Abstract
Hsp70 chaperones assist protein folding by ATP-controlled cycles of substrate binding and release. ATP hydrolysis is the rate-limiting step of the ATPase cycle that causes locking in of substrates into the substrate-binding cavity of Hsp70. This key step is strongly stimulated by DnaJ cochaperones. We show for the Escherichia coli Hsp70 homolog, DnaK, that stimulation by DnaJ requires the linked ATPase and substrate-binding domains of DnaK. Functional interaction with DnaJ is affected by mutations in an exposed channel located in the ATPase domain of DnaK. It is proposed that binding to this channel, possibly involving the J-domain, allows DnaJ to couple substrate binding with ATP hydrolysis by DnaK. Evolutionary conservation of the channel and the J-domain suggests conservation of the mechanism of action of DnaJ proteins.
Keywords: Hsp70, heat shock proteins, protein folding
Hsp70 chaperones provide an essential line of cellular defense against stress by assisting refolding of misfolded proteins, and they are furthermore required for a large variety of protein-folding processes in unstressed cells (1, 2). This diversity in cellular roles, involving promiscuous but also selective substrate recognition (3), requires the activity of DnaJ cochaperones, which target Hsp70 proteins to their substrates (1, 4). DnaJ proteins constitute a large family of multidomain proteins, which share the signature J-domain required for cooperation with Hsp70 partner proteins. They differ with respect to other domains, which presumably permit association with specific Hsp70 target sites (4, 5). The existence of several DnaJ homologs in a cell, in conjunction with the specific substrates associated with them, is thought to be a major determinant for the functional diversity of Hsp70 proteins (1, 6).
DnaJ proteins act by controlling the Hsp70 ATPase activity. Hsp70 alternates between the ATP state characterized by low affinity and fast exchange rates for substrates and the ADP state characterized by high affinity and low exchange rates (7). ATP hydrolysis is the rate-limiting step of the ATPase cycle that locks in substrates in a complex with Hsp70. This control of substrate binding by nucleotide requires communication between the N-terminal ATPase domain and the adjacent substrate-binding domain. The ATP hydrolysis step is the prime target for regulation of Hsp70 proteins, exerted by DnaJ proteins, which stimulate the hydrolysis rate by at least several hundred fold in the case of the Escherichia coli Hsp70, DnaK (1, 6, 8–11). A recent NMR study by using 15N-labeled J-domain of DnaJ revealed that DnaJ interacts via its J-domain with the ATPase domain of DnaK (12) but could not provide information about the binding site(s) on the ATPase domain or possible additional contacts between DnaJ and the substrate-binding domain of DnaK. The molecular basis for the DnaJ interaction with Hsp70 thus remains an open question of central mechanistic importance.
The present work is aimed at identifying interaction sites for DnaJ by mutational analysis of DnaK. We report that DnaJ activity requires the linked ATPase and substrate-binding domains. Mutations in a conserved channel in the ATPase domain of DnaK affect the functional interaction with DnaJ. This channel may have additional roles in the coupling of substrate binding with the nucleotide status of DnaK.
MATERIALS AND METHODS
Purification of DnaK Mutants.
Wild-type (wt) and mutant DnaK were purified as described (13) after overproduction in ΔdnaK52 cells (BB1553) (14, 24). The in vitro functional assays were performed with at least two independently purified protein batches and yielded similar results.
Luciferase-Refolding Assay.
A stock solution of firefly luciferase (64 μM in 1 M glycylglycine, pH 7.4) (Sigma) was 6.4-fold diluted into unfolding buffer (25 mM Hepes-KOH (pH 7.6), 50 mM KCl, 5 mM MgCl2, 5 mM β-mercaptoethanol, and 6 M guanidinium⋅HCl) and denatured (25°C, 10 min), followed by dilution (80 nM, final concentration) into refolding buffer (15) containing DnaJ (160 nM), DnaK and GrpE as indicated in Fig. 3 legend. Aliquots of 1 μl were withdrawn at the indicated times, diluted into 125-μl assay buffer, and analyzed for bioluminescence activity in a Biolumat (Berthold, Bad Wildbad, Germany) as described (15).
