Abstract
The constitutive androstane receptor (CAR) is constitutively activated in immortalized cell lines independent of xenobiotic stimuli. This feature of CAR has limited its use as a sensor for xenobiotic-induced expression of drug-metabolizing enzymes. Recent reports, however, reveal that a splicing variant of human CAR (hCAR3), which contains an insertion of five amino acids (APYLT), exhibits low basal but xenobiotic-inducible activities in cell-based reporter assays. Nonetheless, the underlying mechanisms of this functional shift are not well understood. We have now generated chimeric constructs containing various residues of the five amino acids of hCAR3 and examined their response to typical hCAR activators. Our results showed that the retention of alanine (hCAR1+A) alone is sufficient to confer the constitutively activated hCAR1 to the xenobiotic-sensitive hCAR3. It is noteworthy that hCAR1+A was significantly activated by a series of known hCAR activators, and displayed activation superior to that of hCAR3. Moreover, intracellular localization assays revealed that hCAR1+A exhibits nuclear accumulation upon 6-(4-chlorophenyl) imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl) oxime (CITCO) treatment in COS1 cells, which differs from the spontaneous nuclear distribution of hCAR1 and the nontranslocatable hCAR3. Mammalian two-hybrid and glutathione S-transferase pull-down assays further demonstrated that hCAR1+A interacts with the coactivator SRC-1 and GRIP-1 at low level before activation, while at significantly enhanced level in the presence of CITCO. Thus, the alanine residue in the insertion of hCAR3 seems in charge of the xenobiotic response of hCAR3 through direct and indirect mechanisms. Activation of hCAR1+A may represent a sensitive avenue for the identification of hCAR activators.
Functioning as a xenosensor, human constitutive androstane receptor (hCAR, hCAR1, or NR1I3) regulates numerous hepatic genes that encode phase I oxidation enzymes [e.g., cytochrome P450 (CYP)], phase II conjugation enzymes (e.g., UDP glucuronosyltransferases), and phase III drug transporters (e.g., multidrug resistance MDR1) upon xenobiotic stimulation (Sueyoshi et al., 1999; Honkakoski et al., 2003; Qatanani and Moore, 2005; Stanley et al., 2006). Through induction of these enzymes and transporters, hCAR is also involved in the metabolism and secretion of endogenous signaling molecules such as cholesterol and bilirubin, where bioaccumulation of these endobiotics is associated with disease conditions such as cholestasis and hyperbilirubinemia (Sugatani et al., 2001; Tien and Negishi, 2006). In addition, recent studies also extend the roles of CAR to the regulation of various physiological and pathological processes such as energy homeostasis, cell proliferation/apoptosis, and tumor promotion (Kodama et al., 2004; Maglich et al., 2004; Yamamoto et al., 2004; Huang et al., 2005). Thus, the need for understanding the molecular mechanisms governing CAR activation, and developing novel tools for in vitro identification of hCAR activators has become evident.
Compared with other nuclear receptors, CAR displays several distinctive characteristics in its activation including both direct and indirect mechanisms. In primary cultures of hepatocytes or in vivo, CAR resides in the cytoplasm forming a complex with heat shock protein 90, cytoplasmic CAR retention protein, and membrane-associated subunit of protein phosphatase 1 (PPP1R16A) (Kobayashi et al., 2003; Yoshinari et al., 2003; Sueyoshi et al., 2008). CAR translocates into the nucleus and turns on its target gene expression only after exposure to chemical activators such as 6-(4-chlorophenyl) imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl) oxime (CITCO) a human-specific CAR agonist, and phenobarbital (PB), a universal indirect activator of CAR (Kawamoto et al., 1999; Maglich et al., 2003). In contrast to the observations in primary hepatocytes, expression of CAR in immortalized cell lines, such as HepG2 cells results in spontaneous nuclear accumulation and constitutive activation of this receptor independent of xenobiotic activation (Baes et al., 1994; Zelko et al., 2001; Li et al., 2009). The lack of cell lines that maintain CAR distribution and activation in a physiologically relevant manner has become a major obstacle in investigating the mechanisms of xenobiotic-mediated CAR activation. To date, although several chaperone molecules involving CAR cytoplasmic retention such as cytoplasmic CAR retention protein and PPP1R16A have been identified, the role of CAR protein variants in the constitutive versus chemical-mediated CAR activation remains unclear.
Many naturally occurring alternative splicing variants of hCAR have recently been identified and functionally characterized by several groups (Arnold et al., 2004; Jinno et al., 2004; Auerbach et al., 2005). Among these spliced hCAR transcripts, the hCAR3, which contains an in-frame insertion of five amino acids (APYLT) in the highly conserved region of the ligand-binding domain (LBD), exhibited minimal basal, but potent ligand-induced activities in cell-based reporter assays (Auerbach et al., 2005; Faucette et al., 2007). It is intriguing that CITCO treatment was incapable of facilitating hCAR3 nuclear translocation in COS1 cells or rat primary hepatocytes (Jinno et al., 2004; Auerbach et al., 2005). Given the complexities encountered in studying hCAR activation in vitro, these initial observations of hCAR3 make it an attractive target for illustrating the mechanisms of hCAR activation.
To define the contribution of the five-amino-acid insertion on the functional transformation of hCAR3, we have generated a series of chimeric constructs containing various residues of the five-amino-acid insertion and evaluated their function in response to prototypical hCAR activators. The current studies demonstrate that retention of the alanine residue alone (hCAR1+A) seems sufficient to shift the constitutively activated hCAR1 to the xenobiotic-sensitive hCAR3. The chemical specificities of hCAR1+A activation closely resemble that of the reference hCAR1. Furthermore, our intracellular localization assays revealed that hCAR1+A exhibits nuclear translocation upon CITCO treatment in immortalized cell line. Thus, the unique feature of hCAR1+A may eventually provide a new system that could facilitate our efforts in delineating the mechanisms of hCAR activation.
Materials and Methods
Chemicals and Biological Reagents.
PB, phenytoin (PHN), rifampicin (RIF), artemisinin (ART), carbamezapine (CMZ), WY-14643, 1,4-bis[2-(3,5-dichlorpyridyloxy)]benzene (TCPOBOP), clotrimazole (CLZ), butylated hydroxyanisole (BHA), PK11195, chenodeoxycholic acid (CDCA), 22(R)-hydroxycholesterol (HOC), diazepam (DAP), methadone (MD), 3-methylcholanthrene (3MC), and meclizine (MLZ) were purchased from Sigma-Aldrich (St. Louis, MO). Okadaic acid (OA) was purchased from Calbiochem (San Diego, CA). CITCO was obtained from BIOMOL Research Laboratories (Plymouth Meeting, PA). Efavirenz (EFV) was purchased from Toronto Research Chemicals (Toronto, ON, Canada), and nevirapine (NVP) was purchased from U.S. Pharmacopeia (Rockville, MD). Fluconazole (FLU) and myclobutanil (MCB) were purchased from LKT Laboratories (St. Paul, MN). The Dual-Luciferase Reporter Assay System was purchased through Promega (Madison, WI). FuGENE 6 and Fugene HD transfection reagents were obtained from Roche (Basel, Switzerland). Other cell culture reagents were purchased from Invitrogen (Carlsbad, CA) or Sigma-Aldrich.
