Abstract
To calculate the sample size for a research study it is important to take into account several aspects of the study design. In particular, one needs to take into account the hypotheses being tested, the study design, the sampling design, and the method to be used for the analysis. In this paper we propose a simple method to calculate sample size for clustered continuous data under various scenarios of study design.
1 Introduction
In this paper we focus on sample size calculation for studies leading to clustered continuous data. Study designs that can result in clustered data sets include observational studies involving clusters of observations; cluster-randomized trials, in which a treatment is randomly assigned to all units within a cluster of units; or randomized block design experiments, in which the blocks represent clusters and treatments are assigned to units within blocks. In all of these studies, characteristics of both the clusters and the units forming the clusters are measured. The primary objective of the analysis of clustered data is to quantify the effects of predictors on the dependent variable measured for all units, while accounting for the presence of clusters of units, and for the possible within-cluster correlation of the dependent variable measurements in particular. We propose a simple method to calculate sample size for clustered data under various study design scenarios.
2 Background
2.1 Model specification
The important feature of clustered data is that measurements taken on units within clusters involve two sources of variation, i.e., between and within cluster variation. To properly account for the two sources of variation, mixed-effects models are often used as the analytical method. More specifically, models with two random terms, i.e., a random intercept, to describe the between-cluster variation, and a residual error, to describe the variation within a cluster, are frequently applied.
The model for the j-th unit within the i-th cluster (i = 1,…,n) is specified as
| (1) |
where , , and j = 1,…, ni. The total number of units in all n clusters is equal to
Note that predictors stored in vector xi do not depend on index j, and therefore they characterise clusters themselves, not units within a cluster. Vector β, with p elements, contains the fixed effects to be estimated.
In the context of model (1), the following two important concepts are often defined. The first concept involves an intraclass correlation coefficient for clustered data. The coefficient is defined as . The second concept is a marginal variance-covariance matrix, Vi for cluster i, which results from model (1) after integrating out the random effects ui. Matrix Vi has dimension ni × ni; its diagonal elements are equal to and off-diagonal elements are equal to . This particular structure of Vi is called compound-symmetry or an equicorrelational structure with homogenous variances. Both of these concepts, i.e., ρ and Vi, quantify correlation between any two units, i.e., between yij and yij′ for j ≠ j′, from the same cluster.
2.2 Hypothesis test for fixed effects
In the analysis of clustered data we are typically interested in testing hypotheses about contrasts of fixed effects β. The hypotheses are specified as
| (2) |
where L is a known q × p matrix and Δ0 ≠ 0 is a known vector with q elements. Note that H0 and HA specify the null and alternative hypotheses, respectively.
Null hypothesis H0 can be tested in a variety ways. One possible approach is to use the F-test. The test statistic for H0, constructed based on model (1), can be specified as
| (3) |
where wi = ni/(1 + (ni − 1)ρ) and . Respective estimates , , of β, σc and σe are obtained using likelihood methods. Under H0, statistic F follows approximately the central F distribution with numerator and denominator degrees of freedom equal to rank(L) and N − n, respectively. For details related to a more general case of linear mixed effects models see Verbeke and Molenberghs (2000).
2.3 Statistical Power Calculations
A general method to perform power calculations for linear mixed effects models was proposed in Helms (1992). When applied to model (1), the method implies that, under alternative hypothesis HA, the F-statistic, defined in (3), follows approximately a non-central F distribution, with numerator and denominator degrees of freedom rank(L) and N − p, respectively. The non-centrality parameter, δ, is equal to
| (4) |
Assuming that we are testing hypothesis (2) at significance level α, the calculation of power is performed as follows. We first calculate the (1 − α) quantile F1−α of the central F distribution with rank(L) and N −p numerator and denominator degrees of freedom, respectively. Then we calculate power as 1−F(F1−α, rank(L), N −p, δ), where F(x, d1, d2, δ) is the cumulative distribution function for a non-central F distribution with numerator and denominator degrees of freedom d1 and d2, respectively, and non-centrality parameter δ.
3 Algorithm
In this section we outline a simple algorithm to perform statistical power calculations for clustered data. Based on similarities between the formulae for the F-statistic, specified in (3), and for the non-centrality parameter δ, specified in (4), we propose to perform power calculations in the following three steps:
Step 1: Create an auxiliary dataset.
Step 2: Test hypotheses of interest in the auxiliary data.
Step 3: Calculate statistical power.
Each of these steps is described in more detail in the remainder of this section.
First, we predefine the number of subjects n, and number of units ni per each cluster. Based on pilot data or based on the information from the literature we set and to known values. Then we proceed with Step 1 and prepare an auxiliary dataset containing one row per cluster. The created auxiliary dataset is essentially the same as data commonly used for an analysis using a classical linear model. The data contain the predictor variables needed to calculate the elements of vectors xi and two additional columns containing the pseudo-dependent variable , and weights wi, used in Step 2 of the algorithm. More specifically, values of the pseudo-dependent variable are calculated using the formula , where β* is any value of β fulfilling the equation Lβ = Δ0. The second additional variable contains weights, defined as wi = ni/(1 + (ni − 1)ρ), where ρ is the intraclass correlation coefficient defined in Section 2.1.
