Abstract
Investigating neuronal and photoreceptor regeneration in the retina of zebra fish has begun to yield insights into both the cellular and molecular means by which this lower vertebrate is able to repair its central nervous system. However, knowledge about the signaling molecules in the local microenvironment of a retinal injury and the transcriptional events they activate during neuronal death and regeneration is still lacking. To identify genes involved in photoreceptor regeneration, we combined light-induced photoreceptor lesions, laser-capture microdissection of the outer nuclear layer (ONL) and analysis of gene expression to characterize transcriptional changes for cells in the ONL as photoreceptors die and are regenerated. Using this approach, we were able to characterize aspects of the molecular signature of injured and dying photoreceptors, cone photoreceptor progenitors, and microglia within the ONL. We validated changes in gene expression and characterized the cellular expression for three novel, extracellular signaling molecules that we hypothesize are involved in regulating regenerative events in the retina.
Electronic Supplementary Material
The online version of this article (doi:10.1007/s12177-008-9011-5) contains supplementary material, which is available to authorized users.
Keywords: Regenerative neurogenesis, Retinal stem cells, Laser-capture microdissection, Microarray, Microglia
Introduction
In the teleost retina, extensive neuronal death [20, 30] or the selective death of photoreceptors [35, 38] stimulates intrinsic stem cells to proliferate and give rise to regenerated neurons, which then integrate into existing neural circuits (see [18]). Consequently, the teleost retina is an ideal system to study the intrinsic cellular and molecular mechanisms that allow stem cells to regenerate neurons in the vertebrate central nervous system (CNS). Identifying and testing the molecular determinants of neuronal regeneration in the teleost retina should yield results that will have implications for understanding the regenerative potential of stem cells in the human retina and brain and the potential use of stem-cell-based therapies to treat CNS injuries or disease.
Significant progress has been made in characterizing the cells that are involved in mediating neuronal regeneration of the teleost retina. Injury to photoreceptors was long considered necessary to elicit regeneration [30], although a recent study showed that cell death within the inner nuclear layer (INL) that spares photoreceptors is sufficient [11]. Irrespective of the locus of cell death, Müller glia are the regenerative stem cells [3, 9], and re-expression of developmental regulatory genes and proliferation by Müller glia are required for neuronal regeneration [10, 33]. Although dying neurons and Müller glia are obligatory components of this regenerative neurogenesis, microglia resident within the retina migrate to the site of injury [31, 38] and are also likely critical for neuronal regeneration. Finally, in teleosts, photoreceptors are regenerated in their normal birth order, cones before rods [23, 29, 38]. Following photoreceptor-only death, injury-induced progenitors serve exclusively as “cone progenitors.” Therefore, the teleost retina also provides a model in which to elucidate the molecular mechanisms that govern the selective regeneration of cones.
Since none of the cell types described above is unique to teleost fish [12, 25, 26], a fundamental difference between animals capable of neuronal regeneration and those that are not may be the molecules elicited in response to injury. Previous studies have described the transcriptional changes that occur in the teleost retina in response to either global [6] or photoreceptor-specific injuries [21], and these studies have provided insight into the molecular basis for neuronal regeneration. Particularly revealing are changes in the molecular signature of Müller glia as they adopt the features of neural stem cells [21]. However, knowledge about signaling molecules in the local microenvironment of a retinal injury and the transcriptional events they activate during neuronal death and regeneration is still lacking.
As a means to identify genes necessary for photoreceptor regeneration, we evaluated transcriptional changes for cells in the outer nuclear layer (ONL) as photoreceptors die and are regenerated. To accomplish this, we combined light-induced photoreceptor lesions, laser-capture microdissection (LCM) of the ONL and analysis of gene expression using oligonucleotide arrays. By selectively harvesting cells from the ONL, we were able to limit our analysis to transcriptional changes among cells within the site of injury. By this approach, we hope further to characterize the molecular signatures of injured and dying photoreceptors, cone progenitors, and activated microglia. Further, we are particularly interested in identifying novel extracellular signaling molecules involved in these regenerative events, and here we describe the cellular expression of three such factors.
Materials and methods
Animals and experimental treatments
Wild-type adult zebra fish (3.0–4.0 cm long) were used in this study. Two separate paradigms were employed to induce photoreceptor death. First, for the gene array experiments and analysis of galectin-1-like 2 and galectin-9-like 1 expression, animals were housed in the dark for 7 days then exposed to fluorescent light of moderate intensity (approximately 27,000 lx) for periods between 6 and 72 h. This lesioning paradigm results in the death of photoreceptors in the dorsal retina, while sparing photoreceptors in the ventral retina (see [35, 36]). Second, for in situ hybridization for progranulin-a (see below), animals were exposed to the high-intensity light of a mercury arc lamp (>180,000 lx) for 30 min. This method kills photoreceptors in a narrow horizontally oriented band across the nasal–temporal axis of the retina. To characterize gene expression in the progeny of dividing Müller glia, Tg(gfap:GFP)mi2001 reporter fish [2] were used. In this line, astrocytes throughout the central nervous system and Müller glia within the retina synthesize green fluorescent protein.