Plasmids Constructions.
The pCR3-hCAR1, pEYFP-hCAR1, GST-hCAR1, pcDNA3.1-mSRC-1, and pcDNA3.1-GRIP-1 were provided by Dr. Masahiko Negishi (National Institute of Environmental and Health Sciences, National Institutes of Health, Research Triangle Park, NC). The pCMV2-hCAR3 expression vector and CYP3A4-PXRE/XREM luciferase reporter construct were obtained from Drs. Curtis Omiecinski (Pennsylvania State University, University Park, PA) and Bryan Goodwin (GlaxoSmithKline, Research Triangle Park, NC), respectively. The CYP2B6-PBREM/XREM reporter was described previously (Wang et al., 2003). Plasmids used for mammalian two-hybrid assay, pG5-Luc and pACT, were obtained from Promega. pACT-hCAR1, pM-SRC-1 (621–765), and pM-GRIP-1 were generated as described previously (Ueda et al., 2005). The pEYFP-hCAR3, GST-hCAR3, and pACT-hCAR3, vectors were constructed by subcloning the full-length hCAR3 into the multicloning sites of pEYFP-c1, pGEX4T-3, or pACT, respectively. The pRL-TK Renilla luciferase plasmid used to normalize firefly luciferase activities from Promega.
Generation of hCAR Chimeric Constructs.
Human CAR chimeras (Fig. 1A) were generated by introducing appropriate nucleotides corresponding to the residues of the five-amino-acid insertion of hCAR3 into the pCR3-hCAR1 expression vector by use of the QuikChange Site-directed Mutagenesis System (Stratagene, La Jolla, CA). The mutagenic primers used for constructing the chimeras were summarized in Table 1. The nucleotides GCT reflecting alanine in the hCAR3 insertion were also introduced into hCAR1/pEYFP-c1, GST-hCAR1, and pACT-hCAR1 plasmids for confocal imaging, mammalian two-hybrid, and GST-pull-down assays. All resulted clones were confirmed by sequencing.
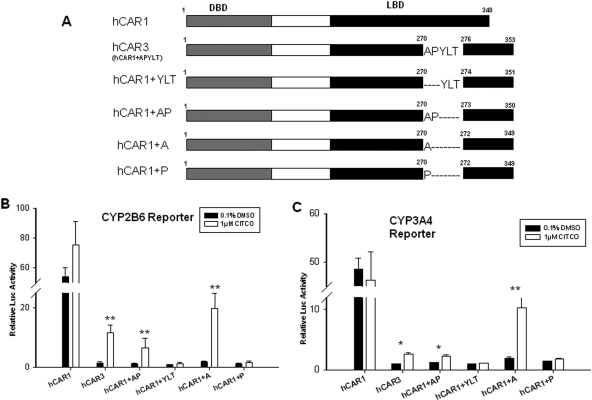
Fig. 1.
Activation of hCAR1, hCAR3, and chimeric constructs in cell-based reporter assays. Schematic structure organization of the reference (hCAR1), splice variant (hCAR3), and chimeric human CAR transcripts (A). HepG2 cells were transfected with CYP2B6-PBREM (B) or CYP3A4 XREM (C) reporter construct in the presence of hCAR1, hCAR3, or hCAR chimeric expression vectors. Transfected cells were treated with vehicle control (0.1% DMSO) or 1 μM CITCO for 24 h. Luciferase activities were determined and expressed relative to hCAR3 vehicle control. Data represent the mean ± S.D. (n = 3) (*, p < 0.05; **, p < 0.01).
Table 1.
Generation of hCAR chimeric constructs
| Vector Names | Mutagenesis Primers | Source Vector | Introduced Amino Acids |
|---|---|---|---|
| hCAR1+YLT | 5′-CATGGCCCTCTTCTCTCCTTATCTTACAGACCGACCTG-3′ | pCR3/hCAR1 | YLT (271–273) |
| 5′-CAGGTCGGTCTGTAAGATAAGGAGAGAAGAGGGCCATG-3′ | |||
| hCAR1+AP | 5′-CTCTTCTCTCCTGCTCCCGACCGACCTGGAGTTACC-3′ | pCR3/hCAR1 | AP (271–272) |
| 5′-GGTAACTCCAGGTCGGTCGGGAGCAGGAGAGAAGAG-3′ | |||
| hCAR1+A | 5′-CCCTCTTCTCTCCTGCTGACCGACCTGGAGTTAC-3′ | pCR3/hCAR1 | A (271) |
| 5′-GTAACTCCAGGTCGGTCAGCAGGAGAGAAGAGGG-3′ | |||
| hCAR1+P | 5′-CCTCTTCTCTCCTCCCGACCGACCTGGAG-3′ | pCR3/hCAR1 | P (271) |
| 5′-CTCCAGGTCGGTCGGGAGGAGAGAAGAGG-3′ |
Transfection Assays in Cell Lines.
HepG2 cells in 24-well plates were transfected with CYP2B6-PBREM/XREM or CYP3A4-PXRE/XREM reporter vector, and control plasmid (pRL-Tk) in the presence of hCAR1, hCAR3, or one of the hCAR chimeric expression constructs (hCAR1+A, hCAR1+P, hCAR1+AP, or hCAR1+YLT) by use of Fugene 6 reagents following the manufacturer's instructions. Eighteen hours after transfection, cells were treated for 24 h with vehicle control (0.1% DMSO), positive control (CITCO), or test compounds at indicated concentrations. Cell lysates were assayed for firefly activities normalized against the activities of cotransfected Renilla by use of Dual-luciferase kit (Promega). Data were represented as mean ± S.D. of three individual transfections.
Intracellular hCAR Localization and Western Blot Assays.
