Abstract
Glioblastomas (GBs) are malignant CNS tumors often associated with devastating symptoms. Patients with GB have a very poor prognosis, and despite treatment, most of them die within 12 months from diagnosis. Several pathways, such as the RAS, tumor protein 53 (TP53), and phosphoinositide kinase 3 (PIK3) pathways, as well as the cell cycle control pathway, have been identified to be disrupted in this tumor. However, emerging data suggest that these aberrations represent only a fraction of the genetic changes involved in gliomagenesis. In this study, we have applied a 32K clone-based genomic array, covering 99% of the current assembly of the human genome, to the detailed genetic profiling of a set of 78 GBs. Complex patterns of aberrations, including high and narrow copy number amplicons, as well as a number of homozygously deleted loci, were identified. Amplicons that varied both in number (three on average) and in size (1.4 Mb on average) were frequently detected (81% of the samples). The loci encompassed not only previously reported oncogenes (EGFR, PDGFRA, MDM2, and CDK4) but also numerous novel oncogenes as GRB10, MKLN1, PPARGC1A, HGF, NAV3, CNTN1, SYT1, and ADAMTSL3. BNC2, PTPLAD2, and PTPRE, on the other hand, represent novel candidate tumor suppressor genes encompassed within homozygously deleted loci. Many of these genes are already linked to several forms of cancer; others represent new candidate genes that may serve as prognostic markers or even as therapeutic targets in the future. The large individual variation observed between the samples demonstrates the underlying complexity of the disease and strengthens the demand for an individualized therapy based on the genetic profile of the patient.
Keywords: amplification, array-CGH, cancer, deletion, glioblastoma
Diffuse astrocytomas are CNS tumors that are thought to be derived from glial precursor or stem cells.1 They are classified and graded according to WHO as diffuse astrocytomas (WHO grade II), anaplastic astrocytomas (WHO grade III), and glioblastomas (GB; WHO grade IV), depending on the prognosis in context of the histological appearance.2 The majority of GBs develop in a short time without clinical evidence of an earlier lower-grade lesion. These GBs are named “de novo” or primary GB (pGB). Most patients with a lower-grade diffuse astrocytoma demonstrate malignant progression to GB. These tumors are termed secondary GBs (sGB). Besides surgical resection, patients with GB are currently treated by adjuvant radiation and chemotherapy. However, the prognosis for GB is poor. Patients without surgical resection have a median survival of only 2.5 months from time of diagnosis. Those that undergo surgical resection demonstrate a median survival of 7.9 months.3 The addition of temozolomide to adjuvant radiotherapy after surgery results in a median survival of 14.6 months.4
Different pathways have already been identified that are disrupted by various structural genetic alterations in GB. The RAS pathway is frequently activated by amplification of growth factor receptor genes such as EGFR (7p11) or PDGFRA (4q12). The tumor protein 53 (TP53) pathway is often disrupted by TP53 mutations (17p13), amplification of MDM2 (12q15), or losses of CDKN2A (9p21). The phosphoinositide kinase 3 (PIK3) pathway is activated by aberrant growth factor signaling and loss of PTEN (10q23) or, more rarely, by PIK3CA mutations (3q26). Disruption of cell cycle control is frequently mediated by loss of CDKN2A (9p21), amplification of CDK4 (12q14), or RB1 alterations (13q14). However, frequently multiple chromosomal alterations are observed (loss of 10p, 10q, 1p, and 22q as well as loss or duplication of 19q) that cannot be linked to a single gene or even a pathway in GB. The frequency of genetic alterations often differs between pGB and sGB and between younger and older patients. Some genetic alterations frequently occur in combination with each other (EGFR amplification and CDKN2A loss) or can be nearly exclusively found in combination (EGFR amplification or TP53 mutations).2
Because of frustrating efforts to optimize therapeutic regimens for patients with GB, the concept of individualized therapy based on the specific genetic profile of the tumor appears promising. For example, screening for MGMT promoter hypermethylation facilitates the identification of patients that benefit from temozolomide therapy,5 and coexpression of epidermal growth factor receptor VIII (EGFRvIII) with phosphatase and tensin homolog (PTEN) is associated with response to EGFR kinase inhibitors.6 However, the genetic heterogeneity of GB mostly prohibits the use of single predictive markers. The design of studies that evaluate such a therapeutic concept not only requires more knowledge about gross chromosomal alterations in GB but also demands a detailed map of gains and losses together with an evaluation of combined alterations.
To generate such a complex map, we comprehensively analyzed a series of 78 GBs by array-based comparative genomic hybridization (aCGH). The technique is based on the assessment of fluorescence ratios between differentially labeled test and reference DNA, competitively hybridized to a microarray of genomic clones spotted onto a glass slide.7–9 Quantitative evaluation of fluorescence intensities allows the identification of altered ratios that are indicative of DNA copy number imbalances in a test versus a reference genome. In this study, we have applied an array composed of 32,396 bacterial artificial chromosomes (BACs) covering 99% of the current assembly of the human genome.10 The high resolution of the platform (average resolution of analysis of 60 kb) allowed us to identify candidate oncogenes and tumor suppressor genes that may serve as prognostic or predictive markers or even as therapeutic targets for more clinically orientated studies in the future.
Materials and Methods
Patient Samples
A total of 78 WHO grade IV GB samples, derived from 50 males and 28 females, was included in the study (average age of onset, 54 and 57 years, respectively). The biopsies were sampled during operations and stored between −80°C and −135°C prior to DNA isolation. Matched peripheral blood samples were also collected for 46 of the cases. Sixty-seven of the samples were obtained from patients treated at the Charité Hospital Berlin and the University of Bonn Medical Center in Germany, seven at the Uppsala University Hospital in Sweden, and four at the Provincial Specialist Hospital in Gdansk, Poland. Tumors were classified and graded by experienced neuropathologists according to the third revised WHO criteria,2 and all cases were further reviewed by a second neuropathologist (A.D., C.H., or T.O.). Peripheral blood–derived DNA obtained from a healthy Caucasian female (F1) was used as the reference control in all hybridization experiments.10 High-molecular- weight DNA was isolated from the tumor and peripheral blood using standard methods.11 The use of DNA from human subjects was approved by the local research ethics committees of the University of Bonn Medical Center, the Charite Humboldt University (Berlin), and the Faculty of Medicine of Uppsala University.
aCGH 32K array
The 32K array was established as described previously.10 In short, the 32K BAC library,12 purchased from BACPAC Resources at Children’s Hospital Oakland Research Institute (Oakland, CA, USA; http://bacpac.chori.org/pHumanMinSet.htm), was amplified using three different degenerate-oligonucleotide–primed PCR primers, designed for the amplification of large insert clones.13 The products from these three amplifications were combined and reamplified with universal primer labeled with an amino group, which allows the attachment of the DNA to a Codelink HD microarray slide (GE HealthCare, Piscataway, NJ, USA). Manufacturing of the 32K BAC arrays was done with a high-throughput microarray printer constructed by the Lawrence Berkeley National Laboratory (Berkeley, CA, USA). After printing, the slides were activated in a humidity chamber overnight, blocked using a sodium borohydrate solution (2.6 g/l) for 5 min, and denatured in boiling water. Validation experiments of the 32K array have been previously reported.10 As an independent verification, the 32K BAC array was supplemented with a different set of clones, used previously in the construction of a chromosome 22-specific array.14 The 32K slides therefore contained two distinct arrays for chromosome 22, named 22 (32K set) and 22_B.14
Labeling and Hybridization
DNA labeling, hybridization, washing, and scanning of arrays were performed as previously described with minor modifications.14 Briefly, 1 μg of test and reference DNA was random prime labeled (BioPrime Array CGH Genomic Labeling Kit; Invitrogen, Carlsbad, CA, USA) with Cy3-dCTP (PA53021, GE HealthCare) and Cy5-dCTP (PA55021, GE HealthCare). Purified DNA probes were combined with 150 μg human Cot-1 DNA (Invitrogen), vacuum evaporated, and resuspended in 50 μl of hybridization solution (50% formamide, 5% sodium dodecyl sulfate [SDS], 10% dextran sulfate [molecular weight 500 kDa], 2× saline–sodium citrate [SSC]). The probe mixture was denatured for 5 min at 95°C, followed by incubation at 45°C for 2 h. Subsequently, the probe mixture was applied to the slide surface, covered with a 22 × 60 mm LifterSlip (Erie Scientific, Ports-mouth, NH, USA), and hybridized at 45°C for 20 h in a slide chamber (Corning Inc., Corning, NY, USA). Slides were washed in 25% formamide, 2× SSC, 0.1% SDS at 45°C for 17 min, followed by 10 min in phosphate-buffered saline at room temperature. The array was finally immersed in distilled H2O and immediately blown dry using pressurized dust-free air.
Array Imaging, Data Analysis, Clustering, and Statistics
Image acquisition was performed using a GenePix 4000B scanner (Axon Instruments Inc., Union City, CA, USA), and hybridization intensity was analyzed using GenePixPro image analysis software (version 6; Axon Instruments). The raw data files were then uploaded to a laboratory information management system database for storage, hosted by the Linnaeus Centre for Bioinformatics (LCB; http://base.lcb.uu.se). LCB also provides tools for filtering and statistical analysis of microarray data within the LCB Data WareHouse (LCB-DWH).15 We applied several filters to the hybridization raw data files. These include removal of oversaturated spots (>5%), spots with low signal-to-noise ratio (<3) in channels, and spots either manually or automatically flagged as bad, absent, or not found in the image analysis program. To remove possible dye bias or spatial effects, we also normalized all data using print-tip locally weighted scatter-plot smoothing.16 Clones were classified as balanced, gained, or deleted using the open-source software SMAP,17 available from Bioconductor (www.bioconductor.org) and LCB-DWH.15 For a specified number of states (six), SMAP iteratively fits a hidden Markov model genomewide to the data and infers the most probable profile of copy numbers for each chromosome using a segmental a posteriori approach until no further improvements can be made. Each individual clone was then assigned a preliminary copy number class (CNC): balanced, CNC = 0; gain, CNC = 1, 2, or 3; and deletion, CNC = −1 or −2. A graphical viewing tool that plots all clones according to their chromosomal positions was used for the visualization of results. Complete hierarchical clustering was used to identify clusters of copy number profiles. Distances between profiles/clusters were calculated using the Euclidean distance at a genomewide level. Five groups of CNCs were considered: homozygous deletion (CNC = −2), heterozygous deletion (CNC = −1), normal (CNC = 0), single copy gain (CNC = 1), and two or more copies gained (CNC ≥2). Clone mapping information was obtained from Ensembl BioMart generic data management system (www.ensembl.org/biomart).
