Abstract
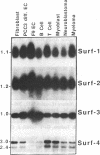
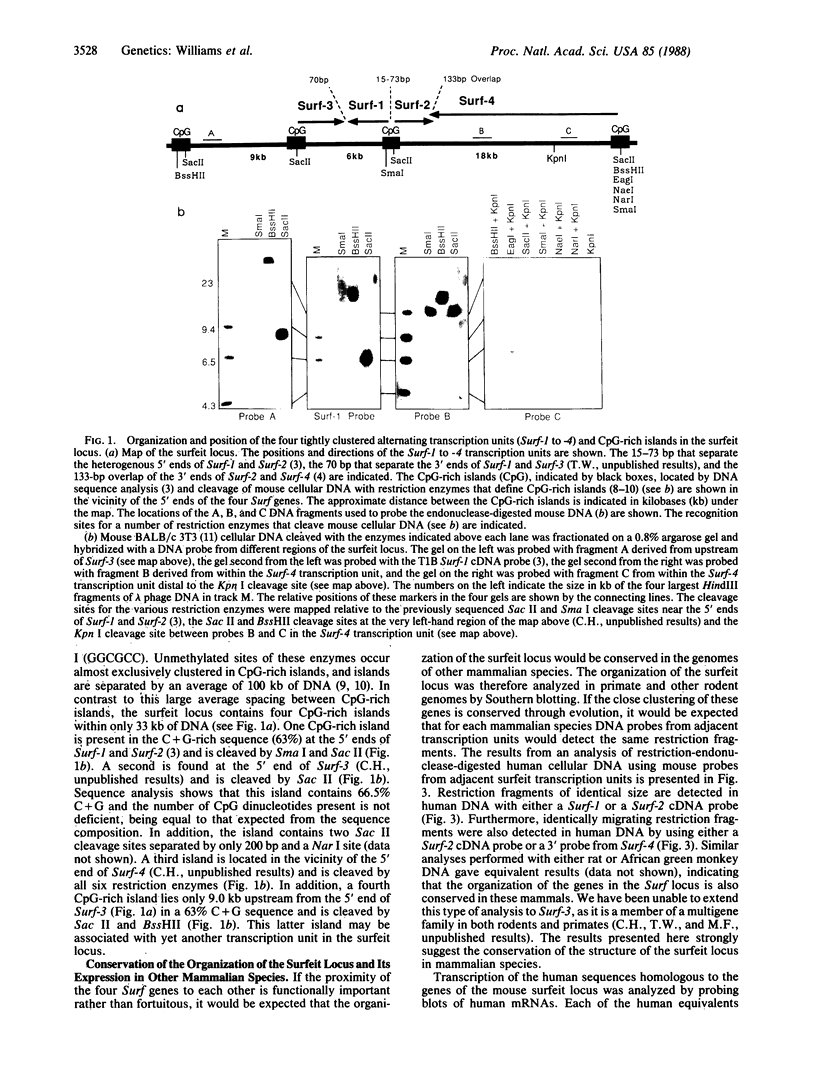
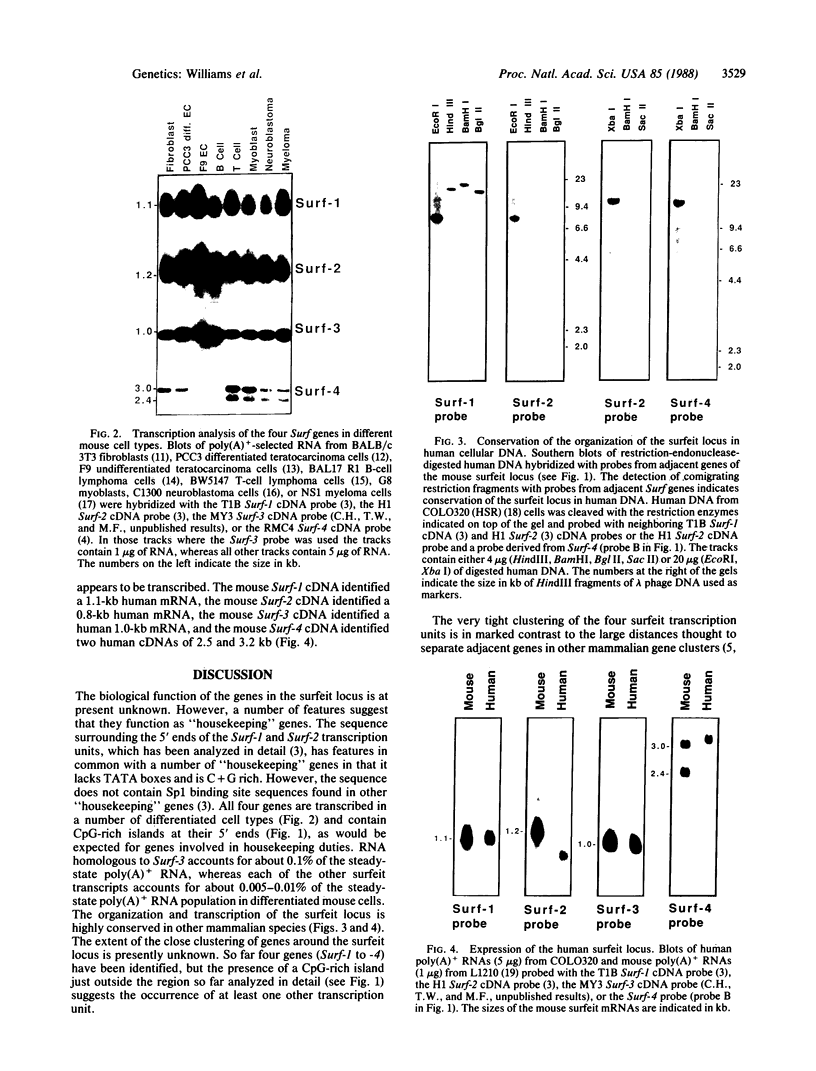
The four identified alternating transcription units (Surf-1 to Surf-4) in the mouse surfeit locus are very tightly clustered, no two neighboring units being separated by more than 73 base pairs and the Surf-2 and Surf-4 transcription units overlapping by 133 base pairs at their 3' ends. All four surfeit genes, which are unrelated by sequence similarity, were found to have the properties of "housekeeping" genes, being expressed in a variety of differentiated mouse cell lines and containing unmethylated CpG-rich islands in the vicinity of their 5' ends. The unusual organization of the four surfeit genes was found not to be unique to the mouse: the same juxtaposition of the genes was found to be conserved in a number of different mammals, including humans. The four human surfeit genes were also found to be transcriptionally active.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aaronson S. A., Todaro G. J. Development of 3T3-like lines from Balb-c mouse embryo cultures: transformation susceptibility to SV40. J Cell Physiol. 1968 Oct;72(2):141–148. doi: 10.1002/jcp.1040720208. [DOI] [PubMed] [Google Scholar]
- Alitalo K., Schwab M., Lin C. C., Varmus H. E., Bishop J. M. Homogeneously staining chromosomal regions contain amplified copies of an abundantly expressed cellular oncogene (c-myc) in malignant neuroendocrine cells from a human colon carcinoma. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1707–1711. doi: 10.1073/pnas.80.6.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augusti-Tocco G., Sato G. Establishment of functional clonal lines of neurons from mouse neuroblastoma. Proc Natl Acad Sci U S A. 1969 Sep;64(1):311–315. doi: 10.1073/pnas.64.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berstine E. G., Hooper M. L., Grandchamp S., Ephrussi B. Alkaline phosphatase activity in mouse teratoma. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3899–3903. doi: 10.1073/pnas.70.12.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985 Jan;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Brown W. R., Bird A. P. Long-range restriction site mapping of mammalian genomic DNA. 1986 Jul 31-Aug 6Nature. 322(6078):477–481. doi: 10.1038/322477a0. [DOI] [PubMed] [Google Scholar]
