Abstract
The endoplasmic reticulum (ER) protein Bap31 associates with nascent class I MHC molecules. It appears to mediate the export of class I MHC molecules from the ER and may also be involved in their quality control. Here we use Förster resonance energy transfer (FRET) and quantitative fluorescence imaging to show that in human, HeLa, cells Bap31 clusters with MHC class I (HLA-A2) molecules in the ER, and traffics via export vesicles to the ER/Golgi intermediate compartment, ERGIC. FRET between Bap31 and HLA-A2 and forward traffic increase when MHC class I molecules are loaded with a pulse of peptide. The increased forward traffic is blocked by over-expression of Bap29, a partner protein for Bap31, which localizes to the ER. Thus, in HeLa cells, Bap31 is involved in the exit of peptide-loaded MHC class I from the ER, and its function is regulated by its interaction with its homolog, Bap29.
Keywords: Human, MHC, trafficking, processing, presentation
Introduction
MHC class I molecules present peptides, derived from intracellular proteins, to T cells. The three components of MHC class I molecules, a transmembrane heavy chain, β2-microglobulin and a peptide, typically 8–10 amino acids, are assembled sequentially in the endoplasmic reticulum (ER) (1-3). When peptide loading is complete, MHC class I molecules are exported from the ER to the cell surface along the secretory pathway.
Although the assembly and peptide loading of MHC class I molecules in the ER is well documented, regulation of the export of assembled molecules is not understood. MHC class I molecules themselves do not contain sequence motifs for association with the coats of ER export or retrieval vesicles. This implies that they either leave the ER by bulk flow, or that they are associated with specific carriers. One candidate for an export carrier is the ER membrane protein, Bap31 (4). Bap31 was defined by its association with the nascent B cell receptor complex immunoprecipitated from cell extracts; complexes of Bap31 and MHC class I have also been found by immunopecipitation (5, 6). These complexes may reflect Bap31 function in regulating the forward trafficking of a number of membrane proteins out of the ER (7-9). Though lack of Bap31 does not abrogate surface expression of MHC class I molecules, it does slow their export from the ER (6). On the other hand, over-expression of Bap31 increases the rate of ER-to-Golgi transport of MHC class I and their stability and so increases the overall level of surface MHC class I (9). Overexpression of a homolog of Bap31, Bap29, reduces surface MHC class I levels (9). If Bap31 transports intact MHC class I molecules out of the ER then we predict that Bap31 will associate more strongly with peptide-loaded MHC class I molecules than with nascent or empty molecules and that it moves with these peptide-loaded MHC class I molecules to post-ER compartments.
Here we use fluorescence resonance energy transfer (FRET), quantitative fluorescence imaging of molecular colocalization and electron microscopy to detect molecular associations and functional relationships between MHC class I molecules, Bap31, and Bap29, as MHC class I molecules traffic from the ER. We find that Bap31 associates with MHC class I molecules in the ER, and that it also forms Bap31 homo-oligomers and Bap31/Bap29 hetero-oligomers. At steady state a significant fraction of Bap31 is found outside the ER, in export vesicles and in the ERGIC. Feeding peptides that bind MHC class I molecules expressed in HeLa cells increases the fraction of Bap31 associated with MHC class I molecules and the fraction of Bap31 localized to the ERGIC. In parallel there is an increase in trafficking of peptide-loaded MHC class I molecules from the ER. This forward traffic is inhibited by over-expression of Bap29, an ER localized partner for Bap31.
Materials and Methods
Antibodies
Mouse mAb for the ERGIC marker G1/93 (ERGIC-53) and for Bap31 were a gift of Dr. Hans-Peter Hauri (University of Basel). Rat CC-4 anti-Bap31 mAb (10) was purchased from Affinity BioReagents (Golden, CO); rabbit polyclonal antibody for Bap29 (11) was a gift of Dr. Gordon C. Shore (McGill University). Anti-calnexin and anti-Sec23 were from Abcam Inc. (Cambridge, MA). mAb W6/32 and KE-2 which detect a monomorphic epitope on all human class I MHC molecules. mAb BB7.2 is specific for HLA-A2. The secondary antibodies used in this study were Alexa 488 and Alexa 633-conjugated goat anti-mouse Ab, Alexa 546-conjugated goat anti-rabbit Ab and Alexa 633-conjugated goat anti-rat Ab (Molecular Probes, Eugene, OR).
Immunolabeling of proteins in HeLa cells
HeLa cells were purchased from the American Type Culture Collection (Manassas, VA) and were maintained in Dulbecco's modified Eagle's medium (Mediatech, Herndon, VA), supplemented with 10% heat-inactivated fetal bovine serum in a humid 5% CO2 atmosphere at 37°C. Cells on cover slips were washed three times in phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (PFA) in PBS for 30 min. The cells were washed three times in 0.25% NH4Cl in PBS and then washed an additional five times in PBS. Next, the cells were permeabilized in 2 ml of PBS containing 1% bovine serum albumin (BSA) and 0.2% saponin at 37°C for 1 h, and incubated with primary antibody in PBS-BSA-saponin at 37°C for 1 h. After washing with PBS-BSA-saponin five times, the cells were incubated with secondary antibodies. After washing five times, cover slips were mounted onto glass slides using the SlowFade Light Antifade Kit (Invitrogen).
Peptides
In experiments on trafficking of Bap31 in response to peptide loading of the endogenous HLA-A68 molecules of HeLa, cells were incubated for 30 minutes in medium containing 25 μM MAGE3.112 peptide, KVAELVHFL which has a measured high affinity for HLA-A68 (12) or with 25 μM Mage3a peptide, EVDPIGHLY, a peptide which we chose to have ∼ ten-fold lower affinity for HLA-A68, using the program SVRMHC (13). This order of magnitude of the relative affinities of the two peptides was also predicted by the program Bimas (14). The predictions were confirmed by comparing the relative abilities of the 2 peptides to restore native MHC class I molecules after their peptides were stripped from the cell surface by brief (90s) treatment at pH 3.2 as described in (15). Peptides were from Sigma Genosys.
In experiments on FRET and trafficking of HLA-A2 molecules, cells transfected with an HLA-A2-YFP plasmid were fed 25μM TAX peptide, LLFGYPVYV (16,17), for 30 minutes.
Plasmid construction
The plasmid encoding Bap31 fused with YFP at the C-terminal end (pBAP31-YFP) was constructed previously (9) but it lacked a C-terminal KKXX motif. To construct a plasmid encoding Bap31-YFP-KKXX, we introduced an oligonucleotide encoding seven amino acids (PMDKKEE) containing the KKEE motif at the end of Bap31-YFP. To construct a BAP31-CFP-KKEE plasmid, the CFP gene was obtained from pHLA-A2-CFP by digesting with BamH I and Not I. The YFP sequence in Bap31-YFP was replaced by the CFP sequence, yielding pBAP31-CFP-KKEE.
To construct a BAP31-YFP-CFP-KKXX tandem plasmid, the following three fragments were assembled in one step using DNA ligation: (i) the large fragment of pBAP31-YFP digested with BsrG I and Not I; (ii) an 80-bp linker fragment of pHLA-A2-YFP-CFP (6) digested with BsrG I and BamH I; and (iii) the CFP-KKXX fragment of pBAP31-CFP-KKEE digested with BamH I and Not I. Construction of a BAP29 plasmid started with HeLa cell RNA and proceeded as described previously for BAP31 (9).