CNC data were used for statistical analysis using Fisher’s exact test within LCB-DWH. The test was used to find statistically significant overrepresentation of changes in tumor tissue compared to blood. p-Values were adjusted using the Benjamini and Hochberg step-up false discovery rate–controlling procedure.18 Spatial correlation between clones in the set of samples was calculated using Spearman’s correlation coefficient. Correlation matrices for the spatial correlation were calculated using R (http://www.r-project.org).19 A Gene Ontology (GO) tool within LCB-DWH was used to test for statistically significant overrepresentation of GO terms of genes resident within aberrant clones with respect to all genes encompassed on the array.15,20
We used publicly available gene expression data from grade IV gliomas, series GSE4412, and from normal brain, series GSE1133, platform GPL96: Affymetrix GeneChip Human Genome U133 Array Set HG-U133A (Affymetrix, Santa Clara, CA, USA) to select differentially expressed genes within minimal overlapping regions of aberrations.
Copy Number Alterations Determined by Quantitative Real-Time PCR
Relative gene copy numbers were determined by quantitative real-time PCR (qPCR) using the Stratagene MX 3005P qPCR system (AH Diagnostics, Århus, Denmark) and SYBR Green JumpStart Taq ReadyMix for High Throughput QPCR (Sigma, St. Louis, MO, USA). The standard curve method described for the ABI PRISM 7700 Sequence Detection System (Applied Biosystems Inc., Foster City, CA, USA; ABI User Bulletin #2, docs.appliedbiosystems.com/pebiodocs/04303859.pdf) was used and the standard curve was generated from human genomic DNA (Applied Biosystems, P/A 4312660). Genomic DNA from 23 tumor samples was analyzed, and we chose the DNAH7 (2q32.3) gene as a reference for all experiments as no copy number changes were observed at this locus in the 32K array analysis. The gene copy number in the tumor DNA was normalized to the reference gene (DNAH7) and calibrated to normal DNA (F1). A subset of candidate genes encompassed within amplicons as well as homozygous deletions were selected for qPCR verification. PCR conditions and primers for all genes are available in Supplementary Table 1 (online only with this article at neuro-oncology.dukejournals.org). PCR reactions were performed in triplicate for each primer pair, and the data were analyzed using the Stratagene MxPro QPCR software (AH Diagnostics).
Results
32K Array Profiling of Copy Number Alterations in GB
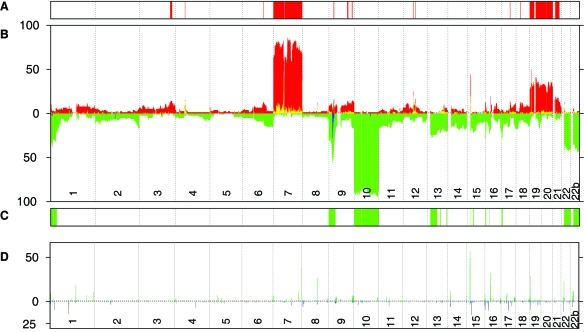
The clinical details of the patients included in this study are summarized in Supplementary Table 2 (online only with this article at neuro-oncology.duke journals.org). To visualize the results, the frequency of copy number changes was calculated for all samples and plotted relative to the position along the chromosome for each clone (Fig. 1). The most common copy number variation involving a whole chromosome was monosomy 10, followed by trisomy 7. The combination of trisomy 7 and monosomy 10 (detected in 83% of the cases) was also the most frequent arrangement of aberrations found in the tumor series. Monosomy 22, trisomy 20, and trisomy 19 were also relatively frequent (33%, 30%, and 25%, respectively). Furthermore, trisomy 20 and trisomy 19 often, but not exclusively, appeared simultaneously (Supplementary Fig. 1 [online only with this article at neuro-oncology.dukejournals.org]). Numerous copy number alterations involving whole p and/or q arms and interstitial and/or terminal gains/deletions were also detected. The most common was the entire or interstitial loss of 9p, identified in 55% of the cases, followed by interstitial deletions of 1p (34%; Fig. 1 and Supplementary Fig. 1). No single chromosome was free from aberrations in the analyzed series. We counted the fraction of the genome involved in DNA copy number alterations for each tumor and found that, on average, 18% of the genome was aberrant in this malignant tumor (Table 1). We also analyzed 46 peripheral blood DNA samples from the patients (Supplementary Fig. 1). Numerous loci affected by gene copy number variation were identified. When compared to our data obtained from the profiling of a series of healthy individuals using the same platform10 and compared to publicly available data (Centre for Applied Genomics, Database of Genomic Variants, http://projects.tcag.ca/variation), we observed that all the alterations detected were previously categorized as disease-unrelated genomic copy number variations. As a result, we concluded that these variations represent only large-scale copy number polymorphisms (data not shown).
Fig. 1.
Frequency of copy number changes in glioblastoma and blood samples calculated for all autosomal clones and plotted relative to the position along the chromosome for each clone, for the 78 tumor samples (B) and for 117 hybridizations run on peripheral blood DNA (D), including 71 samples from healthy subjects10 and 46 blood samples derived from the glioblastoma (GB) patients included in this study. Red bars above the horizontal line indicate the incidence in percent copy number gains (CNC = 1, 2, 3), and yellow bars represent higher copy number gains (CNC = 2, 3). Green bars below the horizontal line indicate the frequency of copy number losses (CNC = −1, −2), and blue bars indicate the incidence of homozygous deletions (CNC = −2). The 32K array was supplemented with 22_B, a different set of clones representing 22q used in our previously reported chromosome 22-specific array,14 indicated on the figure as 22b. The chromosome 22_B clone set was printed on the same slide as the 32K clones, and both sets identified the same frequency of copy number aberrations. To identify tumor-specific aberrations, the array-based comparative genomic hybridization results from the series of 78 GB tumor samples were compared to the data obtained from 117 hybridizations run on peripheral blood DNA. Fisher’s exact test was used to evaluate statistically significant differences between gained and nongained clones as well as between deleted and nondeleted clones in tumor versus blood groups. (A and C) The 3,000 top ranked gained (red) and deleted (green) clones, including clones representing chromosomes 7, 19, 20, and 21 for gains and chromosomes 10, 9, 22, 13, and 1 for deletions.
Table 1.
Summary of array-based comparative genomic hybridization results for 78 glioblastoma samples
| Deletions | Gains | Alterations | Amplicons | Homozygous Deletions | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor ID | Length (Mb) | Length (Mb) | Total [Mb (%a)] | Number | Average Size (Mb) | Number | Average Size (Mb) | EGFR | PDGFRA | CDK4 | CDKN2A/2B | MDM2 |
| G10 | 165.8 | 299.1 | 464.9 (14.5) | 3 | 2.1 | + | + | |||||
| G1143 | 178.2 | 381.4 | 559.5 (17.5) | |||||||||
| G153 | 292.3 | 306.1 | 598.4 (18.7) | 2 | 0.6 | 1 | 4.2 | + | − | |||
| G1596 | 376.3 | 270.8 | 647.1 (20.2) | 1 | 0.6 | 2 | 19.4 | + | − | |||
| G174 | 337.1 | 167.7 | 504.8 (15.8) | 1 | 2.5 | + | ||||||
| G1963 | 152.1 | 161.6 | 313.7 (9.8) | 1 | 0.7 | 1 | 2.7 | + | − | |||
| G20844 | 173.3 | 347.0 | 520.3 (16.3) | 3 | 0.5 | + | ||||||
| G20854 | 139.5 | 286.8 | 426.3 (13.3) | 1 | 0.9 | 2 | 1.9 | + | − | |||
| G20856 | 346.4 | 351.3 | 697.6 (21.8) | |||||||||
| G20890 | 230.9 | 339.3 | 570.2 (17.8) | 3 | 0.8 | 1 | 5.6 | + | + | − | ||
| G20940 | 380.0 | 203.5 | 583.5 (18.2) | 2 | 0.8 | 1 | 6.9 | + | − | |||
| G20944 | 135.9 | 149.4 | 285.3 (8.9) | 1 | 0.4 | + | ||||||
| G20972 | 250.1 | 157.8 | 407.9 (12.7) | 2 | 0.8 | + | ||||||
| G20990 | 294.8 | 151.7 | 446.5 (14.0) | 1 | 0.9 | 2 | 2.6 | + | − | |||
| G21576 | 315.9 | 183.8 | 499.7 (15.6) | 9 | 0.7 | + | + | |||||
| G21602 | 265.4 | 159.1 | 424.5 (13.3) | 2 | 0.5 | + | + | |||||
| G21612 | 450.1 | 166.0 | 616.0 (19.3) | |||||||||
| G21628 | 193.0 | 191.5 | 384.5 (12.0) | 1 | 0.6 | + | ||||||