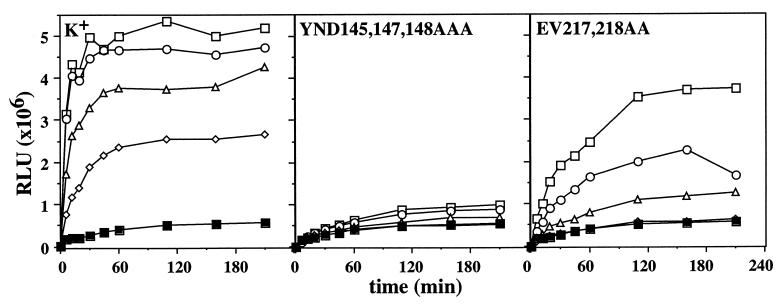
Figure 3.
Activities of DnaK mutants in luciferase refolding. Guanidinium⋅HCl denatured luciferase was diluted into refolding buffer containing DnaJ and GrpE, as well as wt (K+) and mutant DnaK proteins as indicated. The activity of refolded luciferase in relative light units (RLU) was determined as described (25). The final concentrations of luciferase and DnaJ were 80 nM and 160 nM, respectively. The concentrations of DnaK and GrpE were as follows: (■) no DnaK, 800 nM GrpE; (◊) 400 nM DnaK, 200 nM GrpE; (▵) 800 nM DnaK, 400 nM GrpE; (○) 1200 nM DnaK, 600 nM GrpE; (□) 1600 nM DnaK, 800 nM GrpE.
Surface Plasmon Resonance (SPR) Spectroscopy.
SPR analysis of DnaK binding to DnaJ was done by using a BIAcore apparatus (BIAcore, Uppsala) at 30°C in BC buffer [25 mM Hepes, pH 7.6/50 mM KCl/10 mM MgCl2/1 mM EDTA/0.5 mM ZnCl2/0.005% Surfactant P-20 (Pharmacia)] at a flow rate of 20 μl/min according to standard protocols provided by the manufacturer. DnaJ was crosslinked to NHS-LC Biotin (Succinimidyl-6-(biotinamido) hexanoate) crosslinker (Pierce 21335) according to the manufacturer’s protocol. Two thousand response units of crosslinked DnaJ were coupled to a research grade CM5 chip, which contained 5,800 response units of streptavidin, coupled via N-ethyl-N′-(dimethylaminopropyl) carbodiimide N-hydroxy-succinimide (EDC/NHS) crosslinking. DnaK at a concentration of 1.25 μM (in BC buffer) was injected for 3.5 min. Three minutes before injection, a 10-fold excess of ATP in BC buffer was added to DnaK at 4°C. The sensor chip was cleaned of DnaK bound to immobilized DnaJ by injection of 10 μl of 2 M urea.
ATPase Activity Assay.
ATP hydrolysis rates under single turnover conditions were determined as described (8, 9). The final concentration of DnaK–ATP complexes was 250 nM. The peptide substrate used in the single turnover assay was σ32-M195-N207 (22). Hydrolysis was quantified by using the program macbas version 2.5 (Fuji) and fitted to a single exponential decay function by using the program grafit version 3.0 (Erithacus Software, Staines, U.K.).
ATP hydrolysis rates under steady–state conditions were determined as described (8, 9). Reactions were performed at 30°C in mixtures containing buffer HKM (25 mM Hepes-KOH, pH 7.6/50 mM KCl/5 mM MgCl2), 0.69 μM DnaK, 1.4 μM DnaJ, 0.69 μM GrpE, 200 μM ATP, and [α-32P]ATP (0.1 μCi, Amersham). Hydrolysis was quantified by using the program macbas version 2.5 (Fuji) and was fitted by nonlinear regression analysis by using the program grafit version 3.0 (Erithacus Software).
Tryptophan Fluorescence.
Measurements of the intrinsic tryptophan fluorescence of DnaK were performed on a Perkin–Elmer LS50B spectrofluorimeter. The emission spectra of tryptophan fluorescence of DnaK (0.5 μM) in the presence of 50 μM ATP or ADP were recorded at 4°C between 300 and 400 nm at a fixed excitation wavelength of 290 nm (16).
Determination of Substrate Release.