COS1 cells in 12-well plates were transfected with 1 μg of pEYFP-hCAR1, pEYFP-hCAR3, or pEYFP-(hCAR1+A) plasmid by use of Fugene HD reagent following the manufacturer's instruction. Twenty-four hours later, cells were treated with vehicle control (0.1% DMSO) or CITCO (1 μM) for 24 h. Subsequently, cells were fixed in 4% paraformaldehyde and stained with 4,6-diamidino-2-phenylindole for nucleus visualization. Localization of transfected hCARs was examined by use of Confocal Nikon TE2000 as described previously (Li et al., 2009). For each treatment, approximately 100 cells expressing pEYFP-hCARs were counted and classified based on cytosolic, nuclear, or mixed (cytosolic + nuclear) hCAR localizations. In a parallel experiment, COS1 cells in 60-mm dishes were transfected with 5 μg of pEYFP-hCAR1, pEYFP-hCAR3, or pEYFP-(hCAR1+A), and treated with vehicle control or CITCO for 24 h as described above. Preparation of nuclear proteins from these cells were carried out as described previously (Wang et al., 2004; Inoue et al., 2006), and the protein concentrations were determined with the bicinchoninic acid protein assay kit (Pierce Chemical, Rockford, IL). For Western blotting analysis, nuclear proteins (30 μg) were separated on a NuPAGE Novex 4 to 12% Bis-Tris gel (Invitrogen) and transferred on to a polyvinylidene difluoride membrane. The membranes were subsequently probed with specific antibody against hCAR (Perseus Proteomics, Tokyo, Japan) or antibody against transcriptional binding protein (TBP) (Santa Cruz Biotechnology, Santa Cruz, CA) and were incubated with horseradish peroxidase-conjugated anti-rabbit IgG. Protein bands were developed with ECL (GE Healthcare, Little Chalfont, Buckinghamshire, UK).
Chromatin Immunoprecipitation Assay.
HepG2 cells were transfected with pCR3-hCAR1 or pCR3-hCAR1+A expression vector then treated with control (0.1% DMSO) or CITCO (1 μM) for 2 h. Subsequently, cells were cross-linked and processed by use of a ChIP assay kit (Millipore Corporation, Billerica, MA) according to the manufacturer's instructions. Precleared chromatin solution was incubated with 5 μg of anti-hCAR antibody (Perseus Proteomics) or normal rabbit IgG (Santa Cruz Biotechnology) at 4°C overnight, from which immunoprecipitates were collected to purify DNA by use of a QIAQuick PCR purification kit (QIAGEN, Valencia, CA). The PBREM region of the CYP2B6 promoter was amplified by use of the purified DNA as template and the primers, CYP2B6-F: 5′-ctgcaatgagcacccaatctt-3′; and CYP2B6-R: 5′-acacatcctctgacagggtca-3′.
Mammalian Two-Hybrid Assay.
COS1 cells in 24-well plates were transfected with 110 ng of the reporter gene pG5-luc, 40 ng of pM-SRC-1 or pM-GRIP-1, 80 ng of the respective pACT-hCAR, and 20 ng of reference plasmid pRL-TK by use of Fugene 6. Eighteen hours after transfection, the cells were treated with vehicle control (0.1% DMSO) or 1 μM CITCO for 24 h. Luciferase activities were measured as described above. Data were represented as mean ± S.D. of three individual transfections.
GST Pull-Down Assay.
The GST-hCAR1, GST-hCAR3, and GST-(hCAR1+A) were expressed in Escherichia coli BL21(DE3) cells (Stratagene) and purified with Glutathione-Sepharose 4B beads (GE Healthcare). 35S-Labeled SRC-1 and GRIP-1 were produced from pcDNA3.1-mSRC1 and pcDNA3.1-hGRIP1 by use of the TNT T7 Quick Coupled Transcription/Translation System (Promega) along with [35S]methionine. The glutathione-Sepharose 4B beads coupled by equilibrated GST or various GST-hCAR proteins, and [35S]methionine-labeled SRC-1 or GRIP-1 were mixed in the GST interaction buffer containing 2 mg/ml bovine serum albumin, 25 mM HEPES, pH7.6, 100 mM NaCl, 20% glycerol, and 1 mM EDTA. The mixture was rotated overnight at 4°C in the presence of 0.1% DMSO or 2 μM CITCO. The resin was then recovered by centrifugation and washed three times in the same buffer. Proteins were extracted from the resin by heating for 10 min at 70°C in NuPAGE LDS sample buffer (Invitrogen) and separated on 4 to 12% Bis-Tris gel. The gel was then dried under vacuum, and proteins were detected by autoradiography.
Statistical Analysis.
Experimental data are presented as a mean of triplicate determinations ± S.D. unless otherwise noted. Statistical comparisons were made by use of Student's t test and χ2 test. The statistical significance was set at p values of <0.05 (*), or <0.01 (**).
Results
Effects of the Residue(s) of hCAR3 Insertion on the Basal- and Ligand-Mediated Activation.
To determine the roles of different amino acids of the hCAR3 insertion on the activation of the hCAR, a number of chimeric expression vectors have been generated (Fig. 1A). As demonstrated in Fig. 1, B and C, the reference hCAR1 displayed constitutively high basal activity and negligible response to the known hCAR activator (CITCO) in HepG2 cells, which is consistent with previous reports (Kawamoto et al., 1999; Faucette et al., 2006). In contrast, all the generated chimeras (hCAR1+A, hCAR1+P, hCAR1+AP, and hCAR1+YLT) exhibited low basal activity similar to the hCAR3 splicing variant in the absence of CITCO, but only hCAR1+AP and hCAR1+A were activated in the presence of CITCO (1 μM). It is noteworthy that hCAR1+A was activated to 20-fold over vehicle control, whereas hCAR3 and hCAR1+AP were activated to 5- and 3-fold, respectively, in CYP2B6 reporter assays. Similar patterns were observed in CYP3A4 reporter assay, where the activation of hCAR1+A, hCAR1+AP, and hCAR3, by CITCO were also increased to 10-, 3-, and 3-fold over control, respectively. These results suggest that the alanine in the five-amino-acid insertion of hCAR3 is essential for the chemical-mediated activation of hCAR3 in vitro.
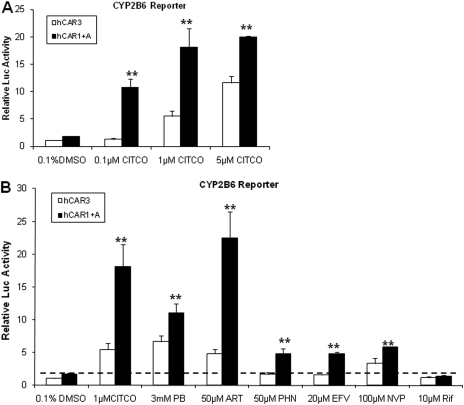
hCAR1+A Exhibits Superior Xenobiotic Response over hCAR3 in Cell-Based Reporter Assays.
