Abstract
The aryl hydrocarbon receptor (AHR) is regarded as an important homeostatic transcriptional regulator within physiological and pathophysiological processes, including xenobiotic metabolism, endocrine function, immunity, and cancer. Agonist activation of the AHR is considered deleterious based on toxicological evidence obtained with environmental pollutants, which mediate toxic effects through AHR. However, a multitude of plant-derived constituents, e.g., polyphenols that exhibit beneficial properties, have also been described as ligands for the AHR. It is conceivable that some of the positive aspects of such compounds can be attributed to suppression of AHR activity through antagonism. Therefore, we conducted a dioxin response element reporter-based screen to assess the AHR activity associated with a range of flavonoid compounds. Our screen identified two flavonoids (5-methoxyflavone and 7,4′-dimethoxyisoflavone) with previously unidentified AHR agonist potential. In addition, we have identified and characterized 6,2′,4′-trimethoxyflavone (TMF) as an AHR ligand that possesses the characteristics of an antagonist having the capacity to compete with agonists, such as 2,3,7,8-tetrachlorodibenzo-p-dioxin and benzo[a]pyrene, thus effectively inhibiting AHR-mediated transactivation of a heterologous reporter and endogenous targets, e.g., CYP1A1, independent of cell lineage or species. Furthermore, TMF displays superior action by virtue of having no partial agonist activity, in contrast to other documented antagonists, e.g., α-napthoflavone, which are partial weak agonists. TMF also exhibits no species or promoter dependence with regard to AHR antagonism. TMF therefore represents an improved tool allowing for more precise dissection of AHR function in the absence of any conflicting agonist activity.
The role of the transcription factor aryl hydrocarbon receptor (AHR) in biology has expanded beyond that of a xenobiotic sensor and regulator of detoxification (Ramadoss et al., 2005). The AHR is documented as affecting numerous physiological and pathophysiological processes; studies investigating the environmental and health impact of persistent pollutants, many of which are AHR agonists, clearly indicate a role of the AHR in modulating endocrine function. Animal knockout models indicate a nonlethal, yet pivotal role for the AHR, as evidenced by reproductive, metabolic, and immunological phenotypes. In the absence of contradictory evidence it is presumed that transactivation function of AHR becomes manifest through activation by endogenous AHR agonists. Despite these phenotypes, pharmacological or environmental activation of the AHR is often regarded as deleterious. Recently, it has been demonstrated that agonists of the AHR play an integral role in T-cell function, promoting a TH2/TH1 switch resulting in a TH1 bias (Negishi et al., 2005). In addition, the AHR aids in stimulating the differentiation of IL17 secreting TH17 cells, thereby generating a potentially proinflammatory autoimmune environment (Veldhoen et al., 2008; Stockinger et al., 2009). It is conceivable therefore that inhibition of AHR activity by antagonists could result in beneficial anti-inflammatory actions. Evidence for such anti-inflammatory effects has recently been identified by use of the AHR antagonist CH-223191, which represses TH17 development in mice leading to diminished levels of the proinflammatory cytokines IL17 and IL22 (Veldhoen et al., 2009).
A number of compounds are described as AHR antagonists, including the flavone derivatives α-napthoflavone (α-NF) (Wilhelmsson et al., 1994), PD98059 (Reiners et al., 1998), and 3′-methoxy-4′-nitroflavone (MNF) (Lu et al., 1995), 6-methoxy-1,3,8-trichlorodibenzofuran (6-MCDF) (Astroff et al., 1988; Harris et al., 1989), 1-amino-3,7,8-trichlorodibenzo-p-dioxin (Luster et al., 1986), and omeprazole sulfide (Gerbal-Chaloin et al., 2006). However, many of these compounds exhibit partial AHR agonist activity and are antagonists only in the sense that they are weakly activating competitive agonists. Furthermore, some display a degree of species specificity and/or AHR-independent activity and thus cannot be considered complete antagonists.
It is interesting that the plant kingdom is rich in AHR ligands predominantly in the form of polyphenolic flavonoid compounds. Many of these compounds exhibit beneficial properties, including antioxidant, antiproliferative, and anti-inflammatory activity, some of which may involve the AHR. However, with the exception of the polyphenol resveratrol, which has been shown to repress AHR-mediated gene expression but also represents an anti-inflammatory estrogen receptor (ER) ligand (Gehm et al., 1997; Casper et al., 1999), it has not been established whether any of the beneficial effects of flavonoids can be attributed to AHR antagonism. We have therefore conducted a screen of a number of substituted flavonoids with the aim of identifying potential bona fide AHR antagonists, which may have therapeutic immunological value in addition to improving the arsenal of antagonists available for investigating the ever-expanding scope of AHR biological function.
Materials and Methods
Materials.
2,3,7,8-Tetrachloroxooxanthrene (TCDD) was a generous gift from Dr. S. Safe (Department of Veterinary Physiology and Pharmacology, Texas A and M, College Station, TX). The following flavonoid compounds were purchased commercially (Indofine Chemicals, Hillsborough, NJ): 2-(4-methoxyphenyl)-4H-chromen-4-one (4′-methoxyflavone); 5-methoxy-2-phenyl-4H-chromen-4-one (5-methoxyflavone); 2-(2,4-dimethoxyphenyl)-6-methoxy-4H-chromen-4-one (6,2′,4′-trimethoxyflavone); 2-phenyl-4H-benzo[h]chromen-4-one (α-naphthoflavone); 3-phenyl-1H-benzo[f]chromen-1-one (β-naphthoflavone); 7-methoxy-3-(4-methoxyphenyl)-4H-chromen-4-one (7,4′-dimethoxyisoflavone); and 1,3,8-trichloro-6-methyldibenzo[b,d]furan (6-methoxy-1,3,8-trichlorodibenzofuran). 2-(3-Methoxy-4-nitrophenyl-4H-chromen-4-one (3′-methoxy-4′-nitroflavone) was a generous gift from Dr. T. Gasiewicz (University of Rochester, Rochester, NY).
Cell Lines and Culture.
The Huh7 human hepatoma cell line was routinely maintained in α-modified essential media (Sigma-Aldrich, St. Louis, MO) supplemented with 8% fetal bovine serum (Hyclone Laboratories, Logan, UT), 100 IU/ml penicillin/100 μg/ml streptomycin (Sigma-Aldrich). The MCF-7 human mammary cell line was maintained in Dulbecco's modified Eagle's media (Sigma-Aldrich) supplemented with 8% fetal bovine serum, penicillin/streptomycin. HepG2 (40/6) human hepatoma stable cell line (Long et al., 1998) containing the stably integrated pGudluc 6.1 luciferase reporter construct under the control of the Cyp1a1 enhancer were cultured under the same conditions as Huh7 cells (Aarts et al., 1995). The Hepa 1.1 mouse hepatoma cell line containing the stably integrated pGudluc 1.1 luciferase reporter construct was originally obtained from Dr. M. Denison (University of California, Davis, CA) and were cultured under the same conditions as HepG2 (40/6). Cells were cultured at 37°C in a humidified atmosphere composed of 95% air and 5% CO2.
Reporter Assays.