Native YFP and CFP may dimerize and so give false positive FRET signals from labeled proteins (18). To avoid this, a codon substitution GCC→AAA, leading to the mutation A206K, was created within the open-reading frames of YFP and CFP, using a QuikChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The substitution was confirmed by DNA sequencing. The monomeric CFP and YFP, mCFP and mYFP, were used to make all of the chimeric proteins described here. Collectively, plasmids constructed and modified in this study were pBAP31-mYFP-KKEE, pBAP31-mCFP-KKEE, pBAP31-mYFP-mCFP, pBAP29-mCFP-KKRL and pHLA-A2-mCFP. In this paper, they are designated pBAP31-YFP, pBAP31-CFP, pBAP31-YFP-CFP, pBAP29-CFP and pHLA-A2-CFP, respectively. Plasmids for N-terminal tagged YFP-Bap31 and YFP-Bap29 and for C-terminal tagged HLA-A2-CFP have been described (9, 19).
Transfection and functional expression of Bap31, Bap29 and HLA-A2 constructs
Cells were trypsinized, placed on coverslips in 6-well plates and were cultured for 1 day before plasmid transfection. For transfection of the cells, 3 μl of FuGENE6 (Roche, Indianapolis, IN) was mixed with 94 μl of OptiMem (Mediatech, Herndon, VA) and the mixture was allowed to stand at room temperature for 5 min. Then, 100–300 ng of plasmid DNA in a volume of < 1 μl was added to the mixture and it was allowed to stand at room temperature for 20 min. The mixture was added to the culture and the cells were grown for another 20–22 h. Because fluorescent protein-fused BAP31 and BAP29 tended to form aggregates in the ER after 30 h of transfection, the cells were fixed before 22 h had elapsed.
Cells were removed from plates using trypsin/collagenase and labeled with either mAb KE-2 or W6/32 to detect all surface HLA class I molecules, or with mAb BB7.2 to detect surface HLA-A2. Secondary antibody was Alexa 633 goat anti-mouse Ig. Effects of transfection of a particular plasmid on HLA expression were then read out as the plot of YFP vs Alexa 633 fluorescence. pBAP31-mYFP-KKEE and pBAP31-mYFP both increased the level of surface MHC class I in proportion to their expression (Supplemental Methods Figure 1) to a level comparable to the increase given by our original, N-terminal-tagged YFP-Bap31 (9). pBap31-mCFP colocalized with YFP-Bap31 in the ER. We could not measure CFP fluorescence in our flow cytometer.
FRET measurements
FRET was measured between endogenous proteins, Bap31 and ERGIC-53, and between transfected, FP-tagged proteins. To measure FRET between Bap31 and ERGIC-53, cells were labeled as described above for fluorescence studies, using directly-conjugated antibodies to Bap31 and ERGIC-53. For FRET between CFP- and YFP-tagged proteins, cells were grown on cover slips for no more than 24 hours after transfection. Cells were washed three times in PBS and fixed with 4% PFA in PBS for 30 min. All procedures were carried out in the dark at room temperature. The cells were washed three times in 0.25% NH4Cl in PBS and then washed an additional five times in PBS. The cover slips were mounted on glass slides using the SlowFade Light Antifade Kit.
Cells were imaged with a LSM510 META confocal laser microscope (Carl Zeiss, Germany) using a Plan Apochromat 63× objective lens (NA = 1.4). For most experiments, we focused on fluorescent regions of the cytoplasm adjacent to the nucleus. This choice of region of interest, together with the optical sectioning by the confocal microscope, largely limited fluorescence to molecules in the ER. This was important when measuring interactions between Bap31 and MHC class I molecules, since a fraction of MHC class I is at the cell surface. YFP and CFP were excited at 514 nm and 458 nm respectively. YFP fluorescence was observed through a 530-nm long-pass filter; CFP fluorescence was observed through a band-pass filter (475–525 nm). Twelve-bit images were collected at 1024 × 1024-pixel resolution. Using cells expressing Bap31-YFP-CFP tandem fusion proteins, we fixed all parameters such as the laser power, detector gain and amplifier offset at individual channels to make the fluorescence intensity of YFP (IYFP) and CFP (ICFP) almost equal. Three to six regions of interest (ROIs) were chosen within the ER of the cell. Each data point represents the calculated FRET efficiency of each ROI as a function of its YFP concentration. For accuracy in calculation we used only ROIs with fluorescence intensities between 500 and 3,000 arbitrary units (a.u.) intensity.
Acceptor (YFP) fluorescence was bleached by 514 nm light to 5–10% of its original level. We found that less complete photobleaching of YFP, for example to 20–30% of initial intensity, significantly increased the fluorescence intensity at the CFP channel excited at 458 nm. This is probably due to photoconversion of the YFP fluorochrome into a CFP-like molecule (20) which causes an artificial increase in FRET efficiency. To further control for this, cells expressing only YFP-tagged protein were prepared for each FRET experiment. The fluorescence intensity of the cells expressing only YFP-tagged protein was measured at the CFP channel before and after photobleaching of YFP (IYFP/prebleach and IYFP/postbleach, respectively), and these values were subtracted from the fluorescence intensity of CFP in cells expressing YFP- and CFP-fused proteins. A corrected % FRET efficiency, E, was calculated as:
E = (1 − [ICFP/prebleach−IYFP/prebleach]/[ICFP/postbleach−IYFP/postbleach]) × 100
Most FRET calculations were done in Excel. The calculations for FRET between Bap31 and HLA-A2 after peptide feeding were done using the FRETcalc v3.0 plugin for ImageJ (21).
When acceptors and donors are clustered on a membrane, E depends upon the acceptor-to-donor ratio, A:D but is independent of the acceptor concentration. When acceptors and donors are randomly distributed, E depends upon the acceptor concentration but is independent of A:D. This difference can distinguish between FRET due to clustering of acceptor and donor and FRET among members of a randomly distributed population of donors and acceptors (19, 22). FRET among clustered molecules is easily recognized in a plot of E vs. A:D.
The extent of colocalization of labeled molecules was determined using the colocalization algorithm of MIPAV, as described previously (9). This calculates both a coefficient of colocalization and the extent to which one label colocalizes with another (% area colocalized). Colocalization coefficients >0.60 were accepted as showing significant colocalization. We have earlier shown that coefficients for markers that are not colocalized (Bap31 and the Golgi marker giantin) were < 0.4 (9). The significance of changes in the extent of colocalization was determined using the built-in statistical functions, t tests and analysis of variance function, of GraphPad Prism (GraphPad Software).
Immuno-gold Electron Microscopy
Approximately 5×105 HeLa cells were incubated at 37°C in serum-free medium containing 25 μM MAGE3.112 peptide. After 30, 60 or 90 min of peptide feeding, cells were fixed in 3% paraformaldehyde + 0.05% glutaraldehyde (Electron Microscopy Sciences, Hatfield PA) in PO4 buffer for 1 hr at room temperature. Cells were pelleted, washed three times in PBS and incubated in 2.3 M sucrose containing 20% polyvinylpyrollidone (10,000 mw) for 2 hours at room temperature. The pellet was mounted on a cyropin and frozen in liquid nitrogen. Ultra-thin sections (∼80 nm to 100 nm thick) were cut on a Leica UCT ultramicrotome equipped with a FCS cryo-stage at -100°C, and collected onto ionized formvar/carbon coated nickel grids. Cryosections were incubated with a mixture of rat anti-Bap31 mAb (CC-4) and mouse anti- ERGIC-53 mAb (G1/93). After washing, the sections were labeled with 6 nm colloidal gold-conjugated anti-rat Ig + 12 nm colloidal gold-conjugated anti-mouse Ig (Jackson ImmunoResearch). Sections were imaged on a Philips EM 420 TEM equipped with a SIS Megaview III CCD digital camera. Colocalization of 6 nm and 12 nm gold labels used the approach of Philimonenko et al. (23). This characterizes proximity of gold particles in terms of a pair cross-correlation function (PCCF), effectively the ratio of the density of 6 nm particles at a specified distance from a 12 nm particle to the average density of 6 nm particles. The distribution of PCCF over distance is constant if 6 nm and 12 nm particles are randomly distributed, but has a peak over a range of distances if the two markers are colocalized. The PCCF was calculated for distances between particles ranging from 10 nm to 300 nm. There was a peak of PCCF at 60-70 nm radius, and this was used to define the distance limit below which 6 nm particles were considered to be clustered with the 12 nm particles.