| G21694 | 721.2 | 667.0 | 1388.2 (43.4) | 1 | 3.1 | − | ||||||
| G21800 | 200.5 | 181.8 | 382.3 (11.9) | 1 | 1.0 | |||||||
| G21804 | 178.3 | 238.0 | 416.4 (13.0) | 3 | 1.4 | 2 | 2.6 | + | − | |||
| G21820 | 194.6 | 301.7 | 496.4 (15.5) | 2 | 2.4 | − | ||||||
| G21828 | 457.0 | 246.6 | 703.6 (22.0) | 2 | 0.2 | + | ||||||
| G21830 | 48.9 | 485.6 | 534.6 (16.7) | 1 | 2.2 | 1 | 0.6 | + | ||||
| G21836 | 180.6 | 296.4 | 477.0 (14.9) | 1 | 0.6 | + | ||||||
| G21864 | 575.7 | 308.4 | 884.0 (27.6) | 16 | 0.5 | + | ||||||
| G22118 | 618.4 | 374.3 | 992.7 (31.0) | 9 | 2.4 | 2 | 2.3 | + | − | |||
| G22334 | 358.5 | 104.2 | 462.6 (14.5) | 3 | 0.6 | 1 | 3.8 | + | − | |||
| G22368 | 248.4 | 211.8 | 460.2 (14.4) | 3 | 1.9 | + | ||||||
| G22370 | 743.6 | 311.6 | 1055.1 (33.0) | |||||||||
| G22520 | 386.1 | 331.8 | 717.9 (22.4) | 5 | 1.3 | 1 | 1.8 | + | ||||
| G22576 | 666.5 | 447.0 | 1113.5 (34.8) | 7 | 1.9 | + | + | + | ||||
| G22616 | 597.3 | 199.7 | 797.0 (24.9) | 1 | 2.0 | 3 | 2.2 | + | − | |||
| G22686 | 407.1 | 521.2 | 928.3 (29.0) | 1 | 0.9 | 3 | 2.4 | + | − | |||
| G22872 | 391.5 | 163.1 | 554.6 (17.3) | |||||||||
| G23120 | 195.7 | 265.9 | 461.7 (14.4) | 1 | 1.2 | + | ||||||
| G23218 | 326.1 | 201.2 | 527.3 (16.5) | 1 | 3.0 | 1 | 1.3 | + | ||||
| G23264 | 826.1 | 543.0 | 1369.1 (42.8) | 2 | 4.1 | − | ||||||
| G23282 | 241.3 | 206.0 | 447.4 (14.0) | 1 | 4.6 | 2 | 0.8 | + | − | |||
| G23304 | 445.8 | 196.2 | 642.0 (20.1) | 1 | 0.7 | 1 | 5.1 | − | + | |||
| G23316 | 328.7 | 304.4 | 633.0 (19.8) | 3 | 0.7 | 1 | 0.5 | + | + | |||
| G23480 | 434.3 | 48.0 | 482.3 (15.1) | 2 | 0.7 | + | + | |||||
| G23786 | 147.0 | 171.9 | 318.8 (10.0) | 1 | 0.9 | 1 | 4.9 | + | ||||
| G23906 | 161.2 | 252.5 | 413.6 (12.9) | 1 | 1.1 | 2 | 1.3 | + | − | |||
| G24014 | 380.4 | 249.1 | 629.5 (19.7) | |||||||||
| G24028 | 304.9 | 248.1 | 553.0 (17.3) | 1 | 2.2 | − | ||||||
| G24064 | 169.4 | 55.0 | 224.4 (7.0) | 14 | 2.3 | + | ||||||
| G24158 | 288.8 | 158.6 | 447.4 (14.0) | 2 | 1.9 | + | + | |||||
| G24178 | 174.5 | 246.7 | 421.2 (13.2) | 1 | 0.9 | + | ||||||
| G24454 | 280.4 | 131.9 | 412.3 (12.9) | 3 | 0.5 | + | ||||||
| G24460 | 202.2 | 254.5 | 456.6 (14.3) | 1 | 0.4 | 1 | 5.3 | + | − | |||
| G24528 | 333.7 | 183.8 | 517.5 (16.2) | 4 | 0.7 | 1 | 7.8 | + | − | |||
| G24596 | 289.3 | 333.3 | 622.6 (19.5) | 1 | 1.2 | 1 | 0.5 | + | ||||
| G24702 | 366.8 | 194.8 | 561.6 (17.5) | 2 | 2.5 | + | ||||||
| G24930 | 391.3 | 307.2 | 698.6 (21.8) | |||||||||
| G25036 | 862.0 | 491.9 | 1354.0 (42.3) | 2 | 0.6 | + | + | |||||
| G251 | 192.5 | 187.9 | 380.4 (11.9) | 1 | 0.7 | 1 | 2.4 | + | − | |||
| G25108 | 316.8 | 282.2 | 599.1 (18.7) | 1 | 5.0 | + | ||||||
| G25488 | 458.2 | 109.4 | 567.6 (17.7) | 1 | 4.7 | + | ||||||
| G25496 | 151.1 | 173.1 | 324.2 (10.1) | 1 | 0.8 | − | ||||||
| G27006 | 453.3 | 235.6 | 688.9 (21.5) | 1 | 0.4 | + | ||||||
| G27030 | 183.4 | 2.5 | 185.9 (5.8) | 1 | 1.4 | 1 | 1.4 | + | − | |||
| G27040 | 260.2 | 145.3 | 405.5 (12.7) | 1 | 0.7 | + | ||||||
| G27614 | 311.1 | 220.3 | 531.4 (16.6) | 1 | 1.3 | − | ||||||
| G27622 | 209.5 | 250.2 | 459.7 (14.4) | 19 | 0.8 | + | + | |||||
| G28882 | 502.5 | 439.3 | 941.7 (29.4) | 2 | 1.0 | |||||||
| G29178 | 210.4 | 271.6 | 482.0 (15.1) | 1 | 1.6 | 1 | 10.0 | + | − | |||
| G29264 | 498.4 | 0.5 | 498.9 (15.6) | 1 | 0.4 | − | ||||||
| G29994 | 298.4 | 155.5 | 453.9 (14.2) | 1 | 2.7 | 1 | 2.2 | + | − | |||
| G30196 | 365.5 | 219.3 | 584.7 (18.3) | 6 | 1.4 | + | + | |||||
| G30276 | 170.9 | 465.4 | 636.3 (19.9) | 1 | 0.2 | 1 | 5.6 | + | − | |||
| G30296 | 586.1 | 590.6 | 1176.8 (36.8) | 1 | 11.3 | |||||||
| G30726 | 334.6 | 157.9 | 492.5 (15.4) | 2 | 0.7 | + | ||||||
| G30758 | 476.4 | 219.3 | 695.7 (21.7) | 4 | 1.7 | + | + | |||||
| G40 | 483.7 | 237.1 | 720.8 (22.5) | 5 | 0.4 | 2 | 2.3 | − | + | |||
| G48 | 273.4 | 130.6 | 404.0 (12.6) | 2 | 1.2 | 1 | 9.1 | + | − | |||
| G50 | 405.4 | 329.5 | 734.9 (23.0) | 4 | 1.9 | + | + | + | ||||
| G8 | 228.8 | 92.4 | 321.2 (10.0) | 3 | 0.7 | 1 | 0.5 | + | ||||
The total number of bases included within deleted or gained regions, as well as the number and average size of amplicons and homozygous deletions are shown for each sample. Amplified, homozygously deleted loci were defined by the presence of at least two neighboring clones with CNC = 3 and CNC = 2. The presence of amplified and deleted loci encompassing genes commonly aberrant in glioblastoma (EGFR, PDGFRA, CDK4, CDKN2A, MDM2) is indicated by “+” for amplicons and “−” for homozygous deletion. Only autosomes are counted.
Percentage of the total genome involved in DNA copy number alterations for each tumor sample.
We used Spearman’s correlation coefficient to calculate the correlation between clones in the series of tumors. A correlation plot was then produced with respect to chromosomal location (Supplementary Fig. 2 [online only with this article at neuro-oncology.dukejournals.org]). We observed a general trend of positive correlation between bins located on the same chromosome/chromosome arm, caused by the presence of long-range gains or deletions (or no alterations). In particular, these wide-range aberrations were found on chromosomes 10, 13, and 14. A remarkable negative correlation was found between chromosomes 7 and 10; this combination of aberrations (trisomy 7 and monosomy 10) is also in agreement with the results obtained from the recurrence analysis (Fig. 1). We also ran Fisher’s exact test to determine frequent and tumor-specific aberrant loci in the GB series. The aCGH results from the 78 tumor samples were compared to data from 117 hybridizations run on peripheral blood DNA, 71 samples from healthy subjects,10 and 46 blood samples from the GB patients included in this study. Clones affected by gains (CNC > 0) or deletions (CNC < 0) were compared in tumor versus blood groups. As much as 35% of the genome, including loci in all chromosomes, appeared significantly affected by gains/deletions (p < 0.001) in the tumor series. The 3,000 top-ranked deleted and gained clones are indicated in Fig. 1 and include clones representing chromosomes 7, 19, 20, and 21 for gains and chromosomes 10, 9, 22, 13, and 1 for deletions. To explore the biological function of these aberrations, we checked for overrepresentation of GO terms within the genes encompassed by these clones. Overrepresented GO biological process terms, found among the genes contained within the 3,000 top-ranked gained clones (>1,000 genes), were related to DNA-dependent regulation of transcription and regulation of cellular and metabolic processes (Supplementary Table 3 [online only with this article at neuro-oncology.dukejournals.org]). The EGFR gene was represented in many of the GO terms. These results are consistent with the current tumor biology model for GB. Within the GO biological process terms represented by genes encompassed within the 3,000 top-ranked deleted clones (>500 genes), processes related to response to virus, cell cycle, and apoptotic programs were found (Supplementary Table 3).
Among the GB samples, 74 were of primary origin, and four were classified as the secondary type (Supplementary Table 2). Representative profiles for a pGB (G20890) and an sGB (G21694) are illustrated in Supplementary Fig. 3 (online only with this article at neuro-oncology.dukejournals.org). Even if only a few samples of the secondary type were studied, clear differences between the groups could be observed. sGB samples presented with a more complex pattern of rearrangements, including whole and partial deletions or gains of multiple chromosomes, including 10 and 7, but complete deletion of chromosome 10 was not detected in sGB (Supplementary Fig. 1). One pGB was derived from a child (G1143). This tumor presented with trisomy 7 and 2, monosomy 6, and a terminal hemizygous deletion of 17p, encompassing the TP53 gene. Monosomy 10 and partial 9p deletion, which were frequently observed in GBs derived from adults, were not present in this sample.