The reporter cell lines Hepa 1.1 and HepG2 (40/6) cells were seeded in six-well plates and cultured to ∼80% confluence. Cells were treated as indicated for 4 h then lysed in 200 μl of lysis buffer [25 mM Tris-phosphate, pH 7.8, 2 mM dithiothreitol, 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% (v/v) glycerol, and 1% (v/v) Triton X-100]. Lysate (20 μl) were combined with 80 μl of Luciferase Reporter Substrate (Promega, Madison, WI), and luciferase activity was measured with a TD-20e luminometer (Turner Designs, Sunnyvale, CA). Luciferase activity was normalized with respect to protein concentration.
RNA Isolation and Reverse Transcription.
Total RNA was isolated from cells cultured in six-well plates using TRIzol (Invitrogen, Carlsbad, CA). RNA concentration was determined via spectrophotometry at λ 260 nm and 280 nm. 2 μg total RNA was reverse transcribed to cDNA by use of a High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA).
Quantitative PCR.
PCR was performed on a MyiQ (Bio-Rad Laboratories, Hercules, CA) system with use of PerfeCTa SYBR Green reagent (Quanta Biosciences, Gaithersburg, MD). Quantitative real-time PCR primers (Integrated DNA Technologies, Coralville, IA) used in this study are listed in Table 1. In all cases, melting point analysis revealed amplification of a single product. Data acquisition and analysis were achieved by use of MyIQ software (Bio-Rad Laboratories).
Table 1.
Nucleotide sequences of oligonucleotides
| Gene | Primer Name | Nucleotide 5′ → 3′ | Species |
|---|---|---|---|
| Ribosomal protein L13a | rL13A_F | CCTGGAGGAGAAGAGGAAAGAGA | Human |
| rL13A_R | GAGGACCTCTGTGTATTTGTCAA | ||
| Cytochrome P450 1A1 | CYP1A1_F | TCTTCCTTCGTCCCCTTCAC | Human |
| CYP1A1_R | TGGTTGATCTGCCACTGGTT | ||
| Cytochrome P450 1A2 | CYP1A2_F | CGGCACTTCGACCCTTACAA | Human |
| CYP1A2_R | GCACATGGCACCAATGACG | ||
| Cytochrome P540 1B1 | CYP1B1_F | TGCCTGTCACTATTCCTCATGCCA | Human |
| CYP1B1_R | ATCAAAGTTCTCCGGGTTAGGCCA | ||
| Ribosomal protein L13a | rL13a_F | TTCGGCTGAAGCCTACCAGAAAGT | Mouse |
| rL13a_R | GCATCTTGGCCTTTTCCTTCCGTT | ||
| Cytochrome P450 1A1 | Cyp1a1_F | CTCTTCCCTGGATGCCTTCAA | Mouse |
| Cyp1a1_R | GGATGTGGCCCTTCTCAAATG |
Photoaffinity Competitive Ligand-Binding Assay.
The AHR photoaffinity ligand; 2-azido-3-[125I]iodo-7,8-dibromodibenzo-p-dioxin (PAL) was synthesized as described previously (Poland et al., 1986). Hepatic cytosol extracts were isolated from B6.Cg-Ahrtm3.1 Bra Tg (Alb-cre, Ttr-AHR)1GHP “Humanized” AHR mice (Flaveny et al., 2009) by homogenization in MENG (25 mM MOPS, 2 mM EDTA, 0.02% NaN3, 10% glycerol, pH 7.4) containing 20 mM sodium molybdate and protease inhibitor cocktail (Sigma-Aldrich) followed by centrifugation at 100,000g for 1 h. All binding experiments were conducted in the dark until photo-cross-linking of the PAL. In brief, a saturating amount of PAL (0.21 pmol, i.e., 8 × 105 cpm/tube) were added to 150 μg of cytosolic protein. Samples were then incubated with α-napthoflavone or 6, 2′,4′-trimethoxyflavone, as indicated for 20 min at room temperature. Samples were photolyzed (402 nm, 8 cm, 4 min), 1% dextran-coated charcoal added, followed by centrifugation at 3000g for 10 min to remove unbound PAL. Labeled samples were resolved on 8% Tricine polyacrylamide gels, transferred to PVDF membrane and visualized by autoradiography. Radio-labeled AHR bands were excised and quantified by γ-counting.
Electromobility Shift Assay.
Gel-shift analyses were performed essentially as described previously (Flaveny et al., 2009). In brief, pCI-AHR and pCI-aryl hydrocarbon receptor nuclear translocator (ARNT) were in vitro translated with the TnT-coupled rabbit reticulocyte lysate system (Promega) supplemented with 1.5 mM sodium molybdate. AHR and ARNT (4 μl) were combined in the presence of 1.5 μl of HEDG buffer (25 mM HEPES, 1 mM EDTA, 10 mM sodium molybdate, and 10% glycerol, pH 7.5) together with indicated treatments for 30 min at room temperature. 32P-labeled dioxin response element (DRE) probe was added to each reaction and incubated for a further 15 min. Lysates were resolved on 6% DNA-retardation gel (Invitrogen) and visualized by autoradiography.
Cytotoxicity Assays.
Short-term cytotoxicity was assessed by use of the MTS assay and is based on the mitochondrial reduction of a substrate by viable cells. Huh7 cells were seeded at 2 × 103 cell/well, after overnight incubation the cells were treated as indicated for a further 48 h. Viability was assessed by adding 40 μl/well MTS reagent and determining the absorbance at 490 nm after 2 h. Data represent viable cell number as a percentage of vehicle (DMSO)-treated cells ± S.E.M. Longer-term cytotoxicity was determined by use of a colony-formation assay. Huh7 cells were seeded at 1 × 103 cells/plate; after overnight incubation, cells were treated as indicated. After 24 h, cells were washed and cultured for an additional 14 days, after which time cells were stained with Coomassie brilliant blue for 2 min and colonies counted. Data represent colony number ± S.E.M. compared with vehicle (DMSO)-treated controls.
Western Immunoblotting.
Huh7 cells were cultured to ∼80% confluence in six-well plates and treated with vehicle or the indicated compounds over increasing time. Cells were lysed with MENG/20 mM sodium molybdate/1% Nonidet P40/1× protease inhibitor cocktail. Lysates were centrifuged (13,000g, 10 min, 4°C) to remove insoluble material. Protein concentrations were determined by use of the bicinchoninic acid kit (Thermo Fisher Scientific, Waltham, MA). Fifty micrograms of protein were resolved on 8% SDS-PAGE gels, transferred to PVDF membrane (Millipore, Billerica, MA). Where indicated, membranes were probed at recommended dilutions for 1 h with the following primary antibodies: anti-AHR mouse IgG (MA1-514; Affinity Bioreagents, Golden, CO), anti-β-actin mouse IgG (sc-47778; Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibody detection was achieved using species-appropriate biotin-conjugated IgG (Jackson ImmunoResearch Laboratories Inc., West Grove, PA). Tertiary detection was achieved through incubation with 0.03 μCi/ml 125I-streptavadin (GE Healthcare, Little Chalfont, Buckinghamshire, UK). Blots were exposed to BioMAX (Eastman Kodak, Rochester, NY) film and developed. Blots were directly quantified by excising 125I-streptavadin-labeled bands and counting in a γ-counter (Iso-data 20/20 series). AHR protein expression was normalized to β-actin and expressed as percentage of vehicle-treated control.
Chromatin Immunoprecipitation.