Results
Clustering of Bap31 with itself, Bap29 and HLA-A2 in the ER
We and others have shown that Bap31 immunoprecipitates with native MHC class I molecules of mouse (5, 6); we also found that human Bap31 co-immunoprecipitates with free MHC class I heavy chains (9). FRET between CFP- and YFP-labeled proteins detected these interactions in situ. FRET was measured in terms of the dequenching of donor (CFP) fluorescence after acceptor (YFP) bleaching. Since labeling Bap31 and Bap29 on the C-terminus could interfere with their ER retention/retrieval, we added retention/retrieval sequences C-terminal to the CFP or YFP sequence: KKEE for Bap31, and KKRL for Bap29. Over-expression of the C-terminally tagged Bap31-YFP increased surface MHC class I levels of HeLa cells similarly to our original construct, N-terminally-tagged Bap31 (9) (Supplemental Methods Figure 1).
Our positive control for FRET was Bap31-YFP linked to CFP with a 25-amino acid linker (Bap31-YFP-CFP in Supplemental Methods Figure 2A). After photobleaching of YFP, the CFP intensity increased by an average of 18% (Supplemental Methods Figure 2B), though FRET in individual ROI ranged from 14-30%. Our negative control for FRET was Bap31 with N-terminal YFP (9), and Bap31 with C-terminal CFP (YFP-BAP31 and BAP31-CFP, respectively in Supplemental Methods Figure 2A). The acceptor and the donor are on opposite sides of the ER membrane, out of FRET range and indeed, no positive FRET was observed between YFP-Bap31 and Bap31-CFP co-expressed in HeLa cells (Supplemental Methods Figure 2C).
Clustering of Bap31 with itself and with Bap29
Biochemical studies show that Bap31 forms homo-oligomers (24). We detected these by FRET when Bap31-YFP and Bap31-CFP were co-expressed in HeLa cells. FRET measurements were made at 16-22 h after transfection. Acceptor concentration in a given ROI was taken as the YFP fluorescence before bleaching. Donor concentration was taken as CFP fluorescence after YFP bleaching. FRET efficiency was independent of the acceptor concentration for given A:D for all levels of acceptor expression, but FRET efficiency did increase with the A:D (Figure 1A). This dependence on A:D but independence of acceptor concentration is the hallmark of FRET between specifically associated proteins (20, 22); Thus, a fraction of Bap31 exists as multimers in the intact ER. Bap31 co-precipitates with Bap29 from extracts of a mouse B cell line J558L (24). To detect this association in our system, Bap31-YFP and Bap29-CFP were co-expressed in HeLa cells. The FRET efficiency between this pair was low, but there was a significant increase in FRET with increasing A:D, implying that a fraction of Bap31 is associated with Bap29 (Figure 1B). As we note below, this association has functional consequences for Bap31.
Figure 1. FRET detects clustering of Bap31-YFP.
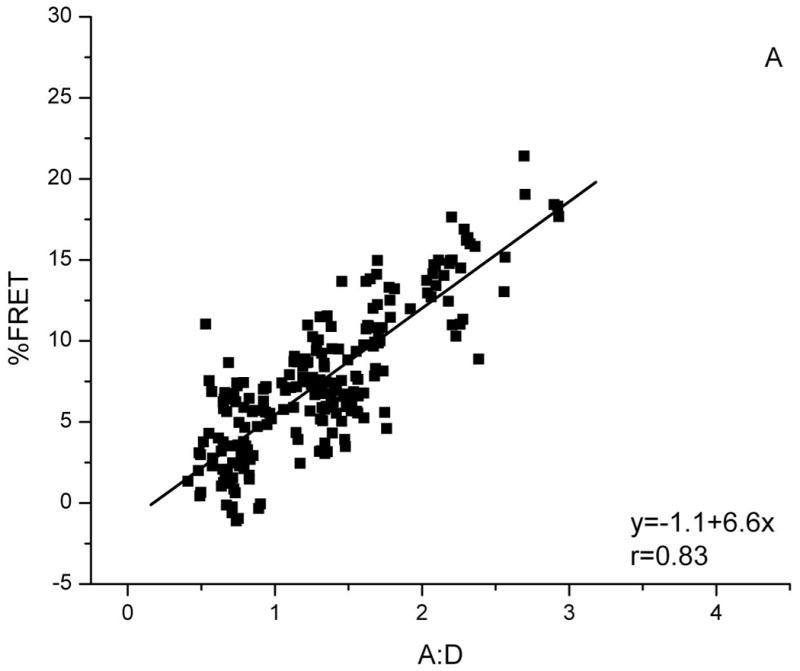
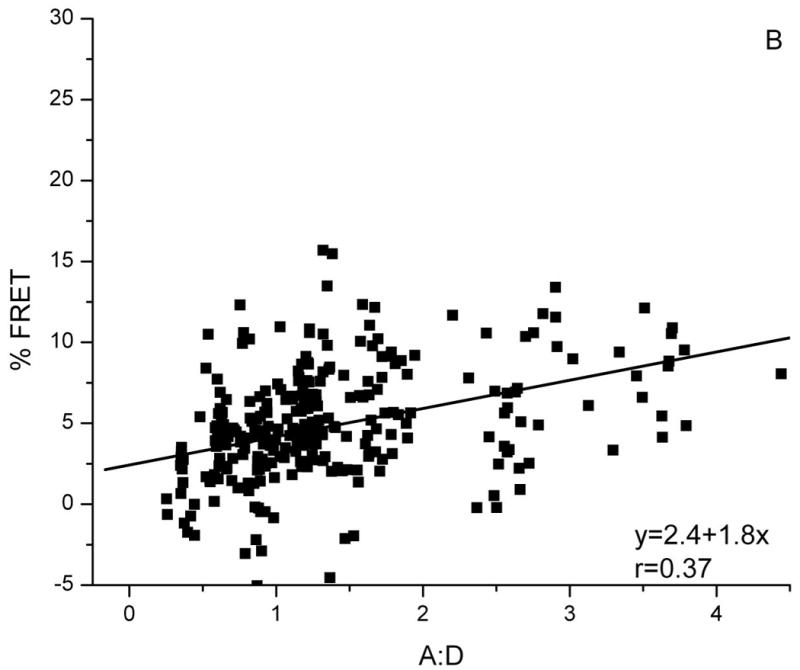
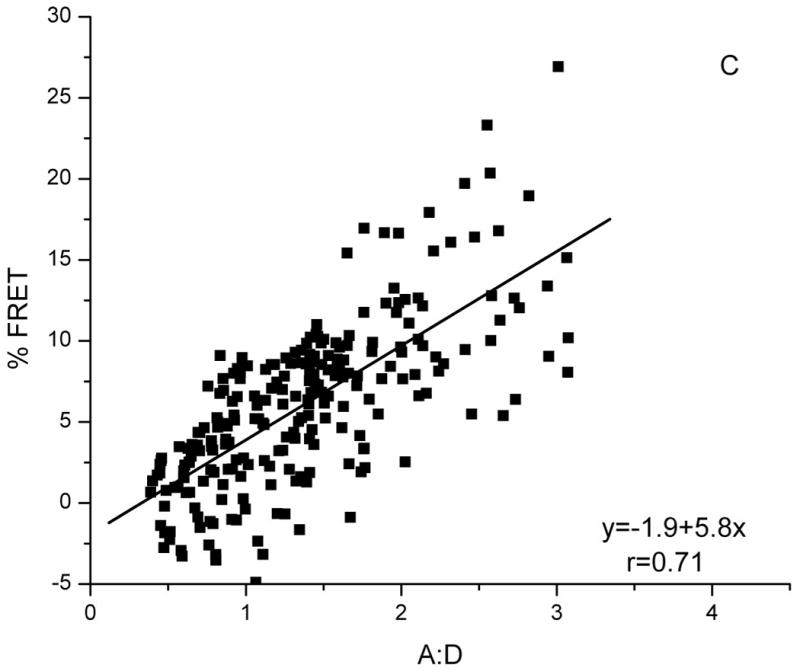
FRET between Bap31-YFP and: A) Bap31-CFP, B) Bap29-CFP and C) HLA-A2-CFP. For all FRET pairs, FRET efficiency increases with increasing A:D ratio (YFP/CFP), a characteristic of FRET between specifically-associated donor and acceptor-tagged molecules. FRET between randomly distributed molecules is independent of A:D (18). The slopes of the plots indicate the FRET efficiency for each pair.