- Fried M., Griffiths M., Davies B., Bjursell G., La Mantia G., Lania L. Isolation of cellular DNA sequences that allow expression of adjacent genes. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2117–2121. doi: 10.1073/pnas.80.8.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian M. J., Hung M. C., Wensink P. C. Independent control elements that determine yolk protein gene expression in alternative Drosophila tissues. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1396–1400. doi: 10.1073/pnas.82.5.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel M. D., Pruitt R. E., Meyerowitz E. M. DNA sequences, gene regulation and modular protein evolution in the Drosophila 68C glue gene cluster. J Mol Biol. 1983 Aug 25;168(4):765–789. doi: 10.1016/s0022-2836(83)80074-6. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Hyman R., Stallings V. Complementation patterns of Thy-1 variants and evidence that antigen loss variants "pre-exist" in the parental population. J Natl Cancer Inst. 1974 Feb;52(2):429–436. doi: 10.1093/jnci/52.2.429. [DOI] [PubMed] [Google Scholar]
- Jakob H., Boon T., Gaillard J., Nicolas J., Jacob F. Tératocarcinome de la spuris: isolement, culture et propriétés de cellules a potentialités multiples. Ann Microbiol (Paris) 1973 Oct;124(3):269–282. [PubMed] [Google Scholar]
- Kim K. J., Weinbaum F. I., Mathieson B. J., McKeever P. E., Asofsky R. Characteristics of BALB/C T cell lymphomas grown as continuous in vitro lines. J Immunol. 1978 Jul;121(1):339–344. [PubMed] [Google Scholar]
- Köhler G., Howe S. C., Milstein C. Fusion between immunoglobulin-secreting and nonsecreting myeloma cell lines. Eur J Immunol. 1976 Apr;6(4):292–295. doi: 10.1002/eji.1830060411. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984 Jul;37(3):969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- Swaroop A., Sun J. W., Paco-Larson M. L., Garen A. Molecular organization and expression of the genetic locus glued in Drosophila melanogaster. Mol Cell Biol. 1986 Mar;6(3):833–841. doi: 10.1128/mcb.6.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. J., Fried M. The MES-1 murine enhancer element is closely associated with the heterogeneous 5' ends of two divergent transcription units. Mol Cell Biol. 1986 Dec;6(12):4558–4569. doi: 10.1128/mcb.6.12.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., Fried M. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3' ends. Nature. 1986 Jul 17;322(6076):275–279. doi: 10.1038/322275a0. [DOI] [PubMed] [Google Scholar]
- von Hoyningen-Huene V., Norbury C., Griffiths M., Fried M. Gene activation properties of a mouse DNA sequence isolated by expression selection. Nucleic Acids Res. 1986 Jul 25;14(14):5615–5627. doi: 10.1093/nar/14.14.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]