HN2095 cells were grown to approximately 90% confluence in 150-cm2 dishes and serum-starved 18 h before treatment. Treatment of cells was done in serum-free media supplemented with 5 mg/ml bovine serum albumin. 6,2′,4′-Trimethoxyflavone (TMF) was added at 10 μM at time 0 and again at 8 h for a total treatment time of 12 h. After treatment, cells were washed once with warm PBS, and chromatin complexes were chemically cross-linked by use of a 1% formaldehyde/PBS solution (final concentration) for 10 min at room temperature. Cross-linking was stopped by addition of glycine solution to a final concentration of 0.125 M. Cells were washed twice with ice-cold PBS and collected in 2 ml of harvest buffer (100 mM Tris, pH 8.3, 10 mM dithiothreitol, 5 mM NaOH). Cells were pelleted, washed in ice-cold PBS, pelleted, and resuspended in 600 μl of lysis buffer (1% SDS, 50 mM Tris-HCl, pH 8.1, 10 mM EDTA). Chromatin was sheared with the Diagenode Bioruptor to an average size of 1 kb. Complexes were precleared with protein A agarose resin (Pierce Chemical, Rockford, IL) and incubated overnight with specific antibodies (AHR rabbit polyclonal or ARNT rabbit polyclonal, Santa Cruz SC-5580). Immunoadsorbed complexes were captured on protein A agarose resin and washed once with TE, pH 8 (10 mM Tris-HCl, pH 8.0, and 0.5 mol/liter EDTA). Resin-bound complexes were then suspended in TE, pH 8, layered on top of a sucrose solution (1 M sucrose, 200 mM NaCl, 1% Nonidet P40) and spun for 3 min through the solution. Resin-bound complexes were then washed once with 0.5× RIPA buffer (10 mM Tris-HCl, pH 8.1, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 1% Triton X-100, 0.10% sodium deoxycholate, 0.10% SDS), followed by four washes with TE8. Samples were eluted off of the resin by use of 200 μl of 100 mM NaHCO3 and 1% SDS, and cross-links were reversed at 65°C overnight. Eluted DNA was isolated, washed, and concentrated by use of the ChIP DNA Clean and Concentrator Kit (ZYMO Research, Orange, CA). Immunoadsorbed DNA was analyzed by PCR.
Data Analysis.
In all cases, studies were performed in triplicate. Statistical analyses of data were performed by use of GraphPad Prism5 graphing and statistical analysis software (Graphpad Software, San Diego, CA). Data were analyzed by use of one-way analysis of variance and Tukey's multiple comparison and Student's t tests. In all cases P values of <0.05 were deemed statistically significant and indicated by an asterisk (*).
Results
Flavonoid Screen for AHR-Driven DRE-Dependent Gene Expression.
In an effort to analyze the potential biological activity of substituted flavones with regard to AHR-dependent signaling, we performed a luciferase reporter-based screen. HepG2 (40/6) cells stably transfected with a reporter construct (pGudluc 6.1) harboring the (−1301/−819 bp) 4 × DRE enhancer region of the mouse Cyp1a1 gene were incubated with 10 nM TCDD or various flavonoid derivatives (Fig. 1), as indicated at a concentration of 10 μM for 4 h and luciferase expression assayed (Fig. 2). TCDD stimulated AHR-driven DRE-dependent reporter activity >10-fold compared with vehicle alone, consistent with its status as a high-affinity prototypical AHR agonist. The flavonoid-based compounds yielded a spectrum of reporter activity, ranging from ∼5-fold induction with 4′-methoxyflavone down to ∼1.5-fold with 7,4′-dimethoxyisoflavone and repression of basal reporter activity with TMF and β-napthoflavone (β-NF). The failure of β-NF to induce reporter activity was initially unexpected, because β-NF is a known and relatively high-affinity AHR agonist (Gillner et al., 1985). Previous work has demonstrated that β-NF is a potent inhibitor of firefly but not Renilla luciferase enzymatic activity (Wang, 2002). We therefore examined the potential of our selected compounds to influence luciferase enzymatic activity, thus limiting the occurrence of false-negative agonists or false-positive antagonists. HepG2 (40/6) cells were incubated with 10 nM TCDD for 4 h to induce luciferase expression; cell lysates were then incubated for 10 min with the selected compounds (10 μM) and luciferase activity was assayed (Supplemental Fig. 1). We observed a total inhibition of luciferase activity with β-NF, ∼25% inhibition with 5-methoxyflavone and 7,4′-dimethoxyisoflavone, but no significant inhibition was observed with any other compound tested.
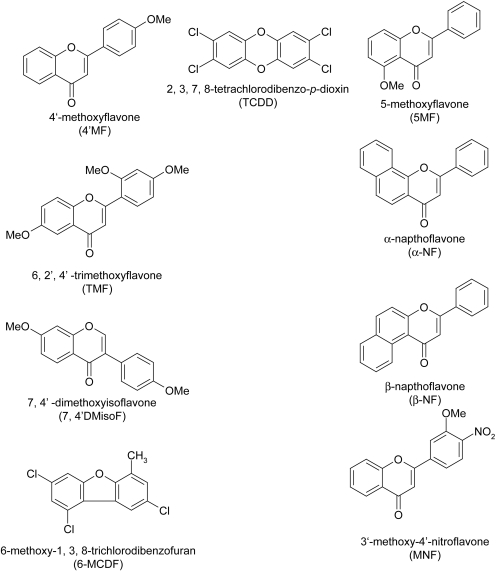
Fig. 1.
Chemical structures. Schematic representations of the chemical structures of AHR agonists and putative antagonists used in this study are depicted (4′-methoxyflavone, 5-methoxyflavone, 6,2′,4′-trimethoxyflavone, α-napthoflavone, β-napthoflavone, 2,3,7,8-tetrachlorodibenzo-p-dioxin, 7,4′-dimethoxyisoflavone, 6-methoxy-1,3,8-trichlorodibenzofuran, 3′-methoxy-4′-nitroflavone).
Fig. 2.
Flavonoid screen for AHR-dependent activity. HepG2 (40/6) cells harboring the AHR-responsive pGudluc 6.1reporter vector were treated with vehicle (DMSO), 10 nM TCDD as a positive control, or the indicated compounds (10 μM) for 6 h; cells were lysed, and luciferase activity was determined. Data represent mean luciferase units ± S.E.M. and were normalized for protein concentration.
Comparison between the Activity of TMF and AHR Antagonists.