To determine the dissociation rate (koff) of DnaK bound peptide in absence of ATP, DnaK (1 μM) was preincubated for 2 hr at 30°C with 2-[4′-(iodoacetamido)anilino]naphthalene-6-sulfonic acid (IAAANS) fluorescence labeled σ32-Q132–Q144-C peptide (17) (0.5 μM). After addition of a 100-fold molar excess of unlabeled peptide, the dissociation of labeled peptide from DnaK was monitored by a decrease in the fluorescence signal. The time-dependent decrease was fitted to a single exponential decay by using grafit. The koff of DnaK bound peptide after addition of ATP was measured by using nucleotide-free DnaK in a stopped flow instrument (Applied Photophysics, Surrey, U.K.). Nucleotide-free DnaK was prepared as described (18), except that DnaK and alkaline phosphatase were separated by Mono Q (Pharmacia) anion exchange chromatography. A mixture of nucleotide-free DnaK (1 μM) and labeled peptide (1 μM) were preincubated at 30°C for 2 hr and mixed 1:1 in a stopped flow apparatus with a solution of 1 mM ATP and 100 μM unlabeled peptide. The decrease of fluorescence was measured for 50 s at 30°C.
RESULTS
Interaction of DnaJ with DnaK Domains.
To identify the domains of DnaK required for DnaJ action, we tested the ability of DnaJ to interact with fragments of DnaK. The tested fragments comprise either the ATPase domain alone (DnaK 1–385) or the ATPase domain linked to the adjacent substrate-binding domain up to the lid-forming α-helix closing the substrate-binding cleft (DnaK 1–538) (19). The DnaK 1–538 protein lacks the 100 C-terminal residues, which have been claimed earlier to be required for DnaJ activity (20). E. coli mutants expressing the chromosomally encoded dnaK mutant allele (dnaK163) that encodes DnaK 1–538 can grow at 40°C but not at 43°C and remain sensitive to lytic infection by bacteriophage λ (data not shown).
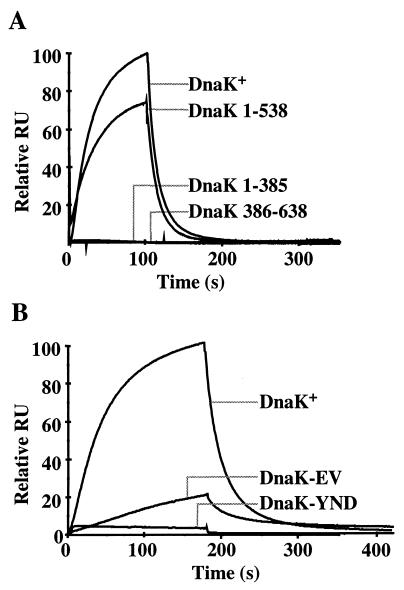
As determined by single turnover ATPase measurements, DnaJ failed to stimulate ATP hydrolysis by DnaK 1–385 whereas it stimulated ATP hydrolysis by DnaK 1–538 100-fold from 0.001 s−1 (unstimulated) to 0.10 s−1 (at 0.5 μM DnaJ and 0.25 μM DnaK; Table 1). The stimulated rate was slightly higher than that determined for wt DnaK (0.06 s−1, Table 1) under these conditions. The ability of DnaJ to physically associate with DnaK fragments was tested by using SPR (Fig. 1A). DnaJ was coupled to the sensor chip, and DnaK was injected into the flow chamber in absence or shortly after addition of ATP. wt DnaK associated with DnaJ in presence but not in absence of ATP, consistent with the role of DnaJ in the ATPase cycle and with earlier findings by using size exclusion chromatography (20). Association and dissociation were rapid but could not be fitted to a single or double exponential function, indicating a high complexity of the DnaK–DnaJ interaction. DnaJ failed to show detectable physical interaction with both, DnaK 1–385 and a fragment comprising the entire C terminus of DnaK including the substrate-binding domain but lacking the ATPase domain (DnaK 386–638) (Fig. 1A). In contrast, DnaJ showed almost normal interaction with DnaK 1–538. These results show that the ATPase domain has to be linked to the substrate-binding domain to allow DnaJ to physically interact with DnaK and to efficiently stimulate its ATPase activity. The C-terminal 100 residues of DnaK are not essential for this DnaJ action.
Table 1.