Protocols for all procedures using animals were approved by the University Committee for Use and Care of Animals at the University of Michigan and conform to National Institutes of Health guidelines.
Histology
Fish were anesthetized in 0.1% methane sulfonate salt (MS222; Sigma Aldridge, St. Louis, MO, USA). Eyecups were then dissected, fixed overnight in 4% paraformaldehyde, cryoprotected by infiltration in 20% sucrose in phosphate buffer, and frozen in OCT (Sakura Finetek, Torrance, CA, USA). Radial sections (8 μm) were cut with a cryostat and mounted on glass slides.
Immunohistochemistry was performed using standard procedures. Briefly, sections were rinsed in phosphate-buffered saline and 0.5% triton X-100 (PBST), incubated with 20% normal goat serum (NGS) in PBST, followed by overnight incubation at 4°C in primary antibody (anti-4C4, gift from Dr. Pamela Raymond; anti-zebra-fish galectin-1-like-2, gift from Geraldo Vasta; rabbit anti-GFP, Invitrogen Corp., Carlsbad, CA, USA) at a concentration of 1:200 diluted in 2% NGS–PBST. After washing with PBST, sections were incubated in fluorescently labeled Alexafluor 555 or Alexa 488 secondary antibodies diluted 1:500 in 2% NGS–PBST (Molecular Probes, Eugene, OR) for 1.5 h at room temperature, washed extensively in PBST, and sealed with mounting media and glass coverslips. Sections were counterstained with 1:1,000 dilution of bisbenzimide to label nuclei. For immunostaining with antibodies against proliferating cell nuclear antigen (PCNA; Sigma clone PC-10, St. Louis, MO, USA), slides were first boiled for 20 min in 10 mM sodium citrate, pH 6.0, with 0.05% Tween 20 prior to the first rinse in PBST.
To identify dying cells in cryosections, terminal deoxynucleotide end labeling (TUNEL) with TUNEL kit (Roche Diagnostics, Indianapolis, IN, USA) was used according to the manufacturer’s protocol.
In situ hybridization was performed on cryosections as previously described [19]. Briefly, sense and antisense riboprobes were synthesized from linearized plasmids, and digoxigenin (DIG)-labeled probes were generated by in vitro transcription using the DIG RNA labeling kit (Roche Diagnostics, Indianapolis, IN, USA). Following prehybridization, 200 ng of probe in 80 μl of hybridization solution was pipetted onto each slide, coverslipped, and hybridized overnight at 55°C. The next day, the sections were washed and digoxigenin was immunolabeled using an alkaline-phosphatase-conjugated antibody. NBT–BCIP or Fast Red (Roche Diagnostics, Indianapolis, IN, USA) served as the enzymatic substrate. After the color reaction, sections were fixed in 4% paraformaldehyde for 15 min before proceeding with immunohistochemistry.
The cDNA encoding galectin-9-like 1 (Accession number BC059573, Open Biosystems, Huntsville, AL, USA) was linearized with EcoRI, and riboprobes were synthesized with T7 polymerase. Similarly, the cDNA encoding progranulin-a (Accession number NM001001949; a gift from Dr. Hugh Bennett) was linearized with SmaI, and riboprobes were synthesized with T3 polymerase. Negative controls were riboprobes encoding the sense strand of the respective cDNAs, which failed to hybridize to retinal sections (data not shown).
Sections were photographed with using a Nikon E300 photomicroscope and a Nikon DMX 1200 digital camera. Images were compiled in Adobe Photoshop CS2 (Adobe, San Jose, CA, USA). Images were resized and occasionally modified for contrast and brightness using the Image-Adjustments-Contrast–Brightness setting. All images within an experiment were manipulated in exactly the same manner.
Laser-capture microdissection
Eyecups were dissected from anesthetized fish and immediately frozen in OCT. Based on the timing of photoreceptor cell death and subsequent retinal stem cell proliferation (see “Results”), five time points were selected to harvest cells from the outer nuclear layers: 0 (control), 6, 12, 24, and 48 h after light onset. Control retinas were from fish removed from the tank and sacrificed immediately prior to turning on the lights. Care was taken throughout to avoid RNase contamination. Eyecups were stored at −80°C until cryosectioning. Sections were cut at 8 μm, collected on silane-coated slides (Sigma, St. Louis, MO, USA), and dried in a desiccator for 45 min. Slides were fixed and dehydrated using HistoGene frozen section staining kit (Arcturus, Molecular Devices Corporation, Sunnyvale, CA, USA) according to the manufacturer’s protocol, except that 0.1% toluidine blue was used in place of the Arcturus staining reagent. Slides were air-dried in a fume hood for 15 min prior to LCM. An AutoPix robot (Arcuturus, Molecular Devices Corporation) was used for all microdissections. Images were captured before and after LCM using the ×4 and ×20 objectives on the AutoPix robot. For each eye, 12 samples were collected from 12 sections, respectively, from the dorsal ONL using the “capture multiple fields” setting. Following each session of LCM, the thermoplastic film was removed from the caps and placed in a microfuge tube. RNA was isolated using PicoPure RNA isolation kit (Arcturus, Molecular Devices Corporation) according to the manufacturer’s protocol. Due to the length of an LCM session and the need to limit variability between sessions, one retina (12 samples) was processed per day, from cryosectioning to RNA isolation.