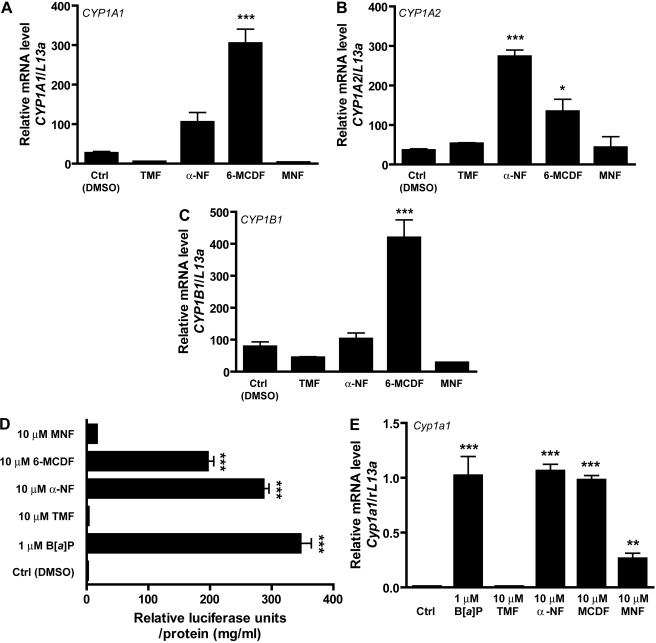
The data from the flavonoid screen for AHR agonist activity indicates that TMF fails to elicit AHR-driven DRE-dependent gene expression but has the capacity to repress basal activity and may function as an AHR antagonist. To eliminate a potential cytotoxic effect, MTS and colony formation assays were performed on Huh7 cells in the presence of 10 μM TMF, α-NF, or β-NF for comparative purposes. The data from the short-term MTS assay revealed a statistically significant cytostatic effect of α-NF, β-NF, and TMF but no difference between each of the treatments. The longer-term colony formation assay indicated no significant cytostatic effects associated with any of the treatment regimes (Supplemental Fig. 2). The repression of basal reporter expression suggests the possibility for TMF to exert an antagonistic effect with regard to AHR. To further explore this notion we compared the activity of TMF against the reported AHR antagonists α-NF, MNF, and 6-MCDF in the context of endogenous AHR-dependent gene expression. Huh7 cells were incubated in the presence of 10 μM TMF, α-NF, MNF, or 6-MCDF for a period of 4 h and the expression of CYP1A1, CYP1A2, and CYP1B1 analyzed by quantitative PCR (Fig. 3, A–C). The data revealed discrepancies with regard to AHR-dependent gene expression after exposure to α-NF or 6-MCDF. α-NF exhibited stimulatory effects on CYP1A family members, prompting a 3-fold induction in both CYP1A1 and A2. In contrast to CYP1A1/2, CYP1B1 expression was not influenced by α-NF. Like α-NF, 6-MCDF exerted a positive influence on CYP1A1 expression, although the magnitude of induction was 2-fold greater than that observed with α-NF. The pattern was reversed with regard to CYP1A2, 6-MCDF induced CYP1A2 expression but to a level that was 50% of the α-NF response. The CYP1B1 response to 6-MCDF was in opposition to the neutral effect of α-NF, and 6-MCDF was able to elicit a 4-fold elevation in CYP1B1 expression. Unlike α-NF and 6-MCDF, neither TMF nor MNF exhibited any agonist potential with regard to CYP1A1/2 or CYP1B1 expression.
Fig. 3.
TMF fails to stimulate AHR-mediated gene expression. Huh7 cells were treated with vehicle (DMSO), or the indicated compounds (TMF, α-NF, 6-MCDF, or MNF) for 4 h. Total RNA was harvested and used to generate cDNA. Expression of AHR-responsive CYP1A1 (A), CYP1A2 (B), and CYP1B1 (C) were determined through quantitative real-time PCR analysis. Data represent relative mRNA levels ± S.E.M. of the indicated target gene normalized against rL13a. D, mouse Hepa 1.1 cells harboring the AHR-responsive pGudluc 6.1 reporter vector were treated with vehicle (DMSO), 1 μM B[a]P as a positive control, or the indicated compounds (TMF, α-NF, 6-MCDF, or MNF) for 6 h; cells were lysed, and luciferase activity was determined. Data represent mean luciferase units ± S.E.M. and were normalized for protein concentration. E, Hepa 1.1 cells were treated with vehicle (DMSO), 1 μM B[a]P as a positive control, or the indicated compounds (TMF, α-NF, 6-MCDF, or MNF) for 4 h. Total RNA was harvested and used to generate cDNA. Expression of AHR-responsive Cyp1a1 was determined through quantitative real-time PCR analysis. Data represent relative mRNA levels ± S.E.M. of the indicated target gene normalized against rL13a.
AHR ligands have been revealed to display a level of species specificity; thus, we examined the capacity of TMF, α-NF, 6-MCDF, and MNF to elicit AHR activity in the mouse Hepa 1.1 reporter cell line (Fig. 3, D and E). As observed with the endogenous CYP1A1/2 gene expression in the human Huh7 cell line both α-NF and 6-MCDF exhibited significant AHR agonist activity (150- and 100-fold, respectively) as assessed by induction of the stably integrated pGudluc 6.1 reporter. In contrast, MNF prompted a marginal increase in reporter activity that proved to be statistically insignificant. In stark contrast to the effect of α-NF, 6-MCDF, and, to a lesser degree, MNF, TMF evoked no increase in reporter expression whatsoever (Fig. 3D). Previous reports have indicated differences in the magnitude of partial agonist effects of compounds such as MNF, depending on the mode of assessment, i.e., reporter assays versus endogenous gene expression (Zhou and Gasiewicz, 2003). Therefore, we also examined the effect of TMF together with the panel of antagonists in Hepa 1.1 cells by use of endogenous Cyp1a1 mRNA expression as a more relevant, less artificial, endpoint (Fig. 3E). As with the reporter-based determination, B[a]P prompted a robust AHR response that was matched by α-NF and 6-MCDF. Unlike the reporter assay, the modest induction observed with MNF proved to be statistically significant, in contrast to TMF, which failed to stimulate Cyp1a1 mRNA expression. The effect of TMF on endogenous gene expression was also examined in a non-hepatocyte-derived cell line to limit the potential of a cell lineage outcome. MCF7 cells showed no elevation in the expression of either CYP1A1 or CYP1A2 in response to TMF, mirroring the Huh7 effect (Supplemental Fig. 3).
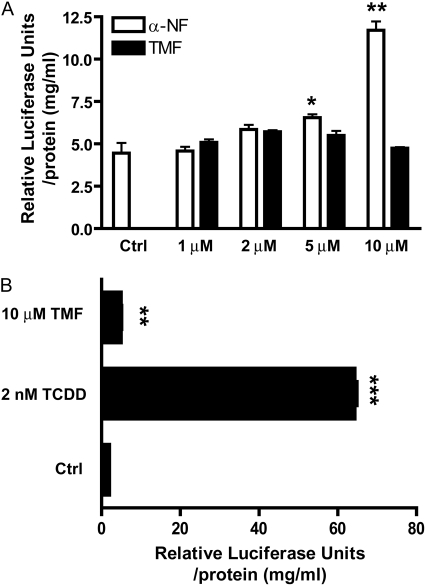
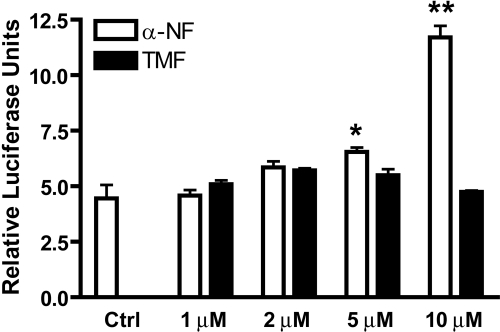
A dose-response comparison between TMF and α-NF, the most widely used AHR antagonist, was performed to examine whether any dose dependence underlies the failure of TMF to induce AHR-mediated gene expression. HepG2 (40/6) cells were exposed to increasing concentrations of TMF or α-NF, as indicated for 6 h, and reporter expression was determined (Fig. 4A). Exposure to increasing α-NF concentrations resulted in a dose-responsive increase in DRE-driven reporter activity, achieving statistical significance at 5 μM. This is in contrast to TMF, which failed to induce reporter activity across all doses tested. To determine whether metabolism of TMF over an extended incubation could yield metabolites with potential AHR agonist activity HepG2 (40/6) cells were exposed to 2 nM TCDD or 10 μM TMF for 24 h, and reporter expression was determined (Fig. 4B). The increased incubation time with TMF resulted in a small (2-fold) but significant increase in reporter activity above vehicle-treated controls, thus indicating a time-dependent generation of agonist activity.