Single turnover ATPase activities of DnaK mutant proteins and their ATPase domain fragments
|
kcat of ATP hydrolysis (s−1)
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Basal
|
+Peptide substrate
|
+DnaJ
|
+ DnaJ/σ32
|
|||||
| Full length | ATPase domain | 100 μM | 50 nM | 0.5 μM | 2.5 μM | 10 μM | 50 nM/2 μM | |
| DnaK+ | 6.3 × 10−4 | 5 × 10−4 | 0.013 | 5.5 × 10−3 | 0.06 | >0.42 | >0.42 | 0.088 |
| YND145,147,148AAA | 2.2 × 10−3 | 6.7 × 10−4 | 0.012 | 3.2 × 10−3 | 8.8 × 10−3 | 0.01 | 9 × 10−3 | 0.011 |
| EV217,218AA | 2.7 × 10−3 | 5 × 10−4 | 0.011 | 3.7 × 10−3 | 6.8 × 10−3 | 0.02 | 0.03 | 5.8 × 10−3 |
| DnaK 1–538 | 1.0 × 10−3 | ND | ND | ND | 0.10 | >0.16 | ND | ND |
ND, not determined
Figure 1.
SPR analysis of the interaction between immobilized DnaJ and DnaK. (A) wt DnaK (DnaK+) and the truncated versions DnaK 1–385, DnaK 1–538, and DnaK 386–638. (B) wt (DnaK+) and mutant proteins YND145,147,148AAA (DnaK-YND) and EV217,218AA (DnaK-EV). The resonance signals given as relative response units (RU) are plotted as a function of time.
Mutagenesis of the ATPase Domain of DnaK.
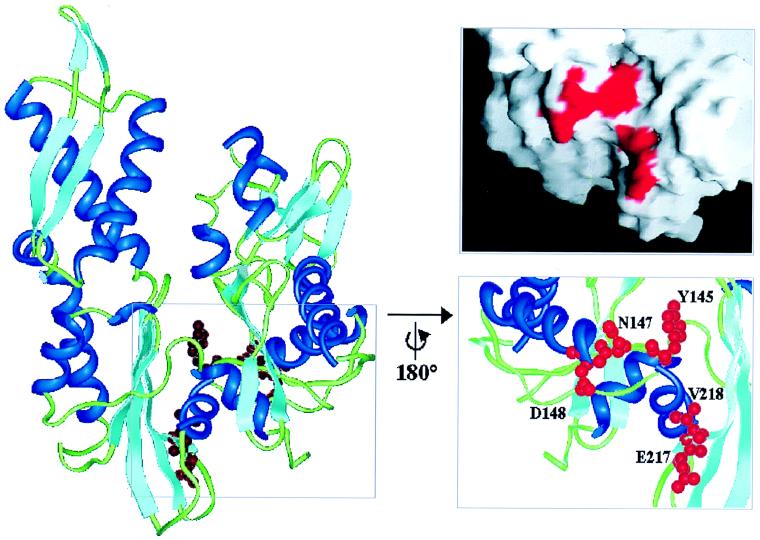
To identify residues in the ATPase domain of DnaK required for DnaJ action we subjected dnaK to site-directed mutagenesis on the basis of the following rationale. The ATPase domain is built of two subdomains connected by two crossed α-helices and separated by a large upper nucleotide-binding cleft and a smaller lower cleft (21, 22) (Fig. 2). Based on analysis of the homologous structures of the Hsc70 ATPase domain, actin and hexokinase, it has been proposed that a nucleotide-dependent opening and closing of the clefts provides the conformational change in the ATPase domain of Hsp70 that is transduced to the substrate-binding domain (23). This proposal is supported by the recently solved structure of the DnaK ATPase domain in complex with the nucleotide exchange factor GrpE, which reveals an opening of the subdomains in the nucleotide-free conformation (21). The residues of the DnaK ATPase domain to be mutated were selected to fulfill the criteria of being (i) surface-exposed, (ii) conserved within the Hsp70 family, (iii) predicted by structure modeling to alter their position in a nucleotide-dependent fashion, and (iv) homologous to residues of actin involved in actin–actin contacts. The only major cluster of residues fulfilling these criteria forms a conserved channel in the lower cleft, located in the back side of the ATPase domain according to the standard view (Fig. 2). To test the function of this channel, we analyzed two DnaK mutant proteins with alanine replacements of two (EV217, 218AA) and three (YND145, 147,148AAA) key residues located at the two sides of the channel (Fig. 2).