We also defined the smallest, tumor-specific overlapping regions of imbalance for each chromosome in the series. A total of 185 regions were identified, involving gain or loss of material, present in at least 3 of the 78 samples. The regions were distributed throughout the genome. Using publically available expression data, we determined the top 10 genes significantly up- or down- regulated within these loci. Many genes known to be involved in glioma pathways as well as novel genes with links to other cancer forms mapped to these loci (Supplementary Table 4 [online only with this article at neuro-oncology.dukejournals.org]). In order to find out in which pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.ad.jp/kegg/pathway.html) these genes could be connected, the list of genes was submitted to the Database for Annotation, Visualization, and Integrated Discovery (DAVID) Functional Annotation Tool (http://david.abcc.ncifcrf.gov).21 The most significantly represented pathway was hsa05214:glioma, with 12 genes recognized in this category (Supplementary Table 5 [online only with this article at neuro-oncology.dukejournals.org]). It is interesting that genes from the list were involved in gap junction, focal adhesion, and tight junction pathways, networks that regulate actin cytoskeleton, cell motility, proliferation, and survival and are known to be usually deregulated in GB.
Narrow Amplicons Pinpoint Novel Candidate Oncogenes in GB
The most striking result from our study is the frequent identification of high and narrow copy number amplicons within the series of GB tumors. Regions involving at least two neighboring clones with CNC ≥ 3 and normalized fluorescence ratio ≥ 2, which represent loci with at least five DNA copies in a diploid tumor, are shown in Table 2. These loci include many previously reported oncogenes involved in GB tumorigenesis (e.g., EGFR, PDGFRA, MDM2, CDK4) as well as numerous novel genes. The detection of previously reported oncogenes stands as an independent validation of our platform. Representative profiles for GB samples displaying EGFR or PDGFRA amplicons are illustrated in Supplementary Figs. 4 and 5 (online only with this article at neuro-oncology.dukejournals.org). A total of 63 cases (81%) presented with amplicons, varying in number from 1 to 19 per sample (three on average), 0.2 to 11.3 Mb in size (1.4 Mb on average; Tables 1, 2). EGFR amplification, detected in 39 samples (48.75%) was the most frequent amplification event identified. Moreover, this was the only amplicon present in 22 of the cases (27.5%). Interestingly, in 19 cases (24%) the normalized ratio for clone CTD-2026N22, within EGFR amplicon, was higher than 10, which is consistent with at least 20 DNA copies of this locus. Distinctly amplified regions, in addition to the EGFR locus, were also detected on 7p. These amplicons encompassed a number of novel genes (NPVF, ZNRF2, CRHR2, LSM5, TBX20, HERPUD2, and GRB10), which probably become coamplified with EGFR and contribute to the tumorigenesis process.
Table 2.
Summary of high-level amplified regions and homozygous deleted regions in glioblastoma samples analyzed by array-based comparative genomic hybridization, involving at least two overlapping consecutive clones on the 32K bacterial artificial chromosome array
| Chromosome | Band | From (Mb) | To (Mb) | Size (Mb) | No. Tumors | Tumor ID | No. Clonesa (Maximum Ratio) | No. Genes in Regionb | Selected Candidate Genesc |
|---|---|---|---|---|---|---|---|---|---|
| Amplifications | |||||||||
| 1 | p36.21 | 13.56 | 13.84 | 0.28 | 1 | G22368 | 2 (15) | 5 | PDPN |
| 1 | p34.2 | 39.84 | 40.34 | 0.50 | 1 | G20844 | 6 (10.6) | 12 | MYCL1 |
| 1 | p34.2 | 42.56 | 43.01 | 0.46 | 1 | G27622 | 3 (16.6) | 10 | SLC2A1 |
| 1 | p13.2 | 115.07 | 115.51 | 0.45 | 1 | G27622 | 3 (3.2) | 5 | NRAS, SYCP1 |
| 1 | q31.1 | 184.73 | 185.51 | 0.78 | 1 | G27622 | 4 (3.5) | 2 | PTGS2, PLA2G4A |
| 1 | q32.1 | 201.24 | 204.18 | 2.94 | 3 | G22368, G48, G30726 | 26 (10.3) | 55 | SOX13, PIK3C2B, MDM444,50–52 |
| 1 | q43, q44 | 241.40 | 242.19 | 0.79 | 1 | G27622 | 8 (3.6) | 3 | AKT331,44,50 |
| 2 | p24.3 | 14.98 | 16.16 | 1.18 | 1 | G27622 | 12 (14.1) | 3 | MYCN44 |
| 2 | p22.1 | 38.47 | 39.92 | 1.45 | 1 | G21576 | 15 (11.1) | 16 | SOS1, CDKL4 |
| 2 | p21 | 42.69 | 43.41 | 0.72 | 1 | G21576 | 7 (13.3) | 4 | MTA3, ZFP36L2 |
| 2 | q14.2, q14.3 | 120.93 | 122.21 | 1.28 | 1 | G27622 | 11 (5.9) | 7 | GLI2, TFCP2L1, CLASP1 |
| 4 | p15.2 | 23.25 | 24.06 | 0.81 | 1 | G23316 | 7 (20.1) | 1 | PPARGC1A |
| 4 | p15.2 | 24.49 | 27.10 | 2.61 | 2 | G30758, G22576 | 25 (11.8) | 16 | PI4K2B, ZCCHC4, ANAPC4, RBPJ, TBC1D19, STIM2 |
| 4 | q11, q12 | 52.35 | 57.36 | 5.01 | 12 | G25108, G21830, G21576, G174, G30758, G27030, G50, G20890, G22576, G30726, G23480, G20944 | 47 (37.7) | 44 | PDGFRA31,44,50,52,53 |
| 5 | p15.33 | 0.35 | 0.56 | 0.20 | 1 | G24528 | 3 (6.6) | 4 | PDCD6 |
| 5 | p15.33 | 1.02 | 1.90 | 0.88 | 3 | G21576, G20844, G24528 | 7 (9.9) | 15 | TERT |
| 5 | q33.3 | 158.57 | 159.26 | 0.69 | 1 | G24528 | 5 (9.1) | 2 | ADRA1B, TTC1 |
| 7 | p15.3, p15.2 | 25.12 | 26.01 | 0.89 | 2 | G22118, G24064 | 8 (12.8) | 7 | NPVF |
| 7 | p15.1, p14.3 | 29.60 | 31.17 | 1.57 | 3 | G24064, G22118, G21804 | 14 (10.6) | 20 | ZNRF2, CRHR2 |
| 7 | p14.3 | 32.41 | 33.32 | 0.91 | 1 | G24064 | 6 (11) | 11 | LSM5 |
| 7 | p14.2 | 35.17 | 35.75 | 0.58 | 2 | G24064, G21804 | 4 (10.8) | 4 | TBX20, HERPUD2 |
| 7 | p12.3, p12.2, p12.1, p11.2, p11.1 | 46.70 | 57.90 | 11.21 | 39 | G22118, G24064, G24454, G23786, G20940, G20890, G10, G25488, G23282, G29994, G23218, G22616, G21804, G24702, G22334, G22520, G20854, G20990, G22368, G23120, G24528, G24596, G153, G24460, G21628, G21836, G22686, G23316, G23906, G24178, G27040, G29178, G48, G1596, G1963, G251, G8, G27006, G30276 | 94 (42.9) | 80 | VWC2, FIGNL1, GRB10, EGFR31,44,50–52,54 |
| 7 | q21.11 | 80.72 | 81.42 | 0.70 | 1 | G24064 | 7 (9.8) | 1 | HGF |
| 7 | q21.2, q21.3 | 91.71 | 95.07 | 3.36 | 1 | G24064 | 22 (15.4) | 35 | CDK644,51,54 |
| 7 | q32.2, q32.3 | 129.53 | 130.95 | 1.42 | 1 | G24064 | 12 (7.9) | 20 | PODXL, MKLN1 |
| 7 | q36.1, q36.2 | 149.96 | 153.39 | 3.43 | 1 | G27622 | 31 (5.8) | 44 | RHEB, NOS3, MLL3, LR8 |
| 9 | p24.3, p24.2 | 1.96 | 4.44 | 2.47 | 1 | G22576 | 25 (3.2) | 9 | GLIS3, RFX3 |
| 9 | q31.3 | 111.36 | 111.74 | 0.38 | 1 | G22334 | 6 (17.8) | 1 | PALM2 |
| 11 | p15.2 | 14.02 | 15.06 | 1.05 | 1 | G21800 | 8 (12.3) | 12 | SPON1, RRAS2 |
| 11 | q23.3 | 117.19 | 118.09 | 0.90 | 1 | G22520 | 9 (4.8) | 25 | TMPRSS4, TMEM25, PHLDB152 |
| 12 | p13.32 | 3.31 | 3.76 | 0.46 | 1 | G30758 | 5 (15.5) | 3 | PRMT852 |
| 12 | q12 | 39.22 | 39.61 | 0.39 | 1 | G40 | 4 (4.7) | 1 | CNTN1 |
| 12 | q13.13 | 52.49 | 53.03 | 0.54 | 1 | G21576 | 7 (7.6) | 16 | HOXC6, HOXC8 |
| 12 | q13.2 | 53.35 | 53.55 | 0.20 | 1 | G21576 | 2 (4.2) | 1 | MUCL1 |
| 12 | q13.2 | 54.48 | 54.74 | 0.26 | 1 | G21828 | 3 (3) | 13 | DGK4, SILV, CDK2, RAB5B |
| 12 | q13.3, q14.1 | 55.93 | 59.15 | 3.22 | 16 | G27622, G50, G20972, G21576, G21864, G23316, G24158, G30196, G20844, G10, G21602, G21828, G22576, G23480, G25036, G30758 | 41 (18.7) | 34 | TSPAN31, CDK4, AVIL31,44,52,55 |
| 12 | q14.1 | 60.08 | 61.16 | 1.08 | 2 | G21576, G22576 | 11 (13.4) | 4 | FAM19A252 |
| 12 | q14.2 q14.3 q15 |
61.85 | 66.02 | 4.17 | 6 | G27622, G24064, G40, G22576, G50, G23304 | 45 (19.2) | 35 | SRGAP1, RASSF3, LEMD3, HMGA2, GRIP1 |
| 12 | q15, q21.1 | 66.22 | 71.61 | 5.38 | 9 | G22576, G50, G27622, G30196, G40, G24158, G25036, G21602, G23304 | 61 (32.6) | 45 | MDM1, NUP107, RAP1B, MDM2, CPSF6, FRS2, CNOT2, RAB2131,44,51,52 |