In Step 2 of the algorithm, we test null hypothesis H0 in the context of the model
| (5) |
Where for i = 1,…n. Note that we use vector as observed values of the dependent variable yi. To find estimates , we use weighted linear regression with weights wi. The reader may note that, in contrast to a classical linear model, the variance of εi term in (5) is known and it is not being estimated from the data. This implies that any software designed for the linear regression models can be used, provided it allows for a known residual variance. In Step 3, we compute the power in the way described in Section 2.3. The idea presented in this section, although similar to that presented in Litell et al (2006), takes advantage of simplifications implied by the specific structure of model (1).
4 Illustration: Michigan Life Science Study
4.1 Study Design and Objectives
The Michigan Life Science (MLS) Corridor Study “Improving muscle power and mobility of elderly men and women” was a four-year study funded by the State of Michigan. The MLS study was designed as a randomised controlled trial, in which healthy young (21–30 years) and older (65–80 years) male and female subjects were randomised into one of two arms of a 12-week progressive resistance training (PRT) exercise intervention. The intervention aimed at strengthening lower extremity muscle strength and power. One group of subjects performed “fast” PRT of the leg muscles using lighter weights, while the other performed the traditional “slow” PRT of the leg muscles using heavier weights. The two study groups were stratified by gender and age (young/old). The training regimen consisted of training three days per week over three months.
The primary outcome was cross-sectional area (CSA) of single muscle fibers obtained from study subjects during biopsy at the end of the study. Data obtained from this study are an example of clustered data, because during each biopsy multiple fibers were obtained and CSA was measured for all the fibers. The primary objective of the study was to test the hypothesis that there was a difference in the effect of the “fast” PRT on CSA as compared to the “slow” PRT.
4.2 Power Calculations
Power calculations are performed assuming the model
where μF and μS are fixed effects for “Fast” and “Slow” PRT study groups. The remaining terms ui and eij are defined in Section 2.1. The null and alternative hypotheses are
| (6) |
To perform the power calculations, we needed an estimate of a clinically and physiologically meaningful intervention effect, Δ, that should be detected. Based on previous experience, the value of Δ was set to 3 μm2. The values of and were estimated from pilot data and were set to 12.4 μm4 and 23.6 μm4, respectively. The corresponding value of ρ is equal to 0.34.
One of the important issues in designing the MLS study was to find an optimal number of muscle fibers needed per biopsy, given budget constraints. We assumed the following cost function:
| (7) |
where C and U are costs associated with a cluster and with a unit within a cluster, respectively. In the MLS study we assumed that the subject-specific cost is equal to C = $2, 860 and the fiber-specific cost is equal to U = $170.
We tentatively set the number of subjects studied in each intervention group to 25. The cost function (7), along with the total (hypothetical) budget that should not exceed $200,000, implies that the number of fibers taken at the biopsy should be equal to six. Assuming α = 0.05, the power is equal to 0.74. In Appendix we present portions of the SAS and R code used to perform calculations.
The reader may note that, for any value of constant c0, all sets of values, c0Δ, c0σc, and c0σe are equivalent, in the sense that they imply the same intraclass correlation coefficient, ρ, and power. Therefore, instead of requesting Δ, , and , to perform power calculations it is sufficient to provide value of ρ and the effect size, say ES, defined as ES = Δ/σ, where . The effect size for our study is equal .
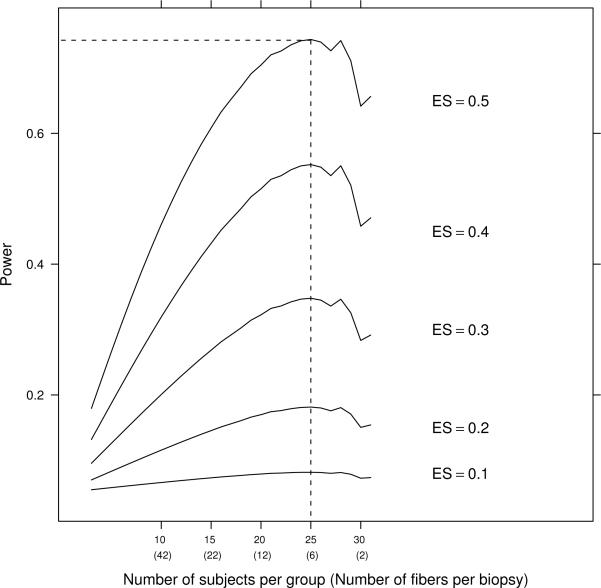
Figure 1 presents the relationship between the power and the number of subjects per study group for different values of ES varying from 0.1 to 0.5, under the assumption that the total budget is equal to $200,000. Note that on the horizontal axis, in addition to the number of subjects, the corresponding number of fibers per subject is given in parentheses. The optimal number of subjects per group is 25 with 6 fibers assessed for every subject. Another combination having power very close to the optimal one is 28 subjects per group and 4 fibers per subject. It is interesting to note that the effect of additional subjects over the optimum number of 25 on statistical power is offset by a lower number of fibers collected for each subject.