Figure 2.
Standard view of the structure of the DnaK ATPase domain within the DnaK–GrpE complex (PDB entry 1DKG) (21), showing the location of the mutated channel residues Y145, N147, D148, E217, and V218. α-helices are shown in dark blue, β-sheets in light blue, loops in green, and the side chains of the mutated residues in red. The marked region was rotated by 180° about the vertical axis. Residues Y145, N147, D148, E217, and V218 are shown as ball and stick models. The figures were prepared with insight II (Micron Separations, San Diego). The surface was created by using the program grasp (29), with mutated residues marked in red.
Chaperone Defects of ATPase Domain Mutants of DnaK.
The in vivo chaperone activities of the DnaK mutant proteins were determined by testing their ability to complement temperature sensitive growth and bacteriophage λ resistance of ΔdnaK52 cells (14, 24) (Table 2). Growth at 40°C was not complemented by the YND145,147,148AAA mutant protein, even when produced to high levels, and was only poorly complemented by the EV217,218AA mutant protein. Lytic growth of λ was only partially complemented by high levels of the YND145,147,148AAA and EV217,218AA mutant proteins. The chaperone activities of the DnaK mutant proteins were determined more precisely in vitro by using guanidinium⋅HCl unfolded firefly luciferase as substrate (25). Refolding of luciferase upon dilution from denaturant required the presence of wt DnaK, DnaJ, and the nucleotide exchange factor GrpE (Fig. 3). The YND145,147,148AAA mutant protein almost completely lacked chaperone activity in this assay. The EV217,218AA mutant protein showed a reduced yield of refolded luciferase and a slower kinetics of refolding. These defects correlate well with the in vivo defects of the mutant proteins.
Table 2.
Functional defects of DnaK channel mutants in vivo
| Growth at 30°C, %
|
Growth at 40°C, %; ΔdnaK52
|
λ plaque formation, %
|
||||||
|---|---|---|---|---|---|---|---|---|
| IPTG, μM | 0–500 | 0 | 100 | 250 | 500 | 0 | 100 | 250 |
| dnaK+ | 100 | 0 | 62 | 100 | 3.8 | 0 | 100 | 100 |
| dnaK-YND145,147,148AAA | 100 | 0 | 0.01 | 0.02 | 0 | 0 | 0 | 61 |
| dnaK-EV217,218AA | 100 | 0 | 0.48 | 7.8 | 0.01 | 0 | 0 | 100 |
Cells of ΔdnaK52 (BB1553) (24) that carry pDMI,1 (lacIq) (30) as well as pUHE21–2fdΔ12(dnaK) (13) encoding the indicated dnaK alleles under control of an IPTG inducible promoter, were tested for growth at the indicated temperatures and for λ plaque formation at 30°C. Formation of colonies and λ plaques (31) were tested on LB/Ap/Km plates containing IPTG at concentrations as indicated in the head row. All values were normalized to the dnaK+ value at 250 μM IPTG at 40°C for colony formation, which was set to 100%. IPTG, isopropyl β-d-thiogalactoside.
Defects of ATPase Domain Mutants in Interaction with DnaJ.
The basis for the functional defects of the two DnaK mutant proteins was examined further. The overall structures of the purified proteins were unaltered as judged by their wt-like proteolysis patterns and their highly similar far UV circular dichroism spectra at 30°C (data not shown). However, both proteins had severe defects in interaction with DnaJ. In single turnover ATPase measurements, the YND145,147,148AAA and EV217,218AA mutant proteins were only poorly stimulated by DnaJ, by 12- and 4-fold at high DnaJ concentrations (10 μM), respectively, whereas wt DnaK was stimulated at these conditions by ≥600-fold (Table 1). The rates for maximally stimulated ATP hydrolysis were at least 13- and 46-fold lower for the YND145,147,148AAA and EV217,218AA mutant proteins, respectively, as compared with wt DnaK. These defects also were observed in the simultaneous presence of DnaJ and protein substrate, σ32 (Table 1). These defects were not caused by strong alterations of the dissociation rates of bound nucleotide (data not shown). Furthermore, we also determined the potential of DnaJ to stimulate the mutant proteins in steady–state ATPase measurements at high ATP concentrations. In these assays, the ATPase activities of the EV217,218AA and the YND145,147,148AAA mutant proteins are only poorly stimulated by DnaJ, both in absence (Table 3) or presence (data not shown) of the σ32 substrate, confirming the results obtained in the single turnover ATPase assays.