Fig. 4.
Lack of TMF agonist activity is independent of dose. A, HepG2 (40/6) cells were treated with vehicle (DMSO) or increasing concentrations of either TMF or α-NF, as indicated for 6 h; cells were lysed, and luciferase activity was determined. Data represent mean luciferase units ± S.E.M. and were normalized for protein concentration. B, HepG2 (40/6) cells were treated with vehicle (DMSO), 2 nM TCDD, or 10 μM TMF, as indicated, for 24 h; cells were lysed, and luciferase activity was determined. Data represent mean luciferase units ± S.E.M.
TMF Fails to Stimulate the Interaction of AHR with Its Cognate Response Element.
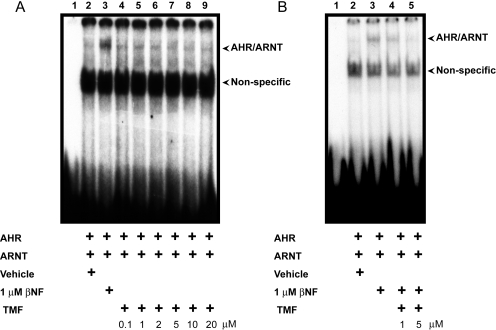
Binding of AHR as a heterodimer in cooperation with the ARNT to its cognate response element, the DRE, is a prerequisite for induction of AHR target genes. We therefore wished to examine the effect of TMF on the association of AHR/ARNT with its response element by use of electromobility shift assays and to determine whether the observed lack of AHR agonist activity is a consequence of diminished AHR/ARNT/DRE complex formation. In vitro translated AHR/ARNT in the absence of additional treatment or with vehicle (DMSO) failed to promote significant binding to the DRE probe. Exposure to 1 μM β-NF yielded a significant probe shift indicative of formation of the AHR/ARNT/DRE complex. Exposure to increasing doses of TMF in the absence of β-NF had a negligible impact on complex formation (Fig. 5A). Such data may suggest that TMF is not a ligand for AHR. To determine whether TMF has the capacity to inhibit the agonist-mediated association of AHR/ARNT with its cognate response element, competitive electromobility shift assays were performed. Coexposure to 1 μM β-NF and TMF at two doses resulted in a decrease in AHR/ARNT/DRE complex formation compared with β-NF alone (Fig. 5B). Such data indicate that TMF can compete with AHR agonists to diminish AHR/DRE binding. The effect of TMF on AHR/DRE binding was also examined in a cellular context by use of chromatin immunoprecipitation assays (Supplemental Fig. 4). HN2095 cells exposed to vehicle or 10 μM TMF for 18 h displayed a decreased signal for both AHR and ARNT at the CYP1A1 proximal promoter compared with vehicle-treated controls. Such data support the inhibitory effect of TMF on the association of AHR with its response element.
Fig. 5.
TMF fails to induce AHR binding to its cognate response element. A, In vitro translated AHR and ARNT were incubated with vehicle (DMSO), 1 μM β-NF or increasing concentrations of TMF (0.1–20 μM) TMF and 32P-labeled DRE probe added. Samples were resolved on 6% nondenaturing retardation gels and visualized through autoradiography. Specific AHR/ARNT/DRE-retarded bands are indicated. B, in vitro translated AHR and ARNT were incubated with vehicle (DMSO), 1 μM β-NF, or 1 μM β-NF together with TMF (1 or 5 μM) and 32P-labeled DRE probe added. Samples were resolved on 6% nondenaturing retardation gels and visualized through autoradiography. Specific AHR/ARNT/DRE-retarded bands are indicated.
TMF Antagonizes AHR-Dependent Gene Expression.
Having established that TMF fails to stimulate AHR binding to its cognate response element together with the complimentary absence of agonist activity suggests that TMF may be neutral with regard to AHR, i.e., is not an AHR ligand. We therefore assessed the ligand-binding status of AHR with regard to TMF by use of a ligand competition approach. HepG2 (40/6) cells were incubated with the AHR agonist B[a]P (1 μM) either in isolation or in conjunction with increasing doses of TMF or α-NF (Fig. 6). Exposure to B[a]P resulted in a robust 8-fold induction of reporter expression, and coexposure to 1 μM α-NF prompted a statistically significant dose-dependent repression of B[a]P-mediated reporter expression. Increasing concentrations of α-NF further repressed reporter expression to ∼50% of the maxima. TMF exhibited a similar dose relationship with regard to repression. Significant repression with TMF was achieved at 2 μM; however, in contrast to α-NF, TMF had the capacity to diminish reporter expression to a greater degree. The competitive nature of TMF both under basal and induced states thus argues in favor of a direct ligand mechanism and against a neutral effect.
Fig. 6.
TMF antagonizes AHR-mediated gene expression. HepG2 (40/6) cells were treated with vehicle (DMSO), 1 μM B[a]P, or B[a]P in conjunction with increasing concentrations of either TMF or α-NF, as indicated, for 6 h; cells were lysed, and luciferase activity was determined. Data represent mean luciferase units ± S.E.M. and were normalized for protein concentration.
TMF Is a Ligand for AHR.
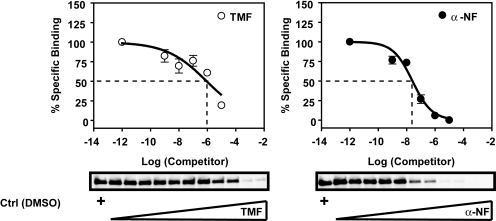
The observations that TMF has the capacity to diminish both basal and agonist-dependent AHR signaling are suggestive but not conclusive evidence that TMF is an antagonistic AHR ligand. To substantiate the potential antagonistic nature of TMF competitive ligand displacement assays were performed against a photoaffinity radioligand by use of α-NF for comparison (Fig. 7). Incubation with increasing concentrations of α-NF effectively displaced the radioligand from AHR yielding an apparent EC50 = 2.5 × 10−8 M. TMF also displayed a capacity to compete for radioligand binding, albeit with reduced efficacy, as evidenced by an apparent EC50 = 9 × 10−7 M, two orders of magnitude greater than that obtained with α-NF. As such, TMF can be considered a direct ligand for the AHR.
Fig. 7.
Competitive ligand binding demonstrates that TMF is an AHR ligand. Cytosolic extracts from “humanized” AHR mice were preincubated with 0.21 pmol if 125I-photoaffinity ligand then treated with increasing concentration of competitor ligand, either TMF or α-NF. After photoaffinity cross-linking samples were resolved through SDS-PAGE and transferred to PVDF membrane. Labeled AHR bands were visualized by autoradiography, excised, and subjected to γ-counting. Data represent percentage of specific binding relative to vehicle (DMSO).