Although hCAR3 has displayed promising features of chemical-induced activation in immortalized cell lines, these responses are predominantly to direct hCAR activators with limited and often muted responses to indirect activators. To compare the chemical response between hCAR3 and hCAR1+A, we examined the effect of hCAR agonist CITCO, and several prototypical hCAR activators, on hCAR3 and hCAR1+A in cell-based reporter assays. Both hCAR3 and hCAR1+A were activated in a concentration-dependent manner by CITCO at 0.1, 1, and 5 μM, where activation of hCAR1+A was significantly greater than that of hCAR3 at each CITCO concentration (Fig. 2A). Furthermore, evaluating the activation profile of each hCAR3 and hCAR1+A with six prototypical hCAR activators, including CITCO, PB, ART, PHN, EFV, and NVP, revealed that hCAR1+A exhibits a greater response than hCAR3 for all the tested activators (Fig. 2B). It is noteworthy that PHN (50 μM) and EFV (20 μM) only demonstrated negligible activation of hCAR3, yet both drugs exhibited potent activation of hCAR1+A in the current experiments. In addition, the selective human PXR agonist, RIF did not activate either hCAR3 or hCAR1+A as expected. These results indicate that hCAR1+A is superior to hCAR3 regarding the sensitivity and magnitude of chemical-mediated activation in immortalized cells.
Fig. 2.
Activation of hCAR3 and hCAR1+A by prototypical hCAR activators. HepG2 cells were transfected with CYP2B6-PBREM reporter, and hCAR3 or hCAR1+A expression vectors as described under Materials and Methods.7, Transfected cells were subsequently treated with vehicle control (0.1% DMSO) or CITCO at the concentration of 0.1, 1.0, and 5.0 μM (A); or with known hCAR activators, including PB, ART, PHN, EFV, and NVP, at indicated concentrations for 24 h (B). RIF (10 μM) was included as non-hCAR activator. After harvesting cell lysates, luciferase activities were determined and expressed relative to hCAR3 vehicle control. Data represent the mean ± S.D. (n = 3) (**, p < 0.01 denotes comparison between hCAR1+A and hCAR3 for each paired group, respectively).
Correlation of the Chemical Spectrum between the Activation of hCAR1+A and hCAR1.
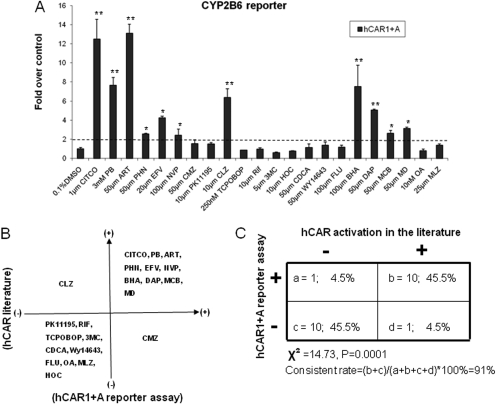
To investigate whether activation of hCAR1+A reflects the chemical selectivities of the reference hCAR1 activation, a series of 22 compounds has been tested in HepG2 cells cotransfected with hCAR1+A and CYP2B6 reporter construct. These compounds include known hCAR activators (CITCO, PB, ART, PHN, EFV, NVP, CMZ, BHA, DAP, MCB, and MD) (Maglich et al., 2003; Wang et al., 2004; Burk et al., 2005; Faucette et al., 2007; Li et al., 2009; Tolson et al., 2009), hCAR deactivators (PK11195, OA and CLZ) (Moore et al., 2000; Stanley et al., 2006; Li et al., 2008), selective rodent CAR activator and/or CYP2B inducers (TCPOBOP, MLZ, and FLU) (Tzameli et al., 2000; Huang et al., 2004), as well as typical activators of other nuclear receptors including: RIF for PXR, CDCA for farnesoid X receptor, HOC for liver X receptor, 3MC for aryl hydrocarbon receptor, and WY-14643 for peroxisome proliferator-activated receptor α. Figure 3A demonstrates that hCAR1+A was significantly activated by 11 of the 22 tested compounds at least 2-fold over the control in cell-based reporter assay. Comparing these data with published literature revealed that 10 of the 11 compounds that showed hCAR1+A-positive responses are known hCAR activators (Fig. 3B), and 10 of 11 compounds that displayed hCAR1+A-negative responses are known hCAR deactivators or selective activators of other nuclear receptors (Fig. 3B). CMZ, which was reported as an hCAR activator previously (Faucette et al., 2007), only demonstrated marginal activation of hCAR1+A in our reporter assay (Fig. 3A). In contrast, the suspected hCAR deactivator CLZ exhibited robust activation of hCAR1+A (Fig. 3A). Statistical analysis of these data showed that the overall consistent rate between hCAR1+A activation and hCAR1 activation in the literature reached 91% (Fig. 3C). Together, these results suggest that activation of hCAR1+A is representative of the reference hCAR with respect to chemical selectivity and sensitivity.
Fig. 3.
Correlation of the chemical specificity between the activation of hCAR1+A and hCAR1. A, HepG2 cells were transfected with CYP2B6-PBREM reporter, and hCAR1+A expression vectors. Transfected cells were then treated with vehicle control (0.1% DMSO), known hCAR activators (CITCO, PB, ART, PHN, EFV, NVP, CMZ, BHA, DAP, MCB, and MD), hCAR deactivators (CLZ, PK11195, OA), selective rodent CAR activator and/or CYP2B inducers (TCPOBOP, MLZ, and FLU), or prototypical activators of other nuclear receptor (RIF, 3MC, HOC, CDCA, and WY-14643) at indicated concentrations for 24 h. Luciferase activities were determined and expressed relative to vehicle control. Data represent the mean ± S.D. (n = 3) (*, p < 0.05; **, p < 0.01). B, hCAR1+A activation data and published hCAR1 data are organized in four-quadrant diagram. C, the correlation between hCAR1+A activation and published findings of hCAR1 activation was analyzed by χ2 test.
Localization and Translocation of hCAR1+A in Immortalized Cell Line.