Clustering of Bap31 and HLA-A2
To detect clustering of HLA-A2 and Bap31, we expressed HLA-A2-CFP and Bap31-YFP in varying proportions. Again FRET efficiency was a function of A:D; HLA-A2-CFP and Bap31-YFP are clustered in the ER. Average FRET efficiency was approximately 5% when the A:D was 1, (Figure 1C), less than that for the tandem positive control, BAP31-YFP-CFP (Supplemental Methods Figure 2B). This is expected since non-fluorescent HLA-A2 and Bap31 are also clustered with Bap31-YFP and HLA-A2-CFP, diluting the interaction of the FRET pair. Also, both HLA-A2 and Bap31 molecules form homo-oligomers (22, 24) as well as clustering with other partners in the ER (25, 26), reducing the fraction of fluorescent HLA-A2 and Bap31 molecules that can interact with one-another to give a FRET signal.
Intracellular localization of Bap31 and Bap29
We have reported that besides colocalizing extensively with calnexin, an ER marker protein, Bap31 also co-localizes with ERGIC-53, a marker for the ER/Golgi intermediate compartment but not with giantin or mannosidase II, markers for the cis- and medial-Golgi complex respectively (9). Consistent with our previous report (9), we found that Bap31 also colocalized with copII, a coat protein of ER export vesicles and with Sec23, a marker of ER exit sites (data not shown), reinforcing our model of Bap31 function in trafficking of MHC class I molecules (9).
To some extent, colocalization of Bap31 with ERGIC-53 reflects the recycling of ERGIC proteins to the ER. As a control for this we compared the fraction of ERGIC marker colocalized with a known ER marker, calnexin, to the fraction localized with Bap31 or with Bap29, at 37°C, with the fraction colocalized at 15°C, a temperature that blocks recycling of the ERGIC marker, to the ER (27). Figure 2A shows representative colocalization images for cells kept at 37°C, as well as the colocalization scatter plot for each image. ERGIC-53 is red and the 3 proteins indicated in the figure are in green. The large number of pixels that are both red and green (along the diagonal in the colocalization scatter plots) is consistent either with traffic of all 3 markers to the ERGIC, or with ERGIC recycling to the ER. These possibilities are distinguished in images of cells fixed and labeled after incubation at 15°C to block recycling of ERGIC components to the ER, Figure 2B. There are few pixels along the diagonal of the colocalization graphs for calnexin and Bap29, but a considerable number of red and green pixels in images of Bap31 and ERGIC-53. The data for a large number of cells are summarized in Figure 3 in terms of the % of ERGIC-53-positive pixels that were also positive for calnexin, Bap29 or Bap31 label. It is clear that the colocalization of ERGIC-53 with calnexin and Bap29 is greatly reduced at 15°C, while that of Bap31 is only slightly reduced. This implies that a significant amount of Bap31 reaches the ERGIC, while Bap29 remains in the ER to the same extent as calnexin, an authentic ER protein. The residual colocalization of ERGIC-53 with calnexin and Bap29 is likely due to the thickness of the optical section analyzed, which results in a flattened, 2-dimensional, image with superposition of labels that are distributed in 3 dimensions.
Figure 2. Colcalization images of ERGIC-53, a marker of the ER/Golgi intermediate compartment with calnexin (an ER marker), Bap29 and Bap31.
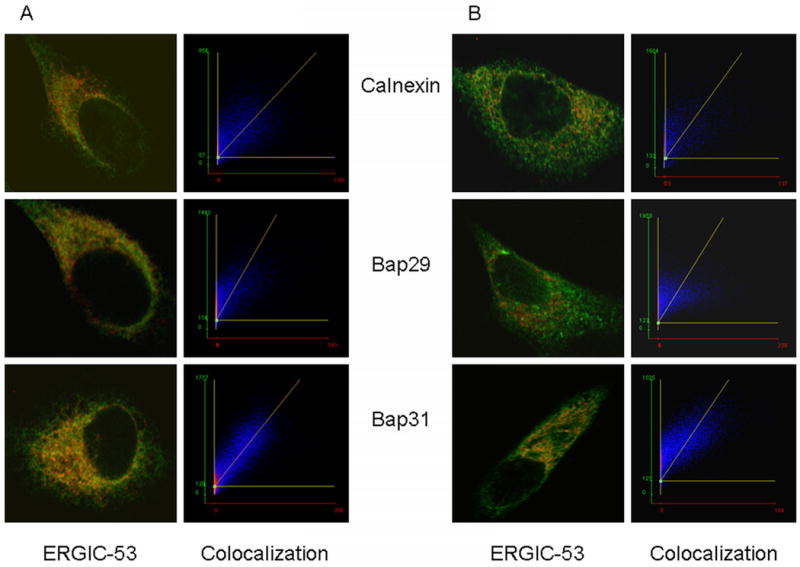
A) 37°C. All proteins were labeled with primary antibodies of different species or isotypes, followed by specific fluorescent anti-Ig. See ‘Methods’ for details. The left panels show the overlays of images of ERGIC-53 in red and the other proteins in green. The right panels are scatter plots of the pixels above a threshold that are both red and green. For ideal colocalization pixels are distributed along a diagonal. The spread away from but parallel to the diagonal is characterized by Pearson's coefficient. Pixels off the diagonal are due to bleed-through of one color into the other channel. It can be seen that there is excellent colocalization of ERGIC-53 and Bap31 (lower panels), and good colocalization of ERGIC-53 with calnexin and Bap29. B) Colocalization after cells were incubated at 15 °C to block recycling of ERGIC to the ER. There is still a significant fraction of Bap31 colocalized with ERGIC-53, indicated by the number of points distributed around the diagonal. There is little colocalization of calnexin or Bap29.
Figure 3. Fraction of ERGIC-53-positive pixels that are also positive for calnexin (Cxn) an authentic ER marker, Bap29 (B29) and Bap31 (B31).
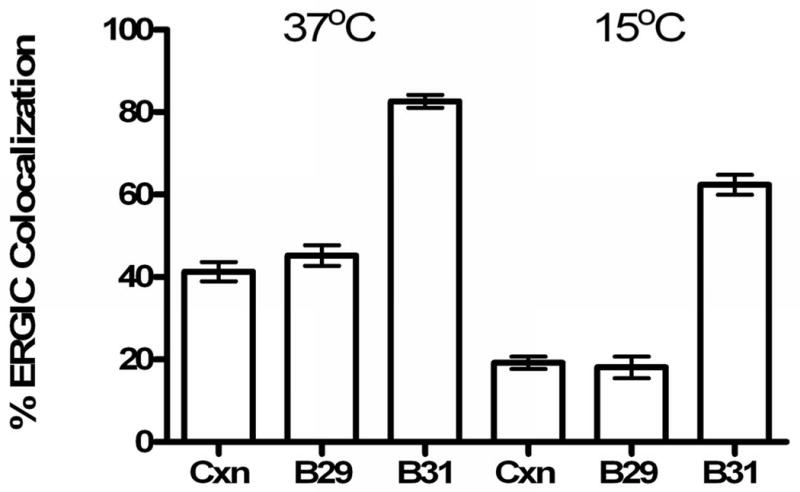
Left bars – apparent fraction of ERGIC marker colocalizing with the three proteins at 37°C. Right bars- apparent fraction of ERGIC marker colocalizing with the three proteins at 15°C. The reduction in colocalization of ERGIC-53 with the other markers at 15°C reflects the temperature-induced block in cycling of ERGIC-53 between the ERGIC and the ER (27). Note that this block affects colocalization of ERGIC-53 and Bap31, much less than it affects the colocalization of ERGIC-53 with calnexin, an ER-resident protein. The result for ERGIC-53 and Bap29 is similar to that for ERGIC-53 and calnexin, which implies that Bap29, like calnexin, is restricted to the ER. The data are averages for 20 (15°C) and 35 (37°C) cells.