TMF Fails to Mediate AHR Protein Degradation.
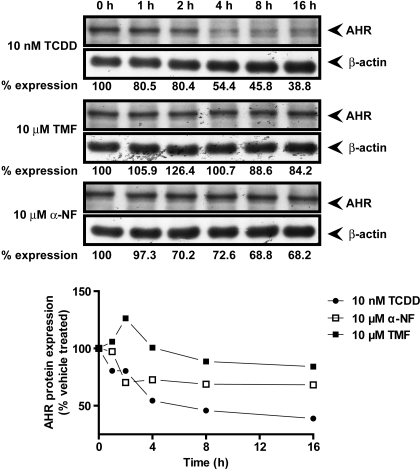
An observed characteristic of AHR ligands is that agonists tend to promote AHR protein turnover, presumably as a mechanism to terminate the stimulatory signal. Conversely, antagonists fail to stimulate protein turnover to the same extent. Examination of Huh7 AHR protein levels revealed a marked 60% reduction in AHR protein levels after 16 h of exposure to 10 nM TCDD compared with vehicle-treated control (Fig. 8). Exposure to 10 μM α-NF also promoted AHR turnover, albeit to a lesser extent than that observed with TCDD. After 16 h of exposure to α-NF, AHR expression was reduced by 40%. AHR expression was least influenced by TMF, after 16 h of incubation AHR levels were diminished by 15% (Fig. 8).
Fig. 8.
TMF fails to stimulate AHR protein degradation. Huh7 cells were treated with vehicle (DMSO), 10 nM TCDD, 10 μM TMF, or 10 μM α-NF for the indicated times. Total protein was harvested and used to assess AHR protein expression. Fifty micrograms of lysate were resolved by SDS-PAGE, transferred to PVDF membrane, and probed for AHR and the loading control β-actin by use of appropriate antibodies. Immune-reactive bands were visualized through autoradiography and quantified by γ-counting. Data represent percentage of AHR expression relative to vehicle-treated control.
Discussion
Despite published accounts of AHR activity occurring in the absence of exogenously added ligand through loss of cell-cell contact (Monk et al., 2001; Cho et al., 2004) or via indirect activators such as omeprazole (Backlund and Ingelman-Sundberg, 2005), it is widely accepted that the majority of the transcriptional activity associated with AHR is a consequence of direct agonist binding. Ligand-mediated transformation of AHR results in the formation of the transcriptionally competent AHR/ARNT heterodimer which is capable of stimulating phase I/II gene expression through association with its cognate DNA response element.
Recent reports have highlighted and expanded the scope of AHR biology beyond that of xenobiotic sensor and phase I/II metabolism. Expansion into the realm of inflammation and adaptive immunity (De Souza et al., 2009; Patel et al., 2009; Stockinger et al., 2009; Veldhoen et al., 2009), raises the notion that inhibition of AHR activity may have therapeutic potential. Attenuation of AHR signaling may be achieved through disruption of the cytoplasmic AHR-chaperone complex with heat-shock protein 90 (HSP90) inhibitors, such as geldanamycin leading to AHR turnover (Song and Pollenz, 2002); however, off-target effects are highly likely because of the ubiquitous chaperone nature of HSP90. Targeted promotion of AHR ubiquitinylation and subsequent proteosomal turnover by use of PROTACS has been proposed as a more specific mode of AHR inhibition (Puppala et al., 2008). While achieving significant inhibition of AHR-dependent gene expression, relatively high doses are required. However, this mechanism does have the advantage of sustained inhibition due to loss of AHR protein. Another approach is the utilization of direct AHR antagonists, ligands capable of AHR binding but rendering it transcriptionally inactive. Such an approach, using the flavone α-NF, has been a mainstay of AHR inhibition. Numerous compounds have demonstratively suppressive effects on AHR-mediated gene expression in the context of exogenous AHR agonists. However, most of these compounds exhibit weak partial agonist activity toward the AHR resulting in modest induction of AHR target genes, and thus, they are considered competitive antagonists (Zhou and Gasiewicz, 2003). These disadvantages led us to screen for compounds that display bona fide AHR antagonist 1 activity.
We therefore conducted an AHR-dependent reporter-based screen of a number of methoxylated flavonoids to assay for AHR ligand activity with an emphasis on antagonism. Our rationale for using methoxyflavone derivatives was 3-fold. First, the flavonoid skeleton adheres to the structural and spatial requirements of the proposed AHR ligand-binding pocket (Waller and McKinney, 1995) as evidenced by the large number of polyphenolic flavonoids that exhibit disparate events on AHR activity (Lu et al., 1996; Ciolino et al., 1999; Flaveny et al., 2009). Second, modification of the flavone ring structure has been shown to alter AHR binding and/or associated agonist potential, depending on the position of the modification, suggesting the possibility of an arrangement that facilitates AHR binding but not receptor transformation and hence agonist activity (Lu et al., 1996). Third, published data indicate higher bioavailability, arising from resistance to P450-dependent metabolism of methoxyflavones relative to their hydroxylated derivatives (Walle, 2004; Wen and Walle, 2006).
The initial reporter-based screen pointed to TMF having suppressive effects with regard to AHR in the context of exogenously added AHR agonists. Cytotoxicity assays identified a modest cytostatic effect of TMF exposure; although statistically significant, the effect is marginal and similar to that obtained with α-NF or β-NF, suggesting that TMF-mediated suppression of the AHR response is not a consequence of overt toxicity. Further examination revealed that exposure to TMF in isolation fails to stimulate AHR-dependent gene expression in the short term, in contrast to AHR antagonists described previously. TMF-mediated inhibition could be attributed to indirect effects, such as through the ER. The ER is known to bind flavone-based compounds and also inhibit AHR activity (Gehm et al., 1997; Beischlag and Perdew, 2005), but our studies were primarily performed in the Huh7 ER negative cell line (unpublished data), thus eliminating such a mechanism. It is plausible that unidentified factors may be similarly influenced by TMF, which, in turn, can negatively affect AHR activity; such a scenario remains to be addressed.
TMF revealed no promoter or species dependence across the AHR target genes examined, which is also in contrast to antagonists described previously (Zhang et al., 2003; Zhou et al., 2003). The reason for the variability observed with other reported antagonists is not established, but it may result from differences in rate of uptake, metabolism, or intrinsic variation in binding affinity for the AHR, or it may reflect differences in how AHR activity is investigated (Wang, 2002; Zhou and Gasiewicz, 2003).
The molecular mechanism underlying TMF-mediated suppression remains to be determined, but it is clear that TMF diminishes the affinity of AHR for its cognate response element rather than perturbing the interaction of DNA-bound AHR with transcriptional cofactors, as evidenced by gel-shift and chromatin immunoprecipitation DNA-binding analyses. It may be speculated that the reduced AHR/DRE interaction is a consequence of a reduction in heterodimerization of AHR with ARNT, a prerequisite for DRE binding. However, the AHR is known to be inherently unstable and liable to proteosomal degradation when dissociated from either the HSP chaperone or ARNT complex (Prokipcak and Okey, 1991; Swanson and Perdew, 1993; Ma and Baldwin, 2000). We were unable to identify any significant loss of AHR protein after exposure to TMF, which could account for such a mode of action. Further studies using coimmunoprecipitation techniques may clarify the effect of TMF on the formation/stability of the AHR/ARNT heterodimer.