Microarray analysis
mRNA was amplified from total RNA to yield double-stranded antisense (or complementary) RNA (aRNA) using RiboAmp HS RNA amplification kit (Arcturus, Molecular Devices Corporation). Biotinylated aRNA was generated by in vitro transcription using Affymetrix GeneChip Expression IVT Labeling Kit (Affymetrix, Santa Clara, CA, USA). Quality of the biotin-labeled aRNA was verified using Agilent Bioanalyzer nanochips. High-quality aRNA was identified as having a smooth curve and peak between 29 and 34 s, indicating an average amplicon size of 500 bp. aRNA was quantified using Quant-iT RiboGreen RNA reagent (Molecular Probes, St. Louis, MO, USA). Five micrograms of labeled aRNA was fragmented and hybridized to Zebra fish Genome Arrays (Affymetrix, Inc., Santa Clara, CA, USA), which contain approximately 15,500 probe sets representing 14,900 genes. The hybridized arrays were scanned using a GeneChip Scanner 3000 (Affymetrix). aRNA from one retina was hybridized to a single array. For each of the five time points, three biological replicates were processed and analyzed. Each biological replicate consisted of RNA collected from a single retina (i.e., samples were not pooled).
CHP files were generated by Affymetrix Microarray Suite v5.0, prior to normalization (quantile method) with RMA software from the R project (http://www.r-project.org). False discovery rate confidence interval was used to identify statistically significant changes in gene expression between the 0 h control retinas and retinas exposed to light for 6, 12, 24, or 48 h [1, 17]. Uncentered correlation and centroid linkage hierarchical clustering was performed using Gene Cluster 3.0, and heat maps were generated using Java TreeView 1.1.1 (http://jtreeview.sourceforge.net). MAIME-compliant files containing the array data are posted at the National Center for Biotechnology Information Gene expression omnibus (http://www.ncbi.nlm.nih.gov.proxy.lib.umich.edu/geo/).
Results
Histological analysis of photoreceptor degeneration in adult pigmented zebra fish
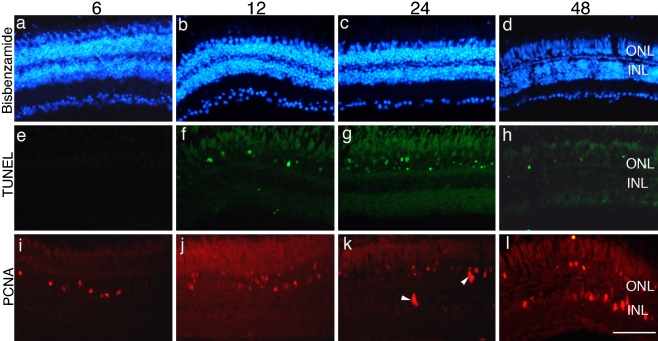
To characterize the time course of the degeneration of photoreceptors, nuclear staining with 4′,6-diamidino-2-phenylindole was used to evaluate the number and organization of nuclei in the ONL; photoreceptor death was evaluated with TUNEL staining, and cell proliferation was evaluated by immunostaining with antibodies against PCNA. After 6-, 12-, and 24-h exposure to moderate light, the three nuclear layers are unremarkable in appearance and little apparent injury occurs to the photoreceptors (Fig. 1a–c). In contrast, 48 h after light onset, the ONL is markedly thinner than normal, and there is some disorganization within the inner nuclear layer (Fig. 1d). TUNEL-positive cells are present in the ONL between 12 and 48 h after light onset (Fig. 1f), with a peak in the number of labeled cells at 24 h after light onset (Fig. 1g). TUNEL-positive cells were present only in the ONL, confirming that cell death was limited to photoreceptors.
Fig. 1.
Light induces photoreceptor-specific cell death and triggers proliferation. Animals were exposed to constant light and retinal sections were prepared from animals after 6 h (a, e, i), 12 h (b, f, j), 24 h (c, g, k), and 48 h (d, h, l). Sections were stained with bisbenzimide to label nuclear layers (a–d). The same retinal sections illustrated in a–d were labeled with TUNEL (arrowheads, e–h). Adjacent sections were immunolabeled with antibodies against PCNA (arrowheads, i–l). Scale bar = 50 μm. ONL, outer nuclear layer; INL, inner nuclear layer
Injury-induced proliferation is first observed at 6 h after light onset and was restricted to cells within the ONL (Fig. 1i). We infer these are rod precursors, mitotically active cells that reside in the ONL. In contrast, at 24 h after light onset, dividing cells are present in the INL (Fig. 1k). These are Müller glia, which have reentered the cell cycle. By 48 h after light onset, clusters of injury-induced progenitors envelope the Müller glia and span the thickness of the INL (Fig. 1l). Over time, the injury-induced progenitors both continue to multiply and migrate to the ONL (data not shown, see [35]).