Our model predicts that Bap31 association with MHC class I and its traffic to the ERGIC should increase when the ER level of peptide-loaded MHC molecules increases. To test this prediction, we first measured FRET between Bap31-mYFP and HLA-A2-mCFP at steady state (when HLA-A2 molecules are loaded with peptides with a range of affinities) and after feeding Tax peptide. Tax binds with high affinity to HLA-A2, but does not bind to the endogenous HLA molecules of HeLa cells. Earlier work has shown that such exogenous peptides reach the ER and so can bind to nascent MHC CLASS I molecules (28, 29), creating a cohort of peptide-loaded MHC CLASS I molecules which exit the ER synchronously. FRET between Bap31 and HLA-A2 increased significantly after feeding Tax peptide. FRET at an A:D of 1:1 was roughly double that of FRET of controls and increased more rapidly than in controls as A:D increased (Figure 4). We also found that this feeding increased the traffic of Bap31 out of the ER. The fraction of ERGIC marker colocalizing with endogenous Bap31 increased significantly from 60% to 80% (p<0.001) (Figure 5A). The fraction of ERGIC marker colocalizing with HLA-A2 also increased from around 65% to nearly 80% (p<0.001). Feeding TAX peptide to the fraction of cells which were not transfected with HLA-A2 did not increase colocalization of ERGIC with Bap31. The extent of ERGIC/Bap31 colocalization at steady state (before peptide feeding) was lower in HLA-A2-expressing cells than in non-transfected cells. This is likely because the increased expression of nascent MHC molecules engages an increased fraction of Bap31 in the ER, and so reduces its association with other molecules moving out of the ER.
Figure 4. FRET between Bap31-YFP and HLA-A2-CFP increases when cells are fed a peptide with high affinity for HLA-A2.
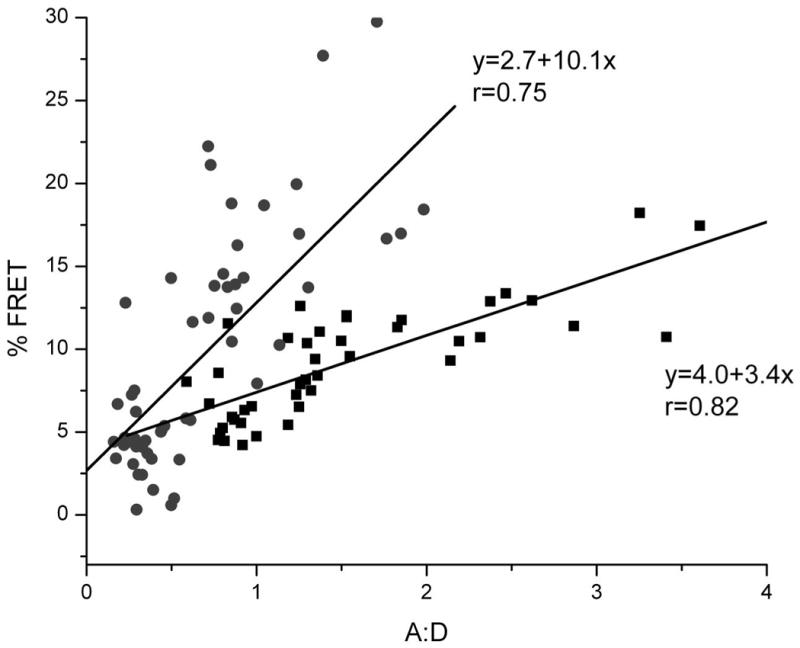
Cells transfected to express Bap31-YFP and HLA-A2-CFP were fed a high-affinity peptide, TAX (LLFGYPVYV), or vehicle (DMSO) blank for 30 minutes. Association of Bap31 and HLA-A2 was detected by FRET measured in terms of increase in donor fluorescence after acceptor photobleaching (19). Regions of interest (ROI) were chosen in the cytoplasm, away from HLA-A2 at the cell surface. FRET increase with increasing A:D, is a hallmark of specific clustering. A:D = acceptor intensity before bleaching/donor intensity after bleaching >90% of the acceptor. ■ + DMSO only, ● + TAX peptide in DMSO.
Figure 5. Feeding peptide to load a bolus of MHC class I molecules increases the amounts of MHC class I and Bap31 colocalized with the ERGIC marker, ERGIC-53.
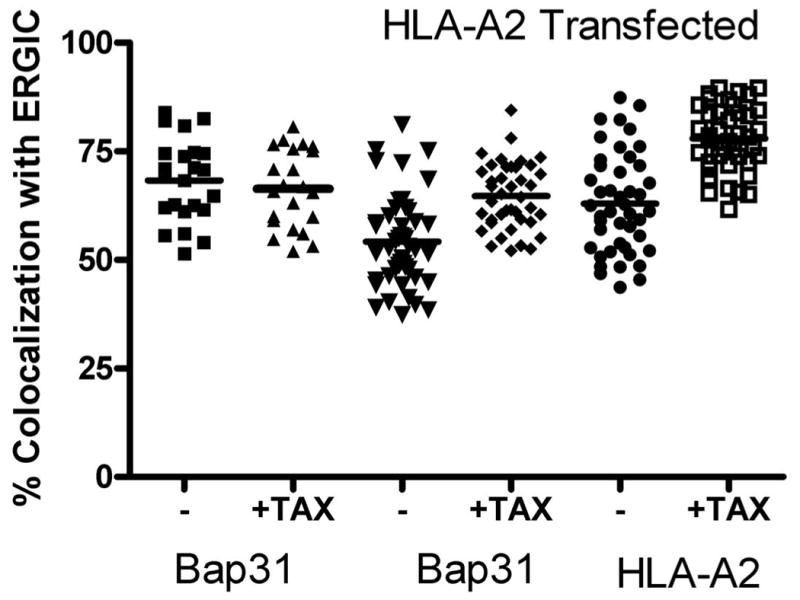

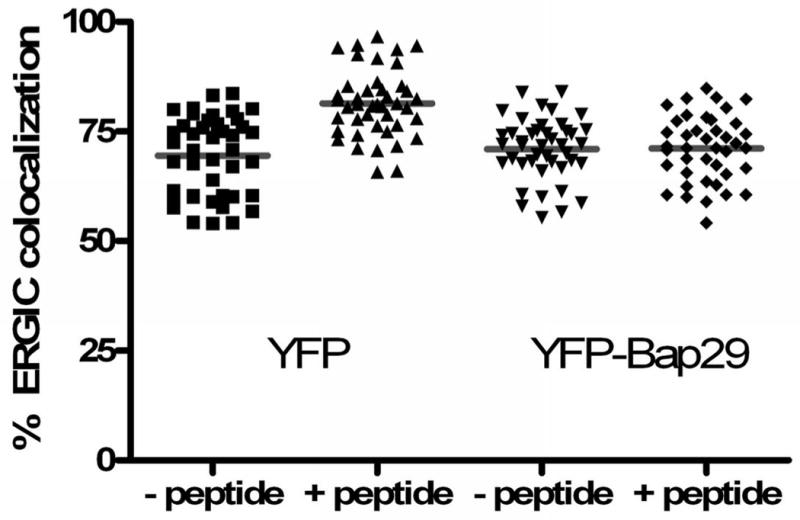
A) Cells were transfected with HLA-A2-YFP. Approximately 20 hours later the cells were fed a high-affinity peptide, TAX (LLFGYPVYV) for 30 minutes. The cells were then permeabilized and endogenous ERGIC-53 and Bap31 were labeled with the appropriate antibodies. Feeding peptide to cells which express HLA-A2-YFP significantly increased the fraction of ERGIC marker colocalized with Bap31 (p<0.001).