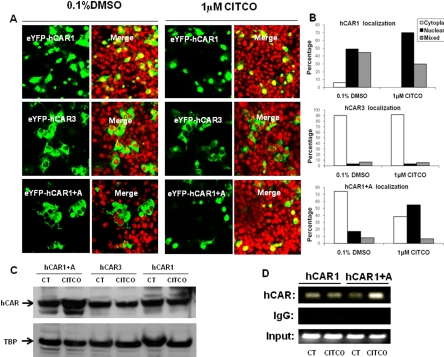
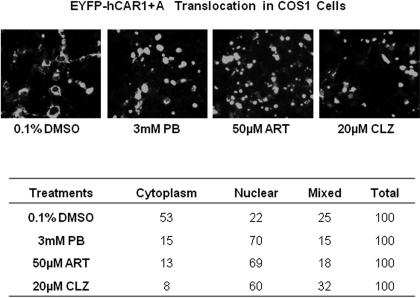
The constitutive activation of hCAR in immortalized cell lines is predominantly attributed to the spontaneous nuclear accumulation of hCAR, regardless of xenobiotic activation (Kawamoto et al., 1999). To examine whether intracellular localization and translocation of hCAR1+A contribute to its robust chemical response, EYFP-tagged hCAR1, hCAR3, or hCAR1+A was transfected in COS1 cells, followed by treatment with control (0.1% DMSO) or CITCO (1 μM). In agreement with previous reports, confocal microscopy analysis showed 94 to 100% of EYFP-hCAR1-expressing cells displayed nuclear or mixed (nuclear + cytoplasm) allocation (Fig. 4, A and B) confirming that the reference hCAR1 constantly accumulates in the nucleus in the absence or presence of CITCO. In contrast, approximately 90% of EYFP-hCAR3 was located in the cytoplasm of COS1 cells, with only 10% showing nuclear or mixed distribution (Fig. 4, A and B) regardless of the CITCO treatment. In the absence of activator, EYFP-(hCAR1+A) displayed a predominantly cytoplasmic distribution of ∼74%, similar to hCAR3. It is noteworthy that, upon the treatment with CITCO, the nuclear and mixed distribution of hCAR1+A increased to approximately 61%, whereas the cytoplasmic allocation dropped to 39% (Fig. 4, A and B). Western blot analysis of nuclear proteins extracted from hCAR1, hCAR3, or hCAR1+A transfected COS1 cells also showed that only hCAR1+A expression was increased after CITCO treatment (Fig. 4C). Additional experiments demonstrated that EYFP-(hCAR1+A) nuclear translocation was clearly increased after the treatment of PB (3 mM), ART (50 μM), or CLZ (20 μM) (Fig. 5A). Of the EYFP-(hCAR1+A)-expressing cells counted, 53 and 22% exhibited cytoplasmic and nuclear localizations, respectively, in the control group, whereas after being treated with the known hCAR activators, 60 to 70% EYFP-(hCAR1+A)-expressing cells demonstrated nuclear distribution, and only 8 to 15% remained in the cytoplasm (Fig. 5B). Overall, these results indicate that hCAR1+A represents a unique hCAR mutant that displays chemical-mediated translocation in immortalized cells.
Fig. 4.
CITCO promotes the nuclear translocation and target gene interaction of hCAR1+A in immortalized cells. COS1 were transfected with 1 μg of EYFP-hCAR1, EYFP-hCAR3, or EYFP-(hCAR1+A) as outlined under Materials and Methods. Transfected cells were then treated with either 0.1% DMSO or 1 μM CITCO for 24 h. A, confocal images illustrate representative localization and translocation of different EYFP-hCAR expression after vehicle control or CITCO treatment. B, one hundred EYFP-hCAR-expressing cells from each group were classified into cytoplasmic, nuclear, or mixed (cytoplasmic + nuclear) distributions. C, nuclear proteins (30 μg) extracted from hCAR1, hCAR3, or hCAR1+A expression vector transfected COS1 cells were subjected to hCAR immunoblot analysis. D, in a separate experiment, HepG2 cells were transfected with hCAR1 or hCAR1+A expression vector for 24 h and subsequently treated with CT (0.1% DMSO) or CITCO (1 μM) for 2 h. Harvested cells were subjected to CHIP assays as described under Materials and Methods.
Fig. 5.
Translocation of EYFP-(hCAR1+A) in COS1 cells after treatment with known hCAR activators. COS1 cells were transfected with EYFP-(hCAR1+A) as described under Materials and Methods, and treated with 0.1% DMSO, PB (3 mM), ART (50 μM), or CLZ (20 μM). After 24 h of treatment, cells were subjected to confocal microscopy analysis. A, representative localization of EYFP-(hCAR1+A) in each treatment group. B, for each treatment, 100 EYFP-(hCAR1+A) expressing cells were calculated and categorized as cytoplasmic, nuclear, or mixed (cytoplasmic + nuclear) localizations.
CITCO Enhances the Recruitment of hCAR1+A to the PBREM Region of CYP2B6.
Although the xenobiotic-induced translocation may represent one of the mechanisms involved in the activation of hCAR1+A in immortalized cells, agonistic ligand of hCAR may also facilitate the interaction between nuclear localized hCAR1+A and the promoter of its target gene to achieve maximal xenobiotic response. To this end, results from CHIP assays indicated that hCAR1+A binding to the PBREM region of CYP2B6 promoter was clearly increased upon the treatment of CITCO, whereas the interaction between hCAR1 and PBREM region of CYP2B6 was rather consistent regardless of the CITCO treatment (Fig. 4D). Thus, recruitment of hCAR1+A to the promoter of CYP2B6 may also contribute to the maximal hCAR1+A activation induced by the selective hCAR activator CITCO.
Protein Interaction between hCAR1+A and Coactivators.
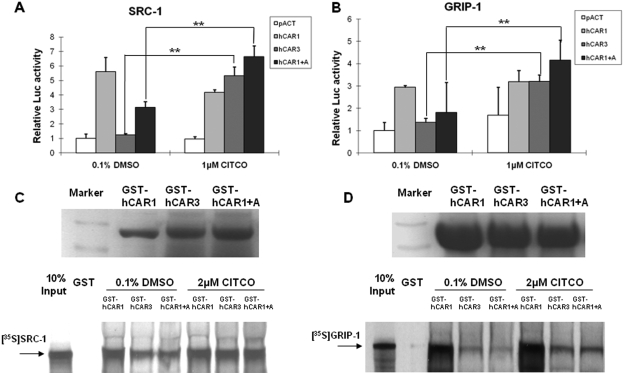
Because reference hCAR was demonstrated to interact constitutively with several coactivators independent of chemical activation (Tzameli et al., 2000), mammalian two-hybrid and GST pull-down assays were performed to further explore the binding specificity of hCAR1, hCAR3, and hCAR1+A with SRC-1 and GRIP-1. As expected, hCAR1 was capable of binding SRC-1 and GRIP-1 constantly in the absence of ligand. In contrast, hCAR3 and hCAR1+A showed minimal binding of SRC-1and GRIP-1 without activation. After treatment with CITCO (1 μM) the binding of hCAR3 and hCAR1+A to SRC-1 or GRIP-1 was significantly enhanced (Fig. 6, A and B). Similar interactions were also observed in the GST pull-down assays. Direct interaction of hCAR3 and hCAR1+A with SRC-1 or GRIP-1 was weaker than that of the constitutive hCAR1 binding under all conditions tested (Fig. 6, C and D). Incubation with 2 μM CITCO enhanced the binding of hCAR3 and hCAR1+A to SRC-1 and GRIP-1, but no change was seen in coactivator interactions with hCAR1, after CITCO treatment. Collectively, these data suggest that direct interaction with coactivators within the nucleus may be an additional factor contributing to the observed robust activation of hCAR1+A in immortalized cells.
Fig. 6.