Microglial cells invade the site of photoreceptor injury
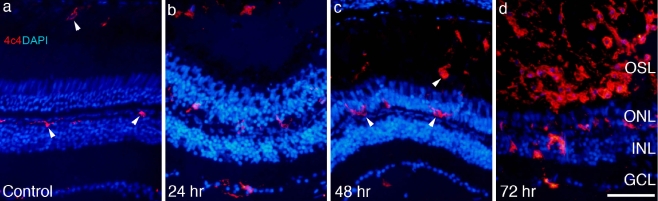
In addition to dead and dying photoreceptors and proliferating cone progenitors, microglia are also present within the outer nuclear (and outer segment) layers (Fig. 2). Microglia, as identified by the 4C4 antibody, normally reside in the optic fiber layer, the plexiform layers, and among cells of the retinal pigment epithelium (RPE; Fig. 2a). At 24 h after light onset, the time of peak cell death, 4C4-positive microglial cells hypertrophy, become polymorphic, and migrate into the ONL (Fig. 2b). At 48 h after light onset, 4C4-positive microglia also invade the field of photoreceptor debris in the outer segment layer, between the ONL and the RPE. These microglia likely originate from those cells that normally reside within the RPE. The number of 4C4-positive microglia within the retina and among the dying photoreceptors increases until at least 72 h after light onset (Fig. 2c,d).
Fig. 2.
As photoreceptors die, resident microglia migrate into the outer nuclear and outer segment layers. Retinal sections were taken from control (a) and experimental retinas exposed to constant light for 24 h (b), 48 h (c), and 72 h (d) and immunolabeled with the microglia-specific antibody, 4C4 (arrowheads), and counterstained with 4′,6-diamidino-2-phenylindole. Scale bar = 50 μm. OSL, outer segment layer; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer
Injury-induced changes in gene expression among cells of the outer nuclear layer
To identify injury-induced changes in gene expression, RNA was isolated from cells within the ONL of light-lesioned retinas using LCM (Fig. 3), and changes in gene expression between control retinas and retinas exposed to light for between 6 and 48 h were identified with oligonucleotide microarrays. Analysis of the microarrays across all time points showed that a total of 1,937 genes showed at least a twofold or greater change in signal intensity (gene expression) on the array (see Supplementary Material). Hierarchical clustering identified the temporal correlation for both increases and decreases in transcript levels (Fig. 4). We categorized the genes into functional groups based on the gene ontology descriptions provided by Affymetrix and by manual classification. Although only approximately 50% of these genes are annotated, we were able to assign a subset of the known genes to the cell types present in the ONL over the time points studied here.
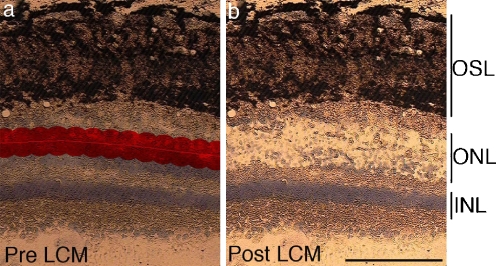
Fig. 3.
Laser-capture microdissection of cells from the ONL. A video image of a section from a control retina is illustrated in a and b. The ONL is highlighted using the LCM robot, identifying the area selected for dissection (a). Panel b illustrates the retinal section in a after LCM. Scale bar = 100 μm. OSL, outer segment layer; ONL, outer nuclear layer; INL, inner nuclear layer
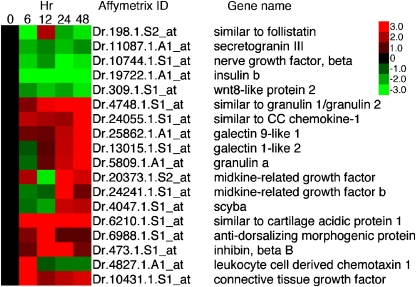
Fig. 4.
Heat map representing the hierarchical clustering of 1,937 genes observed to change in expression in the ONL over the course of photoreceptor injury and regeneration. Uncentered Pearson’s correlation and centroid linkage of genes were used to cluster transcriptional changes. Three clustering differences were observed: genes with expression that increased, genes with expression that decreased, and genes that showed differential expression at the 12-h time point
Transcriptional changes predicted for injured photoreceptors
Based on our cellular analysis, and taking into account that at 0, 6, 12, and 24 h after light onset photoreceptors are the numerically predominant cell type present in the ONL (Fig. 1), we infer that genes that change levels of expression compared to the 0-h control during the first 24 h after light onset will largely be restricted to injured and dying photoreceptors (Fig. 5). To support this assertion, numerous known photoreceptor-specific genes showed decreased expression at early time points, including genes encoding opsins, pde6c (phosphodiesterase 6c), gnat (alpha transducin), and gnb (beta transducin; Fig. 5). Similarly, and as anticipated, at early time points, the expression of numerous stress response genes was dramatically increased (Fig. 6). Genes encoding secreted proteins also show changes at early time points, including insulin b, nerve growth factor b, and wnt-8-like-2 (Fig. 7). Although we are less certain regarding the identities of cells that express genes encoding these growth factors (see below), a first approximation is that some of these genes may be expressed by photoreceptors.