B) Cells were fed peptides predicted to differ by 10-fold in their affinity for HLA-A68, an endogenous HLA molecule of HeLa cells: MAGE3.112, which binds HLA-A-68 with high affinity (12) and MAGE 3a, which is predicted to bind with ∼10-fold lower affinity (13,14). Consistent with this prediction MAGE 3a rescued fewer empty MHC class I molecules from acid-stripped cells (15) than did MAGE 3.112. This was detected by flow cytometry in terms of binding of fluorescent KE-2 IgG. The open symbols and right-hand axis represent mean channel fluorescence intensity +/- 95% confidence interval for control cells, treated with weak acid, pH 3.2, for 90 s, then washed and incubated in buffer at 4°C for 1 hour followed by incubation in buffer at 37°C for 1 hour, and for cells so treated, but incubated with 12μM peptide as indicated. For colocalization studies, peptide was fed for 30 min, the cells were fixed, permeabilized and then stained for Bap31 as described in ‘Materials and Methods’. Both peptides significantly increased the extent of ERGIC colocalization with Bap31 (p<0.05 for MAGE3a and p<0.001 for MAGE 3.112).
C) Expression of Bap29 blocks the peptide-dependent forward traffic of Bap31. Cells were transfected with either free YFP or with YFP-Bap29. Feeding peptide to YFP-expressing control cells had no effect on forward traffic of Bap31 induced by feeding MAGE 3.112 (left bars), but forward traffic was blocked in cells expressing Bap29-YFP (right bars).
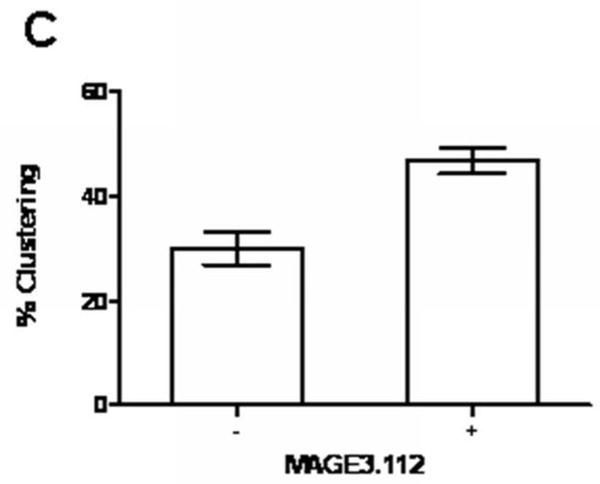
The experiments on Bap31 and HLA-A2-YFP require over-expression of the tagged MHC molecule which could alter trafficking out of the ER. As a control for this we characterized the effects of loading an endogenous HLA molecule of HeLa cells, HLA-A68, with high and low-affinity peptides. At the same concentration, the high-affinity peptide should load a greater fraction of MHC class I molecules that the low affinity peptide. We used the peptide prediction site SVRMHC (13) to specify two nonamer peptides binding to HLA-A68: MAGE3.112 (12), a known high-affinity peptide; and MAGE3a, predicted to have a 10× lower affinity for HLA-A68. The order of magnitude of the SVRMHC prediction was confirmed by a second prediction in Bimas (14). Consistent with the predictions, feeding HeLa cells MAGE3.112 for 12 hours increased surface HLA (measured with a pan-specific antibody) by 50%, while feeding the low-affinity MAGE3a did not measurably increase HLA levels above those of controls (data not shown). We further could show that at a constant concentration MAGE3.112 was more effective than MAGE3a in binding to MHC class I molecules stripped of peptide by mild acid (Figure 5B, open symbols). Feeding either MAGE3.112 peptide or Mage3a significantly increased the apparent fraction of ERGIC marker colocalizing with Bap31, and the increase was significantly larger for MAGE3.112 than for MAGE3a (p<0.05) (Figure 5B); the area of the ERGIC colocalization also increased by about 30% of the total image area (p<0.01). The increase in colocalization of Bap31 and ERGIC-53 was not due to increased recycling of ERGIC-53 to the ER, since peptide feeding did not change the extent of colocalization of ERGIC-53 with Bap29, a protein whose organelle localization matches that of calnexin, an authentic ER protein, indicating that Bap29 is retained in the ER (data not shown). Indeed, over-expression of YFP-Bap29 blocked the increased forward trafficking of Bap31 after peptide feeding (Figure 5C). This implies that the low level FRET detected between Bap31 and Bap29 reflects a functional interaction, as suggested by immunoprecipitation experiments (24).
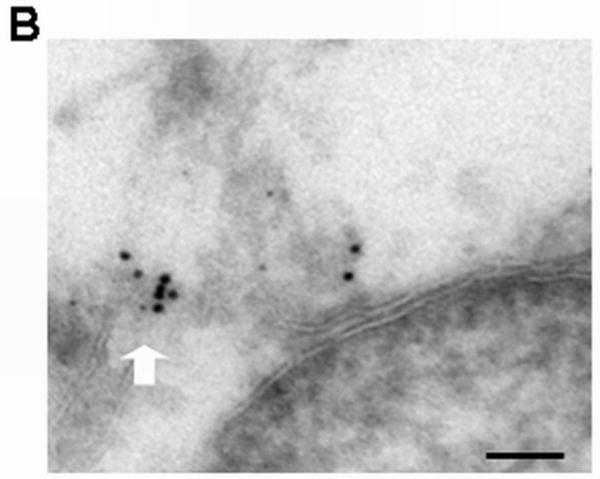
The effects of a cohort of peptide-loaded MHC molecules on traffic of Bap31 to the ERGIC were also detected in terms of increased FRET between Bap31 and ERGIC-53, and by immuno-electron microscopy. HeLa cells were treated with MAGE3.112 peptide as described above, fixed, and stained with Cy5 (acceptor) anti-ERGIC-53 and Alexa 546 (donor) anti-Bap31. FRET between the ERGIC marker and anti-Bap31 increased after feeding peptide for 30 minutes. Surprisingly, this FRET increased with increasing A:D ratio, suggesting some specificity of clustering (Figure 6A). However, this dependence on A:D may be due to the relatively small volume of ERGIC vesicles and consequent high concentration of labeled donors and acceptors. Analysis of electron micrographs of immunolabeled Bap31 and ERGIC-53 also showed that a fraction of the ERGIC-53 and Bap31, respectively defined by 12 nm gold and 6 nm gold particles, clustered in the range 10-70 nm (Figure 6B). The clustered fraction increased significantly after peptide feeding (p<.001) (Figure 6C).
Figure 6. Peptide loading of endogenous HLA-A68 molecules increases the proximity of Bap31 and ERGIC53 resolved by FRET and by immunoelectron microscopy.
