed] [Google Scholar]
- 13.Shioi T, McMullen JR, Kang PM, Douglas PS, Obata T, Franke TF, Cantley LC, Izumo S. Akt/protein kinase B promotes organ growth in transgenic mice. Mol Cell Biol. 2002;22:2799–2809. doi: 10.1128/MCB.22.8.2799-2809.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsui T, Li L, Wu JC, Cook SA, Nagoshi T, Picard MH, Liao R, Rosenzweig A. Phenotypic spectrum caused by transgenic overexpression of activated Akt in the heart. J Biol Chem. 2002;277:22896–22901. doi: 10.1074/jbc.M200347200. [DOI] [PubMed] [Google Scholar]
- 15.Condorelli G, Drusco A, Stassi G, Bellacosa A, Roncarati R, Iaccarino G, Russo MA, Gu Y, Dalton N, Chung C, Latronico MV, Napoli C, Sadoshima J, Croce CM, Ross J., Jr Akt induces enhanced myocardial contractility and cell size in vivo in transgenic mice. Proc Natl Acad Sci U S A. 2002;99:12333–12338. doi: 10.1073/pnas.172376399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antos CL, McKinsey TA, Frey N, Kutschke W, McAnally J, Shelton JM, Richardson JA, Hill JA, Olson EN. Activated glycogen synthase-3 beta suppresses cardiac hypertrophy in vivo. Proc Natl Acad Sci U S A. 2002;99:907–912. doi: 10.1073/pnas.231619298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Astle MV, Seaton G, Davies EM, Fedele CG, Rahman P, Arsala L, Mitchell CA. Regulation of phosphoinositide signaling by the inositol polyphosphate 5-phosphatases. IUBMB Life. 2006;58:451–456. doi: 10.1080/15216540600871159. [DOI] [PubMed] [Google Scholar]
- 18.Hughes WE, Cooke FT, Parker PJ. Sac phosphatase domain proteins. Biochem J. 2000;350(Pt 2):337–352. [PMC free article] [PubMed] [Google Scholar]
- 19.Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- 20.Kearns BG, McGee TP, Mayinger P, Gedvilaite A, Phillips SE, Kagiwada S, Bankaitis VA. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 1997;387:101–105. doi: 10.1038/387101a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong R, Ye ZH. The SAC domain-containing protein gene family in Arabidopsis. Plant Physiol. 2003;132:544–555. doi: 10.1104/pp.103.021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krystal G, Damen JE, Helgason CD, Huber M, Hughes MR, Kalesnikoff J, Lam V, Rosten P, Ware MD, Yew S, Humphries RK. SHIPs ahoy. Int J Biochem Cell Biol. 1999;31:1007–1010. doi: 10.1016/s1357-2725(99)00072-2. [DOI] [PubMed] [Google Scholar]
- 23.Rohrschneider LR, Fuller JF, Wolf I, Liu Y, Lucas DM. Structure, function, and biology of SHIP proteins. Genes Dev. 2000;14:505–520. [PubMed] [Google Scholar]
- 24.Backers K, Blero D, Paternotte N, Zhang J, Erneux C. The termination of PI3K signalling by SHIP1 and SHIP2 inositol 5-phosphatases. Adv Enzyme Regul. 2003;43:15–28. doi: 10.1016/s0065-2571(02)00043-2. [DOI] [PubMed] [Google Scholar]
- 25.Dyson JM, Kong AM, Wiradjaja F, Astle MV, Gurung R, Mitchell CA. The SH2 domain containing inositol polyphosphate 5-phosphatase-2: SHIP2. Int J Biochem Cell Biol. 2005;37:2260–2265. doi: 10.1016/j.biocel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Harris SJ, Parry RV, Westwick J, Ward SG. Phosphoinositide lipid phosphatases: natural regulators of phosphoinositide 3-kinase signaling in T lymphocytes. J Biol Chem. 2008;283:2465–2469. doi: 10.1074/jbc.R700044200. [DOI] [PubMed] [Google Scholar]
- 27.Liu Q, Shalaby F, Jones J, Bouchard D, Dumont DJ. The SH2-containing inositol polyphosphate 5-phosphatase, ship, is expressed during hematopoiesis and spermatogenesis. Blood. 1998;91:2753–2759. [PubMed] [Google Scholar]
- 28.Helgason CD, Damen JE, Rosten P, Grewal R, Sorensen P, Chappel SM, Borowski A, Jirik F, Krystal G, Humphries RK. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sleeman MW, Wortley KE, Lai KM, Gowen LC, Kintner J, Kline WO, Garcia K, Stitt TN, Yancopoulos GD, Wiegand SJ, Glass DJ. Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat Med. 2005;11:199–205. doi: 10.1038/nm1178. [DOI] [PubMed] [Google Scholar]
- 30.Grempler R, Zibrova D, Schoelch C, van Marle A, Rippmann JF, Redemann N. Normalization of prandial blood glucose and improvement of glucose tolerance by liver-specific inhibition of SH2 domain containing inositol phosphatase 2 (SHIP2) in diabetic KKAy mice: SHIP2 inhibition causes insulin-mimetic effects on glycogen metabolism, gluconeogenesis, and glycolysis. Diabetes. 2007;56:2235–2241. doi: 10.2337/db06-1660. [DOI] [PubMed] [Google Scholar]
- 31.Wada T, Sasaoka T, Funaki M, Hori H, Murakami S, Ishiki M, Haruta T, Asano T, Ogawa W, Ishihara H, Kobayashi M. Overexpression of SH2-containing inositol phosphatase 2 results in negative regulation of insulin-induced metabolic actions in 3T3-L1 adipocytes via its 5'-phosphatase catalytic activity. Mol Cell Biol. 2001;21:1633–1646. doi: 10.1128/MCB.21.5.1633-1646.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 33.Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leslie NR, Downes CP. PTEN: The down side of PI 3-kinase signalling. Cell Signal. 2002;14:285–295. doi: 10.1016/s0898-6568(01)00234-0. [DOI] [PubMed] [Google Scholar]
- 35.Brown GE, Stewart MQ, Liu H, Ha VL, Yaffe MB. A novel assay system implicates PtdIns(3,4)P(2), PtdIns(3)P, and PKC delta in intracellular production of reactive oxygen species by the NADPH oxidase. Mol Cell. 2003;11:35–47. doi: 10.1016/s1097-2765(03)00005-4. [DOI] [PubMed] [Google Scholar]
- 36.Takimoto E, Kass DA. Role of oxidative stress in cardiac hypertrophy and remodeling. Hypertension. 2007;49:241–248. doi: 10.1161/01.HYP.0000254415.31362.a7. [DOI] [PubMed] [Google Scholar]
- 37.Ma K, Cheung SM, Marshall AJ, Duronio V. PI(3,4,5)P3 and PI(3,4)P2 levels correlate with PKB/akt phosphorylation at Thr308 and Ser473, respectively; PI(3,4)P2 levels determine PKB activity. Cell Signal. 2008;20:684–694. doi: 10.1016/j.cellsig.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Halstead JR, Jalink K, Divecha N. An emerging role for PtdIns(4,5)P2-mediated signalling in human disease. Trends Pharmacol Sci. 2005;26:654–660. doi: 10.1016/j.tips.2005.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.