Protein interaction between hCAR1+A and coactivators was enhanced by CITCO. Mammalian two-hybrid assays were performed in COS1 cells transiently transfected with indicated expression plasmids encoding VP16-AD/hCAR fusion proteins and GAL4-DBD/coactivators fusion proteins, SRC-1(A) and GRIP-1(B), with the reporter plasmid pG5-Luc. Cells were treated with DMSO (0.1%) or CITCO (1 μM) for 24 h before determination of luciferase activities. Data represent mean ± S.D. from three independent transfections (**, p < 0.01). In the GST pull-down assay, bacterially expressed GST-fused hCAR1, hCAR3, and hCAR1+A were equilibrated for loading, and then incubated with in vitro-translated 35S-labeled SRC-1 (C) and 35S-labeled GRIP-1 (D) in the presence of 0.1% DMSO, or 2 μM CITCO as detailed under Materials and Methods.
Discussion
The orphan nuclear receptor, CAR, has evolved in mammals to function as a “stress” sensor dictating both xenobiotic and endobiotic signals. CAR requires nuclear localization and activation to be a fully functional receptor. Unlike other nuclear receptors, activation of hCAR may occur by direct ligand binding or indirect mechanisms. Currently, one of the major drawbacks in studying the mechanisms underlying CAR activation is the spontaneous activation of this receptor in immortalized cells independent of xenobiotic stimuli (Kawamoto et al., 1999; Qatanani and Moore, 2005). Functional characterization of alternatively spliced variants of hCAR, however, revealed that hCAR3 exhibits low basal but ligand-induced activation in several cell lines, though the exact mechanisms remain elusive. Here, we delineate the functional importance of the five-amino-acid insertion of hCAR3 and have identified that the insertion of alanine alone is sufficient to switch the constitutively activated reference hCAR1 to the xenobiotic-responsive hCAR3. The hCAR1+A construct displayed robust responsiveness to over 90% of known hCAR activators tested. Moreover, hCAR1+A exhibited xenobiotic-dependent nuclear translocation in COS1 cells. To our knowledge, hCAR1+A represents the first hCAR mutant that translocates to the nuclei of immortalized cells upon chemical stimulation. Although many mechanistic questions remain unanswered at this point, this deletion mutant may provide an experimental model for further investigating the mechanisms governing hCAR nuclear translocation and xenobiotic activation.
Human CAR3 is one of the prominent hCAR splice variants incorporating 15 nucleotides in intron 7, which resulted in a five-amino-acid (APYLT) in-frame insertion in the highly conserved loop between helices α8 and α9 of the LBD (Auerbach et al., 2003; Arnold et al., 2004; Jinno et al., 2004). Previous computational modeling analysis purported that the bulged extension of the loop 8–9 by the APYLT insertion is responsible for the unique activation property of hCAR3 (Auerbach et al., 2003). Nevertheless, our present study shows that the low basal activities were well maintained in all mutants retaining one or more amino acids from the insertion (+YLT, +AP, +P, +A), but only the constructs containing the +AP or +A residues were capable of responding to ligand-stimulated activation. These results indicate that the specific amino acid properties not the number of amino acid residues in this particular region is crucial for restoring the distinctive feature of hCAR3. It is interesting that replacement of the aliphatic alanine with proline, which usually disrupts the α-helix structure, led to the total loss of xenobiotic responsiveness in the receptor construct. This raises an intriguing question as to whether any of the other 20 amino acids could have an alanine- or proline-like effect on the hCAR3 activation. Solving this question will shed light on the effort of dissecting the molecular mechanisms underlying the chemical-mediated activation of hCAR.
Splicing variants of hCAR protein characterized with unique structural changes in the LBD are associated with different ligand specificities (Savkur et al., 2003; Arnold et al., 2004; Lamba et al., 2004). Recently, DeKeyser et al. reported the common plasticizer, di(2-ethylhexyl) phthalate, known to be a ligand for PXR but not an activator of reference hCAR, is a highly potent and selective activator of hCAR2, another prominent hCAR splicing variant (DeKeyser et al., 2009). Molecular modeling analysis suggested that hCAR3 is the only differentially spliced hCAR variant with an unaltered ligand-binding pocket (Auerbach et al., 2003). Alterations in the hCAR LBD may affect the outcome of direct ligand activation, but the consequences to indirect activation pathways are unknown. To determine the chemical specificities in activation of hCAR1+A versus the reference hCAR1, the current study has further evaluated a series of 22 compounds, including known hCAR activators, deactivators, prototypical activators of other nuclear receptors, and selective rodent CAR activators. Collectively, more than 90% of known hCAR activators resulted in at least 2-fold activation of hCAR1+A, whereas more than 90% of non-hCAR activators failed to activate hCAR1+A in the cell-based reporter assays. It is noteworthy that one of the known hCAR deactivators, CLZ significantly induced the activity of hCAR1+A, which seems to conflict with the previous observation that CLZ antagonized the constitutive activity of hCAR by 50% in HepG2 cells (Moore et al., 2000). Nevertheless, our current results are in agreement with several reports that challenge the deactivating characteristic of CLZ. Instead of deactivation, CLZ increased the hCAR activation in a human embryonic kidney 293 cell transfection assay (Honkakoski et al., 2001), and enhanced the activation of hCAR3 in transfected COS1 cells (Auerbach et al., 2005). However, PK11195, a potent deactivator of hCAR identified previously by this laboratory, displayed negligible activation of hCAR1+A, and hCAR3, yet consistent deactivation of the reference hCAR1 in multiple cell lines (Li et al., 2008). Although both CLZ and PK11195 are capable of translocating hCAR to the nucleus in primary hepatocytes (Wang and Tompkins, 2008; Li et al., 2009), the dramatic differences in their activation of hCAR1+A and hCAR3 led to the speculation that these two compounds may exert their antagonistic effects through binding and interacting with distinct regions of the nuclear localized hCAR. Compared with hCAR3, the newly generated hCAR1+A demonstrated a higher activation in response to all known hCAR activators tested. In particular, CAR1+A was significantly responsive to indirect hCAR activators such as PB, PHN, and EFV, to which hCAR3 was nonresponsive in most cases. Together, current evidence suggests that the hCAR1+A construct exhibits robust xenobiotic responses that correlate well with the activation profile of the reference hCAR.