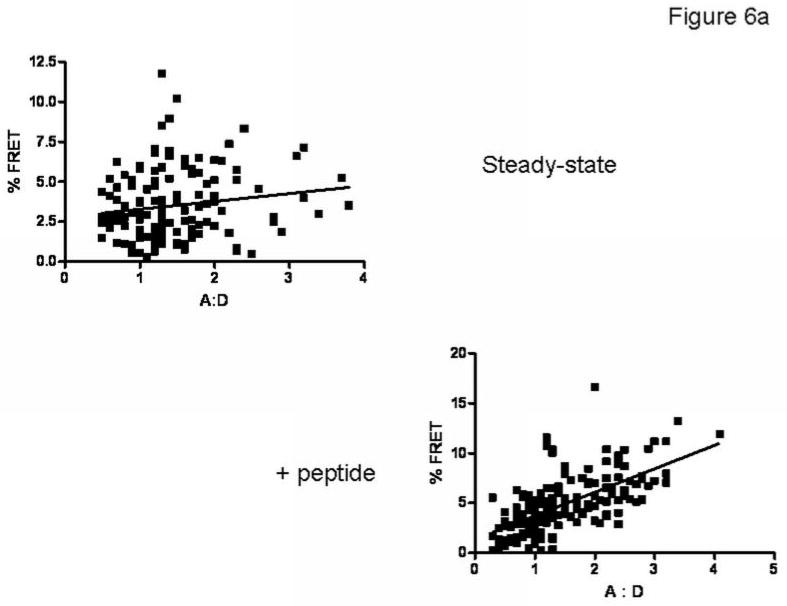
A) Peptide feeding increases FRET between antibody-labeled proteins. At steady-state FRET is independent of A:D. The data are fit by y=3+.5x. The slope is not significantly different from 0. After peptide feeding y=1+2.4x, a slope significantly > 0. The dependence of FRET on A:D after peptide feeding suggests specific clustering of Bap31 and ERGIC-53. However, this may also reflect the limited membrane area of ERGIC vesicles.
B) Proximity of immuno-gold labeled ERGIC-53 and Bap31 after feeding peptide. Bap31 labeled with 6 nm colloidal gold appears to be within some 10's of nm of ERGIC-53 labeled with 12 nm gold (white arrow). Scale bar = 100 nm.
C) Statistics of 6 nm and 12 nm gold within 60 nm of one another before and after feeding MAGE3.112 peptide to create a bolus of peptide-loaded MHC class I molecules ready for export from the ER. The difference in extent of clustering is statistically significant, p<0.001.
Discussion
We have proposed that the ER membrane protein Bap31 traffics peptide-loaded MHC class I molecules out of the ER, moving with the MHC class I molecules to the ERGIC (9). This model originated in our findings that peptide-loaded MHC class I molecules were not synchronously released from the ER, implying that they required specific carriers and that Bap31 co-precipitated with mouse (5) and human (9) MHC class I molecules. In the present study, we used FRET measurements and quantitative imaging to show that Bap31 is associated with HLA-A2 molecules in the ER of intact cells, and that it associates with peptide-loaded MHC class I molecules to a greater extent than with nascent molecules. Another consequence of peptide loading is that there is an increase in Bap31 traffic to the ERGIC. We also showed a regulatory interaction of Bap31 with its homolog, Bap29.
The difference in the extent of Bap31 traffic to the ERGIC after HeLa cells were fed high- or low-affinity peptides could reflect retention of empty MHC class I molecules or free heavy chains in the ER, perhaps by association of Bap31/MHC class I with Bap29. Alternately, it could reflect enhanced recycling of empty MHC class I molecules and free heavy chains from the ERGIC to the ER. A given concentration of low-affinity peptide would load a smaller fraction of nascent MHC class I molecules than the same concentration of high-affinity peptide, leaving more empty MHC class I molecules to be retained in the ER. On the other hand, a fraction of MHC class I molecules loaded with low-affinity peptide could lose this peptide in a slightly acid environment in the ERGIC (30), and recycle back to the ER complexed with Bap31. Whatever the mechanism, the requirement for Bap31 or other carriers limits the rate of MHC class I export from the ER and a population of peptide-loaded molecules persists there for some time after peptide loading (5). Further work on the pH of the ERGIC, and its influence on the stability of MHC/peptide binding is needed to further elucidate Bap31 function in the quality control of MHC class I peptide complexes.
Though it is clear that there is selection for stable MHC class I molecules, those that have bound peptides with high affinity, the mechanisms that ensure this, so-called quality-control mechanisms, are not well defined and are controversial. A specialized ER chaperone, tapasin, has been implicated in MHC class I quality control (experiments summarized in (31)). There appear to be parallels between tapasin action on nascent MHC class I molecules and the action of the known quality control protein for the homologous MHC class II proteins (32). However, in our hands tapasin does not appear to affect the stability of MHC class I molecules that reach the cell surface of B lymphoblasts (31), though it does enhance stability of MHC class I molecules in HeLa cells (9). This inconsistency may reflect the fact that different quality-control mechanisms are used in different cell types. Indeed, Bap31 itself does not reach the ERGIC in some MHC class I-expressing cells (32), and is not required for ER export and trafficking of stable MHC class I molecules in cells lacking Bap31(9). There may well be multiple redundant mechanisms of MHC class I quality control.
Besides interacting with MHC class I molecules, Bap31 also self-associated and associated with its homolog, Bap29. Unlike Bap31, Bap29 was confined to the ER where it interacted with a fraction of Bap31; this interaction blocked the forward traffic of Bap31 after peptide feeding. We earlier showed that over-expressing Bap29 reduced surface MHC class I expression (9). From the present result it appears that the effect of Bap29 on MHC class I expression is due to its restriction of Bap31 traffic out of the ER. Hence it appears that in cells in which Bap31 functions as a major ER export carrier for MHC class I molecules; this function, as recently suggested (24) is modulated by associations with its partner protein, Bap29.
Supplementary Material
Acknowledgments
We thank Dr. Hans-Peter Hauri (University of Basel) for providing antibodies for Bap31 and ERGIC-53; Dr. Gordon C. Shore (McGill University) for providing Bap29 antibody; Dr. J. Michael McCaffery, Michelle Husein and Ned Perkins (Integrated Imaging Center, Johns Hopkins University) for technical support; and Sarah Boyle, Maya Everett, David Fooksman and Saame R. Shaikh (Edidin Laboratory, Johns Hopkins University) for technical advice and valuable comments.
Abbreviations used in this paper
- CFP
cyan fluorescent protein
- ER
endoplasmic reticulum
- ERGIC
ER/Golgi intermediate compartment
- FRET
Förster resonance energy transfer
- YFP
yellow fluorescent protein
- A:D
acceptor-to-donor ratio
Footnotes
This work was supported by a grant from the National Institutes of Health, AI-14584 (M.E.).
