Abstract
Mechanisms regulating CYP4F genes remain under investigation, although characterization of CYP4F regulatory modalities would facilitate the discovery of new drug targets. This present study shows that all-trans- and 9-cis-retinoic acids can inhibit CYP4F11 expression in human keratinocyte-derived HaCaT cells. Transrepression of many genes by retinoic acids is mediated by interactions between retinoid receptors and the activator protein 1 (AP-1) complex. Proinflammatory cytokines tumor necrosis factor α (TNF-α) and interleukin 1β, which can activate the AP-1 complex, induce CYP4F11 transcription in HaCaT cells. The c-Jun N-terminal kinase (JNK)-specific inhibitor 1,9-pyrazoloanthrone (SP600125) blocked the induction of CYP4F11 by both cytokines, indicating involvement of the JNK pathway. Furthermore, TNF-α failed to induce CYP4F11 transcription when HaCaT cells were preincubated with retinoic acids. Retinoic acids are ligands for the retinoic acid receptors (RARs) and the retinoid X receptors (RXRs). The RXR agonist 6-(1(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)cyclopropyl) nicotinic acid (LG268) greatly induced CYP4F11 transcription, whereas the RAR agonist 4-(2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl)benzoic acid (TTNPB) markedly inhibited CYP4F11 transcription, indicating that down-regulation of CYP4F11 transcription by retinoic acid is mediated by RARs and may also be related to ligand competition for RXRs. Thus, the CYP4F11 gene is positively regulated by multiple signaling pathways in HaCaT keratinocytes, including RXR and JNK signaling pathways.
Cytochrome P450 4F enzymes are involved in cellular protection and metabolism of numerous small molecules, including drugs, toxins, and eicosanoids. To date, studies have been focused on CYP4F functions rather than transcriptional regulatory mechanisms. This approach is partially because of the apparent complexity of CYP4F gene regulation and differences observed among the various model systems studied.
Despite the seeming complexity of CYP4F regulation, some research groups have made substantive contributions to this puzzle. Zhang and colleagues showed that retinoic acids and peroxisome proliferators can regulate CYP4F2 gene activities in HepG2 cells, and that retinoid X receptor (RXR) α heterodimers stimulated and retinoic acid receptor (RAR) α repressed CYP4F2 expression (Zhang and Hardwick, 2000; Zhang et al., 2000; Hsu et al., 2007) showed that statins induced CYP4F2 in primary human hepatocytes and HepG2 cells. We recently showed that retinoic acids induced the expression of CYP4F2, CYP4F3A, CYP4F3B, and CYP4F11 in primary human epidermal keratinocytes, and that RXR (but not RAR) nuclear receptors mediate retinoic acid-induced up-regulation of CYP4F2 and CYP4F3A (Kalsotra et al., 2008). In a preliminary study (data not shown) using a transformed immortal human keratinocyte cell line (HaCaT), a different story unfolded. Lovastatin did not affect HaCaT cell CYP4F2 expression, which is in contrast to its up-regulatory effects on CYP4F2 in human hepatic cells (Hsu et al., 2007). In addition, 9-cis-retinoic acid failed to induce CYP4F11 in HaCaT cells; rather, it significantly decreased CYP4F11 transcript levels.
Retinoic acids are derived from retinol, the animal form of vitamin A. Retinoic acids play a role in gene regulation via two groups of nuclear receptors (NRs), RARs and RXRs (Mangelsdorf et al., 1995), each of which consists of three isotypes: α, β, and γ (Mangelsdorf et al., 1990; Leid et al., 1993). Retinoic acids can up-regulate or down-regulate certain genes by three mechanisms: transactivation, transrepression, and competition with other NRs for heterodimerization with RXR. Transrepression by retinoic acids is the cross-talk between retinoid receptors and the activator protein 1 (AP-1) complex (Saatcioglu et al., 1994; Chen et al., 1995). Cytokines, oncogenes, and growth factors that activate protein kinase C modulate the activities of the AP-1 complex composed of c-Jun and c-Fos proteins (Zhang and Pfahl, 1993). Active RAR-RXR heterodimers interact with AP-1 complexes, thereby inhibiting the transcription of genes regulated by AP-1 (Fisher et al., 1998). Transrepression of AP-1 is a major mechanism of the pharmacological anti-inflammatory and antiproliferative effects of retinoids (Fisher and Voorhees, 1996).
RXR can form heterodimers with other NRs, including peroxisome proliferator-activated receptors (Green, 1995; Qi et al., 2000; Lazar, 2005), farnesoid X receptor (Laffitte et al., 2000; Desvergne, 2007), liver X receptors (Willy et al., 1995; Willy and Mangelsdorf, 1997; Desvergne, 2007), thyroid receptor (Apriletti et al., 1998; Barra et al., 2004), and NR-related 1 receptor (Perlmann and Jansson, 1995; Perlmann and Wallén-Mackenzie, 2004). RXRs can also form homodimers under certain conditions (Zhang and Pfahl, 1993). Thus, when an RAR-specific agonist stimulates the interaction of RXR with RARs, it can act to negatively regulate the activity of other NRs that partner with RXR in favor of RAR-mediated responses (Heinz Nau, 2007). The relative abundance of classic retinoid receptors and other RXR-interacting NRs, which have varying affinities for RXR, influences RXR dimerization and thereby gene regulation mediated by these NRs (Fisher and Voorhees, 1996). The aim of the current study is to clarify the mechanism and factors responsible for the down-regulation of HaCaT CYP4F11 expression by retinoic acids.
Materials and Methods
Chemicals.
The 9-cis- and all-trans-retinoic acids, 4-(2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl)benzoic acid (TTNPB), and 12-O-tetradecanoyl-phorbol-13-acetate (TPA) were from Sigma-Aldrich (St. Louis, MO). 6-(1(3,5,5,8,8-Pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl) cyclopropyl) nicotinic acid (LG268) was a gift from Dr. Peter J. A. Davies (University of Texas-Houston). Human tumor necrosis factor α (TNF-α) and interleukin 1β (IL-1β) were from Invitrogen (Carlsbad, CA). SP600125 and SB203580 were produced by Calbiochem (San Diego, CA). Optimal concentrations of drugs were chosen according to literature citations. Polyclonal anti-CYP4F11 antibody was from Proteintech Group, Inc. (Chicago, IL). Polyclonal antibodies against human RARs and RXRs were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Monoclonal c-Jun and phosphor-c-Jun antibodies were from Cell Signaling Technology Inc. (Danvers, MA). All of the other chemicals used were of reagent grade quality or higher.