Fig. 5.
Heat map of selected phototransduction genes that change over the course of the light injury. The majority of photoreceptor-specific genes decreased their levels of expression, consistent with photoreceptor loss due to cell death
Fig. 6.
Heat map of selected stress response genes that change in expression over the course of light injury. The majority of these genes showed marked increase in expression during the early phases of the light-induced injury
Fig. 7.
Heat map of selected secreted proteins that change over the course of the light injury
Transcriptional changes predicted to occur in cone progenitors and microglia
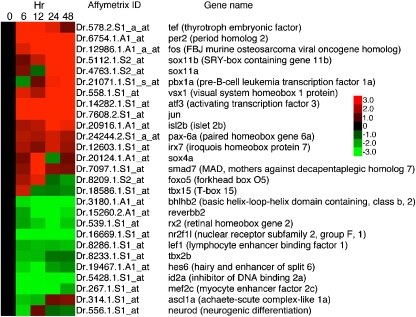
Following a similar logic, we predict that transcripts with levels that change at the 24- and 48-h time points compared to the control retinas will be expressed in cells that migrate to the ONL, cone progenitors, and microglia (see Figs. 1 and 2). Genes at the 24- and 48-h time points were among those that showed the greatest magnitude of change. Included in these genes are those encoding the transcription factors sox11b, fos, jun, and pax6a (Figs. 8 and 9), and the secreted proteins, lymphocyte cytosolic plastin 1 (l-plastin, see [16]), CC chemokine, members of the galectin family, members of the progranulin family, and midkines a and b (Fig. 7).
Fig. 8.
Heat map of selected transcription factors that change over the course of light injury
Fig. 9.
Heat map of selected genes that showed a marked increase in transcription at the 48-h time point
From the genes encoding growth factors, three were selected to study further: two members of the galectin family, galectin-9 like-1 (lgals9l1) and galectin-1 like-2 (lgals 1l2), and progranulin-a (pgrna). In situ hybridization or immunocytochemistry were used both to independently validate the changes in the expression of these genes and to characterize their cellular patterns of expression. Lgals9l1 and Lgals1l2 are members of a large family of secreted carbohydrate-binding proteins that bind β-galactosides on the surface of glycoproteins such as laminin, integrins, osteopontin, and fibronectin [5]. Galectins are believed to mediate interactions between cells and the extracellular matrix [27]. In situ hybridization showed that lgals9l1 is expressed by a sparse population of cells confined to a narrow strata at the interface of the ONL and outer plexiform layer, a location that is suggestive of microglia (Fig. 10; see [37]). This inference was confirmed by combining in situ hybridization with immunolabeling with the microglia-specific antibody, 4c4 (Fig. 10). In contrast, immunostaining with antibodies against Lgals1l2 labeled both radial columns of cells spanning the INL, suggestive of Müller glia and/or cone progenitors, and microglia. Retinal sections taken from the Tg(gfap:GFP)mi2001 line and immunostained with the Lgals1l2 antibodies confirmed that cells immunopositive for Lgals1l2 originate from Müller glia (Fig. 11a–c), and immunostaining with 4c4 identified GFP-negative, Lgals1l2-positive microglia.
Fig. 10.
Expression of Lgals9l1 mRNA in microglia in the ONL. Panels a–c illustrate a retinal section from an animal exposed to constant light for 12 h, then labeled by in situ hybridization with probes against lgals9l1 mRNA (a), immunolabeled for microglia with 4C4 antibody (b). Panel c is the digital overlay of a and b. Panels d–f illustrate a retina section from an animal exposed to constant light for 48 h, then labeled by in situ hybridization with probes against lgals9l1 mRNA (d), immunolabeled for microglia with 4C4 antibody (e). Panel f is the digital overlay of d and e. Arrowheads indicate the same cells in a–c and d–f. ONL, outer nuclear layer; INL, inner nuclear layer. Scale bar = 50 μm
Fig. 11.