References
- 1.York IA, Rock KL. Antigen processing and presentation by the class I major histocompatibility complex. Annu Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Pamer E, Cresswell P. Mechanisms of MHC class I--restricted antigen processing. Annu Rev Immunol. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Williams DB. Assembly of MHC class I molecules within the endoplasmic reticulum. Immunol Res. 2006;35:151–162. doi: 10.1385/IR:35:1:151. [DOI] [PubMed] [Google Scholar]
- 4.Kim KM, Adachi T, Nielsen PJ, Terashima M, Lamers MC, Kohler G, Reth M. Two new proteins preferentially associated with membrane immunoglobulin D. EMBO J. 1994;13:3793–3800. doi: 10.1002/j.1460-2075.1994.tb06690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spiliotis ET, Manley H, Osorio M, Zuniga MC, Edidin M. Selective export of MHC class I molecules from the ER after their dissociation from TAP. Immunity. 2000;13:841–851. doi: 10.1016/s1074-7613(00)00081-9. [DOI] [PubMed] [Google Scholar]
- 6.Paquet ME, Cohen-Doyle M, Shore GC, Williams DB. Bap29/31 influences the intracellular traffic of MHC class I molecules. J Immunol. 2004;172:7548–7555. doi: 10.4049/jimmunol.172.12.7548. [DOI] [PubMed] [Google Scholar]
- 7.Annaert WG, Becker B, Kistner U, Reth M, Jahn R. Export of cellubrevin from the endoplasmic reticulum is controlled by BAP31. J Cell Biol. 1997;139:1397–1410. doi: 10.1083/jcb.139.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert G, Becker B, Schreiber R, Boucherot A, Reth M, Kunzelmann K. Control of cystic fibrosis transmembrane conductance regulator expression by BAP31. J Biol Chem. 2001;276:20340–20345. doi: 10.1074/jbc.M011209200. [DOI] [PubMed] [Google Scholar]
- 9.Ladasky JJ, Boyle S, Seth M, Li H, Pentcheva T, Abe F, Steinberg SJ, Edidin M. Bap31 enhances the endoplasmic reticulum export and quality control of human class I MHC molecules. J Immunol. 2006;177:6172–6181. doi: 10.4049/jimmunol.177.9.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manley HA, Lennon VA. Endoplasmic reticulum membrane-sorting protein of lymphocytes (BAP31) is highly expressed in neurons and discrete endocrine cells. J Histochem Cytochem. 2001;49:1235–1243. doi: 10.1177/002215540104901005. [DOI] [PubMed] [Google Scholar]
- 11.Ng FW, Nguyen M, Kwan T, Branton PE, Nicholson DW, Cromlish JA, Shore GC. p28 Bap31, a Bcl-2/Bcl-XL- and procaspase-8-associated protein in the endoplasmic reticulum. J Cell Biol. 1997;139:327–338. doi: 10.1083/jcb.139.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawashima I, Hudson SJ, Tsai V, Southwood S, Takesako K, Appella E, Sette A, Celis E. The multi-epitope approach for immunotherapy for cancer: identification of several CTL epitopes from various tumor-associated antigens expressed on solid epithelial tumors. Hum Immunol. 1998;59:1–14. doi: 10.1016/s0198-8859(97)00255-3. [DOI] [PubMed] [Google Scholar]
- 13.Wan J, Liu W, Xu Q, Ren Y, Flower DR, Li T. SVRMHC prediction server for MHC-binding peptides. BMC Bioinformatics. 2006;7:463. doi: 10.1186/1471-2105-7-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 15.Van der Burg SH, Ras E, Drijfhourt JW, Benckhuijsen WE, Bremers AJA, Melief CJ, Kast WM. An HLA Class 1 Peptide-Binding Assay Based on Competition for Binding to Class Molecules on Intact Human B cells Identification of Conserved HIV-1 Polymerase Peptides Binding to HLA-A*0301. Hum Immunol. 1995;44:189–198. doi: 10.1016/0198-8859(95)00105-0. [DOI] [PubMed] [Google Scholar]
- 16.Kannagi M, Shida H, Igarashi H, Kuruma K, Murai H, Aono Y, Maruyama I, Osame M, Hattori T, Inoko H, Harada S. Target epitope in the Tax protein of human T-cell leukemia virus type I recognized by class I major histocompatibility complex-restricted cytotoxic T cells. J Virol. 1992;66:2928–2933. doi: 10.1128/jvi.66.5.2928-2933.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauvau G, Kakimi K, Niedermann G, Ostankovitch M, Yotnda P, Firat H, Chisari FV, van Endert PM. Human transporters associated with antigen processing (TAPs) select epitope precursor peptides for processing in the endoplasmic reticulum and presentation to T cells. J Exp Med. 1999;190:1227–1240. doi: 10.1084/jem.190.9.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 19.Kenworthy AK, Edidin M. Distribution of a glycosylphosphatidylinositol anchored protein at the apical surface of MDCK cells examined at a resolution of <100 A using imaging fluorescence resonance energy transfer. J Cell Biol. 1998;142:69–84. doi: 10.1083/jcb.142.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valentin G, Verheggen C, Piolot T, Neel H, Coppey-Moisan M, Bertrand E. Photoconversion of YFP into a CFP-like species during acceptor photobleaching FRET experiments. Nat Methods. 2005;2:801. doi: 10.1038/nmeth1105-801. [DOI] [PubMed] [Google Scholar]
- 21.Stepensky D. FRETcalc plugin for calculation of FRET in non-continuous intracellular compartments. Biochem Biophys Res Commun. 2007;359:752. doi: 10.1016/j.bbrc.2007.05.180. [DOI] [PubMed] [Google Scholar]
- 22.Pentcheva T, Edidin M. Clustering of peptide-loaded MHC class I molecules for endoplasmic reticulum export imaged by fluorescence resonance energy transfer. J Immunol. 2001;166:6625–6632. doi: 10.4049/jimmunol.166.11.6625. [DOI] [PubMed] [Google Scholar]
- 23.Philimonenko AA, Janacek J, Hozak P. Statistical evaluation of colocalization patterns in immunogold labeling experiments. J Struct Biol. 2000;132:201–210. doi: 10.1006/jsbi.2000.4326. [DOI] [PubMed] [Google Scholar]
- 24.Schamel WW, Kuppig S, Becker B, Gimborn K, Hauri HP, Reth M. A high-molecular-weight complex of membrane proteins BAP29/BAP31 is involved in the retention of membrane-bound IgD in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2003;100:9861–9866. doi: 10.1073/pnas.1633363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pentcheva T, Spiliotis ET, Edidin M. Tapasin is retained in the endoplasmic reticulum by dynamic clustering and exclusion from endoplasmic reticulum exit sites. J Immunol. 2002;168:1538–1541. doi: 10.4049/jimmunol.168.4.1538. [DOI] [PubMed] [Google Scholar]
- 26.Szczesna-Skorupa E, Kemper B. BAP31 is involved in the retention of cytochrome P450 2C2 in the endoplasmic reticulum. J Biol Chem. 2005;281:4142–8. doi: 10.1074/jbc.M509522200. [DOI] [PubMed] [Google Scholar]
- 27.Saraste J, Svensson K. Distribution of the intermediate elements oper ating in ER to Golgi transport. J Cell Sci. 1991;100:415–430. doi: 10.1242/jcs.100.3.415. [DOI] [PubMed] [Google Scholar]
- 28.Day PM, Yewdell JW, Porgador A, Germain RN, Bennink JR. Direct delivery of exogenous MHC class I molecule-binding oligopeptides to the endoplasmic reticulum of viable cells. Proc Natl Acad Sci U S A. 1997;94:8064–8069. doi: 10.1073/pnas.94.15.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marguet D, Spiliotis ET, Pentcheva T, Lebowitz M, Schneck J, Edidin M. Lateral diffusion of GFP-tagged H2Ld molecules and of GFP-TAP1 reports on the assembly and retention of these molecules in the endoplasmic reticulum. Immunity. 1999;11:231–240. doi: 10.1016/s1074-7613(00)80098-9. [DOI] [PubMed] [Google Scholar]
- 30.Palokangas H, Ying M, Väänänen K, Saraste J. Retrograde transport from the pre-Golgi intermediate compartment and the Golgi complex is affected by the vacuolar H+-ATPase inhibitor bafilomycin A1. Mol Biol Cell. 1998;9:3561–3578. doi: 10.1091/mbc.9.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Everett MW, Edidin M. Tapasin increases efficiency of MHC CLASS I assembly in the endoplasmic reticulum but does not affect MHC CLASS I stability at the cell surface. J Immunol. 2007;179:7646–7652. doi: 10.4049/jimmunol.179.11.7646. [DOI] [PubMed] [Google Scholar]
- 32.Chen M, Bouvier M. Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J. 2007;26:1681–1690. doi: 10.1038/sj.emboj.7601624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breuza L, Halbeisen R, Jeno P, Otte S, Barlowe C, Hong W, Hauri HP. Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J Biol Chem. 2004;279:47242–47253. doi: 10.1074/jbc.M406644200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