Cell Culture.
The HaCaT cell line was from CLS–Cell Line Services (Heidelberg, Germany) (Boukamp et al., 1988). Cells were grown at 37°C in a humidified incubator with 5% CO2 in high glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Atlas Biologicals, Fort Collins, CO), 1% l-glutamine (Invitrogen), 1% sodium pyruvate (Invitrogen), and 1% antibiotic-antimycotic (Invitrogen). Cells were passaged at a 1:10 ratio after they had been 100% confluent for 6 to 7 days and used for experiments at 70 to 80% confluency.
Protein Isolation and Western Blot Analyses.
Cells were washed once with cold phosphate-buffered saline and then quickly detached by gently scraping. Cell pellets were collected by centrifugation for 2 min at 4500 rpm and stored at −80°C. Whole protein lysate buffer was prepared using 50 mM Tris-HCl buffer, pH 6.8, with 2% SDS. Cell pellets were dissolved in the lysate buffer by pipetting the suspension up and down and then boiled at 95°C for 5 min. The cell lysate was cleared by centrifugation at 13,000 rpm for 2 min. Protein concentrations of the clear supernatant fractions were measured using the bicinchoninic acid assay kit (Pierce, Rockford, IL) and subjected to Western blot assays.
Protein samples were boiled in Laemmli buffer and resolved electrophoretically on 4 to 15% gradient Tris-glycine-SDS-polyacrylamide gels (Bio-Rad Laboratories, Hercules, CA). Proteins were transferred to polyvinylidene fluoride membranes (wet transfer apparatus; Bio-Rad Laboratories). Membranes were blocked for 1 h at room temperature with 5% dried nonfat milk followed by incubation for 3 h at room temperature with antibodies raised against CYP4F11 (1:1000 dilution) or c-Jun (1:500 dilution). Membranes were then washed and incubated at room temperature with horseradish peroxidase-conjugated secondary antibody (1:2000 dilution) for 1 h. Immunoreactivity was visualized using a horseradish peroxidase chemiluminescence system (Pierce).
Quantitative Real-Time Polymerase Chain Reaction.
Cells were washed with phosphate-buffered saline, and total RNA was isolated using TRIzol reagent (Invitrogen). All the samples were treated with DNase I (Invitrogen). Aliquots (200 ng) of total RNA were reverse-transcribed by SuperScript II Reverse Transcriptase (Invitrogen) in triplicate, including a reverse transcription blank to evaluate presence of contaminating genomic DNA. Amplification was performed using Taq DNA Polymerase (Invitrogen) with an ABI Prism 7700 (Applied Biosystems, Foster City, CA) at 95°C for 1 min, followed by 40 cycles at 95°C for 12 s and 60°C for 30 s. CYP4F11 mRNA levels were measured using standard curves generated by plotting Ct versus the log of the amount of purified amplicon for CYP4F11 (custom synthesis by Integrated DNA Technologies, Inc., Coralville, IA) (200 ag–2 pg). Abundance of human 18S RNA was used as the internal control. Primers and fluorescent probe sequences for CYP4F11 and 18S RNA were reported (Kalsotra et al., 2008).
Reverse Transcriptase-Polymerase Chain Reaction.
Aliquots (200 ng) of total RNA were reverse-transcribed and amplified using Superscript One-Step reverse transcriptase-polymerase chain reaction (RT-PCR) kit (Invitrogen) and transcript-specific oligonucleotides designed using Primer3 (version 0.4.0; http://frodo.wi.mit.edu/primer3/) (Table 1). For RAR and RXR, the reverse-transcription reaction (50°C, 30 min) was followed by 40 cycles of amplification (94°C, 15 s; 60°C, 30 s; and 72°C, 1 min). For CYP4F11, the reverse-transcription reaction (60°C, 30 min) was followed by 40 cycles of amplification (94°C, 1 min and 65°C, 1.5 min). Products were resolved on 1.5% agarose gels and visualized with ethidium bromide.
TABLE 1.
Sequences of primers for RARs, RXRs, and CYP4F11 for RT-PCR
| Transcript | Oligonucleotide Sequences (5′-3′) | Product Size/Annealing Temperature |
|---|---|---|
| RAR-α | Forward GGCATGTCCAAGGAGTCTGT | 420 bp/60°C |
| Reverse GTCCGAGAAGGTCATGGTGT | ||
| RAR-β | Forward GAGAGGTGGCATTGATCCAGG | 435 bp/62°C |
| Reverse GGCCTGGGCCAGCCTGACCTC | ||
| RAR-γ | Forward TCGAGATGCTGAGCCCTAGCTTCC | 351 bp/58°C |
| Reverse CATGCCCACTTCGAAGCACTTCTGT | ||
| RXR-α | Forward AATGAGGTGGAGTCGACCAG | 400 bp/60°C |
| Reverse TCAGCACCCTGTCAAAGATG | ||
| RXR-β | Forward GGACAGAAGCTCAGGCAAAC | 540 bp/60°C |
| Reverse AATGGATCGGTGTGAGAAGG | ||
| RXR-γ | Forward TGTGGTCAACAGTGTCAGCA | 392 bp/60°C |
| Reverse TCTTGCACAGCTTCCCTCTT | ||
| CYP4F11 | Forward GCCTCAGGATCCCACCCTCCAT | 290 bp/65°C |
| Reverse ATGTGGTCACCAGCTGGGTCAATGT |
bp, base pair.
Plasmids.
To construct pGL3-CYP4F11 plasmid, CYP4F11 promoter was cloned by PCR using human genomic DNA as a template with the forward primer 5′-TCT TAC GCG TGC TAG CTA CCC AGC ACC CAG AGT AGG-3′ and the reverse primer 5′-CCG GAA TGC CAA GCT TGC TCC AAG GAC AGT GGA AAG-3′. The resulting 1729-base pair product was the promoter region of CYP4F11 (GenBank accession no. NG_0008335) including exon 1 as the coding start site in exon 2. The PCR product was then cloned into HindIII- and NheI-digested linear pGL3 Basic luciferase vector (Promega, Madison, WI) using the infusion Advantage PCR Cloning Kit (Clontech, Mountain View, CA).