| 12 | q21.2 | 76.38 | 77.15 | 0.76 | 1 | G27622 | 4 (9.4) | 1 | NAV3 |
| 12 | q21.2 | 77.87 | 78.11 | 0.24 | 1 | G27622 | 4 (9.1) | 1 | SYT1 |
| 13 | q22.3 | 76.69 | 76.93 | 0.24 | 1 | G22520 | 2 (3.1) | 1 | MYCBP2 |
| 13 | q31.1, q31.2, q31.3 | 85.01 | 96.35 | 11.35 | 4 | G30296, G22520, G24702 | 102 (7.1) | 29 | MIRH1, DCT, TGDS, LOC144874, ABCC4, DZIP1, UGCGL2 |
| 15 | q25.2 | 82.15 | 82.51 | 0.36 | 1 | G40 | 2 (6.6) | 1 | ADAMTSL3 |
| 15 | q26.1 | 88.09 | 90.46 | 2.37 | 1 | G22118 | 18 (3.4) | 25 | SEMA4B, NGRN, IQGAP1, FES, PRC156–58 |
| 15 | q26.2, q26.3 | 95.97 | 97.80 | 1.83 | 1 | G22118 | 18 (3.4) | 10 | IGF1R, DMN |
| 15 | q26.3 | 98.25 | 100.31 | 2.06 | 1 | G22118 | 23 (3.3) | 28 | ASB7, TM2D331 |
| 16 | p13.3 | 4.44 | 4.95 | 0.51 | 1 | G30196 | 4 (2.7) | 18 | NUDT16L1 |
| 17 | q21.33 | 45.86 | 46.77 | 0.91 | 1 | G20972 | 8 (5.4) | 19 | ABCC3, CROP, TOB1, NME1, NME2 |
| 19 | q13.31 | 49.34 | 50.18 | 0.84 | 1 | G10 | 8 (4.3) | 23 | CEACAM20/19/16, BCL3, BCAM |
| 21 | q22.13 | 38.08 | 38.57 | 0.49 | 1 | G21576 | 5 (13.1) | 5 | DSCR4/8 |
| 22 | q11.1, q11.21 | 16.30 | 16.59 | 0.30 | 1 | G21864 | 2 (2.9) | 3 | CECR227 |
| 22 | q11.21 | 17.82 | 18.67 | 0.85 | 2 | G21864, G153 | 4 (4.6) | 19 | CDC45L, CLDN5, TBX1, TXNRD2, RANBP127 |
| 22 | q11.21 | 19.05 | 19.64 | 0.58 | 1 | G21864 | 8 (19.2) | 12 | PI4KA, CRKL27 |
| 22 | q11.21, q11.22 | 19.84 | 20.66 | 0.82 | 1 | G21864 | 6 (8.1) | 13 | MAPK1 |
| 22 | q12.1 | 26.69 | 26.86 | 0.17 | 1 | G21864 | 2 (10.4) | 1 | TTC28 |
| 22 | q12.2 | 28.13 | 28.86 | 0.73 | 1 | G21864 | 4 (5) | 13 | RFPL1, MTMR3 |
| 22 | q12.2 | 29.09 | 30.03 | 0.95 | 1 | G21864 | 8 (9.9) | 27 | GAL3ST1, PES127 |
| 22 | q12.3 | 30.65 | 31.21 | 0.57 | 1 | G21864 | 5 (4) | 13 | RFPL2, RFPL3, FBXO727 |
| 22 | q12.3 | 31.79 | 32.61 | 0.82 | 1 | G21864 | 2 (9.9) | 1 | LARGE27 |
| 22 | q12.3 | 34.38 | 34.93 | 0.55 | 1 | G21864 | 5 (10.6) | 4 | RBM927 |
| 22 | q13.2 | 41.62 | 42.19 | 0.58 | 1 | G21864 | 4 (7) | 9 | TSPO, SCUBE127 |
| 22 | q13.31 | 42.87 | 44.57 | 1.70 | 1 | G21864 | 10 (6.2) | 16 | PHF21B, FBLN127 |
| Homozygous deletions | |||||||||
| 1 | p36.23, p36.22 | 7.31 | 9.98 | 2.68 | 3 | G22616, G20990, G21804 | 27 (0.27) | 28 | CAMTA1, PER3, TNFRSF9, ERRFI1, DNB5, RERE, ENO130,44 |
| 3 | q21.1, q21.2 | 123.26 | 126.82 | 3.56 | 1 | G22686 | 39 (0.39) | 32 | CSTA, DIRC2, SEMA5B, KALRN, MUC13, SNX4 |
| 3 | q21.3 | 127.83 | 128.48 | 0.65 | 1 | G22686 | 8 (0.37) | 5 | CHCHD6, PLXNA1 |
| 6 | q27 | 168.94 | 169.35 | 0.41 | 1 | G22616 | 3 (0.43) | 1 | THBS244 |
| 9 | p23 | 9.19 | 9.87 | 0.68 | 1 | G22370 | 3 (0.48) | 1 | PTPRD |
| 9 | p22.3 | 15.87 | 16.44 | 0.57 | 1 | G23282 | 7 (0.40) | 2 | BNC2 |
| 9 | p22.2, p22.1 | 17.98 | 18.88 | 0.89 | 1 | G20854 | 9 (0.52) | 1 | ADAMTSL1 |
| 9 | p22.1, p21.3, p21.2, p21.1 |
19.43 | 30.42 | 10.99 | 32 | G20990, G22616, G1596, G251, G40, G20940, G29994, G22686, G23906, G48, G153, G29178, G20854, G27030, G21694, G22334, G23282, G20890, G21804, G21820, G22118, G27614, G30276, G24460, G23304, G24528, G1963, G25496, G23264, G28882, G29264, G24028 | 113 (0.27) | 57 | MLLT3, KIAA1797, PTPLAD2, KLHL9, CDKN2A/2B, ELAVL244,59 |
| 9 | q22.32 | 96.31 | 97.20 | 0.89 | 1 | G40 | 6 (0.50) | 5 | FANCC |
| 10 | q23.2, q23.31 | 89.40 | 91.27 | 1.87 | 4 | G21830, G23316, G21820, G22118 | 16 (0.24) | 22 | PTEN44 |
| 10 | q26.2, q26.3 | 129.37 | 131.18 | 1.81 | 1 | G22520 | 18 (0.38) | 5 | PTPRE, MGMT44 |
| X | p21.1, p11.4 | 37.05 | 38.13 | 1.07 | 1 | G28882 | 14 (0.23) | 11 | SYTL5, DYNLT3, PRGR |
| X | q25 | 123.09 | 128.99 | 31.67 | 3 | G1596, G23218, G8 | 65 (0.38) | 16 | OCRL, XPNPEP2, ZDHHC9, BCORL1 |
Total number of genomic clones in the array representing the loci.
Total number of Ensembl annotated and predicted genes mapped to the region.
Candidate genes encompassed within aberrant loci were selected within the minimum overlapping region and/or top of the amplicons, after reported involvement in cancer and control of cell growth/proliferation. Clone and gene mapping information were obtained from Ensembl (http://ensembl.org/biomart, Homo sapiens genes, National Center for Biotechnology Information Build 36). For loci reported to be aberrant in glioblastoma, at least one reference is listed.
Chromosome 12 was also of particular interest, as many distinctly amplified regions were detected on this autosome. In total, 19 (24%) of the samples presented with high copy number gains on 12q11–21 (Supplementary Fig. 6 and Supplementary Table 7 [online only with this article at neuro-oncology.dukejournals.org]). Amplification of CDK4 locus, identified in 16 cases, was the second most frequent amplification event detected in GB samples. However, amplification of this locus was never identified as a solitary amplicon. In seven cases, simultaneous amplification of CDK4 and MDM2 loci was determined, and in an additional five samples, the CDK4 locus was coamplified with PDG-FRA locus. In two cases, simultaneous amplification of CDK4 and EGFR loci was determined. Furthermore, CDK4 amplicon in the absence of EGFR, PDGFRA, or MDM2 loci amplification was detected in four cases. Several candidate loci were identified in these four cases. In sample G20844, two coamplified loci were detected, one encompassing the MYCL1 gene on 1p34.2, and the other the TERT gene on 5p15.33 (Table 2). In samples G20972 and G21828, only one additional amplicon was present in combination with the CDK4 amplicon. One of them mapped to a gene-rich region on 17q21.33 (sample G20972), including several interesting genes, among them TOB1, which acts as transducer of ERBB2. The other amplicon (sample G21828) was located on 12q13.2 and included DGK4, SILV, CDK2, and RAB5B genes (Table 2). The fourth case, G21864, presented a complex chromosome 22 profile, with multiple amplicons in addition to the CDK4 amplicon. PDGFRA and MDM2 loci amplifications were also relatively common incidents, detected in 11 (13.75%) and 9 (11.25%) cases, respectively. PDGFRA amplification was found both as a solitary amplification event and in combination with other amplicons (Tables 1, 2). In one of the pGB tumors, amplification of PDGFRA locus was detected simultaneously with EGFR amplification. The MDM2 locus was coamplified with CDK4 and/or PDGFRA loci, but not in combination with EGFR locus amplification or with 17p13 deletion. Exclusive amplification of MDM2 locus was not found.
The presence of a unique amplicon per sample, different from EGFR or PDGFRA, was detected in two cases (samples G30296 and G21800). These loci deserve special attention as they represent interesting candidate areas. The region amplified in sample G30296 overlaps with amplifications present in three additional tumors and defined a minimal candidate region on 13q31.3, containing several genes, among them MIRH1, GP6, DCT, TGDS, GPR180, LOC144874, ABCC4, DZIP1, and UGCGL2 (Table 2). Further aberrations of trisomy 1, 7, 9, and 21 as well as monosomy 10, 13, 14, and 22 were determined in this sample. Noteworthy was also the profile exhibited by sample G21800 (Supplementary Fig. 7 [online only with this article at neuro-oncology.dukejournals.org]), in which in addition to trisomy 7 and monosomy 10, a single high copy number amplicon, mapping to 11p15.2 and only 1 Mb in size, was identified. A few genes are located in this region, but SPON1 and RRAS2 represent attractive candidate oncogenes. SPON1 (spondin 1) is an extracellular matrix protein, aberrantly expressed in ovarian carcinoma. RRAS2 is a protooncogene that belongs to the RAB subfamily of small GTPases.