Competitive DNA-binding analysis identified the capacity of TMF to compete with β-NF-mediated AHR/DRE binding, suggesting that TMF is a direct ligand for the AHR. The status of TMF acting directly on AHR was confirmed through competitive ligand-binding assays. In the absence of a crystal structure for the AHR, the molecular events underlying ligand-mediated AHR transformation are poorly understood and the nature of antagonism even less so. It is likely that partial agonists, such as α-NF and MNF, or bona fide antagonists, such as TMF, adopt a conformation in the AHR ligand-binding pocket that either fails to make complete contact with critical residues that facilitate receptor transformation or interact with different residues to introduce an inhibitory conformation.
Despite suppressing AHR activity in the short-term, we were able to identify a modest increase in AHR-dependent gene expression after extended exposure to TMF. Such an observation may be attributable to metabolism of TMF yielding weak agonist potential, as has been described with other flavone derivatives (Androutsopoulos et al., 2008). Nonetheless, TMF seems to have a better antagonist profile compared with described previously compounds and may suggest that improved TMF-derivatives more resistant to metabolism could be developed.
In summary, we have documented for the first time the identification and characterization of the commercially available synthetic substituted flavonoid TMF as an AHR ligand. Furthermore, TMF exhibits the functional characteristics of an AHR antagonist, possessing the capacity to effectively compete with AHR agonists thus repressing AHR-mediated gene induction. The inhibitory mode of TMF action remains to be fully determined but evidence suggests that AHR suppression is a consequence of reduced association with its cognate response element. Moreover, unlike current AHR antagonists, TMF exhibits no AHR agonist activity in the short term. It is noteworthy that TMF treatment of various cell types reveals no promoter or species dependence with regard to antagonism; thus, TMF represents a true antagonist of the AHR with limited cytotoxicity and may have applications both therapeutically and for the further investigation of AHR function.
Supplementary Material
Acknowledgments
We thank Drs. S. Safe and T. Gasiewicz, Texas A&M University and University of Rochester School of Medicine & Dentistry, for the generous gifts of TCDD and 3′-methoxy-4′-nitroflavone, respectively, and Kelly Wagner (The Pennsylvania State University) for generating some of the PCR data.
This work was supported by the National Institutes of Health National Institute of Environmental Health Sciences [Grant ES04869].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.109.158261
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
1 An antagonist in our context represents an inhibitor of DRE-mediated gene expression in the absence of residual partial agonist activity.
- AHR
- aryl hydrocarbon receptor
- ARNT
- aryl hydrocarbon receptor nuclear translocator
- TMF
- 6,2′,4′-trimethoxyflavone
- α-NF
- α-napthoflavone
- β-NF
- β-napthoflavone
- MNF
- 3′-methoxy-4′-nitroflavone
- 6-MCDF
- 6-methoxy-1,3,8-trichlorodibenzofuran
- ER
- estrogen receptor
- B[a]P
- benzo[a]pyrene
- TCDD
- 2,3,7,8-tetrachlorodibenzo-p-dioxin
- DRE
- dioxin response element
- PAL
- 2-azido-3-[125I]iodo-7,8-dibromodibenzo-p-dioxin
- CH-223191
- 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazophenyl)-amide
- PD98059
- 5′-methoxy-6′-aminoflavone
- DMSO
- dimethyl sulfoxide
- PVDF
- polyvinylidene difluoride
- PAGE
- polyacrylamide gel electrophoresis
- PBS
- phosphate-buffered saline
- PCR
- polymerase chain reaction
- MOPS
- 4-morpholinepropanesulfonic acid
- HSP90
- heat-shock protein 90.
References
- Aarts JM, Denison MS, Cox MA, Schalk MA, Garrison PM, Tullis K, de Haan LH, Brouwer A. (1995) Species-specific antagonism of Ah receptor action by 2,2′,5,5′-tetrachloro- and 2,2′,3,3′4,4′-hexachlorobiphenyl. Eur J Pharmacol 293:463–474 [DOI] [PubMed] [Google Scholar]
- Androutsopoulos V, Arroo RR, Hall JF, Surichan S, Potter GA. (2008) Antiproliferative and cytostatic effects of the natural product eupatorin on MDA-MB-468 human breast cancer cells due to CYP1-mediated metabolism. Breast Cancer Res 10:R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astroff B, Zacharewski T, Safe S, Arlotto MP, Parkinson A, Thomas P, Levin W. (1988) 6-Methyl-1,3,8-trichlorodibenzofuran as a 2,3,7,8-tetrachlorodibenzo-p-dioxin antagonist: inhibition of the induction of rat cytochrome P-450 isozymes and related monooxygenase activities. Mol Pharmacol 33:231–236 [PubMed] [Google Scholar]
- Backlund M, Ingelman-Sundberg M. (2005) Regulation of aryl hydrocarbon receptor signal transduction by protein tyrosine kinases. Cell Signal 17:39–48 [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Perdew GH. (2005) ER alpha-AHR-ARNT protein-protein interactions mediate estradiol-dependent transrepression of dioxin-inducible gene transcription. J Biol Chem 280:21607–21611 [DOI] [PubMed] [Google Scholar]
- Casper RF, Quesne M, Rogers IM, Shirota T, Jolivet A, Milgrom E, Savouret JF. (1999) Resveratrol has antagonist activity on the aryl hydrocarbon receptor: implications for prevention of dioxin toxicity. Mol Pharmacol 56:784–790 [PubMed] [Google Scholar]
- Cho YC, Zheng W, Jefcoate CR. (2004) Disruption of cell-cell contact maximally but transiently activates AhR-mediated transcription in 10T1/2 fibroblasts. Toxicol Appl Pharmacol 199:220–238 [DOI] [PubMed] [Google Scholar]
- Ciolino HP, Daschner PJ, Yeh GC. (1999) Dietary flavonols quercetin and kaempferol are ligands of the aryl hydrocarbon receptor that affect CYP1A1 transcription differentially. Biochem J 340:715–722 [PMC free article] [PubMed] [Google Scholar]
- De Souza VR, Cabrera WK, Galvan A, Ribeiro OG, De Franco M, Vorraro F, Starobinas N, Massa S, Dragani TA, Ibañez OM. (2009) Aryl hydrocarbon receptor polymorphism modulates DMBA-induced inflammation and carcinogenesis in phenotypically selected mice. Int J Cancer 124:1478–1482 [DOI] [PubMed] [Google Scholar]
- Flaveny CA, Murray IA, Chiaro CR, Perdew GH. (2009) Ligand selectivity and gene regulation by the human aryl hydrocarbon receptor in transgenic mice. Mol Pharmacol 75:1412–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehm BD, McAndrews JM, Chien PY, Jameson JL. (1997) Resveratrol, a polyphenolic compound found in grapes and wine, is an agonist for the estrogen receptor. Proc Natl Acad Sci U S A 94:14138–14143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbal-Chaloin S, Pichard-Garcia L, Fabre JM, Sa-Cunha A, Poellinger L, Maurel P, Daujat-Chavanieu M. (2006) Role of CYP3A4 in the regulation of the aryl hydrocarbon receptor by omeprazole sulphide. Cell Signal 18:740–750 [DOI] [PubMed] [Google Scholar]