Transfection and Luciferase Assays.
Cells were transfected by Attractene Transfection Reagent (QIAGEN, Valencia, CA) according to the manufacturer's protocol. Cells were seeded onto six-well plates (3 × 105 cells/well). Immediately after the seeding, cells were transfected with 4.5 μl of Attractene/well and 1.2 μg of DNA/well. Transfection efficiency was monitored by simultaneous cotransfection with a β-galactosidase reporter construct at the concentration of 20 ng/well. The culture medium was changed at 6 h post-transfection. Twenty-four hours after transfection, cells were treated with agonists for RARs or RXRs with or without TNF-α. After the treatment, cells were lysed in passive lysis buffer (Promega). Luciferase activity was measured for 10 s in a luminometer. β-Galactosidase activity was measured to normalize for variations in transfection efficiency. Promoter activity of each construct was expressed as firefly luciferase/β-galactosidase activity. Each experiment was independently performed on three separate occasions with at least triplicate wells in each experiment.
Statistical Analysis.
Data are presented as mean ± S.E. One-way analysis of variance followed by Tukey's multiple comparison test was used for the statistical analysis. Statistical differences were considered significant if P < 0.05.
Database Sequence Analysis.
The TRANSFAC database (BIOBASE, Beverly, MA) was searched using AliBaba 2.1 (BIOBASE) for putative AP-1 binding sites and hormone responsive elements on the 5′ flanking sequence (2000 base pairs from the ATG start codon) of CYP4F11 gene.
Results
Expression of Retinoid Receptors in HaCaT Cells and Inhibitory Effects of Retinoic Acids on CYP4F11 Expression.
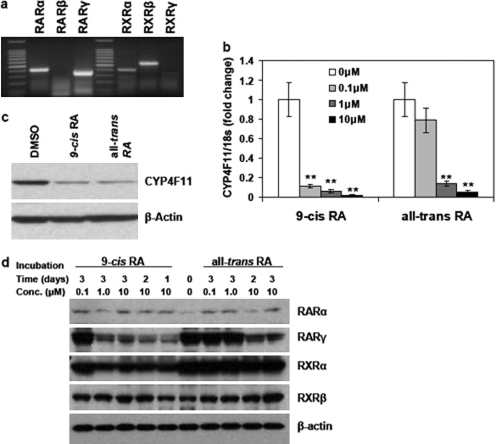
We first determined which RXR and RAR isoforms are expressed in HaCaT cells. Results by RT-PCR show that HaCaT cells express RARα, RARγ, RXRα, and RXRβ, whereas neither RARβ nor RXRγ was detectable (Fig. 1a). To elucidate mechanisms by which retinoic acids regulate CYP4F11 expression, we treated HaCaT cells with 9-cis- and all-trans-retinoic acids for 3 days at 0.1, 1, and 10 μM concentrations or vehicle [dimethyl sulfoxide (DMSO)] alone. Both retinoic acids resulted in dose-dependent suppression of CYP4F11 transcript levels in HaCaT cells, measured by quantitative real-time polymerase chain reaction (qRT-PCR) (Fig. 1b). The reductions in CYP4F11 mRNA at the 0.1, 1, and 10 μM dose levels were 89.1, 93.9, and 98.3% for 9-cis-retinoic acid and 21.1, 86.5, and 94.8% for all-trans-retinoic acid, respectively. Data from luciferase assay indicated that at the concentration of 1 μM, 9-cis- and all-trans-retinoic acids were able to decrease CYP4F11 promoter activity by 83.0 and 70.9%, respectively (Supplemental Fig. 1). Furthermore, Western blot analyses showed the down-regulation of CYP4F11 protein by both retinoic acids at 10 μM (Fig. 1c). Western blots against RARα, RARγ, RXRα, and RXRβ were performed to study the potential effects of retinoic acids on the expression levels of retinoid receptors. As shown in Fig. 1d, cells treated with high concentrations (1 and 10 μM) of 9-cis-retinoic acid had reduced levels of RARγ and RXRα. Cells treated with 10 μM all-trans-retinoic acid showed a lower level of RARγ. In contrast, 3-day treatment of 0.1 μM 9-cis-retinoic acid, 0.1 μM all-trans-retinoic acid, or 1 μM all-trans-retinoic acid did not affect the protein level of RARγ and RXRα. The data also showed that retinoic acid treatments could not decrease the protein levels of RARα and RXRβ.
Fig. 1.
Expression of RARs and RXRs in HaCaT cells and inhibition of CYP4F11 expression by retinoic acids. a, expression of RARs and RXRs in HaCaT cells. Total RNA was isolated from HaCaT cells, and an aliquot of 200 ng of the total RNA was used to run RT-PCR assay with primers specific for each RAR and RXR isoform. The size of each PCR product is indicated in Table 1. b, inhibition of CYP4F11 transcripts by retinoic acids. Cells were treated with 0.1, 1, or 10 μM 9-cis- or all-trans-retinoic acid for 3 days. Cells treated with 0.1% DMSO were used as the vehicle control. The expression of CYP4F11 was quantitated by qRT-PCR. Each data point represents n = 5. ∗∗, P < 0.01 compared with the relative control group. c, inhibition of CYP4F11 protein expression by retinoic acids. Cells were treated with 10 μM 9-cis- or all-trans-retinoic acid for 3 days. Whole cell lysates were isolated, and totally 30 μg of protein from each sample was subjected to Western blot assay using CYP4F11 antibody. The amount of β-actin expression from each sample was used as the loading control. d, time course and dose effects of retinoic acids on the protein expression of retinoid receptors. Cells were treated with 0.1, 1, or 10 μM 9-cis- or all-trans-retinoic acid and collected at different time points as indicated in the figure. Cells treated with 0.1% DMSO were used as the vehicle control. Whole cell lysates were isolated, and totally 20 μg of protein from each sample was subjected to Western blot assay using antibodies specific for RARα, RARγ, RXRα, and RXRβ, respectively. The amount of β-actin expression from each sample was used as the loading control.