Figure 1.
MLS Study: Power plotted versus number of subjects per group for different values of effect size (ES). (Note: Total budget = $200, 000, ρ = 0.34, α = 0.05)
5 Discussion
The method most commonly used to perform power calculations for clustered data is based on a correction of the sample size that involves the design effect defined as 1 + ρ(k − 1), where k is the average cluster size. The method has some drawbacks. For example, it does not allow one to take into account the presence of covariates, and it does not allow one to properly take into account a varying cluster size. The method presented in this paper allows one to overcome these shortcomings. It can be directly used in designing a study or in simulations.
Acknowledgments
We gratefully acknowledge support from the Claude Pepper Center Grants P30-AG08808 and AG024824 from the National Institute on Aging and the financial support from the IAP Research Network P6/03 of the Belgian Government (Belgian Science Policy).
Appendix
In the Appendix we present portions of the SAS and R code used to perform calculations.
In Step 1 of the algorithm, described in Section 3, we created data named dt that contained 50 rows and three variables named recno, grp, and y. Selected rows from these data are given for illustration below
| recno | grp | y |
| 1 | 1 | 0 |
| … | ||
| 25 | 1 | 0 |
| 26 | 2 | 3 |
| … | ||
| 50 | 2 | 3 |
Variables recno, grp, and y indicate row number, study group and value of variable, respectively. Variable wght (not shown) is created separately during the execution.
Calculations using SAS
In the code below we demonstrate how to use SAS to perform Steps 2 and 3 of the algorithm. To fit linear model in Step 2, we use PROC MIXED. Statement PARMS is used to specify σ2. Options NOPROFILE and NOITER in PARMS statement are used to keep it constant during calculations. ODS OUTPUT statement is used to save the F-statistic in data named contrasts. In the DATA step power is calculated, and Step 3 of the algorithm is completed.
| Title “Power calculations: Step 2”; |
| ods output contrasts=contrasts; |
| proc mixed data=dt noprofile; |
| class grp; |
| weight wght; |
| model y=grp; |
| parms (36)/ noiter; |
| contrast “grp” grp 1 −1 ; |
| quit; |
| ods output close; |
| Title “Power calculations: Step 3”; | |
| data power; | |
| set contrasts; | |
| alpha = 0.05; | |
| n = 50; | * Number of subjects; |
| ni = 6; | * Number of fibers per subject |
| P = 2; | * Number of fixed effects |
| dendf0 = n*ni − n; | |
| qf = finv(1−alpha, numdf, dendf0); | |
| dendfA = n*ni − p; | |
| power = 1 − probf(qf, numdf, dendfA, Fvalue*numdf); | |
| put power=; | * Equal to 0.7436023; |
| run; | |
Calculations using R language
The code below demonstrates key commands used in R language to perform power calculations. Note that, in contrast to PROC MIXED in SAS, function lm() in R does not allow to specify arbitrary value of σ. Instead, the {lm()} function estimates σ from data and, not surprisingly, obtains a value of sd0 very close to 0. To obtain the desired F-statistic, we calculated scalar sc and rescaled the F-statistic. Although the procedure of “rescaling” appears to work fine, some caution is required, because it involves division by sd0 having value very close to zero.
| # Step 2 using R: | |
| lm0 | <- lm(ym ~ grp, data = dt, weights = wght) |
| sd0 | <- summary(lm0)$sigma |
| sc | <- sqrt(36)/sd0 |
| Fvalue | <- anova(lm0)[“grp”,“F value”]/(sc*sc) # Rescale F stat |
| # Step 3 | |
| alpha | <- 0.05 |
| n | <- 50 |
| ni | <- 6 |
| P | <- 2 |
| numdf | <- 1 |
| dendf0 | <- n*ni − n |
| qF | <- qf(1−alpha, numdf,dendf0) |
| dendfA | <- n*ni − p |
| (power | <- 1− pf(qF,numdf,dendfA, numdf*Fvalue)) |
| [1] 0.7436023 | |
References
- [1].Verbeke G, Molenberghs G. Linear Mixed Models for Longitudinal Data. Springer; 2000. [Google Scholar]
- [2].Helms RW. Intentionally incomplete longitudinal designs: I. Methodology and comparison of some full span designs. Statistics in Medicine. 1992;11:1889–1913. doi: 10.1002/sim.4780111411. [DOI] [PubMed] [Google Scholar]
- [3].Litell RC, Milliken GA, Stroup WW, Wolfinger RD, Schabenberger O. SAS for Mixed Models. SAS Publishing; 2006. [Google Scholar]