- Gillner M, Bergman J, Cambillau C, Fernström B, Gustafsson JA. (1985) Interactions of indoles with specific binding sites for 2,3,7,8-tetrachlorodibenzo-p-dioxin in rat liver. Mol Pharmacol 28:357–363 [PubMed] [Google Scholar]
- Harris M, Zacharewski T, Astroff B, Safe S. (1989) Partial antagonism of 2,3,7,8-tetrachlorodibenzo-p-dioxin-mediated induction of aryl hydrocarbon hydroxylase by 6-methyl-1,3,8-trichlorodibenzofuran: mechanistic studies. Mol Pharmacol 35:729–735 [PubMed] [Google Scholar]
- Long WP, Pray-Grant M, Tsai JC, Perdew GH. (1998) Protein kinase C activity is required for aryl hydrocarbon receptor pathway-mediated signal transduction. Mol Pharmacol 53:691–700 [DOI] [PubMed] [Google Scholar]
- Lu YF, Santostefano M, Cunningham BD, Threadgill MD, Safe S. (1995) Identification of 3′-methoxy-4′-nitroflavone as a pure aryl hydrocarbon (Ah) receptor antagonist and evidence for more than one form of the nuclear Ah receptor in MCF-7 human breast cancer cells. Arch Biochem Biophys 316:470–477 [DOI] [PubMed] [Google Scholar]
- Lu YF, Santostefano M, Cunningham BD, Threadgill MD, Safe S. (1996) Substituted flavones as aryl hydrocarbon (Ah) receptor agonists and antagonists. Biochem Pharmacol 51:1077–1087 [DOI] [PubMed] [Google Scholar]
- Luster MI, Hong LH, Osborne R, Blank JA, Clark G, Silver MT, Boorman GA, Greenlee WF. (1986) 1-amino-3,7,8-trichlorodibenzo-p-dioxin: a specific antagonist for TCDD-induced myelotoxicity. Biochem Biophys Res Commun 139:747–756 [DOI] [PubMed] [Google Scholar]
- Ma Q, Baldwin KT. (2000) 2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced degradation of aryl hydrocarbon receptor (AhR) by the ubiquitin-proteasome pathway. Role of the transcription activaton and DNA binding of AhR. J Biol Chem 275:8432–8438 [DOI] [PubMed] [Google Scholar]
- Monk SA, Denison MS, Rice RH. (2001) Transient expression of CYP1A1 in rat epithelial cells cultured in suspension. Arch Biochem Biophys 393:154–162 [DOI] [PubMed] [Google Scholar]
- Negishi T, Kato Y, Ooneda O, Mimura J, Takada T, Mochizuki H, Yamamoto M, Fujii-Kuriyama Y, Furusako S. (2005) Effects of aryl hydrocarbon receptor signaling on the modulation of TH1/TH2 balance. J Immunol 175:7348–7356 [DOI] [PubMed] [Google Scholar]
- Patel RD, Murray IA, Flaveny CA, Kusnadi A, Perdew GH. (2009) Ah receptor represses acute-phase response gene expression without binding to its cognate response element. Lab Invest 89:695–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland A, Glover E, Ebetino FH, Kende AS. (1986) Photoaffinity labeling of the Ah receptor. J Biol Chem 261:6352–6365 [PubMed] [Google Scholar]
- Prokipcak RD, Okey AB. (1991) Downregulation of the Ah receptor in mouse hepatoma cells treated in culture with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Can J Physiol Pharmacol 69:1204–1210 [DOI] [PubMed] [Google Scholar]
- Puppala D, Lee H, Kim KB, Swanson HI. (2008) Development of an aryl hydrocarbon receptor antagonist using the proteolysis-targeting chimeric molecules approach: a potential tool for chemoprevention. Mol Pharmacol 73:1064–1071 [DOI] [PubMed] [Google Scholar]
- Ramadoss P, Marcus C, Perdew GH. (2005) Role of the aryl hydrocarbon receptor in drug metabolism. Expert Opin Drug Metab Toxicol 1:9–21 [DOI] [PubMed] [Google Scholar]
- Reiners JJ, Jr, Lee JY, Clift RE, Dudley DT, Myrand SP. (1998) PD98059 is an equipotent antagonist of the aryl hydrocarbon receptor and inhibitor of mitogen-activated protein kinase kinase. Mol Pharmacol 53:438–445 [DOI] [PubMed] [Google Scholar]
- Song Z, Pollenz RS. (2002) Ligand-dependent and independent modulation of aryl hydrocarbon receptor localization, degradation, and gene regulation. Mol Pharmacol 62:806–816 [DOI] [PubMed] [Google Scholar]
- Stockinger B, Veldhoen M, Hirota K. (2009) Modulation of Th17 development and function by activation of the aryl hydrocarbon receptor–the role of endogenous ligands. Eur J Immunol 39:652–654 [DOI] [PubMed] [Google Scholar]
- Swanson HI, Perdew GH. (1993) Half-life of aryl hydrocarbon receptor in Hepa 1 cells: evidence for ligand-dependent alterations in cytosolic receptor levels. Arch Biochem Biophys 302:167–174 [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Christensen J, O'Garra A, Stockinger B. (2009) Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med 206:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. (2008) The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453:106–109 [DOI] [PubMed] [Google Scholar]
- Walle T. (2004) Absorption and metabolism of flavonoids. Free Radic Biol Med 36:829–837 [DOI] [PubMed] [Google Scholar]
- Waller CL, McKinney JD. (1995) Three-dimensional quantitative structure-activity relationships of dioxins and dioxin-like compounds: model validation and Ah receptor characterization. Chem Res Toxicol 8:847–858 [DOI] [PubMed] [Google Scholar]
- Wang TT. (2002) β-Naphthoflavone, an inducer of xenobiotic metabolizing enzymes, inhibits firefly luciferase activity. Anal Biochem 304:122–126 [DOI] [PubMed] [Google Scholar]
- Wen X, Walle T. (2006) Methylation protects dietary flavonoids from rapid hepatic metabolism. Xenobiotica 36:387–397 [DOI] [PubMed] [Google Scholar]
- Wilhelmsson A, Whitelaw ML, Gustafsson JA, Poellinger L. (1994) Agonistic and antagonistic effects of alpha-naphthoflavone on dioxin receptor function. Role of the basic region helix-loop-helix dioxin receptor partner factor Arnt. J Biol Chem 269:19028–19033 [PubMed] [Google Scholar]
- Zhang S, Qin C, Safe SH. (2003) Flavonoids as aryl hydrocarbon receptor agonists/antagonists: effects of structure and cell context. Environ Health Perspect 111:1877–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gasiewicz TA. (2003) 3′-Methoxy-4′-nitroflavone, a reported aryl hydrocarbon receptor antagonist, enhances Cyp1a1 transcription by a dioxin responsive element-dependent mechanism. Arch Biochem Biophys 416:68–80 [DOI] [PubMed] [Google Scholar]
- Zhou JG, Henry EC, Palermo CM, Dertinger SD, Gasiewicz TA. (2003) Species-specific transcriptional activity of synthetic flavonoids in guinea pig and mouse cells as a result of differential activation of the aryl hydrocarbon receptor to interact with dioxin-responsive elements. Mol Pharmacol 63:915–924 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.