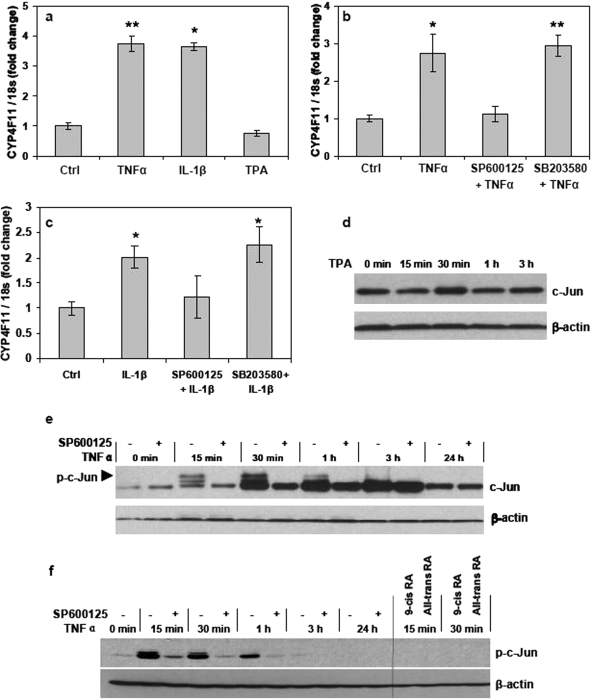
TNF-α and IL-1β, but Not TPA, Can Induce CYP4F11 mRNA Levels in HaCaT Cells through the c-Jun N-Terminal Kinase Pathway.
We investigated the ability of TNF-α and IL-1β or TPA to induce CYP4F transcription in HaCaT cells through mechanisms involving AP-1, as reported for other cell systems (Lamph et al., 1988; Sassone-Corsi et al., 1990; Chedid et al., 1991; Kyriakis, 1999; Yu et al., 2008). We treated HaCaT cells with TNF-α (10 ng/ml) and IL-1β (10 ng/ml) or TPA (100 ng/ml) for 24 h. Both cytokines induced CYP4F11 mRNA levels significantly, whereas TPA had no effect compared with the vehicle control (Fig. 2a). Luciferase assay also indicated the induction of CYP4F11 promoter activity by the treatment of 10 ng/ml TNF-α for 24 h (Supplemental Fig. 1). Cytokines and many other factors activate the AP-1 complex through multiple signaling pathways such as the c-Jun N-terminal kinase (JNK), P38, and extracellular signal-regulated kinase pathways (Whitmarsh and Davis, 1996). Hence, we preincubated HaCaT cells with the JNK inhibitor SP600125 or the p38 inhibitor SB203580 for 4 h before administering TNF-α or IL-1β. Inhibiting JNK completely blocked the cytokine-induced CYP4F11 transcription (Fig. 2, b and c). Inhibiting p38 had no effect on CYP4F11 expression. After administering TNF-α, phosphorylated c-Jun was observed within 15 min; it was and remained elevated for at least 3 h (Fig. 2, e and f). Inhibiting JNK by SP600125 pretreatment markedly blocked the phosphorylation of c-Jun protein in HaCaT cells (Fig. 2, e and f). In contrast, TPA (100 ng/ml) had no effect on c-Jun phosphorylation (Fig. 2d). Cells were also preincubated with 1 μM 9-cis- or all-trans-retinoic acid for 24 h and then treated with TNF-α. As shown in Fig. 2f, both retinoic acids completely blocked the phosphorylation of c-Jun at 15 and 30 min post-TNF-α administration.
Fig. 2.
Induction of CYP4F11 by cytokines through the JNK pathway. a, effects of cytokines and TPA on CYP4F11 transcripts in HaCaT cells. Cells were treated with 10 ng/ml TNF-α, 10 ng/ml IL-1β, or 100 ng/ml TPA for 24 h. Cells treated with 0.1% DMSO or 0.1% bovine serum albumin were used as the vehicle control. The expression of CYP4F11 was quantitated by qRT-PCR. Each data point represents n = 5. ∗, P < 0.05, ∗∗, P < 0.01 compared with the control group. b and c, effects of pathway inhibitors on the induction of CYP4F11 by cytokines. HaCaT cells preincubated with or without SP600125 (JNK inhibitor) or SB203580 (p38 inhibitor) were treated with 10 ng/ml TNF-α (b) or 10 ng/ml IL-1β (c). At 24 h post-treatment, total RNA was isolated, and the expression of CYP4F11 was quantitated by qRT-PCR. Cells preincubated with 0.1% DMSO and then treated with 0.1% bovine serum albumin were used as the vehicle control. Each data point represents n = 5. ∗∗, P < 0.01 compared with the relative control group. d, TPA had no effect on c-Jun phosphorylation in HaCaT cells. Cells were treated with 100 ng/ml TPA and collected at different time points as indicated in the figure. Whole cell lysate was isolated from each sample, and 20 μg of proteins from each sample was loaded for Western blot assay. β-Actin was used as the loading control. e, effects of TNF-α on c-Jun phosphorylation in HaCaT cells. Cells preincubated with or without 20 μM SP600125 were treated with 10 ng/ml TNF-α and collected at different time points as indicated in the figure. Whole cell lysate was isolated from each sample, and 15 μg of proteins from each sample was loaded for Western blot assay. β-Actin was used as the loading control. f, inhibition effects of SP600125, 9-cis-retinoic acid, and all-trans-retinoic acid on TNF-α-induced c-Jun phosphorylation in HaCaT cells. Cells preincubated with or without 20 μM SP600125, 1 μM 9-cis-retinoic acid, or 1 μM all-trans-retinoic acid were treated with 10 ng/ml TNF-α and collected at different time points as indicated in the figure. Whole cell lysate was isolated from each sample, and 15 μg of proteins from each sample was loaded for Western blot assay. β-Actin was used as the loading control.
JNK Pathway Is Not the Sole Mechanism Mediating CYP4F11 Regulation.