Table 3.
Effects of DnaJ and GrpE on the steady–state ATPase activity of DnaK channel mutants
| Stimulation factors and kcat of ATP hydrolysis, s−1
|
|||
|---|---|---|---|
| Basal | +DnaJ | +DnaJ/GrpE | |
| DnaK+ | 1 (7.2 × 10−4) | 20 | 240 |
| YND145,147,148AAA | 1 (1.8 × 10−3) | 2 | 24 |
| EV217,218AA | 1 (2.5 × 10−3) | 2 | 12 |
The stimulation factors are relative to the basal ATP hydrolysis rates, which are given in parentheses.
We determined the ability of the mutant proteins to physically interact with DnaJ by SPR analysis. The YND145,147,148AAA and EV217,218AA mutant proteins had strongly reduced apparent affinities for the coupled DnaJ, with the YND145,147,148AAA mutant protein showing a stronger reduction (>20-fold) than the EV217,218AA mutant protein (5-fold) (Fig. 1B). In addition, both DnaK mutant proteins had altered apparent association and dissociation behavior as compared with wt DnaK, as judged by the altered shapes of the response curves.
Interdomain Communication in ATPase Domain Mutants.
The single turnover ATPase measurements also revealed that the basal rates of ATP hydrolysis by the two mutant proteins are slightly increased, 4-fold for EV217,218AA and 3.5-fold for YND145,147,148AAA, as compared with wt (Table 1). Such relatively small increases in the rate of ATP hydrolysis are typically observed in wt DnaK upon binding of chaperone substrate (8) and reflect the coupling between substrate binding and ATPase activity. The two mutant proteins may thus have either defects in the interdomain coupling or structural changes in the ATPase domain directly affecting ATP hydrolysis. To differentiate between these possibilities, we generated the 44-kDa ATPase domain fragments of the wt and mutant DnaK proteins by papain digestion followed by their purification (26) and tested their ATPase activities. The ATPase domain fragments of both mutant proteins had low basal ATPase activities in single turnover assays, similar to that of the corresponding fragment of wt DnaK (Table 1), demonstrating that the increased basal ATPase activities of the full length mutant proteins were caused by altered interdomain communication.
We investigated the functional consequences of these alterations by several criteria. First, single turnover assays revealed that the ATPase activities of the two mutant proteins were further stimulated by addition of protein substrate (σ32; not shown) and peptide substrate (σ32-M195-N207) (17) to the same level as wt DnaK (Table 1). Second, the intrinsic tryptophan fluorescence of DnaK, known to undergo an ATP-induced blueshift indicative of the interdomain coupling (16), showed only minor changes for the two mutant proteins compared with wt (Table 4). Third, the mutant proteins showed ATP induced alterations in the kinetics and pattern of proteolysis by trypsin similar to the changes found for wt DnaK (16) (data not shown). However, these assays may not allow to elucidate potential kinetic alterations in the coupling process. We therefore determined the kinetics by which ATP stimulates the dissociation rate constant (koff) of bound peptide substrate in stopped flow measurements, using nucleotide-free DnaK preparations. For these experiments, it was instrumental to generate nucleotide-free DnaK because for nucleotide (ADP) containing DnaK the koff for peptide is limited by the koff for bound ADP (18). For wt DnaK, ATP stimulated the koff of bound peptide substrate ≈2,000-fold to 2.31 s−1 (Table 4). For the YND145,147,148AAA and EV217,218AA mutant proteins, ATP stimulated the koff by ≈700-fold and 200-fold to 1.23 s−1 and 0.24 s−1, respectively. Together with the other assays described above, these findings indicate only minor coupling defects in the YND145,147,148AAA mutant protein and slightly more severe coupling defects in the EV217,218AA mutant protein.
Table 4.