Nuclear translocation triggered by direct ligand binding is a common mechanism required for the activation of most steroid hormone receptors (Walker et al., 1999). In contrast, the nuclear translocation of CAR does not require direct ligand binding, and actually the majority of identified CAR activators activate CAR through a PB-like indirect mechanism. Moreover, although CAR demonstrates xenobiotic-mediated translocation and activation in primary cultured hepatocytes, this characteristic of CAR was lost entirely in all transformed cell lines interfering with investigations of CAR activation in vitro. Recently, several studies revealed that a number of hCAR splice variants including hCAR3 displayed mixed cellular distribution in hepatocytes and cell lines with a majority of the proteins localizing in the cytoplasm (Jinno et al., 2004; Auerbach et al., 2005). Nevertheless, typical hCAR activators cannot trigger a translocation of these CAR proteins to the nucleus, indicating that the CITCO-mediated activation of hCAR3 targets only those CAR proteins already localized to the nucleus. Our current results unexpectedly showed that, although hCAR1+A displayed a mixed cellular distribution similar to that of hCAR3 in the absence of treatment, the nuclear localization of this mutant was clearly increased after treatment with several known hCAR activators. It is noteworthy that this alanine residue is not located to the leucine-rich peptide (L/MXXLXXL) region termed xenobiotic response signal (XRS), which was involved in dictating nuclear translocation of CAR in response to PB in mouse liver (Zelko et al., 2001). However, because the XRS contains multiple amino acid residues responsible for inter- and intramolecular interactions, it is possible that the alanine insertion may interact with the XRS and affect the translocation of hCAR thereafter.
The unique feature of nuclear translocation of hCAR1+A seems to contribute to its xenobiotic activation in immortalized cells. Nevertheless, it is difficult to explain the robust activation of hCAR1+A entirely by this relatively moderate nuclear accumulation. Results from chromatin immunoprecipitation assay indicated that the enhanced recruitment of hCAR1+A to the promoter region of CYP2B6 gene upon CITCO treatment may also account for the optimal xenobiotic response of hCAR1+A in vitro. In addition, ligand-independent coactivator assembly has been established as a foundation for the intrinsically high activity of CAR (Tzameli et al., 2000; Li et al., 2008). In contrast, the relatively low basal interaction between hCAR3 and SRC-1 was significantly enhanced in the presence of CITCO (Auerbach et al., 2005). Our mammalian two-hybrid and GST pull-down assays showed that, similarly to hCAR3, the hCAR1+A also exhibits low basal and high CITCO-inducible interactions with SRC-1 and GRIP-1, suggesting alanine alone seems adequate to confer this distinctive nature of protein-protein interaction to hCAR3.
Taken together, we demonstrate in this report that a single amino acid residue, alanine, within the unique hCAR3 insert is sufficient to convert the constitutively active hCAR1 to the xenobiotic-responsive hCAR3, while maintaining the chemical specificities correlate to the reference hCAR1. Moreover, in contrast to known hCAR splicing variants and mutants, hCAR1+A exhibits xenobiotic-induced nuclear translocation in immortalized cell lines. However, we do realize that, although both increased nuclear translocation and coactivator recruitment contribute to the xenobiotic activation of hCAR1+A, these data alone cannot explain entirely the functional switch conferred by hCAR1+A. In particular, future structure-function analysis is required for the better understanding of the mechanisms underlying this functional shift of CAR. Overall, hCAR1+A may offer an unique and sensitive model for investigating hCAR translocation and activation, as well as screening hCAR activators in vitro.
Acknowledgments
We thank Dr. Masahiko Negishi (National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC) for providing multiple CAR expression vectors; Drs. Curtis Omiecinski (The Pennsylvania State University, University Park, PA) and Bryan Goodwin (GlaxoSmithKline, Research Triangle Park, NC) for providing the CMV2-hCAR3 expression vector and the CYP3A4-PXRE/XREM reporter vector, respectively; Bo Feng for assistance with the Western blot analysis; and John Cottrell (University of Maryland Medical Center, Baltimore, MD) for aid in procuring human liver tissues.
This work was supported by the National Institutes of Health National Institute of Diabetes Digestive and Kidney Diseases [Grant DK061652].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.109.159210
- CAR
- constitutive androstane receptor
- hCAR
- human CAR
- ART
- artemisinin
- BHA
- butylated hydroxyanisole
- CMZ
- carbamezapine
- CLZ
- clotrimazole
- CITCO
- 6-(4-chlorophenyl) imidazo[2,1-b][1,3]thiazole-5-carbaldehyde-O-(3,4-dichlorobenzyl) oxime
- CYP
- cytochrome P450
- DMSO
- dimethyl sulfoxide
- CDCA
- chenodeoxycholic acid
- DAP
- diazepam
- EFV
- efavirenz
- EYFP
- enhanced yellow fluorescent protein
- FLU
- fluconazole
- GRIP-1
- glucocorticoid receptor-interacting protein-1
- GST
- glutathione S-transferase
- HOC
- 22(R)-hydroxycholesterol
- LBD
- ligand-binding domain
- MLZ
- meclizine
- 3MC
- 3-methylcholanthrene
- MCB
- myclobutanil
- MD
- methadone
- NVP
- nevirapine
- OA
- okadaic acid
- PB
- phenobarbital
- PHN
- phenytoin
- PXR
- pregnane X receptor
- PK11195
- 1-(2-chlorophenyl-nmethylpropyl)-3-isoquinolinecarboxamide
- PBREM
- phenobarbital-responsive enhancer module
- XREM
- xenobiotic-responsive enhancer module
- RXR
- retinoic acid receptor
- RIF
- rifampicin
- TCPOBOP
- 1,4-bis[2-(3,5-dichlorpyridyloxy)]benzene
- SRC-1
- steroid receptor coactivator-1
- XRS
- xenobiotic response signal
- WY-14643
- 4-chloro-6-(2,3-xylidino)-2-pyrimidinylthioacetic acid.