A number of amplicons encompassing a single gene were identified in the tumor set, and most of them are novel (Table 2). Some of the genes mapping to these regions are PPARGC1A (peroxisome proliferative activated receptor, gamma, coactivator 1 alpha) on 4p15.2, HGF (hepatocyte growth factor) on 7q21.1, PALM2 (paralemmin 2) on 9q31.3, CNTN1 (contactin 1) on 12q12 (encoding a neuronal cell adhesion molecule, involved in initiating a NOTCH/DTX1 signaling pathway that promotes oligodendrocyte maturation and myelination),22 NAV3 (neuron navigator 3) on 12q21.2 (gene belonging to the neuron navigator family, encoding a protein involved in the process of neuron growth and regeneration, as well as in neural tumorigenesis),23,24 SYT1 (synaptotagmin I) on 12q21.2 (an integral membrane protein of synaptic vesicles presenting with two protein kinase C-homologous repeats), MUCL1 (mucin-like 1) on 12q13.2, MYCBP2 (MYC binding protein 2) on 13q22.3, ADAMTSL3 (ADAMTS-like 3 precursor) on 15q25.2, TTC28 (tetratricopeptide repeat domain 28) on 22q12.1, and LARGE (like-glycosyltransferase) on 22q12.3. Furthermore, numerous additional amplicons encompassing more than a single gene were determined. Most often, genes reported to be involved in cancer and control of cell growth/proliferation were located at these loci (Table 2). To confirm the copy number ratio observed from aCGH experiments, a subset of candidate genes encompassed within these narrow amplicons was analyzed by qPCR. In general, there was good agreement between relative gene copy number values obtained from 32K array and qPCR (Supplementary Table 6 [online only with this article at neuro-oncology.dukejournals.org]).
Complex Amplifier Genotype Target-Specific Chromosomes in GB
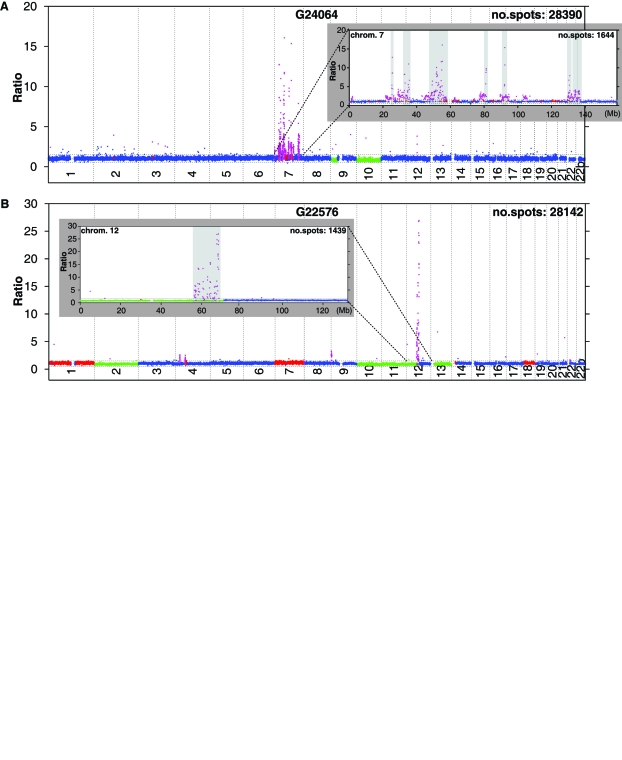
The presence of multiple, narrow, and high copy number level amplicons was an original and recurrent finding in the tumor set. We also discovered specific chromosomes displaying a complex aCGH profile. An outstanding case, in which several independent high copy number amplicons were identified on chromosome 7, in addition to EGFR, is represented in Fig. 2A. The maximum ratio observed in this sample was 16.05, which theoretically indicates 32 gene copies of EGFR locus. The coamplified regions in this tumor mapped to gene rich loci; one of them encompassed one single oncogene, HGF (7q21.11). HGF is a multifunctional growth factor, involved in invasive tumor growth,25 and its overexpression has been associated with poor prognosis of malignant gliomas.26 Other candidate oncogenes include CDK6 (7q21.2), PODXL (7q32.3), and MKLN1(7q32.3). This type of complex amplifier genotype picture, with numerous high and low copy number amplicons, was not exclusive for chromosome 7. In G22576, a complex rearrangement pattern was identified on chromosome 12 (Fig. 2B). The maximum ratio observed in this case was approximately 27, which indicates at least 54 gene copies. This locus encompassed, among others, the MDM2 gene. The same sample presented with several additional amplicons encompassing novel genes on chromosome 12, as well as amplification of CDK4 locus (Table 2). Finally a third tumor, G21864, presented with a similar and complex amplifier genotype, involving chromosome 22; this profile is in agreement with our previous results derived from the analysis of the sample using a chromosome 22–specific genomic array.27 The size and boundaries of overlapping amplified regions detected in these tumors, as well as selected genes contained within the loci, are summarized in Table 2.
Fig. 2.
Identification of multiple, narrow, and high copy number amplicons on chromosomes 7 and 12 in glioblastoma using 32K array. (A) Whole-genome profile of tumor G24064 identifies several narrow amplicons on chromosome 7, in addition to 9p deletion and monosomy 10. (Inset) Chromosome 7 profile reveals several highly amplified regions encompassing candidate oncogene loci. (B) Whole-genome profile of tumor G22576 identifies several independent amplicons on chromosome 12; a narrow amplicon on 9p; two amplified regions on chromosome 4; trisomy 1, 7, and 18; monosomy 2, 10, 11, 13; and partial deletion of 12. (Inset) Chromosome 12 profile reveals several different amplicons encompassing amplified gene loci, including CDK4, RASSF3, and MDM2. Amplicons are indicated in gray bars. The x-axis shows the clone positions for each chromosome, and the y-axis depicts fluorescence ratios. The samples were hybridized against a healthy female reference (F1). Clone mapping information was obtained from Ensembl, and coordinates from human genome assembly of March 2006 (National Center for Biotechnology Information Build 36, hg18) were used.
Novel Homozygously Deleted Loci in GB
aCGH can distinguish between hemizygous (one copy loss) and homozygous (total loss) deletions; the latter are particularly interesting as they can provide an important resource for identifying candidate tumor suppressor genes. Homozygously deleted loci, defined by the presence of at least two consecutive clones with CNC = −2, identified in our tumor series are shown in Table 2. These regions included several previously reported tumor suppressor genes involved in GB tumorigenesis as well as novel genes. The most frequent homozygously deleted locus, detected in 32 samples (41%), mapped to 9p21.3 (Supplementary Fig. 8 [online only with this article at neuro-oncology.dukejournals.org]). The size of the homozygous deletion varied considerably among the tumors (0.53–10.99 Mb), yet in all but one of these samples, the biallelic 9p21.3 deletion encompassed the CDKN2A gene. The only sample presenting with a single homozygous deletion on 9p21.3, not including the CDKN2A gene, was an sGB, G28882. The locus was also encompassed within several larger biallelic deletions found on 9p21-p22 in other cases and was homozygously deleted in one other sGB, sample G21694 (Supplementary Fig. 8). A unique candidate tumor suppressor gene, ELAVL2, resides within this novel region. ELAVL2 encodes a nervous-system–specific RNA- binding protein, implicated in the control of stabilization, nuclear export, and/or translation of specific mRNAs. We also discovered a few additional homozygous deletions on 9p21-p22. In case G20854, the deleted locus encompassed only one gene, ADAMTSL1. The product of this gene belongs to a family of zinc extracellular metalloproteases, with a thrombospondin type 1 motif. Additional tumors presenting with extra homozygously deleted loci on 9p21-p22 were G23282, G153, and G23906, and possible candidate genes mapping to these regions include MLLT3, KIAA1797, PTPLAD2, and KLHL9 (Table 2, Supplementary Table 8, Supplementary Fig. 8). These cases provide strong indications for the presence of additional tumor suppressor genes (other than CDKN2A) at this location.
Hemizygous deletion of chromosome 10 was the most frequent event detected in the series of GBs, while biallelic deletions within this chromosome were not common. Four samples presented with a minimal homozygous deletion on 10q23.31, which encompassed the PTEN gene. The profile of one of these tumors, sample G21830, is shown in Supplementary Fig. 5. PTEN is a tumor suppressor, known to be inactivated in GB and in multiple advanced cancers, involved in the PIK3/AKT pathway and highly implicated in cell death and/or cell cycle arrest. A second tumor suppressor gene locus on chromosome 10 was identified in sample G22520, encompassing the PTPRE and the MGMT genes (Supplementary Fig. 4). Homozygous deletion of this region, 10q26, has not been previously reported. PTPRE is a novel protein tyrosine phosphatase (PTP) located in the plasma membrane. Studies in mice show that PTPRE may have a regulatory role in RAS-related signal transduction pathways and inhibits the mitogen-activated protein kinase cascade.28,29 MGMT is a methylguanine-DNA methyltransferase involved in repairing alkylated guanine in DNA. Promoter methylation and silencing of the MGMT gene compromise DNA repair in the tumor cell and have been associated with longer survival in patients with GB who receive alkylating agents.5 Additional novel homozygous deleted regions were identified on chromosomes 1, 3, 6, 9, and X (Table 2). Three samples displayed homozygous deletion on 1p36. This region encompassed several candidate genes and has been reported as a preferential target region for deletions in astrocytic tumors.30 The loci detected on chromosomes 3 and X are novel and contained a few candidate genes. Loci mapped to 9p23 and 6q27 enclosed one gene each (Table 2). PTPRD at chr9:8.304–10.603 Mb encodes a tyrosine phosphatase, located at the plasma membrane (sample G22370; Supplementary Fig. 8). THBS2 at chr6:169.358–169.396 Mb encodes a polypeptide with a procollagen homology domain and several type I and type III thrombospondin domains. Noteworthy, the telomeric breakpoint of five additional interstitial hemizygous deletions present in other samples mapped to the THBS2 locus. A subset of the candidate tumor suppressor genes (BNC2, ADAMTSL1, PTPLAD2, and PTPRE) encompassed within the homozygously deleted regions was selected for qPCR validation. The relative gene copy number values were confirmed with good agreement (Supplementary Table 6).