Lgals1l2 protein is expressed by proliferating Müller glial stem cells and their progeny and microglia in the ONL of injured retina. Panels a–c illustrate a retinal section from a transgenic (gfap:GFP)mi2001 fish exposed to constant light for 48 h followed by immunolabeling for Lgals1l2 (a) and GFP (b). Panel c is a digital overlay of a and b. Panels d–f illustrate a retinal section from an animal exposed to constant light for 48 h and immunolabeled for Lgals1l2 (d) and 4C4 (e). Panel f is a digital overlay of d and e. Arrowheads indicate the same cells in a–c and d–f. Scale bar = 50 μm. ONL, outer nuclear layer; INL, inner nuclear layer
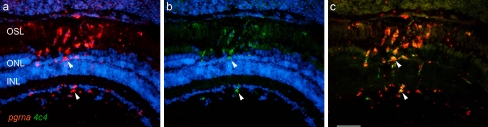
Progranulin-a (a.k.a., granulin-epithelin precursor, proepithelin, PC cell-derived growth factor, acrogranin) is one of the five granulin paralogs expressed in zebra fish [4] that encode proteins with multiple functions, including mediating tissue repair [15]. In situ hybridization with probes encoding pgrna labeled cells within the retina that are in close association with the lesioned photoreceptors and, most prominently, cells within the ONL and the overlying outer segment layer (Fig. 12a). Combining in situ hybridization with immunostaining with the 4c4 antibody showed that pgrna is expressed exclusively by microglia (Fig. 12b,c).
Fig. 12.
Progranulin-a is expressed by microglia at the site of photoreceptor injury. Panel a illustrates an in situ hybridization showing pgrna at the site of photoreceptor injury. Panel b is the same section as in a but immunostained with the 4c4 antibody. Panel c is the digital overlay of panels a and b. Arrowheads identify the same cells in a–c. Scale bar = 50 μm. OSL, outer segment layer; ONL, outer nuclear layer; INL, inner nuclear layer
Transcriptional changes at 12 h after light onset may identify genes regulated by light or the circadian rhythm
Fortuitously, the design of our experiment also allowed us to identify transcriptional changes that are likely regulated either by light or the circadian rhythm. Cells harvested at 0- (control), 24-, and 48-h time points were collected in the morning shortly after the room lights were switched on, whereas the 12-h time points were collected shortly before the room lights were switched off. Therefore, genes that are differentially expressed at 12- vs. 0-, 24-, or 48-h time points may encode proteins that are synthesized in response to light or regulated by the circadian rhythm [13]. As evidence for this, we detected numerous genes that were differentially expressed at the 12-h time point (Figs. 13 and 14).
Fig. 13.
Heat map highlighting selected genes that increased expression at the 12-h time point
Fig. 14.
Heat map highlighting selected genes that decreased in expression at the 12-h time point
Discussion
The dataset described in this report provides the vision community with a resource to mine for transcriptional changes that regulate the first stages of photoreceptor regeneration, from death to the specification of photoreceptor progenitors. Early transcriptional changes were observed in genes likely expressed by injured photoreceptors. Later transcriptional changes allowed us to identify novel genes expressed in Müller glia and cone progenitors, which give rise to regenerated photoreceptors, and microglia, which may also play a crucial role in photoreceptor regeneration. More broadly, this dataset provides information about the molecular mechanisms that regulate injury-induced neurogenesis in an adult nervous system and thus has utility for neuroscience in general.
By combining LCM of the ONL with a time course of injury-induced gene expression, we were able nominally to assign genes to each of the three cell types the cellular analysis identified within the lesioned ONL. We predicted that changes in gene expression that occur during the first 24 h are largely attributable to the injured photoreceptors. Thus, as anticipated, we observed transcriptional changes in known injury-induced genes at the early time points, including the heat shock proteins. Similarly, we detected a decrease in photoreceptor-specific genes, including genes encoding opsins and phototransduction proteins. The decrease in the expression of these genes is most easily explained by the apoptotic loss of photoreceptors. Nonetheless, the observed decrease in photoreceptor-specific genes serves to validate the logic behind our approach to assign early gene changes to photoreceptors. In the mammalian retina, injury to photoreceptors results in increased transcription of endothelin2, which encodes a secreted protein that signals through endothelin B receptors on Müller glia [28]. We anticipate that similar signaling events occur in the regenerating retina of teleosts (although no changes in endothelin 1 or 2 were observed), and genes encoding molecules that signal from photoreceptors to Müller glia are represented in the dataset. One category of genes worth investigating further are the stress response genes. Recent studies have suggested that heat shock proteins may be secreted [8] and thus may serve as extracellular signaling molecules during the early events of photoreceptor regeneration.
In addition, we anticipated identifying known and novel genes expressed by cone progenitors in the ONL at the 48-h time point. It should be noted, however, that the changes in the expression of these genes, and in contrast to the loss of photoreceptors, are most easily explained by the accretion of cone progenitors migrating from the INL to the ONL. Nonetheless, our approach allowed us to identify putative cone-progenitor-specific genes, and these data will provide insights into the molecular mechanisms that govern the terminal steps of cone specification and differentiation. In support of this hypothesis, we detected an increase in the expression of lgals1l2, which encodes a secreted protein, and demonstrated that Lgals1l2 is expressed by putative cone progenitors (and microglia and Müller glia).