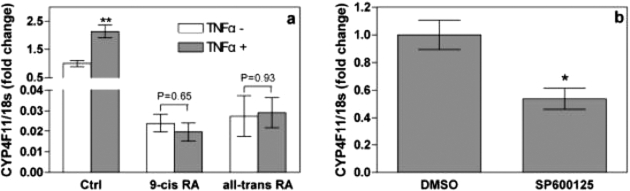
To determine whether the JNK pathway is the primary mechanism regulating CYP4F11 expression, we treated HaCaT cells with cytokine TNF-α (10 ng/ml) for 24 h after a 3-day pretreatment with 9-cis- or all-trans-retinoic acid at 10 μM concentration. TNF-α alone increased CYP4F11 mRNA levels, and pretreatment with either retinoic acid blocked this induction (Fig. 3a). Hence, TNF-α could not reverse the inhibitory effects of retinoic acids on CYP4F11 transcription. When we treated HaCaT cells with the JNK inhibitor SP600125 alone (20 μM for 24 h), CYP4F11 mRNA levels decreased by only 50% (Fig. 3b). At this concentration, SP600125 completely blocked the TNF-α-induced CYP4F11 transcription (Fig. 2b). Inhibiting the JNK pathway did not totally suppress the transcription of CYP4F11, indicating other signaling pathways positively regulate CYP4F11 expression in HaCaT cells.
Fig. 3.
JNK pathway is not the sole mechanism mediating CYP4F11 regulation. a, effects of TNF-α on cells treated with retinoic acids. Cells preincubated with or without 10 μM 9-cis- or all-trans-retinoic acid were treated with 10 ng/ml TNF-α for 24 h. The expression of CYP4F11 was quantitated by qRT-PCR. Each data point represents n = 5. ∗, P < 0.05 compared with the negative control. b, effects of JNK inhibitor SP600125 on CYP4F11 transcription in HaCaT cells. Cells were treated with 0.1% DMSO (vehicle control) or 20 μM SP600125 for 24 h. The expression of CYP4F11 was quantitated by qRT-PCR. Each data point represents n = 6. ∗, P < 0.05 compared with the control group.
Opposing Effects of RAR and RXR Agonists on CYP4F11 Expression in HaCaT Cells.
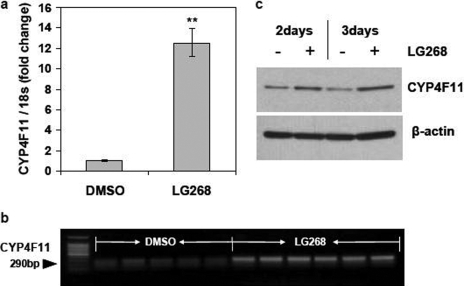
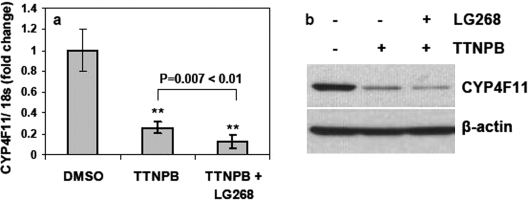
Because retinoic acids activate both RARs and RXRs, we investigated which NR(s) mediate the inhibitory effects of retinoic acids on CYP4F11 expression. We treated HaCaT cells for 2 or 3 days with the pan agonists for RXR (1 μM LG268, also known as LG100268) and RAR (10 μM TTNPB). The RXR pan agonist LG268 increased CYP4F11 mRNA levels 12-fold versus vehicle (Fig. 4a). We confirmed this induction by visualizing RT-PCR products that were amplified using different primer sequences (Fig. 4b) and by Western blot analyses (Fig. 4c). CYP4F11 protein levels were greater in cells treated with LG268 for 2 and 3 days compared with vehicle control. In HaCaT cells treated with the RAR pan agonist TTNPB, CYP4F11 mRNA levels were greatly decreased versus vehicle, and combined treatment with both pan agonists for 3 days decreased CYP4F11 mRNA levels even further (Fig. 5a). Western blot analyses confirmed the markedly decreased CYP4F11 protein levels elicited by TTNPB treatment with or without LG268 (Fig. 5b).
Fig. 4.
Effects of LG268 on CYP4F11 expression in HaCaT cells. a and b, effects of LG268 on the transcription of CYP4F11. HaCaT cells were treated with DMSO (vehicle control, n = 5) or 1 μM LG268 (n = 6) for 3 days. The CYP4F11 mRNA level was quantitated by qRT-PCR (a). ∗∗, P < 0.01 compared with the control group. Aliquots of 200 ng of total RNA from each sample were used to run RT-PCR (b) with primers specific for CYP4F11. c, effects of LG268 on expression of CYP4F11 protein. HaCaT cells were treated with or without 1 μM LG268 for 2 or 3 days. Cells treated with 0.1% DMSO were used as the negative control. Whole cell lysate was isolated, and 20 μg of total protein from each sample was loaded to run Western blot assay. β-Actin was used as the loading control.
Fig. 5.
Effects of TTNPB on CYP4F11 expression in HaCaT cells. HaCaT cells were treated with 0.1% DMSO, 1 μM TTNPB, or 1 μM TTNPB plus 1 μM LG268 for 3 days. a, the CYP4F11 mRNA level was quantitated by qRT-PCR. Each data point represents n = 6. ∗∗, P < 0.01 compared with the control group. b, whole cell lysate was isolated, and 30 μg of total protein from each sample was loaded to run Western blot assay. β-Actin was used as the loading control.