Discussion
Regions of gain and loss of genomic DNA occur in many cancers and induce and promote the disease. Using a whole human genome clone-based array, we found a large number of DNA copy number aberrations present in GB. From the recurrence plot and Fisher test, it is obvious that copy number aberrations affecting a whole chromosome (e.g., 10, 7, 22, 19, 20, and 13) as well as partial chromosomal gains or deletions (e.g., 9p, 1p, and 21q) are frequent events. Gain of chromosome 7 and deletion of chromosome 10 were the most common aberrations detected in the series of tumors, results that are in agreement with previous publications.31 Our results also fit the hypothesis that molecular mechanisms target whole or parts of chromosomes in tumors.32,33 This co-occurrence was also indicated by the Spearman’s test. The identification of narrow regions with aberrant DNA copy number is of special interest as the genes mapping within amplicons/homozygously deleted regions represent candidate oncogenes/tumor suppressor genes. Under continuous selection pressure, aberrant regions can become narrower and focused on the genes under selection.34 Owing to the high resolution of the platform, we were able to identify numerous small homozygous deletions as well as amplifications, the majority of which are novel. The presence of numerous and independent amplicons as well as more than one homozygously deleted locus within a single chromosome from the same sample was also observed. aCGH profiles with extremely complex amplifier genotypes were specifically found for chromosomes 7, 12, and 22 (e.g., tumors G24064, G22576, and G21864),27 and several different homozygously deleted loci were detected on chromosome 9 (e.g., tumors G21694, G20854, G23906, and G23282). These findings suggest that local genetic instability also plays an important role in genetic selection of GB.
High copy number amplicons were repeatedly found in GB (81% of cases). The most frequently gained region was EGFR locus (50% of samples), identified as the sole amplification event or coamplified with other loci in the same sample. EGFR amplification is known to occur more frequently in pGB, and mutations of TP53 are more common in sGBs.2 In our series of tumors, we observed amplification of EGFR locus in 37 of 74 pGBs, but also in two of four sGB samples. Furthermore, hemizygous deletion of TP53 locus at 17p13.1 was observed in five samples in addition to the tumor derived from a child (G1143; Supplementary Table 4). Interestingly, all the samples presenting with deletion of 17p13.1 were classified as pGB. Moreover, in one of them amplification of EGFR locus was also observed, which points to the difficulties in differentiating these groups of patients. Highly complex amplifications were also frequent on chromosome 12, results that are in agreement with recent data indicating the presence of stable amplicons on 12q13-21 in glioma.35 MDM2, for example, a powerful oncogene known to be involved in GB development, was highly amplified in nine cases of this series but was never coamplified with the EGFR locus. The MDM2 locus was often coamplified with CDK4, yet the latter aberration was not detected in combination with homozygous deletion of the CDKN2A locus. This reflects the fact that different combinations of aberrations confer the most favorable growth advantage to the individual tumors.
Consistent with the hypothesis that more than one tumor suppressor gene is located on 9p,36,37 we discovered several different loci affected by biallelic deletion on this autosome. Biallelic deletion of ELAVL2, which has previously been reported in pediatric gliomas,38 was detected in 16 cases (Supplementary Fig. 8). ELAVL2 encodes a predicted protein with significant similarity to the product of the Drosophila ELAV gene. Absence of the ELAV gene in Drosophila causes multiple structural defects and hypotrophy of the CNS of the fly. Our results strengthen the notion that ELAV2 may represent a candidate tumor suppressor gene in glioma progression. The identification of several PTPs resident within homozygously deleted loci (PTPRD on 9p23, PTPLAD2 on 9p21, and PTPRE on 10q26.2) was a captivating discovery. PTPs are known to be signaling molecules that regulate a variety of cellular processes, including cell growth, differentiation, mitotic cycle, and oncogenic transformation. Activation of tyrosine kinases is a common feature in several cancer forms, including GB, and recent evidence shows that PTPs can function as tumor suppressors, negatively regulating tyrosine phosphorylation.29 The function of PTPRD, PTPLAD2, and PTPRE has not been completely determined, but they may regulate neurite growth and control the oncogenic activation of tyrosine kinases and RAS-related signal transduction pathways. Additional reports indicate that PTPs not only can serve as tumor suppressors but also can positively regulate the signaling of growth factor receptors.29 Further understanding of how these enzymes function and how they are regulated might help us to develop new anticancer drugs.
Deregulation of proteins of the extracellular matrix was also observed. ADAMTSL3 and ADAMTSL1 were located at amplified (15q25.2) and homozygous deleted (21q22.1-q22) loci, respectively. The proteins encoded by these genes present strong similarity to members of the ADAMTS family (a disintegrin and metalloproteinase with thrombospondin motif). ADAMTS proteins are involved in the regulation of the cleavage of EGF family signal protein precursors, including EGF and tumor growth factor-α (TGF-α), and in the destruction of components of the extracellular matrix, which facilitate metastasis. ADAMTS4 and ADAMTS5, for example, have been reported to be expressed in human GBs.39 ADAMTSL3 and ADAMTSL1 lack the metalloproteinase and disintegrin-like domains typical for ADAMTS family but contain other important motifs. The function of these proteins has not been determined, but they may have important functions in the extracellular matrix. ADAMTSL3, for instance, is a secreted glycoprotein that possesses a PLAC (protease and lacunin) domain, a motif usually found in extracellular matrix protein convertases.40 Moreover, the presence of frequent mutations of ADAMTSL3 has recently been identified in colorectal cancer.41 ADAMTSL1 encodes a secreted protein that contains a thrombospondin type 1 motif. ADAMTSL1 could play a role in the activation of TGF-β, which is usually stored as a latent inactive complex in the extra-cellular matrix, where it gets activated by thrombospondin. THBS2 represents an additional candidate tumor suppressor gene with thrombospondin domains. It was the only gene mapped to a homozygous deletion detected on 6q27. Interestingly, THBS2 has been shown to function as a potent endogenous inhibitor of tumor growth and angiogenesis.42
Recently, two comprehensive genomic analysis of GB have been performed.43,44 The authors identified frequent amplification of EGFR and CDK4, as well as homozygously deletion of CDKN2A and PTEN genes, which is in agreement with our present results as well as previous results.43–45 Rare focal amplifications of MDM4 (mouse double minute 4 homolog), CDK6 (cyclin-dependent kinase 6), MYCN (v-myc myelocytomatosis viral related oncogenes), and AKT3 (v-akt murine thymoma viral oncogene homolog 3) loci, as well as low frequency of biallelic deletion at 1p36.23, 6q27, 9p23, and 10q26.2, q26.3, were also found,44 which represent an independent validation of our data. Recurrent alterations of NF1 (neurofibromin 1) not observed in our study were reported, but they were mainly point mutations.43,44 Alterations in PIK3CA (phosphoinositide-3-kinase, catalytic, alpha) were detected as well.43,44 Interestingly, although the locus was not amplified in the series of GB analyzed here, the gene mapped to an overlapping region of gain on 3q26.32, detected in nine (11.5%) samples. In addition, one of these studies reported mutations in IDH1 (isocitrate dehydrogenase 1) gene in a large fraction of young patients.43 However, copy number aberrations at this gene locus, 2q33.3, were not observed in our study. This discrepancy may be explained by the difference in average age between the patient cohorts.
In conclusion, we have conducted a detailed profiling of a large series of GB samples and provide evidence for the presence of numerous gains and losses of chromosomal regions in this tumor, aberrations that most likely contribute to tumorigenesis by altering gene expression. Recurrent regions affected by copy number alterations were observed; however, numerous and novel unique events that target loci encompassing genes with clear links to cancer were also identified. Moreover, not a single pair of tumors presented with identical genomic profiles, which demonstrates the underlying complexity of the disease. Profiling of tumors is a valuable tool in the identification of specific patients that could benefit from particular treatments. It is clear that a better understanding of the biological significance of genetic differences is crucial in order to improve treatment strategies for these patients.
Supplementary Information
Acknowledgments
We thank Stefan Enroth, Linnaeus Centre for Bioinformatics, for assistance regarding the GO tool within the LCB-DWH and for adaptation to aCGH data.
T.D.S. was supported by the Swedish Children’s Cancer Foundation, the Swedish Cancer Society, and Uppsala University. J.P.D. was supported by the Swedish Cancer Society, the Swedish Children’s Cancer Foundation, and the U.S. Army Medical Research and Materiel Command, award W81XWH-04-1-0269. C.H. and A.D. were supported by the German National Genome Research Network and the German Research Foundation.