Interdomain communication of DnaK channel mutants
|
koff of DnaK bound peptide, s−1
|
Tryptophan fluorescence, λmax [nm]
|
|||
|---|---|---|---|---|
| −ATP | +ATP | ADP | ATP | |
| DnaK+ | 1.17 × 10−3 | 2.31 (2,000-fold) | 344.6 | 341.1 |
| YND145,147,148AAA | 1.83 × 10−3 | 1.23 (700-fold) | 345.4 | 342.6 |
| EV217,218AA | 1.17 × 10−3 | 0.24 (200-fold) | 344.6 | 342.5 |
DISCUSSION
The results of our study indicate that the channel located at the lower back side of the ATPase domain of DnaK (Fig. 2) is an important site for interaction with DnaJ. The difference in the strength of the phenotypes of the two mutants may reflect differences in the importance of the two sides of the channel for functional interaction with DnaJ. The side of the channel where the residues 145,147,148 are located seems more important for the functional interaction with DnaJ as judged by the more severe defects of the YND145,147,148AAA mutant protein. However, the criterium of severity of defects is complicated as for the koff of chaperone-bound peptide in the presence of ATP, the EV217,218AA mutant protein has a slightly more severe defect as compared with the YND145,147,148AAA mutant protein. Obviously, the maximally stimulated koff observed with wt is not strictly required for residual chaperone activity as seen with the EV217,218AA mutant protein. The most plausible explanation of our data is that DnaJ interacts directly with residues located within this channel. A different genetic approach described in the accompanying study came to the same conclusion (27). Binding of DnaJ to this channel may involve the J-domain, in accordance with the demonstration by NMR experiments that helix 2 and the HPD motif of the J-domain contact the ATPase domain of DnaK (12). This possibility is further supported by the presence of negatively charged, exposed residues in the channel, which may electrostatically interact with the positively charged residues of the J-domain helix 2. Our failure to detect interaction of DnaJ with the ATPase domain fragment of wt DnaK by SPR spectroscopy indicates that this interaction exhibits a high dissociation rate that is outside of the kinetic range of this method (kd 10−5 − 10−2 s−1, ka 103 − 5 × 106 M−1 s−1).
The slight defects of the analyzed mutant proteins in interdomain communication suggest an additional role of this channel or its surrounding in the coupling of ATPase activity with substrate binding. This role is further indicated by a mutation localized in the close neighborhood of this channel, which causes a complete uncoupling phenotype (unpublished results). Such neighboring or overlapping interaction sites would be in accordance with the intimate relationship between the mechanism of interdomain coupling and DnaJ activity.
For physical interaction of DnaJ with the ATPase domain of DnaK further contacts are required to allow DnaJ to stimulate ATP hydrolysis. This is manifested by our findings that physical and functional DnaK–DnaJ interactions were detected only when the ATPase domain is linked to the substrate-binding domain of DnaK. In addition to the interaction of the J-domain with DnaK a second signal has been proposed to be binding of substrate to the substrate-binding cleft of DnaK (1, 28). This proposal is supported by our findings (Tables 1, 3, and unpublished results) that DnaJ requires presence of substrates to efficiently stimulate the ATPase activity of DnaK and that mutations in DnaK blocking the central hydrophobic substrate-binding pocket disturb the interaction with DnaJ (unpublished results). A possible scenario is that ATP dependent association of the J-domain with the ATPase channel of DnaK, along with further contacts to the substrate-binding domain of DnaK, positions the DnaJ bound substrate in close vicinity of the DnaK substrate-binding cleft. This allows efficient substrate transfer into the substrate-binding cleft of DnaK, thereby stimulating ATP hydrolysis by DnaK. These orchestrated interactions would allow DnaJ to tightly couple ATP hydrolysis with substrate binding by DnaK. The high evolutionary conservation of the involved structures of DnaJ (the J-domain) and DnaK (ATPase and substrate-binding domains) within the DnaJ and Hsp70 protein families strongly suggests that the mechanism of action of DnaJ proteins in the functional cycle of Hsp70 partner proteins is conserved as well.
Acknowledgments
We thank Klaus Paal for excellent performance of BIAcore experiments, Stefan Rüdiger for critical reading of the manuscript, and Jochen Reinstein for helpful advice concerning the off-rate determinations. This work was supported by a grant of the Deutsche Forschungsgemeinschaft to B.B. (SFB388).