Discussion
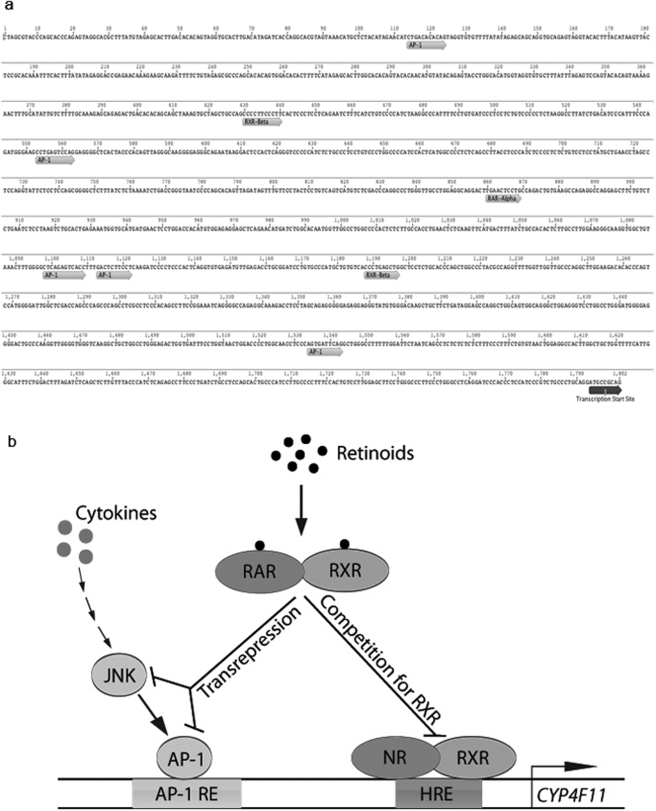
CYP4Fs catalyze the metabolism of both endogenous and exogenous molecules (Kalsotra and Strobel, 2006). For example, they inactivate the leukotriene and prostaglandin prompts for the inflammation cascade playing an anti-inflammatory role (Kikuta et al., 2002), and they also catalyze the metabolism of many drugs (Hashizume et al., 2002; Kalsotra et al., 2004). Among the human CYP4F enzymes, CYP4F11 is most active in metabolizing therapeutic drugs. To elucidate the role of CYP4F11, it is necessary to decipher the mechanism of CYP4F11 gene regulation under various physiological and pathological conditions. The results of our present study have shown that several distinct but interacting pathways of gene regulation impinge on the regulatory region of the CYP4F11 gene in HaCaT cells. We report for the first time that two possible pathways, the JNK/AP-1 pathway and the RXR-mediated pathway, are involved in the regulation of CYP4F11 in HaCaT cells. These pathways and their interactions are depicted in the scheme as shown in Fig. 6b.
Fig. 6.
Scheme for CYP4F11 regulation. a, putative hormone responsive element and AP-1 binding sites in the promoter region of CYP4F11 gene. Sites shown were predicted using AliBaba2.1 (BIOBASE). Figure was generated using Biomatters Geneious Pro 4.5.3 software (http://www.biomatters.com/). b, CYP4F11 regulation model. The JNK/AP-1 pathway and the RAR/RXR pathway modulate the transcription of CYP4F11 in HaCaT cells. Adobe (San Jose, CA) Illustrator CS4 was used to draw this model.
Our present study shows that retinoids down-regulate CYP4F11 expression in HaCaT cells. The anti-AP-1 activity of retinoids supports the positive regulation of CYP4F11 through the AP-1 complex by inflammatory cytokines TNF-α and IL-1β. Moreover, this induction of CYP4F11 was shown to be mediated by the JNK pathway as indicated by the loss of cytokine induction of CYP4F11, as well as c-Jun phosphorylation in the presence of JNK inhibitor SP600125. TPA had no effect on CYP4F11 mRNA levels in HaCaT, which is consistent with previous findings that TPA did not induce JNK activity in HaCaT cells (Zhou et al., 2000). CYP4F11 expression was strongly induced by addition of the cytokines TNF-α and IL-1β but not TPA, indicating the efficacy of AP-1 sites in regulation of CYP4F11 mRNA expression. The blockage of TNF-α-induced CYP4F11 expression and c-Jun phosphorylation by 9-cis- or all-trans-retinoic acid confirms the role of AP-1 transrepression in down-regulating CYP4F11. Furthermore, five AP-1 sites were found using AliBaba2.1 program (BIOBASE) within the first 2000 bases upstream of the start site for CYP4F11 (Fig. 6a). These results support transrepression of AP-1 complexes as a mechanism whereby retinoic acids suppress CYP4F11 transcription, as shown for other gene systems (Saatcioglu et al., 1994; Chen et al., 1995).
The transrepression mechanism proposed here is not adequate to explain the findings that 9-cis- or all-trans-retinoic acid almost completely eliminated CYP4F11 expression, whereas SP600125 alone only showed moderate inhibition, although SP600125 preincubation could completely block the induction of CYP4F11 by TNF-α. It appears that the inducible regulation is different from the basal regulation through JNK activity. As also proposed in the scheme of Fig. 6b, retinoic acids bind to retinoid receptors and activate the formation of RAR/RXR heterodimers. RXRs, in conjunction with one or more other NRs, are proposed to stimulate CYP4F11 transcription by acting through the hormone responsive element. On the other hand, the formation of RAR/RXR heterodimers reduces the number of RXRs available for interaction with NRs, thereby suppressing the transcription of CYP4F11 through competition for RXRs. Thus, retinoid treatment of proliferating HaCaT cells leads to reduced expression of CYP4F11 through transrepression of the AP-1 pathway and/or by competition for RXRs. RAR/RXR heterodimers lead to down-regulation, whereas NR/RXR heterodimers would lead to up-regulation of CYP4F11 expression.
Our results indicate that simultaneous activation of RARs and RXRs intensifies the inhibition of CYP4F11 in HaCaT, which is consistent with data in Fig. 1b, given the fact that 9-cis-retinoic acid is able to activate both RARs and RXRs, whereas all-trans-retinoic acid only activates RARs (Leid et al., 1993; Fisher and Voorhees, 1996). It is worth noting that retinoids are able to activate retinoic acid 4-hydroxylase within hours (Marikar et al., 1998). Retinoic acid 4-hydroxylase hydroxylates all-trans- but not 9-cis-retinoic acid. Therefore, the activation of retinoic acid 4-hydroxylase may result in the reduced level of all-trans-retinoic acid, which may make all-trans-retinoic acid less effective than 9-cis-retinoic acid in the inhibition of CYP4F11. However, the distinct effects of TTNPB and LG268 on CYP4F11 expression increase the possibility of a competition mechanism and allow for the potential existence of an NR that can directly activate CYP4F11 transcription. It seems unlikely that RXR homodimers are involved in the induction of CYP4F11 because 9-cis-retinoic acid, which is able to promote the formation of RXR homodimers (Zhang and Pfahl, 1993), inhibited CYP4F11 to a greater extent than the specific RAR ligand all-trans-retinoic acid did. In addition, it has been reported that endogenous RAR/RXR heterodimers are the major functional forms regulating retinoid-responsive elements in human keratinocytes (Xiao et al., 1995), which further seems to exclude RXR homodimers as primary regulator for CYP4F11 expression in HaCaT cells. Therefore, the partner of RXR in the up-regulation of CYP4F11 could be other NRs that were not tested in this study or some unknown orphan NRs whose interaction with RXRs has not yet been clarified. It is worth noting that data from the luciferase assay (Supplemental Fig. 1) showed the down-regulation of CYP4F11 promoter activity by retinoic acids and RAR agonist TTNPB, whereas RXR agonist LG268 was not able to induce the promoter activity. This promoter may be ascribed to the limited length of the promoter sequence we used in this assay. Two putative RXR-β binding sites were found in this promoter region (Fig. 6a). However, it is possible that other untested or unknown RXR binding sites beyond the current sequence are essential for the regulation of CYP4F11 by RXR agonists.