References
- Arnold KA, Eichelbaum M, Burk O. (2004) Alternative splicing affects the function and tissue-specific expression of the human constitutive androstane receptor. Nucl Recept 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Ramsden R, Stoner MA, Verlinde C, Hassett C, Omiecinski CJ. (2003) Alternatively spliced isoforms of the human constitutive androstane receptor. Nucleic Acids Res 31:3194–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Stoner MA, Su S, Omiecinski CJ. (2005) Retinoid X receptor-alpha-dependent transactivation by a naturally occurring structural variant of human constitutive androstane receptor (NR1I3). Mol Pharmacol 68:1239–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. (1994) A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol 14:1544–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk O, Arnold KA, Nussler AK, Schaeffeler E, Efimova E, Avery BA, Avery MA, Fromm MF, Eichelbaum M. (2005) Antimalarial artemisinin drugs induce cytochrome P450 and MDR1 expression by activation of xenosensors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol 67:1954–1965 [DOI] [PubMed] [Google Scholar]
- DeKeyser JG, Stagliano MC, Auerbach SS, Prabhu KS, Jones AD, Omiecinski CJ. (2009) Di(2-ethylhexyl) phthalate is a highly potent agonist for the human constitutive androstane receptor splice variant CAR2. Mol Pharmacol 75:1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucette SR, Sueyoshi T, Smith CM, Negishi M, Lecluyse EL, Wang H. (2006) Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther 317:1200–1209 [DOI] [PubMed] [Google Scholar]
- Faucette SR, Zhang TC, Moore R, Sueyoshi T, Omiecinski CJ, LeCluyse EL, Negishi M, Wang H. (2007) Relative activation of human pregnane X receptor versus constitutive androstane receptor defines distinct classes of CYP2B6 and CYP3A4 inducers. J Pharmacol Exp Ther 320:72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P, Jaaskelainen I, Kortelahti M, Urtti A. (2001) A novel drug-regulated gene expression system based on the nuclear receptor constitutive androstane receptor (CAR). Pharmacol Res 18:146–150 [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Sueyoshi T, Negishi M. (2003) Drug-activated nuclear receptors CAR and PXR. Ann Med 35:172–182 [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, Moore DD. (2005) Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol Endocrinol 19:1646–1653 [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Wei P, Schrader WT, Moore DD. (2004) Meclizine is an agonist ligand for mouse constitutive androstane receptor (CAR) and an inverse agonist for human CAR. Mol Endocrinol 18:2402–2408 [DOI] [PubMed] [Google Scholar]
- Inoue K, Borchers CH, Negishi M. (2006) Cohesin protein SMC1 represses the nuclear receptor CAR-mediated synergistic activation of a human P450 gene by xenobiotics. Biochem J 398:125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinno H, Tanaka-Kagawa T, Hanioka N, Ishida S, Saeki M, Soyama A, Itoda M, Nishimura T, Saito Y, Ozawa S, et al. (2004) Identification of novel alternative splice variants of human constitutive androstane receptor and characterization of their expression in the liver. Mol Pharmacol 65:496–502 [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. (1999) Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol 19:6318–6322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Sueyoshi T, Inoue K, Moore R, Negishi M. (2003) Cytoplasmic accumulation of the nuclear receptor CAR by a tetratricopeptide repeat protein in HepG2 cells. Mol Pharmacol 64:1069–1075 [DOI] [PubMed] [Google Scholar]
- Kodama S, Koike C, Negishi M, Yamamoto Y. (2004) Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol 24:7931–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba JK, Lamba V, Yasuda K, Lin YS, Assem M, Thompson E, Strom S, Schuetz E. (2004) Expression of constitutive androstane receptor splice variants in human tissues and their functional consequences. J Pharmacol Exp Ther 311:811–821 [DOI] [PubMed] [Google Scholar]
- Li H, Chen T, Cottrell J, Wang H. (2009) Nuclear translocation of Ad/EYFP-hCAR: a novel tool for screening human CAR activators in human primary hepatocytes. Drug Metab Dispos 37:1098–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chen T, Stanton JD, Sueyoshi T, Negishi M, Wang H. (2008) The peripheral benzodiazepine receptor ligand 1-(2-chlorophenyl-methylpropyl)-3-isoquinoline-carboxamide is a novel antagonist of human constitutive androstane receptor. Mol Pharmacol 74:443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, et al. (2003) Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem 278:17277–17283 [DOI] [PubMed] [Google Scholar]
- Maglich JM, Watson J, McMillen PJ, Goodwin B, Willson TM, Moore JT. (2004) The nuclear receptor CAR is a regulator of thyroid hormone metabolism during caloric restriction. J Biol Chem 279:19832–19838 [DOI] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, et al. (2000) Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem 275:15122–15127 [DOI] [PubMed] [Google Scholar]
- Qatanani M, Moore DD. (2005) CAR, the continuously advancing receptor, in drug metabolism and disease. Curr Drug Metab 6:329–339 [DOI] [PubMed] [Google Scholar]
- Savkur RS, Wu Y, Bramlett KS, Wang M, Yao S, Perkins D, Totten M, Searfoss G, 3rd, Ryan TP, Su EW, et al. (2003) Alternative splicing within the ligand binding domain of the human constitutive androstane receptor. Mol Genet Metab 80:216–226 [DOI] [PubMed] [Google Scholar]
- Stanley LA, Horsburgh BC, Ross J, Scheer N, Wolf CR. (2006) PXR and CAR: nuclear receptors which play a pivotal role in drug disposition and chemical toxicity. Drug Metab Rev 38:515–597 [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. (1999) The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem 274:6043–6046 [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Moore R, Sugatani J, Matsumura Y, Negishi M. (2008) PPP1R16A, the membrane subunit of protein phosphatase 1beta, signals nuclear translocation of the nuclear receptor constitutive active/androstane receptor. Mol Pharmacol 73:1113–1121 [DOI] [PubMed] [Google Scholar]
- Sugatani J, Kojima H, Ueda A, Kakizaki S, Yoshinari K, Gong QH, Owens IS, Negishi M, Sueyoshi T. (2001) The phenobarbital response enhancer module in the human bilirubin UDP-glucuronosyltransferase UGT1A1 gene and regulation by the nuclear receptor CAR. Hepatology 33:1232–1238 [DOI] [PubMed] [Google Scholar]
- Tien ES, Negishi M. (2006) Nuclear receptors CAR and PXR in the regulation of hepatic metabolism. Xenobiotica 36:1152–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolson AH, Li H, Eddington ND, Wang H. (2009) Methadone Induces the Expression of Hepatic Drug-Metabolizing Enzymes Through the Activation of Pregnane X Receptor and Constitutive Androstane Receptor. Drug Metab Dispos [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. (2000) The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol 20:2951–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Matsui K, Yamamoto Y, Pedersen LC, Sueyoshi T, Negishi M. (2005) Thr176 regulates the activity of the mouse nuclear receptor CAR and is conserved in the NR1I subfamily members PXR and VDR. Biochem J 388:623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D, Htun H, Hager GL. (1999) Using inducible vectors to study intracellular trafficking of GFP-tagged steroid/nuclear receptors in living cells. Methods 19:386–393 [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Moore R, Sueyoshi T, Negishi M, LeCluyse E. (2004) Human constitutive androstane receptor mediates induction of CYP2B6 gene expression by phenytoin. J Biol Chem 279:29295–29301 [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, LeCluyse EL. (2003) A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem 278:14146–14152 [DOI] [PubMed] [Google Scholar]
- Wang H, Tompkins LM. (2008) CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab 9:598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. (2004) The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res 64:7197–7200 [DOI] [PubMed] [Google Scholar]
- Yoshinari K, Kobayashi K, Moore R, Kawamoto T, Negishi M. (2003) Identification of the nuclear receptor CAR:HSP90 complex in mouse liver and recruitment of protein phosphatase 2A in response to phenobarbital. FEBS Lett 548:17–20 [DOI] [PubMed] [Google Scholar]
- Zelko I, Sueyoshi T, Kawamoto T, Moore R, Negishi M. (2001) The peptide near the C terminus regulates receptor CAR nuclear translocation induced by xenochemicals in mouse liver. Mol Cell Biol 21:2838–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]