Based on our cellular analysis, we also anticipated identifying genes expressed by reactive microglia that migrate to the damaged ONL. Microglia are a little studied cell type but one whose importance in various developmental and disease states is becoming recognized [14, 24]. Of note, each of the genes encoding the secreted proteins lgals1l2, lgals9l1, and prgna is expressed by resident microglia after injury to photoreceptors. Microglia are phagocytic cells that can secrete both proinflammatory and anti-inflammatory signals to either inhibit or promote neuronal repair and regeneration [14]. Therefore, microglial-derived secreted signals are likely important regulators of cone photoreceptor regeneration. Functional evaluation of microglial-specific genes should provide further insights into the mechanisms that regulate photoreceptor (and neuronal) regeneration.
Gene array technologies have been used previously to profile transcriptional changes in zebra fish during regeneration of the fin [32], heart [22], and retina [6, 21]. Recently, LCM and gene arrays were combined to identify transcriptional changes in retinal ganglion cells during axonal regeneration [34]. Of note, some of the gene coding for secreted factors identified in our dataset are also upregulated during heart and fin regeneration [22, 32]. The injury-induced upregulation of these genes suggest that in zebra fish common molecular mechanisms may regulate all regenerative processes. However, further comparisons of datasets from zebra fish arrays should also reveal molecular signatures that are specific to retinal stem cells and the regeneration of cone photoreceptors. Finally, although the annotation of the zebra fish genome does not yet match that for mammals, comparing transcriptional changes in the injured and regenerating retina of zebra fish with similar studies conducted in mammals [7, 28] may begin to identify molecular mechanisms that are essential to control the fates of stem cells in the mammalian retina and brain.
Electronic Supplementary Material
Below is the link to the Electronic Supplementary Material.
(XLS 464 KB)
Acknowledgements
We thank Drs. Pamela Raymond for the transgenic (gfap:GFP)mi2001 reporter fish and the 4C4 antibody, Geraldo Vasta for the anti-zebra-fish Galectin-1-like-2 antibody and Hugh Bennett for the progranulin-a cDNA. We also thank Dr. David Reed, Matthew Brooks, and Ritu Khanna for their help in conducting and analyzing the microarray experiments and Erica Dawsey and Laura Kakuk-Atkins for technical assistance. This work was supported by the National Institutes of Health Grants R01-EY007060 and P30-EY07003 (PFH) and T32-EY13934 (S.E.L.C.) and Research to Prevent Blindness. S.E.L.C. was also supported by a postdoctoral fellowship from the Foundation Fighting Blindness-Canada.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Benjamini Y, Yekutieli DJ. Am Stat Assoc. 2005;100:71–80. doi: 10.1198/016214504000001907. [DOI] [Google Scholar]
- 2.Bernardos RL, Raymond PA. GFAP transgenic zebra fish. Gene Expr Patterns. 2006;6:1007–1013. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadieux B, Chitramuthu BP, Baranowski D, Bennett HP. The zebra fish progranulin gene family and antisense transcripts. BMC Genomics. 2005;6:156. doi: 10.1186/1471-2164-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camby I, Le Mercier M, Lefranc F, Kiss R. Galectin-1: a small protein with major functions. Glycobiology. 2006;16:137R–157R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- 6.Cameron DA, Gentile KL, Middleton FA, Yurco P. Gene expression profiles of intact and regenerating zebra fish retina. Mol Vis. 2005;11:775–791. [PubMed] [Google Scholar]
- 7.Chen L, Wu W, Dentchev T, Zeng Y, Wang J, Tsui I, Tobias JW, Bennett J, Baldwin D, Dunaief JL. Light damage induced changes in mouse retinal gene expression. Exp Eye Res. 2004;79:239–247. doi: 10.1016/j.exer.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Eustace BK, Sakurai T, Stewart JK, Yimlamai D, Unger C, Zehetmeier C, Lain B, Torella C, Henning SW, Beste G, Scroggins BT, Neckers L, Ilag LL, Jay DG. Functional proteomic screens reveal an essential extracellular role for hsp90 alpha in cancer cell invasiveness. Nat Cell Biol. 2004;6:507–514. doi: 10.1038/ncb1131. [DOI] [PubMed] [Google Scholar]
- 9.Fausett BV, Goldman D. A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebra fish retina. J Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fausett BV, Gumerson JD, Goldman D. The proneural basic helix–loop–helix gene ascl1a is required for retina regeneration. J Neurosci. 2008;28:1109–1117. doi: 10.1523/JNEUROSCI.4853-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fimbel SM, Montgomery JE, Burket CT, Hyde DR. Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebra fish. J Neurosci. 2007;27:1712–1724. doi: 10.1523/JNEUROSCI.5317-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer AJ, Reh TA. Müller glia are a potential source of neural regeneration in the postnatal chicken retina. Nat Neurosci. 2001;4:247–252. doi: 10.1038/85090. [DOI] [PubMed] [Google Scholar]
- 13.Green CB, Besharse JC. Retinal circadian clocks and control of retinal physiology. J Biol Rhythms. 2004;19:91–102. doi: 10.1177/0748730404263002. [DOI] [PubMed] [Google Scholar]