It is also worth noting that differences in cell type and cell status may greatly affect the regulation pattern of CYP4F11. We have previously reported that retinoid treatment of differentiated human primary keratinocytes induced the expression of CYP4F11 and other CYP4F subfamily members, and this induction response seems to be mediated by RAR/RXR heterodimers (Kalsotra et al., 2008). These differences in response to retinoid treatments are likely attributable to the fact that HaCaT cells are a spontaneously arising cell line derived from keratinocytes. Cultures state whether proliferating or differentiated plays a significant role in the type of response to retinoid treatment, although the expression pattern of retinoid receptors in HaCaT cells (Fig. 1a) appears to be the same as primary human keratinocytes (Elder et al., 1992). Such differences are also seen in the comparison of expression levels of CYP4Fs among various tissues. For example, in rat lung CYP4F1 is absent and CYP4F6 comprises 95% of CYP4F expression, whereas in kidney CYP4F1 expression comprises 90% of CYP4F expression and CYP4F6 expression accounts for only 8% (Kalsotra and Strobel, 2006).
Retinoic acids are important for skin development, and they are also therapeutic agents against many skin diseases such as inflammation and cancer. Therefore, the regulation of CYP4F11 by retinoic acids may result in the drug-drug interaction between retinoic acids and CYP4F11-targeted drugs. In addition, inflammation and cancer, which stimulate JNK/AP-1 pathway, may also affect CYP4F11 drug-metabolizing activity. Although CYP4F11 is the most active drug metabolizer in the CYP4F subfamily, no primary endogenous substrate for CYP4F11 has been uncovered. As the functions of CYP4F11 in normal physiological conditions and under disease conditions are clarified, CYP4F11 regulatory mechanisms will be known more completely, and their significance will become clearer.
Supplementary Material
Acknowledgments.
We thank Dr. Peter J. A. Davies for provision of chemicals.
This work was supported in part by the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS44174]; and the National Institutes of Health National Institute of Arthritis and Musculoskeletal and Skin Diseases [Grant AR45603].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.109.029025
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- RXR
- retinoid X receptor
- RAR
- retinoic acid receptor
- NR
- nuclear receptor
- AP-1
- activator protein 1
- TTNPB
- 4-(2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl)benzoic acid
- TPA
- 12-O-tetradecanoyl-phorbol-13-acetate
- LG268
- 6-(1(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)cyclopropyl) nicotinic acid
- TNF-α
- tumor necrosis factor α
- IL-1β
- interleukin 1β
- SP600125
- 1,9-pyrazoloanthrone
- SB203580
- 4-(4-fluorophenyl)-2-(4-methylsulfinylphenyl)-5-(4-pyridyl)1H-imidazole
- RT-PCR
- reverse transcriptase-polymerase chain reaction
- DMSO
- dimethyl sulfoxide
- qRT-PCR
- quantitative real-time polymerase chain reaction
- JNK
- c-Jun N-terminal kinase.
References
- Apriletti JW, Ribeiro RC, Wagner RL, Feng W, Webb P, Kushner PJ, West BL, Nilsson S, Scanlan TS, Fletterick RJ, Baxter JD. (1998) Molecular and structural biology of thyroid hormone receptors. Clin Exp Pharmacol Physiol Suppl 25:S2–S11 [DOI] [PubMed] [Google Scholar]
- Barra GB, Velasco LF, Pessanha RP, Campos AM, Moura FN, Dias SM, Polikarpov I, Ribeiro RC, Simeoni LA, Neves FA. (2004) [Molecular mechanism of thyroid hormone action]. Arq Bras Endocrinol Metabol 48:25–39 [DOI] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. (1988) Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol 106:761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedid M, Yoza BK, Brooks JW, Mizel SB. (1991) Activation of AP-1 by IL-1 and phorbol esters in T cells. Role of protein kinase A and protein phosphatases. J Immunol 147:867–873 [PubMed] [Google Scholar]
- Chen JY, Penco S, Ostrowski J, Balaguer P, Pons M, Starrett JE, Reczek P, Chambon P, Gronemeyer H. (1995) RAR-specific agonist/antagonists which dissociate transactivation and AP1 transrepression inhibit anchorage-independent cell proliferation. EMBO J 14:1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvergne B. (2007) RXR: from partnership to leadership in metabolic regulations. Vitam Horm 75:1–32 [DOI] [PubMed] [Google Scholar]
- Elder JT, Aström A, Pettersson U, Tavakkol A, Griffiths CE, Krust A, Kastner P, Chambon P, Voorhees JJ. (1992) Differential regulation of retinoic acid receptors and binding proteins in human skin. J Invest Dermatol 98:673–679 [DOI] [PubMed] [Google Scholar]
- Fisher GJ, Talwar HS, Lin J, Lin P, McPhillips F, Wang Z, Li X, Wan Y, Kang S, Voorhees JJ. (1998) Retinoic acid inhibits induction of c-Jun protein by ultraviolet radiation that occurs subsequent to activation of mitogen-activated protein kinase pathways in human skin in vivo. J Clin Invest 101:1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GJ, Voorhees JJ. (1996) Molecular mechanisms of retinoid actions in skin. FASEB J 10:1002–1013 [DOI] [PubMed] [Google Scholar]
- Green S. (1995) PPAR: a mediator of peroxisome proliferator action. Mutat Res 333:101–109 [DOI] [PubMed] [Google Scholar]
- Hashizume T, Imaoka S, Mise M, Terauchi Y, Fujii T, Miyazaki H, Kamataki T, Funae Y. (2002) Involvement of CYP2J2 and CYP4F12 in the metabolism of ebastine in human intestinal microsomes. J Pharmacol Exp Ther 300:298–304 [DOI] [PubMed] [Google Scholar]