References
- 1.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, et al. WHO Classification of Tumours of the Central Nervous System. Lyon, France: International Agency for Research on Cancer; 2007. [Google Scholar]
- 3.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 6.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 7.Mantripragada KK, Buckley PG, Díaz de Ståhl T, Dumanski JP. Genomic microarrays in the spotlight. Trends Genet. 2004;20:87–94. doi: 10.1016/j.tig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 9.Solinas-Toldo S, Lampel S, Stilgenbauer S, et al. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- 10.de Ståhl TD, Sandgren J, Piotrowski A, et al. Profiling of copy number variations (CNVs) in healthy individuals from three ethnic groups using a human genome 32 K BAC-clone-based array. Hum Mutat. 2008;29:398–408. doi: 10.1002/humu.20659. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 12.Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nature Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 13.Fiegler H, Carr P, Douglas EJ, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 14.Buckley PG, Mantripragada KK, Benetkiewicz M, et al. A full- coverage, high-resolution human chromosome 22 genomic microarray for clinical and research applications. Hum Mol Genet. 2002;11:3221–3229. doi: 10.1093/hmg/11.25.3221. [DOI] [PubMed] [Google Scholar]
- 15.Ameur A, Yankovski V, Enroth S, Spjuth O, Komorowski J. The LCB Data Warehouse. Bioinformatics. 2006;22:1024–1026. doi: 10.1093/bioinformatics/btl036. [DOI] [PubMed] [Google Scholar]
- 16.Yang YH, Dudoit S, Luu P, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson R, Bruder CE, Piotrowski A, et al. A segmental maximum a posteriori approach to genome-wide copy number profiling. Bioinformatics. 2008;24:751–758. doi: 10.1093/bioinformatics/btn003. [DOI] [PubMed] [Google Scholar]
- 18.Benjimini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 19.Team RDC. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2006. [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman BT, Huang da W, Tan Q, et al. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu QD, Ang BT, Karsak M, et al. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. 2003;115:163–175. doi: 10.1016/s0092-8674(03)00810-9. [DOI] [PubMed] [Google Scholar]
- 23.Coy JF, Wiemann S, Bechmann I, et al. Pore membrane and/or filament interacting like protein 1 (POMFIL1) is predominantly expressed in the nervous system and encodes different protein isoforms. Gene. 2002;290:73–94. doi: 10.1016/s0378-1119(02)00567-x. [DOI] [PubMed] [Google Scholar]
- 24.Maes T, Barcelo A, Buesa C. Neuron navigator: a human gene family with homology to unc-53, a cell guidance gene from Caenorhabditis elegans. Genomics. 2002;80:21–30. doi: 10.1006/geno.2002.6799. [DOI] [PubMed] [Google Scholar]
- 25.Uchinokura S, Miyata S, Fukushima T, et al. Role of hepatocyte growth factor activator (HGF activator) in invasive growth of human glioblastoma cells in vivo. Int J Cancer. 2006;118:583–592. doi: 10.1002/ijc.21362. [DOI] [PubMed] [Google Scholar]
- 26.Arrieta O, Garcia E, Guevara P, et al. Hepatocyte growth factor is associated with poor prognosis of malignant gliomas and is a predictor for recurrence of meningioma. Cancer. 2002;94:3210–3218. doi: 10.1002/cncr.10594. [DOI] [PubMed] [Google Scholar]
- 27.de Ståhl TD, Hartmann C, de Bustos C, et al. Chromosome 22 tiling-path array-CGH analysis identifies germ-line- and tumor-specific aberrations in patients with glioblastoma multiforme. Genes Chromosomes Cancer. 2005;44:161–169. doi: 10.1002/gcc.20226. [DOI] [PubMed] [Google Scholar]
- 28.Wabakken T, Hauge H, Funderud S, Aasheim HC. Characterization, expression and functional aspects of a novel protein tyrosine phosphatase epsilon isoform. Scand J Immunol. 2002;56:276–285. doi: 10.1046/j.1365-3083.2002.01127.x. [DOI] [PubMed] [Google Scholar]
- 29.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 30.Ichimura K, Vogazianou AP, Liu L, et al. 1p36 is a preferential target of chromosome 1 deletions in astrocytic tumours and homozygously deleted in a subset of glioblastomas. Oncogene. 2007;27:2097–2108. doi: 10.1038/sj.onc.1210848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maher EA, Brennan C, Wen PY, et al. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res. 2006;66:11502–11513. doi: 10.1158/0008-5472.CAN-06-2072. [DOI] [PubMed] [Google Scholar]
- 32.Charames GS, Bapat B. Genomic instability and cancer. Curr Mol Med. 2003;3:589–596. doi: 10.2174/1566524033479456. [DOI] [PubMed] [Google Scholar]
- 33.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 34.Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Fischer U, Keller A, Leidinger P, et al. A different view on DNA amplifications indicates frequent, highly complex, and stable amplicons on 12q13–21 in glioma. Mol Cancer Res. 2008;6:576–584. doi: 10.1158/1541-7786.MCR-07-0283. [DOI] [PubMed] [Google Scholar]
- 36.Perinchery G, Bukurov N, Nakajima K, Chang J, Li LC, Dahiya R. High frequency of deletion on chromosome 9p21 may harbor several tumor-suppressor genes in human prostate cancer. Int J Cancer. 1999;83:610–614. doi: 10.1002/(sici)1097-0215(19991126)83:5<610::aid-ijc7>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Grady B, Goharderakhshan R, Chang J, et al. Frequently deleted loci on chromosome 9 may harbor several tumor suppressor genes in human renal cell carcinoma. J Urol. 2001;166:1088–1092. [PubMed] [Google Scholar]
- 38.Wong KK, Tsang YT, Chang YM, et al. Genome-wide allelic imbalance analysis of pediatric gliomas by single nucleotide polymorphic allele array. Cancer Res. 2006;66:11172–11178. doi: 10.1158/0008-5472.CAN-06-2438. [DOI] [PubMed] [Google Scholar]
- 39.Held-Feindt J, Paredes EB, Blomer U, et al. Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. Int J Cancer. 2006;118:55–61. doi: 10.1002/ijc.21258. [DOI] [PubMed] [Google Scholar]
- 40.Nardi JB, Martos R, Walden KK, Lampe DJ, Robertson HM. Expression of lacunin, a large multidomain extracellular matrix protein, accompanies morphogenesis of epithelial monolayers in Manduca sexta. Insect Biochem Mol Biol. 1999;29:883–897. doi: 10.1016/s0965-1748(99)00064-8. [DOI] [PubMed] [Google Scholar]
- 41.Koo BH, Hurskainen T, Mielke K, et al. ADAMTSL3/punctin-2, a gene frequently mutated in colorectal tumors, is widely expressed in normal and malignant epithelial cells, vascular endothelial cells and other cell types, and its mRNA is reduced in colon cancer. Int J Cancer. 2007;121:1710–1716. doi: 10.1002/ijc.22882. [DOI] [PubMed] [Google Scholar]
- 42.Streit M, Riccardi L, Velasco P, et al. Thrombospondin-2: a potent endogenous inhibitor of tumor growth and angiogenesis. Proc Natl Acad Sci U S A. 1999;96:14888–14893. doi: 10.1073/pnas.96.26.14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mischel PS, Nelson SF, Cloughesy TF. Molecular analysis of glioblastoma: pathway profiling and its implications for patient therapy. Cancer Biol Ther. 2003;2:242–247. doi: 10.4161/cbt.2.3.369. [DOI] [PubMed] [Google Scholar]
- 46.Nigro JM, Misra A, Zhang L, et al. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res. 2005;65:1678–186. doi: 10.1158/0008-5472.CAN-04-2921. [DOI] [PubMed] [Google Scholar]
- 47.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 48.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. article 3. [DOI] [PubMed] [Google Scholar]
- 49.Lonnstedt I, Britton T. Hierarchical Bayes models for cDNA microarray gene expression. Biostatistics. 2005;6:279–291. doi: 10.1093/biostatistics/kxi009. [DOI] [PubMed] [Google Scholar]
- 50.Mulholland PJ, Fiegler H, Mazzanti C, et al. Genomic profiling identifies discrete deletions associated with translocations in glioblastoma multiforme. Cell Cycle. 2006;5:783–691. doi: 10.4161/cc.5.7.2631. [DOI] [PubMed] [Google Scholar]
- 51.Misra A, Pellarin M, Nigro J, et al. Array comparative genomic hybridization identifies genetic subgroups in grade 4 human astrocytoma. Clin Cancer Res. 2005;11:2907–2918. doi: 10.1158/1078-0432.CCR-04-0708. [DOI] [PubMed] [Google Scholar]
- 52.Lo KC, Rossi MR, LaDuca J, et al. Candidate glioblastoma development gene identification using concordance between copy number abnormalities and gene expression level changes. Genes Chromosomes Cancer. 2007;46:875–894. doi: 10.1002/gcc.20474. [DOI] [PubMed] [Google Scholar]
- 53.Liu F, Park PJ, Lai W, et al. A genome-wide screen reveals functional gene clusters in the cancer genome and identifies EphA2 as a mitogen in glioblastoma. Cancer Res. 2006;66:10815–10823. doi: 10.1158/0008-5472.CAN-06-1408. [DOI] [PubMed] [Google Scholar]
- 54.Korshunov A, Sycheva R, Golanov A. Genetically distinct and clinically relevant subtypes of glioblastoma defined by array-based comparative genomic hybridization (array-CGH) Acta Neuropathol (Berl) 2006;111:465–474. doi: 10.1007/s00401-006-0057-9. [DOI] [PubMed] [Google Scholar]
- 55.Lukashova-v Zangen I, Kneitz S, Monoranu CM, et al. Ependymoma gene expression profiles associated with histological subtype, proliferation, and patient survival. Acta Neuropathol. 2007;113:325–337. doi: 10.1007/s00401-006-0190-5. [DOI] [PubMed] [Google Scholar]
- 56.Balenci L, Clarke ID, Dirks PB, et al. IQGAP1 protein specifies amplifying cancer cells in glioblastoma multiforme. Cancer Res. 2006;66:9074–9082. doi: 10.1158/0008-5472.CAN-06-0761. [DOI] [PubMed] [Google Scholar]
- 57.McDonald KL, O’Sullivan MG, Parkinson JF, et al. IQGAP1 and IGFBP2: valuable biomarkers for determining prognosis in glioma patients. J Neuropathol Exp Neurol. 2007;66:405–417. doi: 10.1097/nen.0b013e31804567d7. [DOI] [PubMed] [Google Scholar]
- 58.Marie SK, Okamoto OK, Uno M, et al. Maternal embryonic leucine zipper kinase transcript abundance correlates with malignancy grade in human astrocytomas. Int J Cancer. 2007;122:807–815. doi: 10.1002/ijc.23189. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki T, Maruno M, Wada K, et al. Genetic analysis of human glioblastomas using a genomic microarray system. Brain Tumor Pathol. 2004;21:27–34. doi: 10.1007/BF02482174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.