- 14.Hanisch UK, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci. 2007;10:1387–1394. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- 15.He Z, Ong CH, Halper J, Bateman A. Progranulin is a mediator of the wound response. Nat. Med. 2003;9:225–229. doi: 10.1038/nm816. [DOI] [PubMed] [Google Scholar]
- 16.Herbomel P, Thisse B, Thisse C. Zebra fish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. 2001;238:274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- 17.Hero AO, Fleury G, Mears AJ, Swaroop A. Multicriteria gene screening for analysis of differential expression with DNA microarrays. EURASIP JASP. 2004;2004:43–52. [Google Scholar]
- 18.Hitchcock PF. Tracer coupling among regenerated amacrine cells in the retina of the goldfish. Vis Neurosci. 1997;14:463–472. doi: 10.1017/s095252380001213x. [DOI] [PubMed] [Google Scholar]
- 19.Hitchcock P, Kakuk-Atkins L. The basic helix-loop-helix transcription factor neuroD is expressed in the rod lineage of the teleost retina. J Comp Neurol. 2004;477:108–117. doi: 10.1002/cne.20244. [DOI] [PubMed] [Google Scholar]
- 20.Hitchcock PF, Lindsey Myhr KJ, Easter SS, Jr, Mangione-Smith R, Jones DD. Local regeneration in the retina of the goldfish. J Neurobiol. 1992;23:187–203. doi: 10.1002/neu.480230209. [DOI] [PubMed] [Google Scholar]
- 21.Kassen SC, Ramanan V, Montgomery JE, Burket CT, Liu CG, Vihtelic TS, Hyde DR. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebra fish. Dev Neurobiol. 2007;67:1009–1031. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- 22.Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebra fish heart regeneration. PLoS Biol. 2006;4:e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mader MM, Cameron DA. Photoreceptor differentiation during retinal development, growth, and regeneration in a metamorphic vertebrate. J Neurosci. 2004;24:11463–11472. doi: 10.1523/JNEUROSCI.3343-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallat M, Marín-Teva JL, Chéret C. Phagocytosis in the developing CNS: more than clearing the corpses. Curr Opin Neurobiol. 2005;15:101–107. doi: 10.1016/j.conb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Ooto S, Akagi T, Kageyama R, Akita J, Mandai M, Honda Y, Takahashi M. Potential for neural regeneration after neurotoxic injury in the adult mammalian retina. Proc Natl Acad Sci U S A. 2004;101:13654–13659. doi: 10.1073/pnas.0402129101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osakada F, Ooto S, Akagi T, Mandai M, Akaike A, Takahashi M. Wnt signaling promotes regeneration in the retina of adult mammals. J Neurosci. 2007;27:4210–4219. doi: 10.1523/JNEUROSCI.4193-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabinovich GA, Toscano MA, Jackson SS, Vasta GR. Functions of cell surface galectin–glycoprotein lattices. Curr. Opin. Struct. Biol. 2007;17:513–520. doi: 10.1016/j.sbi.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rattner A, Nathans J. The genomic response to retinal disease and injury: evidence for endothelin signaling from photoreceptors to glia. J Neurosci. 2005;25:4540–4549. doi: 10.1523/JNEUROSCI.0492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raymond PA. Retinal regeneration in teleost fish. Ciba Found Symp. 1991;160:171–186. doi: 10.1002/9780470514122.ch9. [DOI] [PubMed] [Google Scholar]
- 30.Raymond PA, Reifler MJ, Rivlin PK. Regeneration of goldfish retina: rod precursors are a likely source of regenerated cells. J Neurobiol. 1988;19:431–463. doi: 10.1002/neu.480190504. [DOI] [PubMed] [Google Scholar]
- 31.Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebra fish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schebesta M, Lien CL, Engel FB, Keating MT. Transcriptional profiling of caudal fin regeneration in zebra fish. Scientific World Journal. 2006;6(Suppl 1):38–54. doi: 10.1100/tsw.2006.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thummel R, Kassen SC, Montgomery JE, Enright JM, Hyde DR. Inhibition of Müller glial cell division blocks regeneration of the light-damaged zebra fish retina. Dev Neurobiol. 2008;68:392–408. doi: 10.1002/dneu.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebra fish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebra fish (Danio rerio) retina. J Neurobiol. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::AID-NEU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 36.Vihtelic TS, Soverly JE, Kassen SC, Hyde DR. Retinal regional differences in photoreceptor cell death and regeneration in light-lesioned albino zebra fish. Exp Eye Res. 2006;82:558–575. doi: 10.1016/j.exer.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 37.Wagner EC, Raymond PA. Müller glial cells of the goldfish retina are phagocytic in vitro but not in vivo. Exp Eye Res. 1991;53:583–589. doi: 10.1016/0014-4835(91)90216-2. [DOI] [PubMed] [Google Scholar]
- 38.Wu DM, Schneiderman T, Burgett J, Gokhale P, Barthel L, Raymond PA. Cones regenerate from retinal stem cells sequestered in the inner nuclear layer of adult goldfish retina. Invest Ophthalmol Vis Sci. 2001;42:2115–2124. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS 464 KB)