ABBREVIATIONS
- SPR
surface plasmon resonance
- wt
wild type
References
- 1.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 2.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 3.Rüdiger S, Buchberger A, Bukau B. Nat Struct Biol. 1997;4:342–349. doi: 10.1038/nsb0597-342. [DOI] [PubMed] [Google Scholar]
- 4.Silver P A, Way J C. Cell. 1993;74:5–6. doi: 10.1016/0092-8674(93)90287-z. [DOI] [PubMed] [Google Scholar]
- 5.Laufen T, Zuber U, Buchberger A, Bukau B. In: Molecular Chaperones in Proteins: Structure, Function, and Mode of Action. Fink A L, Goto Y, editors. New York: Marcel Dekker; 1998. pp. 241–274. [Google Scholar]
- 6.Mayer M, Bukau B. Biol Chem. 1998;379:261–268. [PubMed] [Google Scholar]
- 7.Schmid D, Baici A, Gehring H, Christen P. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 8.McCarty J S, Buchberger A, Reinstein J, Bukau B. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 9.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierpaoli E V, Sandmeier E, Schönfeld H-J, Christen P. J Biol Chem. 1998;273:6643–6649. doi: 10.1074/jbc.273.12.6643. [DOI] [PubMed] [Google Scholar]
- 11.Ziegelhoffer T, Lopez-Buesa P, Craig E. J Biol Chem. 1995;270:10412–10419. doi: 10.1074/jbc.270.18.10412. [DOI] [PubMed] [Google Scholar]
- 12.Greene M K, Maskos K, Landry S J. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchberger A, Schröder H, Büttner M, Valencia A, Bukau B. Nat Struct Biol. 1994;1:95–101. doi: 10.1038/nsb0294-95. [DOI] [PubMed] [Google Scholar]
- 14.Bukau B, Walker G C. J Bacteriol. 1989;171:2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schröder H, Langer T, Hartl F-U, Bukau B. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchberger A, Theyssen H, Schröder H, McCarty J S, Virgallita G, Milkereit P, Reinstein J, Bukau B. J Biol Chem. 1995;270:16903–16910. doi: 10.1074/jbc.270.28.16903. [DOI] [PubMed] [Google Scholar]
- 17.McCarty J S, Rüdiger S, Schönfeld H-J, Schneider-Mergener J, Nakahigashi K, Yura T, Bukau B. J Mol Biol. 1996;256:829–837. doi: 10.1006/jmbi.1996.0129. [DOI] [PubMed] [Google Scholar]
- 18.Theyssen H, Schuster H-P, Bukau B, Reinstein J. J Mol Biol. 1996;263:657–670. doi: 10.1006/jmbi.1996.0606. [DOI] [PubMed] [Google Scholar]
- 19.Zhu X, Zhao X, Burkholder W F, Gragerov A, Ogata C M, Gottesman M, Hendrickson W A. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wawrzyn A, Zylicz M. J Biol Chem. 1995;270:19300–19306. doi: 10.1074/jbc.270.33.19300. [DOI] [PubMed] [Google Scholar]
- 21.Harrison C J, Hayer-Hartl M, Di Liberto M, Hartl F-U, Kuriyan J. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 22.Flaherty K M, Deluca-Flaherty C, McKay D B. Nature (London) 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 23.Holmes K C, Sander C, Valencia A. Trends Cell Biol. 1993;3:53–59. doi: 10.1016/0962-8924(93)90161-s. [DOI] [PubMed] [Google Scholar]
- 24.Bukau B, Walker G. EMBO J. 1990;9:4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szabo A, Langer T, Schröder H, Flanagan J, Bukau B, Hartl F U. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montgomery D, Jordan R, McMacken R, Freire E. J Mol Biol. 1993;232:680–692. doi: 10.1006/jmbi.1993.1418. [DOI] [PubMed] [Google Scholar]
- 27.Suh W-C, Burkholder W F, Lu C Z, Zhao X, Gottesman M E, Gross C A. Proc Natl Acad Sci USA. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karzai A W, McMacken R. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- 29.Nicholls A, Sharp K A, Honig B J. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 30.Lanzer M. Ph.D. thesis. Heidelberg, Germany: University of Heidelberg; 1988. [Google Scholar]
- 31.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]