- Heinz Nau WSB. (2007) Retinoids: The Biochemical and Molecular Basis of Vitamin A and Retinoid Action. Springer-Verlag, Berlin, Heidelberg, New York: [Google Scholar]
- Hsu MH, Savas U, Griffin KJ, Johnson EF. (2007) Regulation of human cytochrome P450 4F2 expression by sterol regulatory element-binding protein and lovastatin. J Biol Chem 282:5225–5236 [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Du L, Wang Y, Ladd PA, Kikuta Y, Duvic M, Boyd AS, Keeney DS, Strobel HW. (2008) Inflammation resolved by retinoid X receptor-mediated inactivation of leukotriene signaling pathways. FASEB J 22:538–547 [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Strobel HW. (2006) Cytochrome P450 4F subfamily: at the crossroads of eicosanoid and drug metabolism. Pharmacol Ther 112:589–611 [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Turman CM, Kikuta Y, Strobel HW. (2004) Expression and characterization of human cytochrome P450 4F11: putative role in the metabolism of therapeutic drugs and eicosanoids. Toxicol Appl Pharmacol 199:295–304 [DOI] [PubMed] [Google Scholar]
- Kikuta Y, Kusunose E, Kusunose M. (2002) Prostaglandin and leukotriene omega-hydroxylases. Prostaglandins Other Lipid Mediat 68–69:345–362 [DOI] [PubMed] [Google Scholar]
- Kyriakis JM. (1999) Activation of the AP-1 transcription factor by inflammatory cytokines of the TNF family. Gene Expr 7:217–231 [PMC free article] [PubMed] [Google Scholar]
- Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD, Edwards PA. (2000) Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J Biol Chem 275:10638–10647 [DOI] [PubMed] [Google Scholar]
- Lamph WW, Wamsley P, Sassone-Corsi P, Verma IM. (1988) Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature 334:629–631 [DOI] [PubMed] [Google Scholar]
- Lazar MA. (2005) PPAR gamma, 10 years later. Biochimie 87:9–13 [DOI] [PubMed] [Google Scholar]
- Leid M, Kastner P, Durand B, Krust A, Leroy P, Lyons R, Mendelsohn C, Nagpal S, Nakshatri H, Reibel C. (1993) Retinoic acid signal transduction pathways. Ann N Y Acad Sci 684:19–34 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Ong ES, Dyck JA, Evans RM. (1990) Nuclear receptor that identifies a novel retinoic acid response pathway. Nature 345:224–229 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. (1995) The nuclear receptor superfamily: the second decade. Cell 83:835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikar Y, Wang Z, Duell EA, Petkovich M, Voorhees JJ, Fisher GJ. (1998) Retinoic acid receptors regulate expression of retinoic acid 4-hydroxylase that specifically inactivates all-trans retinoic acid in human keratinocyte HaCaT cells. J Invest Dermatol 111:434–439 [DOI] [PubMed] [Google Scholar]
- Perlmann T, Jansson L. (1995) A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev 9:769–782 [DOI] [PubMed] [Google Scholar]
- Perlmann T, Wallén-Mackenzie A. (2004) Nurr1, an orphan nuclear receptor with essential functions in developing dopamine cells. Cell Tissue Res 318:45–52 [DOI] [PubMed] [Google Scholar]
- Qi C, Zhu Y, Reddy JK. (2000) Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem Biophys 32:187–204 [DOI] [PubMed] [Google Scholar]
- Saatcioglu F, Claret FX, Karin M. (1994) Negative transcriptional regulation by nuclear receptors. Semin Cancer Biol 5:347–359 [PubMed] [Google Scholar]
- Sassone-Corsi P, Ransone LJ, Verma IM. (1990) Cross-talk in signal transduction: TPA-inducible factor jun/AP-1 activates cAMP-responsive enhancer elements. Oncogene 5:427–431 [PubMed] [Google Scholar]
- Whitmarsh AJ, Davis RJ. (1996) Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J Mol Med 74:589–607 [DOI] [PubMed] [Google Scholar]
- Willy PJ, Mangelsdorf DJ. (1997) Unique requirements for retinoid-dependent transcriptional activation by the orphan receptor LXR. Genes Dev 11:289–298 [DOI] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. (1995) LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev 9:1033–1045 [DOI] [PubMed] [Google Scholar]
- Xiao JH, Durand B, Chambon P, Voorhees JJ. (1995) Endogenous retinoic acid receptor (RAR)-retinoid X receptor (RXR) heterodimers are the major functional forms regulating retinoid-responsive elements in adult human keratinocytes. Binding of ligands to RAR only is sufficient for RAR-RXR heterodimers to confer ligand-dependent activation of hRAR beta 2/RARE (DR5). J Biol Chem 270:3001–3011 [DOI] [PubMed] [Google Scholar]
- Yu Y, Ge N, Xie M, Sun W, Burlingame S, Pass AK, Nuchtern JG, Zhang D, Fu S, Schneider MD, et al. (2008) Phosphorylation of Thr-178 and Thr-184 in the TAK1 T-loop is required for interleukin (IL)-1-mediated optimal NFkappaB and AP-1 activation as well as IL-6 gene expression. J Biol Chem 283:24497–24505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen L, Hardwick JP. (2000) Promoter activity and regulation of the CYP4F2 leukotriene B(4) omega-hydroxylase gene by peroxisomal proliferators and retinoic acid in HepG2 cells. Arch Biochem Biophys 378:364–376 [DOI] [PubMed] [Google Scholar]
- Zhang X, Hardwick JP. (2000) Regulation of CYP4F2 leukotriene B4 omega-hydroxylase by retinoic acids in HepG2 cells. Biochem Biophys Res Commun 279:864–871 [DOI] [PubMed] [Google Scholar]
- Zhang XK, Pfahl M. (1993) Hetero- and homodimeric receptors in thyroid hormone and vitamin A action. Receptor 3:183–191 [PubMed] [Google Scholar]
- Zhou H, Lin A, Gu Z, Chen S, Park NH, Chiu R. (2000) 12-O-Tetradecanoylphorbol-13-acetate (TPA)-induced c-Jun N-terminal kinase (JNK) phosphatase renders immortalized or transformed epithelial cells refractory to TPA-inducible JNK activity. J Biol Chem 275:22868–22875 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.